Abstract
Pleosporales is the largest and most morphologically diverse order in Dothideomycetes, including a large proportion of saprobic fungi. During the investigation of microfungi from decaying wood in Sichuan Province, several novel fungal taxa of asexual and sexual morphs were collected, identified, and well-described. Phylogenetic analyses based on SSU, ITS, LSU, RPB2 and TEF1α gene sequences suggested that these new taxa were related to Pleosporales and distributed in five families, viz. Amorosiaceae, Bambusicolaceae, Lophiostomataceae, Occultibambusaceae and Tetraplosphaeriaceae. The morphological comparison and molecular phylogeny evidence justify the establishment of six new taxa, namely Bambusicola guttulata sp. nov., Flabellascoma sichuanense sp. nov., Neoangustimassarina sichuanensis gen. et sp. nov., Occultibambusa sichuanensis sp. nov. and Pseudotetraploa bambusicola sp. nov. Among them, Neoangustimassarina was introduced as the second sexual morph genus in Amorosiaceae; Bambusicola guttulata, O. sichuanensis and P. bambusicola were isolated from bamboos, which contributed to the diversity of bambusicolous fungi. The detailed, illustrated descriptions and notes for each new taxon are provided, as well as a brief note for each family. The potential richness of fungal diversity in Sichuan Province is also discussed.
Keywords: 6 new taxa, Dothideomycetes, multi-gene, phylogeny, taxonomy
1. Introduction
Pleosporales is the largest order in the class Dothideomycetes [1], and its members are found worldwide on a variety of host plants as epiphytes, endophytes, saprobes and parasites [2,3,4]. In addition, they are commonly found in terrestrial, marine and freshwater habitats [5,6,7]. Some of them produce secondary metabolites that can serve as a basis for developing new antimicrobials, agrochemical pesticides and other useful compounds [8].
The Pleosporales was invalidly introduced by Luttrell [9], and later revised by Barr [10], based on the family Pleosporaceae with the type species Pleospora herbarum [11]. Members of Pleosporales usually have perithecioid ascomata, cellular pseudoparaphyses, bitunicate and fissitunicate asci, and various shaped, aseptate or septate ascospores, with or without a gelatinous sheath [12,13,14], and their asexual morphs are coelomycetes and hyphomycetes [12,13]. For example, asexual morphs of Bambusicola and Pseudotetraploa are the most common forms of coelomycetes and hyphomycetes in Pleosporales, respectively. Zhang et al. listed 26 families in Pleosporales [12], while Hyde et al. revised Pleosporales and accepted 41 families [13]. Hongsanan et al. redefined the families of Dothideomycetes and accepted 91 families in Pleosporales based on morphology and multigene analysis [15]. Currently, Pleosporales consists of approximately 91 families and 655 genera (including 41 genera incertae sedis) [16].
Fungi have a broad geographical distribution and diversity comparable to plants and other organisms [17,18]. However, the fungal kingdom, in general, is less well-documented than the plant kingdom in terms of the number of species [19]. As one of the biodiversity hotspots in China, Sichuan Province (located in southwestern China) has a variety of complex topography (mountains, hills, plains, basins and plateaus) and climate conditions, and these are important factors contributing to the biodiversity [20]. However, little research on fungi has been carried out in this area; meanwhile, the highly variable climate and lush vegetation are shown to have an important influence on fungal diversity. Therefore, Sichuan Province is believed to have a large amount of hidden fungal diversity to be explored and discovered [21,22].
During a survey of micro-fungi from decomposing wood in Sichuan Province, China, a series of interesting asexual and sexual fungi were collected. In this study, we aim to describe these new findings and contribute fungal diversity to China. The multi-gene phylogeny integrated with morphological comparison was carried out to determine the classification of these new collections. One new genus and five new species are introduced, and the establishment of these new taxa is justified by morphology and phylogenetic evidences.
2. Materials and Methods
2.1. Isolation and Morphological Examination
Fungi associated with decaying wood were collected from Sichuan Province, China in 2021. Specimens were placed in envelopes and taken to the laboratory. Fungal colonies and fruiting bodies were observed using Motic SMZ 168-B. Fungal structures were examined and photographed by using a Nikon ECLIPSE Ni-U compound microscope fitted with a Nikon DS-Ri2 digital camera. The detailed morphological examination approaches used in this paper were generally based on Senanayake et al. [23]. Single spore isolations were made following the method in Senanayake et al. [23]. Measurements were made with the Tarosoft (R) Image Framework program v. 0.9.7, following the procedures outlined by Liu et al. [24]. Photo plates representing fungal structures were processed in Adobe Photoshop CS6 software (Adobe Systems Inc., San Jose, CA, USA). Herbarium specimens (dry branches with fungal material) were deposited in the herbarium of Cryptogams, Kunming Institute of Botany Academia Sinica (HKAS), Kunming, China and the herbarium of the University of Electronic Science and Technology (HUEST), Chengdu, China. The isolates obtained in this study were deposited in China General Microbiological Culture Collection Center (CGMCC), Beijing, China and the University of Electronic Science and Technology Culture Collection (UESTCC), Chengdu, China. The names of the new taxa were registered in MycoBank [25].
2.2. DNA Extraction, PCR Amplification and Sequencing
A Trelief TM Plant Genomic DNA Kit (Beijing TsingKe Biotech Co., Ltd., Beijing, China) was used to extract total genomic DNA from fresh mycelia, according to the manufacturer’s instructions. DNA amplification was performed by polymerase chain reaction (PCR). SSU, ITS, LSU, RPB2 and TEF1α gene regions were amplified using the primer pairs NS1/NS4, ITS5/ITS4, LR0R/LR5, fRPB2-5F/fRPB2-7cR and 983F/2218R, respectively, [26,27,28,29]. The amplification reactions were performed in 25 μL PCR mixtures containing 22 μL PCR MasterMix (Green) (TsingKe Co., Beijing, China), 1 μL DNA template and 1 μL of each primer (10 µM/L). The PCR thermal cycle program for SSU, ITS, LSU, RPB2 and TEF1α amplification were listed in Table 1. PCR products were checked on 1% agarose electrophoresis gels stained with Gel Red. The sequencing reactions were carried out with primers, mentioned above, by Beijing Tsingke Biotechnology Co., Ltd., Chengdu, China.
Table 1.
PCR thermal cycles for SSU, ITS, LSU, RPB2 and TEF1α amplification.
| Step | SSU | ITS, LSU, RPB2 | TEF1α | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Temperature | Time | Cycles | Temperature | Time | Cycles | Temperature | Time | Cycles | |
| Initial Denaturation | 98 °C | 2 min | 1 | 98 °C | 2 min | 1 | 98 °C | 2 min | 1 |
| Denaturation | 98 °C | 10 s | 35 | 98 °C | 10 s | 35 | 98 °C | 10 s | 35 |
| Annealing | 47 °C | 10 s | 56 °C | 10 s | 61.7 °C | 10 s | |||
| Extension | 72 °C | 10 s | 72 °C | 10 s | 72 °C | 10 s | |||
| Final Extension | 72 °C | 5 min | 1 | 72 °C | 5 min | 1 | 72 °C | 5 min | 1 |
| Hold | 4 °C | - | - | 4 °C | - | - | 4 °C | - | - |
2.3. Phylogenetic Analyses
The chromatograms of the new sequences obtained in this study were viewed in Finch TV Version 1.4.0 (https://digitalworldbiology.com/FinchTV (accessed on 22 September 2021)). The BLAST searches were performed for finding similar sequences that match our data. A concatenated dataset of the SSU, ITS, LSU, RPB2 and TEF1α sequences were used for phylogenetic analyses with the inclusion of reference taxa from GenBank (Table 2). The sequences were aligned by using the online multiple-alignment program MAFFT v.7 (http://mafft.cbrc.jp/alignment/server/ (accessed on 5 January 2022)) [30], and the alignment was manually optimized in BioEdit v.7.0.9 [31]. Each gene dataset was concatenated by Mesquite v. 3.11 (http://www.mesquiteproject.org/ (accessed on 15 April 2022)) for multi-gene phylogenetic analyses. Maximum likelihood (ML) and bayesian inference (BI) were carried out as detailed in Dissanayake et al. [32]. The programs used in this study are RAxMLGUI v. 1.0 [33], PAUP v.4.0b10 [34], Mr Modeltest 2.3 [35] and MrBayes v. 3.1.2 [36,37]. The phylogenetic tree was visualized by FigTree v.1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 April 2022)).
Table 2.
Taxa used in the phylogenetic analyses and their GenBank accession numbers. Newly generated sequences are indicated with * and the ex-type strains are in bold.
| Species | Voucher/Strain/Isolate | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|
| SSU | ITS | LSU | RPB2 | TEF1α | ||
| Alfoldia vorosii | CBS 145501 | MK589346 | JN859336 | MK589354 | N/A | MK599320 |
| Alfoldia vorosii | REF113 | MK589345 | JN859333 | MK589353 | N/A | MK599319 |
| Alfoldia vorosii | REF117 | MK589347 | JN859337 | MK589355 | N/A | MK599321 |
| Amorocoelophoma camelliae | NTUCC 18-097-1 | MT071230 | MT112303 | MT071279 | MT459143 | MT743271 |
| Amorocoelophoma camelliae | NTUCC 18-097-2 | MT071231 | MT112304 | MT071280 | MT459141 | MT743272 |
| Amorocoelophoma camelliae | NTUCC 18-097-3 | MT071232 | MT112305 | MT071281 | MT459142 | MT743273 |
| Amorocoelophoma cassiae | MFLUCC 17-2283 | MK347847 | MK347739 | MK347956 | MK434894 | MK360041 |
| Amorocoelophoma neoregeliae | CBS 146820 | N/A | MZ064410 | MZ064467 | MZ078193 | MZ078247 |
| Amorosia littoralis | CBS 120399 | AM292056 | AM292047 | AM292055 | N/A | N/A |
| Angustimassarina acerina | MFLUCC 14-0505 | KP899123 | KP899132 | KP888637 | N/A | KR075168 |
| Angustimassarina arezzoensis | MFLUCC 13-0578 | KY501113 | KY496743 | KY496722 | N/A | KY514392 |
| Angustimassarina camporesii | MFLU 18-0057 | MN244173 | MN244197 | MN244167 | N/A | N/A |
| Angustimassarina italica | MFLUCC 15-0082 | KY501124 | KY496756 | KY496736 | N/A | KY514400 |
| Angustimassarina lonicerae | MFLUCC 15-0087 | N/A | KY496759 | KY496724 | N/A | N/A |
| Angustimassarina populi | MFLUCC 13-0034 | KP899128 | KP899137 | KP888642 | N/A | KR075164 |
| Angustimassarina premilcurensis | MFLUCC 15-0074 | N/A | KY496745 | KY496725 | KY514404 | N/A |
| Angustimassarina quercicola | MFLUCC 14-0506 | KP899124 | KP899133 | KP888638 | N/A | KR075169 |
| Angustimassarina rosarum | MFLUCC 15-0080 | N/A | MG828869 | MG828985 | N/A | N/A |
| Angustimassarina sylvatica | MFLUCC 18-0550 | MK314097 | MK307843 | MK307844 | N/A | MK360181 |
| Angustimassarina alni | MFLUCC 15-0184 | KY548098 | KY548099 | KY548097 | N/A | N/A |
| Angustimassarina coryli | MFLUCC 14-0981 | N/A | MF167431 | MF167432 | N/A | MF167433 |
| Aquatisphaeria thailandica | MFLUCC 21-0025 | MW890967 | MW890969 | MW890763 | N/A | N/A |
| Bambusicola aquatica | MFLUCC 18-1031 | MT864293 | MT627729 | MN913710 | MT878462 | MT954392 |
| Bambusicola bambusae | MFLUCC 11-0614 | JX442039 | JX442031 | JX442035 | KP761718 | KP761722 |
| Bambusicola didymospora | MFLUCC 10-0557 | KU872110 | KU940116 | KU863105 | KU940163 | KU940188 |
| Bambusicola dimorpha | MFLUCC 13-0282 | KY038354 | KY026582 | KY000661 | KY056663 | N/A |
| Bambusicola ficuum | MFLUCC 17-0872 | MT215581 | N/A | MT215580 | N/A | MT199326 |
| Bambusicola fusispora | MFLUCC 20-0149 | MW076529 | MW076532 | MW076531 | MW034589 | N/A |
| Bambusicola guttulata * | CGMCC 3.20935 | ON332919 | ON332909 | ON332927 | ON383985 | ON381177 |
| Bambusicola guttulata * | UESTCC 22.0002 | ON332920 | ON332910 | ON332928 | ON383986 | ON381178 |
| Bambusicola irregulispora | MFLUCC 11-0437 | JX442040 | JX442032 | JX442036 | KP761719 | KP761723 |
| Bambusicola loculata | MFLUCC 13-0856 | KP761735 | KP761732 | KP761729 | KP761715 | KP761724 |
| Bambusicola massarinia | MFLUCC 11-0389 | JX442041 | JX442033 | JX442037 | KP761716 | KP761725 |
| Bambusicola pustulata | MFLUCC 15-0190 | KU872112 | KU940118 | KU863107 | KU940165 | KU940190 |
| Bambusicola sichuanensis | SICAUCC 16-0002 | MK253528 | MK253473 | MK253532 | MK262830 | MK262828 |
| Bambusicola splendida | MFLUCC 11-0439 | JX442042 | JX442034 | JX442038 | KP761717 | KP761726 |
| Bambusicola subthailandica | SICAU 16-0005 | MK253529 | MK253474 | MK253533 | MK262831 | MK262829 |
| Bambusicola thailandica | MFLUCC 11-0147 | N/A | KU940119 | KU863108 | KU940166 | KU940191 |
| Bambusicola triseptatispora | MFLUCC 11-0166 | N/A | KU940120 | KU863109 | KU940167 | N/A |
| Brunneofusispora clematidis | MFLUCC 17-2070 | MT226685 | MT310615 | MT214570 | MT394692 | MT394629 |
| Brunneofusispora inclinatiostiola | CGMCC 3.20403 | MZ964884 | MZ964866 | MZ964875 | OK061075 | OK061069 |
| Brunneofusispora sinensis | KUMCC 17-0030 | MH393556 | MH393558 | MH393557 | N/A | MH395329 |
| Corylicola italica | MFLU 19-0500 | MT554923 | MT554925 | MT554926 | MT590776 | N/A |
| Corylicola italica | MFLUCC 20-0111 | MT633084 | MT633085 | MT626713 | MT635596 | MT590777 |
| Crassiclypeus aquaticus | CBS 143641 | LC312470 | LC312499 | LC312528 | LC312586 | LC312557 |
| Crassiclypeus aquaticus | CBS 143643 | LC312472 | LC312501 | LC312530 | LC312588 | LC312559 |
| Ernakulamia krabiensis | MFLUCC 18–0237 | MK347880 | MK347773 | MK347990 | N/A | N/A |
| Ernakulamia tanakae | NFCCI 4615 | N/A | MN937229 | MN937211 | N/A | N/A |
| Ernakulamia xishuangbannaensis | KUMCC 17-0187 | MH260354 | MH275080 | MH260314 | N/A | N/A |
| Exosporium stylobatum | CBS 160.30 | N/A | JQ044428 | JQ044447 | N/A | N/A |
| Flabellascoma aquaticum | KUMCC 15-0258 | MN304832 | MN304827 | MN274564 | MN328895 | MN328898 |
| Flabellascoma cycadicola | CBS 143644 | LC312473 | LC312502 | LC312531 | LC312589 | LC312560 |
| Flabellascoma fusiforme | MFLUCC 18-1584 | N/A | MN304830 | MN274567 | N/A | MN328902 |
| Flabellascoma minimum | CBS 143645 | LC312474 | LC312503 | LC312532 | LC312590 | LC312561 |
| Flabellascoma minimum | CBS 143646 | LC312475 | LC312504 | LC312533 | LC312591 | LC312562 |
| Flabellascoma sichuanense * | CGMCC 3.20936 | ON332921 | ON332911 | ON332929 | ON383987 | ON381179 |
| Flabellascoma sichuanense * | UESTCC 22.0003 | ON332922 | ON332912 | ON332930 | ON383988 | ON381180 |
| Hysterium rhizophorae | NFCCI-4250 | MG844280 | MG844284 | MG844276 | MG968956 | N/A |
| Lentistoma bipolare | CBS 115375 | LC312477 | LC312506 | LC312535 | LC312593 | LC312564 |
| Lentistoma bipolare | KT 3056 | LC312484 | LC312513 | LC312542 | LC312600 | LC312571 |
| Leptoparies palmarum | CBS 143653 | LC312485 | LC312514 | LC312543 | LC312601 | LC312572 |
| Leucaenicola aseptata | MFLUCC 17-2423 | MK347853 | MK347746 | MK347963 | MK434891 | MK360059 |
| Leucaenicola camelliae | NTUCC 18-093-4 | MT071229 | MT112302 | MT071278 | MT743283 | MT374091 |
| Leucaenicola phraeana | MFLUCC 18-0472 | MK347892 | MK347785 | MK348003 | MK434867 | MK360060 |
| Lophiomurispora hongheensis | KUMCC 20-0216 | MW264227 | MW264218 | MW264197 | MW256810 | MW256819 |
| Lophiomurispora hongheensis | KUMCC 20-0217 | MW264225 | MW264216 | MW264195 | MW256808 | MW256817 |
| Lophiostoma macrostomum | KT 709/HHUF 27293 | AB521732 | AB433276 | AB433274 | JN993493 | LC001753 |
| Lophiostoma pseudodictyosporium | MFLUCC 13-0451 | N/A | KR025858 | KR025862 | N/A | N/A |
| Lophiostoma ravennicum | MFLUCC 14-0005 | KP698415 | KP698413 | KP698414 | N/A | N/A |
| Massarina corticola | CBS 154.93 | FJ795491 | N/A | FJ795448 | FJ795465 | N/A |
| Neoangustimassarina sichuanensis * | CGMCC 3.20937 | ON332917 | ON332907 | ON332925 | ON383983 | ON381175 |
| Neoangustimassarina sichuanensis * | UESTCC 22.0001 | ON332918 | ON332908 | ON332926 | ON383984 | ON381176 |
| Neooccultibambusa chiangraiensis | MFLUCC 12-0559 | KU712458 | KU712442 | KU764699 | N/A | KU872761 |
| Neooccultibambusa kaiyangensis | CGMCC 3.20404 | MZ964886 | MZ964868 | MZ964877 | OK061077 | OK061071 |
| Neooccultibambusa trachycarpi | CGMCC 3.20405 | MZ964888 | MZ964870 | MZ964879 | OK061079 | OK061073 |
| Neothyrostroma encephalarti | CBS 146037 | N/A | MN562104 | MN567612 | N/A | MN556830 |
| Neothyrostroma encephalarti | CPC 35999 | N/A | MN562105 | MN567613 | N/A | MN556831 |
| Neovaginatispora clematidis | MFLUCC 17–2156 | MT226676 | MT310606 | MT214559 | N/A | MT394738 |
| Neovaginatispora fuckelii | KH 161 | AB618689 | LC001731 | AB619008 | N/A | LC001749 |
| Neovaginatispora fuckelii | KT 634 | AB618690 | LC001732 | AB619009 | N/A | LC001750 |
| Occultibambusa aquatica | MFLUCC 11-0006 | KX698112 | KX698114 | KX698110 | N/A | N/A |
| Occultibambusa bambusae | MFLUCC 13-0855 | KU872116 | KU940123 | KU863112 | KU940170 | KU940193 |
| Occultibambusa chiangraiensis | MFLUCC 16-0380 | KX655551 | N/A | KX655546 | KX655566 | KX655561 |
| Occultibambusa fusispora | MFLUCC 11-0127 | N/A | KU940125 | KU863114 | KU940172 | KU940195 |
| Occultibambusa hongheensis | KUMCC 21-0020 | MZ329029 | MZ329037 | MZ329033 | N/A | MZ325467 |
| Occultibambusa jonesii | GZCC 16-0117 | KY628324 | N/A | KY628322 | KY814758 | KY814756 |
| Occultibambusa kunmingensis | HKAS 102151 | MT864342 | MT627716 | MN913733 | MT878453 | MT954407 |
| Occultibambusa maolanensis | GZCC 16-0116 | KY628325 | N/A | KY628323 | KY814759 | KY814757 |
| Occultibambusa pustula | MFLUCC 11-0502 | KU872118 | KU940126 | KU863115 | N/A | N/A |
| Occultibambusa sichuanensis * | CGMCC 3.20938 | N/A | ON332913 | ON332931 | ON383989 | ON381181 |
| Occultibambusa sichuanensis * | UESTCC 22.0004 | N/A | ON332914 | ON332932 | ON383990 | ON381182 |
| Palmiascoma gregariascomum | MFLUCC 11-0175 | KP753958 | KP744452 | KP744495 | KP998466 | N/A |
| Palmiascoma qujingense | KUMCC 19-0201 | MT477186 | MT477183 | MT477185 | MT495782 | N/A |
| Parapaucispora pseudoarmatispora | KT 2237 | LC100018 | LC100021 | LC100026 | N/A | LC100030 |
| Paucispora quadrispora | KT 843 | AB618692 | LC001734 | AB619011 | N/A | LC001755 |
| Paucispora versicolor | KH 110 | LC001721 | AB918731 | AB918732 | N/A | LC001760 |
| Podocarpomyces knysnanus | CBS 146076 | N/A | MN562155 | MN567662 | MN556816 | MN556836 |
| Polyplosphaeria fusca | KT1616 | AB524463 | AB524789 | AB524604 | N/A | N/A |
| Pseudotetraploa curviappendiculata | JCM 12852 | AB524467 | AB524792 | AB524608 | N/A | N/A |
| Pseudotetraploa javanica | JCM 12854 | AB524470 | AB524795 | AB524611 | N/A | N/A |
| Pseudotetraploa longissima | JCM 12853 | AB524471 | AB524796 | AB524612 | N/A | N/A |
| Pseudotetraploa rajmachiensis | NFCCI 4618 | N/A | MN937222 | MN937204 | N/A | N/A |
| Pseudotetraploa bambusicola * | CGMCC 3.20939 | ON332923 | ON332915 | ON332933 | ON383991 | ON381183 |
| Pseudotetraploa bambusicola * | UESTCC 22.0005 | ON332924 | ON332916 | ON332934 | ON383992 | ON381184 |
| Quadricrura bicornis | CBS 125427 | AB524472 | AB524797 | AB524613 | N/A | N/A |
| Quadricrura meridionalis | CBS 125684 | AB524473 | AB524798 | AB524614 | N/A | N/A |
| Seriascoma bambusae | KUMCC 21-0021 | MZ329031 | MZ329039 | MZ329035 | MZ325470 | MZ325468 |
| Seriascoma didymosporum | MFLUCC 11-0179 | KU872119 | KU940127 | KU863116 | KU940173 | KU940196 |
| Seriascoma yunnanense | MFLU 19-0690 | MN174694 | N/A | MN174695 | MN210324 | MN381858 |
| Shrungabeeja aquatica | MFLUCC 18-0664 | N/A | MT627722 | MT627663 | N/A | N/A |
| Shrungabeeja longiappendiculata | BCC 76463 | KT376471 | KT376474 | KT376472 | N/A | N/A |
| Shrungabeeja vadirajensis | MFLUCC 17-2362 | N/A | MT627681 | MN913685 | N/A | N/A |
| Tetraploa aquatica | MFLU 19-0995 | N/A | MT530448 | MT530452 | N/A | N/A |
| Tetraploa aristata | CBS 996.70 | AB524486 | AB524805 | AB524627 | N/A | N/A |
| Tetraploa dwibahubeeja | NFCCI 4621 | N/A | MN937226 | MN937208 | N/A | N/A |
| Tetraploa nagasakiensis | JCM 13168 | AB524489 | AB524806 | AB524630 | N/A | N/A |
| Triplosphaeria acuta | JCM 13171 | AB524492 | AB524809 | AB524633 | N/A | N/A |
| Triplosphaeria maxima | JCM 13172 | AB524496 | AB524812 | AB524637 | N/A | N/A |
| Triplosphaeria yezoensis | CBS 125436 | AB524497 | AB524813 | AB524638 | N/A | N/A |
| Vaginatispora amygdali | CBS 143662 | LC312495 | LC312524 | LC312553 | LC312611 | LC312582 |
| Vaginatispora appendiculata | MFLUCC 16-0314 | KU743219 | KU743217 | KU743218 | N/A | KU743220 |
| Vaginatispora aquatica | MFLUCC 11-0083 | KJ591575 | KJ591577 | KJ591576 | N/A | N/A |
| Versicolorisporium triseptatum | JCM 14775 | AB524501 | AB365596 | AB330081 | N/A | N/A |
| Versicolorisporium triseptatum | UESTCC 21.0016 = NMX1222 | OL741381 | OL741378 | OL741318 | N/A | N/A |
3. Results
3.1. Phylogenetic Analyses
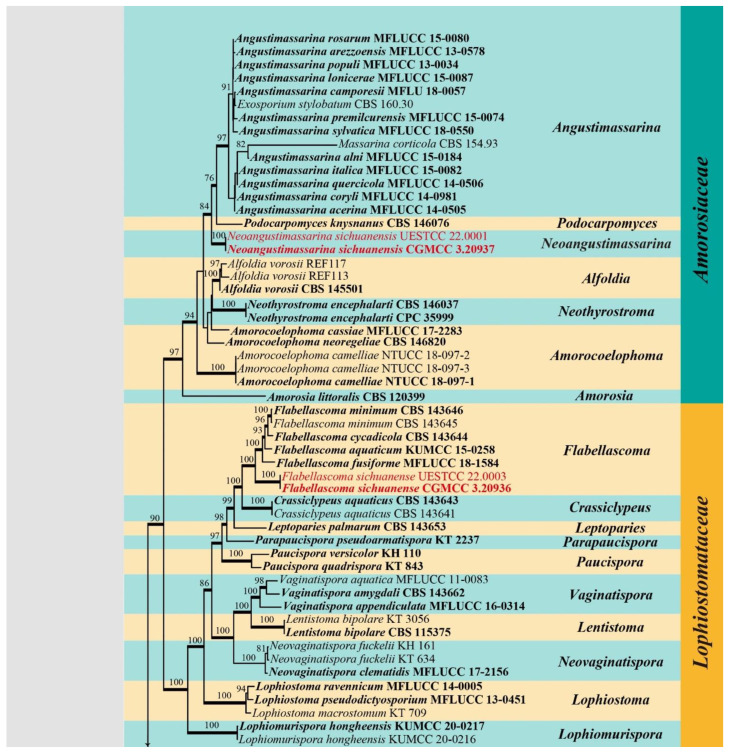
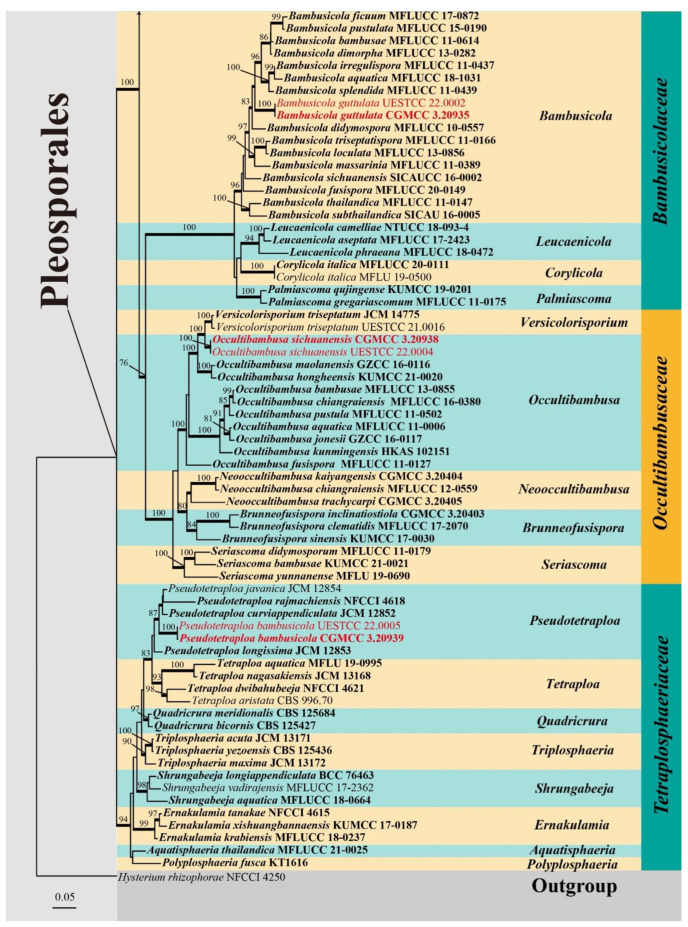
Five gene loci SSU, ITS, LSU, RPB2 and TEF1α were used to determine the phylogenetic placement of the new collections. The concatenated matrix comprised 124 taxa with a total of 4633 characters (SSU: 1021 bp; ITS: 684 bp; LSU: 904 bp; RPB2: 1031 bp; TEF1α: 993 bp) including gaps. Maximum likelihood (ML) and Bayesian inference (BI) analyses were carried out to infer phylogenetic relationships. The best scoring ML tree (Figure 1) was selected to represent the relationships among taxa, in which a final likelihood value of –51844.747390 is presented. The matrix had 2418 distinct alignment patterns. Estimated base frequencies were as follows: A = 0.245989, C = 0.250069, G = 0.270993, T = 0.232949; substitution rates AC = 1.588007, AG = 3.646881, AT = 1.331328, CG = 1.142338, CT = 7.560715, GT = 1.000000. GTR + I + G is the best-fit model selected by AIC in MrModeltest based on each gene (SSU, ITS, LSU, RPB2 and TEF1α), which was used for maximum likelihood and Bayesian analysis. Six simultaneous Markov chains were run for 1,970,000 generations and trees were sampled every 1000 generations and 1970 trees were obtained. The first 394 trees representing the burn-in phase of the analyses were discarded, while the remaining 1576 trees were used for calculating posterior probabilities in the majority rule consensus tree (critical value for the topological convergence diagnostic is 0.01).
Figure 1.
RAxML tree generated from combined SSU, ITS, LSU, RPB2 and TEF1α sequence data of targeted five families (Amorosiaceae, Bambusicolaceae, Lophiostomataceae, Occultibambusaceae, and Tetraplosphaeriaceae) in Pleosporales. Bootstrap values for ML equal to or greater than 75% are placed above the branches. Branches with Bayesian posterior probabilities (PP) from MCMC analysis equal to or greater than 0.95 are in bold. The tree was rooted with Hysterium rhizophorae (NFCCI 4250). The ex-type strains are indicated in bold and newly generated sequences are indicated in red.
The newly obtained isolates were grouped with pleosporalean families of Amorosiaceae, Bambusicolaceae, Lophiostomataceae, Occultibambusaceae and Tetraplosphaeriaceae. Two isolates of Neoangustimassarina sichuanensis (CGMCC 3.20937 and UESTCC 22.0001) formed a distinct, well-supported clade (84% ML/1.00 BYPP) in Amorosiaceae. Two isolates of Bambusicola guttulata (CGMCC 3.20935 and UESTCC 22.0002) were nested in the genus Bambusicola in Bambusicolaceae. Two isolates of Flabellascoma sichuanense (CGMCC 3.20936 and UESTCC 22.0003) clustered with Flabellascoma in Lophiostomataceae. Two strains of Occultibambusa sichuanensis (CGMCC 3.20938 and UESTCC 22.0004) formed a distinct branch within Occultibambusaceae, which was closely related to Occultibambusa hongheensis (KUMCC 21-0020), O. maolanensis (GZCC 16-0116), and Versicolorisporium triseptatum (JCM 14775 and NMX1222) with statistical support (100% ML/1.00 BYPP). The other two strains of Pseudotetraploa bambusicola (CGMCC 3.20939 and UESTCC 22.0005) grouped with Pseudotetraploa species in Tetraplosphaeriaceae.
3.2. Taxonomy
Pleosporales Luttr. ex M.E. Barr, Prodromus to class Loculoascomycetes: 67 (1987)
Amorosiaceae Thambug and K.D. Hyde, Fungal Diversity 74: 252 (2015)
Notes: Amorosiaceae was established by Thambugala et al. [38] and typified by Amorosia Mantle and D. Hawksw., which is characterized by micronematous to semi-macronematous conidiophores, integrated, terminal, or intercalary, monoblastic conidiogenous cells, elongate-clavate and 3–4-septate conidia [39]. Six genera were accepted in the family, viz. Alfoldia D.G. Knapp, Imrefi and Kovács, Amorosia Mantle and D. Hawksw., Amorocoelophoma Jayasiri, E.B.G. Jones and K.D. Hyde, Angustimassarina Thambugala, Kaz. Tanaka and K.D. Hyde, Neothyrostroma Crous and Podocarpomyces Crous [38,39,40,41,42]. Angustimassarina is the only genus in the family that represents the sexual morph [38]. Herein, we introduce the second genus with a sexual morph to Amorosiaceae.
Neoangustimassarina X.D. Yu and Jian K. Liu, gen. nov.
Type species: Neoangustimassarina sichuanensis X.D. Yu, S.N. Zhang and Jian K. Liu
MycoBank: MB 843716
Etymology: The name refers to the similarity to the genus Angustimassarina.
Saprobic on dead wood in terrestrial habitat. Sexual morph: Ascomata solitary, scattered, immersed, visible as pale brown, circular cap with a small central black dot, subglobose, uniloculate. Peridium composed of several layers of hyaline to brown cells of textura angularis. Hamathecium hyphae-like, pseudoparaphyses, embedded in a gelatinous matrix. Asci 8-spored, bitunicate, fissitunicate, broad clavate to cylindric-clavate, short pedicellate. Ascospores biseriate, fusiform with obtuse ends, hyaline, 1-septate, guttulate, smooth-walled, surrounded by a mucilaginous sheath. Asexual morph: Undetermined.
Notes: The monotypic genus was introduced to accommodate Neoangustimassarina sichuanensis, which formed a distinct clade within Amorosiaceae (Figure 1). Neoangustimassarina resembles Angustimassarina in forming globose to subglobose ascomata, hyaline, and septate ascospores surrounded by mucilaginous sheaths [38]. However, Neoangustimassarina differs from the latter in having immersed ascomata without a pore opening, broader asci (broad clavate to cylindric-clavate vs. cylindrical to cylindric-clavate), and the septa of the ascospores (1-septate vs. 1–3-septate). We, hereby, introduce the new genus based on the distinctiveness of morphology and multi-gene phylogeny.
Neoangustimassarina sichuanensis X.D. Yu, S.N. Zhang and Jian K. Liu, sp. nov., Figure 2
Figure 2.
Neoangustimassarina sichuanensis (HKAS 123092, holotype) (a–c) Ascomata on host substrate. (d–f) Vertical section of ascoma. (g–k) Asci. (l,m) Structure of peridium. (n) Pseudoparaphyses. (o–s) Ascospores. (t) Germinated ascospore. (u,v) Colonies on PDA, above (u) and below (v). Scale bars: (d–f) = 100 μm, (g–t) = 20 μm.
MycoBank: MB 843717
Etymology: The epithet refers to Sichuan Province where the fungus was collected.
Holotype: HKAS 123092
Saprobic on dead wood in terrestrial habitat. Sexual morph: Ascomata solitary, scattered, immersed, visible as circular, pale brown to nearly white flat cap, with a small black dot in the center, in vertical section 135–235 μm high, 190–260 μm diam., subglobose, uniloculate, ostiolate. Peridium 10–23 μm wide, composed of several layers of hyaline to brown cells of textura angularis. Hamathecium 1.9–2.9 µm wide, hyphae-like, pseudoparaphyses, embedded in a gelatinous matrix. Asci 78–125 × 20–30 µm ( = 92 × 25 µm, n = 30), 8-spored, bitunicate, fissitunicate, broad clavate to cylindric-clavate, straight or slightly curved, short pedicellate to subsessile, rounded at the apex, with an ocular chamber. Ascospores 23–35 × 6.5–10.5 µm ( = 30 × 9 µm, n = 30), overlapping biseriate, fusiform with obtuse ends, hyaline, 1-septate, constricted at the septum, the upper cell slightly wider than the lower cell, guttulate when young, smooth-walled, surrounded by a wide mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics: Colonies on PDA reaching 50–60 mm after 7 weeks at 25 °C, circular, dry, the mycelium sparse at the margin, greyish-brown, reverse dark brown.
Material examined: CHINA, Sichuan Province, Chengdu City, Pengzhou County, Huilonggou Scenic Area, 31°14′21″ N, 103°47′28″ E, 1135 m Elevation, on dead wood, 28 July 2021, X.D. Yu, HLG3 (HKAS 123092, holotype); ex-holotype living culture CGMCC 3.20937; ibid., HUEST 22.0001, isotype, ex-isotype living culture UESTCC 22.0001.
Bambusicolaceae D.Q. Dai and K.D. Hyde, Fungal Diversity 63: 49 (2013)
Notes: Bambusicolaceae was established by Hyde et al. [13] to accommodate Bambusicola, D.Q. Dai and K.D. Hyde [43]. Four genera were accepted in this family, viz. Bambusicola [43], Corylicola [44], Leucaenicola [41] and Palmiascoma [45]. Most Bambusicola species are parasites or saprobes and have been found on Bamboos (Poaceae) in terrestrial habitats [43,46,47,48,49,50], except B. aquatica (from a freshwater habitat) and B. ficuum (on Ficus sp., Moraceae) [51,52]. In this study, a coelomycetous Bambusicola species is introduced.
Bambusicola D.Q. Dai and K.D. Hyde, Cryptogamie Mycologie 33: 367 (2012)
Bambusicola guttulata X.D. Yu, S.N. Zhang and Jian K. Liu, sp. nov., Figure 3.
Figure 3.
Bambusicola guttulata (HKAS 123091, holotype) (a–c) Conidiomata on surface of dead bamboo culms. (d,e) Vertical section of conidioma. (f) Wall of conidioma. (g–j) Conidiogenous cells bearing conidia (the arrows indicated how the conidiogenous cells produce conidia). (k–o) Conidia. (p) Germinating conidia. (q,r) Colonies on PDA, above and below. Scale bars: (d,e) = 50 μm, (f,g,p) = 20 μm, (h–o) = 10 μm.
MycoBank: MB 843718
Etymology: Referring to the conidia with large guttules.
Holotype: HKAS 123091
Saprobic on dead branches of bamboo. Sexual morph: Undetermined. Asexual morph: Conidiomata 100–170 μm high, 130–250 μm diam., dark brown to black, pycnidial, usually forming in a linear series on the host surface, solitary, closed when young, becoming erumpent, stromatic, irregular subglobose in section, immersed or semi-immersed, unilocular, thick-walled. Conidiomatal wall 25–55 μm wide, composed of thick-walled, subhyaline to brown cells of textura angularis. Conidiophores hyaline, cylindrical, branched, straight or slightly flexuous, septate, and occasionally reduced to conidiogenous cells. Conidiogenous cells 6–16 × 3–5 μm, holoblastic, hyaline, cylindrical to subcylindrical, determinate, terminal, smooth-walled. Conidia 14–21 × 4–6 μm ( = 17 × 5 μm; n = 30), hyaline to pale brown, unicellular when young, becoming 1-septate at maturity, cylindrical to subcylindrical, sometimes with a narrow and truncate base, straight or slightly curved, smooth-walled, guttulate.
Culture characteristics: Colonies on PDA reaching 30–40 mm after 7 weeks at 25 °C, irregular, raised to umbonate, surface rough, dense, edge undulate, greyish-yellow, dry, reverse dark brown.
Material examined: CHINA, Sichuan Province, Chengdu City, Tianfu New Area, Dalin Village, 30°16′43″ N, 104°6′44″ E, 500 m Elevation, on dead branches of bamboo, 24 July 2021, X.D. Yu, B2 (HKAS 123091, holotype); ex-holotype living culture CGMCC 3.20935; ibid., HUEST 22.0002, isotype, ex-isotype living culture UESTCC 22.0002.
Notes: Two isolates of Bambusicola guttulata formed a distinct lineage in Bambusicola (Figure 1). Morphologically, B. guttulata is most similar to the asexual morph of the generic type B. massarinia compared to the anamorphic species in the genus Bambusicola [43]. However, B. guttulata has broader conidia than that of B. massarinia (14–21 × 4–6 μm vs. 14–20 × 2–3 μm). The establishment of the new species B. guttulata is justified by morphological and phylogenetic evidence.
Lophiostomataceae Sacc., Sylloge Fungorum 2: 672 (1883)
Notes: Nitschke [53] introduced “Lophiostomeae” based on the type species of Lophiostoma macrostomum (Tode) Ces. and De Not. Saccardo formally established the family Lophiostomataceae and placed “Lophiostomeae” in the order Pleosporales [54]. Members of this family have crest-like ostioles in most cases and easily to be recognized. They are characterized by immersed to erumpent ascomata, mostly clavate asci, hyaline to dark brown ascospores with appendages or mucilaginous sheaths [38,55]. With the continuous increase of new members, the family currently comprises 30 genera [16]. A new species added to the genus Flabellascoma is identified and described.
Flabellascoma A. Hashim., K. Hiray. and Kaz. Tanaka, Studies in Mycology 90: 167 (2018)
Flabellascoma sichuanense X.D. Yu, S.N. Zhang and Jian K. Liu, sp. nov., Figure 4.
Figure 4.
Flabellascoma sichuanense (HKAS 123094, holotype) (a–c) Ascomata on host substrate. (d) Vertical section of ascoma. (e) Ostiole, showing periphyses. (f) Structure of peridium. (g) Pseudoparaphyses. (h–k) Asci. (l–o) Ascospores. (p) Germinated ascospore. (q,r) Colonies on PDA, above and below. Scale bars: (d,e) = 50 μm, (f,p) = 20 μm, (g–o) = 10 μm.
MycoBank: MB 843719
Etymology: The epithet refers to Sichuan Province where the fungus was collected.
Holotype: HKAS 123094
Saprobic on dead branches of Eriobotrya sp. (Rosaceae). Sexual morph: Ascomata solitary, scattered, rarely clustered, immersed to erumpent, visible as black, crest-like ostiolar neck on the substrate, in vertical section 150–350 μm high, 190–280 μm diam., subglobose, uniloculate. Ostiole central, laterally compressed, periphysate. Peridium 25–35 μm wide, composed of several layers of brown, thick-walled cells of textura angularis. Hamathecium 1.5–3.5 µm wide, hyphae-like, pseudoparaphyses, embedded in a gelatinous matrix. Asci 55–75 × 9.5–13 µm ( = 64 × 11 µm, n = 30), 8-spored, bitunicate, fissitunicate, cylindric-clavate, straight or slightly curved, shortly pedicellate, rounded at the apex, with an ocular chamber. Ascospores 15–18 × 5–7 µm ( = 16.5 × 5.5 µm, n = 30), overlapping biseriate, fusiform, hyaline, 1-septate, constricted at the septum, the upper cell slightly wider than the lower cell, guttulate, smooth-walled, with a narrow bipolar sheath. Sheath 3.0–7.0 μm long, 1.5–2.5 μm wide, drawn-out at both ends, with an internal chamber at both ends of ascospores (Figure 4l). Asexual morph: Undetermined.
Culture characteristics: Colonies on PDA reaching 40–50 mm after 7 weeks at 25 °C, circular, with dense mycelium on the surface, dark grayish of the inner ring, and brown of the outer ring; in reverse black of the inner ring, and brown of the outer ring.
Material examined: CHINA, Sichuan Province, Chengdu City, Tianfu New Area, Dalin Village, 30°16′43″ N, 104°6′44″ E, 500 m Elevation, on dead branches of Eriobotrya sp. (Rosaceae), 24 July 2021, X.D. Yu, L4 (HKAS 123094, holotype); ex-holotype living culture CGMCC 3.20936; ibid., HUEST 22.0003, isotype, ex-isotype living culture UESTCC 22.0003.
Notes: The phylogenetic result based on SSU, ITS, LSU, RPB2 and TEF1α sequence data showed that the new collection Flabellascoma sichuanense nested in Flabellascoma (Figure 1) and formed a distinct lineage. Morphologically, it fits well with the genus Flabellascoma in having immersed ascomata, bitunicate, fissitunicate, cylindrical, clavate asci and fusiform, hyaline, 1-septate ascospores with a narrow bipolar sheath [56]. However, the dimensions of asci and ascospores distinguish F. sichuanense from other species (Table 3).
Table 3.
Morphological comparative data of Flabellascoma species.
| Taxa | Ascomata (μm) | Hamathecium (μm) | Asci (μm) | Ascospores (μm) | Sheath (μm) | References |
|---|---|---|---|---|---|---|
| F. aquaticum | 280–440 × 260–390 | 1.2–2.0 | 48.0–72.0 × 8–9 | 16–18 × 4.3–5.3 | 4.7–7.0 μm wide | [57] |
| F. cycadicola | 490–530 × 600–620 | 1.0–3.0 | 67.5–88.0 × 9–12 | 17–23 × 4.5–7.0 | 7.0–10 μm long | [56] |
| F. fusiforme | 310–420 × 320–380 | 1.5–3.0 | 66.0–80.0 × 10–12 | 15–18 × 4.0–5.0 | 5.4–8.0 μm wide | [57] |
| F. minimum T | 250–320 × 350–500 | 1.5–3.0 | 45.0–77.5 × 7.5–12 | 12–17.5 × 3.5–5 | 5.5–8.0 μm long | [56] |
| F. sichuanense | 150–350 × 190–280 | 1.5–3.5 | 55.0–75.0 × 9.5–13 | 15–18 × 5.0–7.0 | 3.0–7.0 μm long, 1.5–2.5 μm wide | This study |
Occultibambusaceae D.Q. Dai and K.D. Hyde, Fungal Diversity 82: 25 (2017)
Notes: Species of Occultibambusaceae are mostly saprobic and frequently found on monocotyledons or hardwood trees in terrestrial and aquatic habitats [47,58]. Dai et al. [47] established this family to accommodate Neooccultibambusa, Occultibambusa, Seriascoma and Versicolorisporium. Brunneofusispora, typified by Brunneofusispora sinensis, was subsequently introduced to this family by Phookamsak et al. [59]. Phylogenetically, the coelomycetous genus Versicolorisporium appeared to be a close relationship with Occultibambusa in previous studies [21,52,60,61]. However, they continue to be treated as two distinct genera because the known asexual morph of Occultibambusa is different from Versicolorisporium [60]. In this study, a new Occultibambusa species is introduced.
Occultibambusa D.Q. Dai and K.D. Hyde, Fungal Diversity 82: 25 (2017)
Occultibambusa sichuanensis X.D. Yu, S.N. Zhang and Jian K. Liu, sp. nov., Figure 5.
Figure 5.
Occultibambusa sichuanensis (HKAS 123093, holotype) (a–c) Ascomata on host substrate. (d,e) Vertical section of ascoma. (f–j) Asci. (k) Structure of peridium. (l) Pseudoparaphyses. (s) Ascospores. (t) Germinated ascospore. Colonies on PDA, above and below. Scale bars: (d,e) = 50 μm, (f–k,m–t) = 20 μm, (l) = 10 μm.
MycoBank: MB 843720
Etymology: The epithet refers to Sichuan Province where the fungus was collected.
Holotype: HKAS 123093
Saprobic on dead branches of Bamboo. Sexual morph: Ascomata solitary to gregarious, semi-immersed, visible as black domes on the substrate, in vertical section 130–180 μm high, 340–440 μm diam., subglobose, coriaceous, uniloculate. Peridium 25–90 μm wide, composed of several layers of brown, thick-walled cells of textura angularis. Hamathecium 2.8–3.3 µm wide, hyphae-like, cellular pseudoparaphyses, embedded in a gelatinous matrix. Asci 70–100 × 22–27 µm ( = 85 × 24 µm, n = 30), 8-spored, bitunicate, fissitunicate, obovoid to pyriform, straight or slightly curved, shortly pedicellate, rounded at the apex, with an ocular chamber. Ascospores 31–41 × 6.5–10 µm ( = 36 × 8 µm, n = 30), 3-seriate, fusiform, straight to somewhat curved, brown, 1-septate, constricted at the septum, guttulate, smooth-walled, surrounded by a mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics: Colonies on PDA reaching 40–50 mm after 7 weeks at 25 °C, circular, with sparse mycelium on the surface, light gray of the inner ring, and brown of the outer ring; in reverse olive green.
Material examined: CHINA, Sichuan Province, Chengdu City, Pengzhou County, Huilonggou Scenic Area, 31°14′21″ N, 103°47′28″ E, 1135 m Elevation, on dead branches of bamboo in a terrestrial environment, 28 July 2021, X.D. Yu, HLG8 (HKAS 123093, holotype); ex-holotype living culture CGMCC 3.20938; ibid., HUEST 22.0004, isotype, ex-isotype living culture UESTCC 22.0004.
Notes: The blast search based on LSU sequence data of our new collection showed that the closest hits were Versicolorisporium triseptatum (HHUF 28815 = JCM14775, identity 99.18%; NMX1222, identity 99.03%), and Occultibambusa bambusae (MFLUCC 11-0394, identity 98.01%); the closest hits based on ITS sequence were Versicolorisporium triseptatum (JCM 14775, identity 93.95%; NMX1222, identity 93.72%), and Occultibambusa hongheensis (KUMCC 21-0020, identity 90.95%); the closest hits based on TEF1α sequence were Occultibambusa hongheensis (KUMCC 21-0020, identity 97.02%), and O. maolanensis (KUMCC 21-0020, identity 96.91%). Multi-gene phylogeny showed that the new collection grouped with Occultibambusa and Versicolorisporium. It formed a sister clade with V. triseptatum with high statistical support (100% ML/1.00 BYPP, Figure 1). However, the morphology of our collection fits well with Occultibambusa. Further morphological evidence of its association with Versicolorisporium is somewhat difficult due to the lack of asexual morph in our collection. Therefore, we recognize our new collection as a new species of Occultibambusa, namely, O. sichuanensis. The morphological comparison of Occultibambusa species was listed in Table 4.
Table 4.
Morphological comparative data of Occultibambusa.
| Taxa | Ascomata (μm) | Asci | Ascospores | References | ||
|---|---|---|---|---|---|---|
| Morphology | Size (μm) | Morphology | Size (μm) | |||
| O. aquatica | 100–250 × 180–280 | Clavate | 73–86 × 9–13 | Narrowly fusiform, 1-septate, brownish, with a sheath | 19–25 × 3.5–6.5 | [62] |
| O. bambusae T | 150–200 × 400–550 | Broadly cylindrical | (50–)60–80(−90) × (9.5–)11.5–14.5(−15) | Slightly broad and fusiform, 1-septate, dark brown, with a sheath | (22–)23.5–27.5× 4.5–7 | [47] |
| O. chiangraiensis | 195–295 × 352–520 | Clavate-oblong | 47–92 × 12–16 | Fusiform, (1–)3-septate, hyaline when immature, pale brown to red-brown at maturity, without a sheath | 16–24 × 5–7 | [62] |
| O. fusispora | 135–185 × 240–275 | Clavate to cylindric-clavate | (60–)65–90(−110) × (11–)12–14(−15)(−16) | Fusiform, mostly 1-septate, rarely 2–3-septate, light brown, without a sheath | (20–)22–25(−26) × 5–6(−6.5) | [47] |
| O. hongheensis | 180–340 × 400–550 | Cylindric-clavate to clavate | (78–)80–130(–137) × (18–)19–23(–25) | Fusiform, 1-septate, hyaline when young and becoming pale brown when mature, with a sheath | (25–)27–30 × (5.5–)8–9(–10) | [60] |
| O. jonesii | 196–236 × 200–260 | Broadly cylindrical to clavate | (65–)75–89(–105) × 13.5–19 | Inequilateral-fusiform, 2-celled, hyaline when young and becoming brown to grayish when mature, without a sheath | 27–33.5 × 5.5–6.5 | [63] |
| O. kunmingensis | 110–150 × 220–260 | Clavate or cylindric-clavate | 110–140(–160) × 13–16.5 | Fusiform, 1-septate, brown, without a sheath | 32–40 × 5–6.5 | [52] |
| O. maolanensis | 544–600 diameter | Broadly cylindrical to clavate | (66–)77–85(–94) × 17–20(–24) | Inequality-fusiform, 2-celled, hyaline when young and become light brown when mature, without a sheath | 25–31 × 8–10 | [63] |
| O. pustula | 150–200 × 200–300 | Cylindrical | 80–105 × 8–12 | Slightly broad-fusiform, 1-septate, hyaline to pale brown, with a sheath | 22–25 × 5–5.5 | [47] |
| O. sichuanensis | 130–180 × 340–440 | Obovoid to pyriform | 70–100 × 22–27 | Fusiform, 1-septate, brown, with a sheath | 31–41 × 6.5–10 | This study |
Tetraplosphaeriaceae Kaz. Tanaka and K. Hiray, Studies in Mycology 64: 177 (2009)
Notes: Tetraplosphaeriaceae was introduced by Tanaka et al. [64], and typified by Tetraplosphaeria. The latest taxonomic treatment of the family contains nine genera [1]. Pseudotetraploa is a genus with only known asexual forms, which were commonly associated with Poaceae (Dendrocalamus stocksii, Pleioblastus chino, Pleioblastus chino, Sasa kurilensis) distributed in Japan or India [64,65]. In this study, a new Pseudotetraploa species associated with bamboos from China is introduced.
Pseudotetraploa Kaz. Tanaka and K. Hiray, Studies in Mycology 64: 193 (2009)
Pseudotetraploa bambusicola X.D. Yu, S.N. Zhang and Jian K. Liu, sp. nov., Figure 6.
Figure 6.
Pseudotetraploa bambusicola (HKAS 123095, holotype) (a–c) Colonies on natural substratum. (d–l) Conidia. (m) Germinating conidium. (n) Colony on PDA from above and below. Scale bars: (d–m) = 20 μm.
MycoBank: MB 843721
Etymology: Refers to the bamboo host.
Holotype: HKAS 123095
Saprobic on dead branches of Bamboo. Sexual morph: Undetermined. Asexual morph: Colonies effuse, black. Mycelium superficial. Conidiophores absent. Conidiogenous cells monoblastic, integrated, usually indistinguishable from superficial hyphae. Conidia 23–41 × 14–24 µm ( = 32 × 19 µm, n = 50), solitary, amygdaliform to ovoid, or obovoid, dark brown to black, pseudoseptate, consisting of 3–4 columns, with 0–4 setose appendages. Appendages 8.85–95 × 2.5–4.5 µm ( = 36 × 3.5 µm, n = 50), 0–4-septate, dark brown, smooth, unbranched, straight or curved.
Culture characteristics: Colonies on PDA reaching 40–50 mm after 7 weeks at 25 °C, circular, dry, with dense mycelium, raised, entire at the edge, grayish brown, reverse dark brown.
Material examined: CHINA, Sichuan Province, Chengdu City, Longquanyi District, Longquan Mountain Scenic Area, 30°32′47″ N, 104°19′11″ E, 800 m Elevation, on dead branches of bamboo in a terrestrial environment, 13 August 2021, X.D. Yu, THGL14 (HKAS 123095, holotype); ex-holotype living culture CGMCC 3.20939; ibid., HUEST 22.0005, isotype, ex-isotype living culture UESTCC 22.0005.
Notes: The phylogenetic result (Figure 1) showed that Pseudotetraploa bambusicola formed a distinct lineage within Pseudotetraploa [64]. Morphologically, Pseudotetraploa bambusicola resembles P. curviappendiculata, P. javanica, P. longissimi and P. rajmachiensis in having monoblastic conidiogenous cells. However, they can be distinguished by the shape of conidia (obclavate to narrowly obpyriform in P. curviappendiculata and P. longissimi; ovoid in P. javanica, ovoid to obclavate or obpyriform in P. rajmachiensis; amygdaliform to ovoid, or obovoid in P. bambusicola) [64].
4. Discussion
The genus Flabellascoma was introduced by Hashimoto et al. [56] to accommodate two terrestrial species F. cycadicola A. Hashim., K. Hiray and Kaz. Tanaka and F. minimum. Subsequently, two species F. aquaticum D.F. Bao, Z.L. Luo, K.D. Hyde and H.Y. Su and F. fusiforme D.F. Bao, Z.L. Luo, K.D. Hyde and H.Y. Su from freshwater habitats were introduced based on multi-gene phylogeny [57]. Members of Flabellascoma have similar morphological features [56,57] and it is difficult to distinguish Flabellascoma species by the size and shape of asci and ascospores [57]. Bao et al. [57] proposed that the ascomatal features appear to be remarkable features to distinguish taxa in this genus. Molecular data were found to be more supportive for the identification of the new species in this study, and we believed that DNA data provided more objective evidence for the species distinction of Flabellascoma.
In previous studies, the relationship between Occultibambusa and Versicolorisporium has not been well resolved due to the asexual morphs of Occultibambusa and Versicolorisporium being inconsistent [60]. In our phylogenetic tree, however, the genus Occultibambusa is not monophyletic (Figure 1), of which O. fusispora formed an independent lineage in Occultibambusaceae; this is consistent with recent relevant studies [60,61]. Occultibambusa fusispora is the only species in the genus reported with its holomorph [47]; we cannot solve the problem between Occultibambusa and Versicolorisporium due to the type-of-species issue, although O. fusispora has an asexual morph. Therefore, further studies are needed to provide sexual and asexual links of the type of species of O. bambusae and V. triseptatum towards the classification of Occultibambusa and Versicolorisporium with more sampling and taxa population included in the analysis.
During the investigation of microfungi in Sichuan Province, we randomly sampled three times in the vicinity of Chengdu city from July to August 2021. Morphological and phylogenetic results showed that these newly collected interesting taxa were distributed in five different pleosporalean families. It is worth noting that three new species found on bamboo are typical bambusicolous fungi. Bamboo is a gramineous plant with economic and ornamental value, and its culms and leaves are abundant in saprobic fungi [44,66,67,68]. China has the richest bamboo resources, with a total of 861 species distributes in 43 genera [69]. Among them, Sichuan has a large area of bamboo forests, with an area of 592,800 ha, ranking fifth in the country after Fujian, Jiangxi, Zhejiang and Hunan [69]. In recent years, an increasing number of new species of bambusicolous fungi have been reported and discovered in China [60,67,70,71]. Therefore, the unique natural conditions in Sichuan are of great potential for the excavation and identification of bamboo fungi.
Acknowledgments
Sajeewa Maharachchikumbura is thanked for his helps with sample collection.
Author Contributions
Conceptualization, X.-D.Y., S.-N.Z. and J.-K.L.; methodology, X.-D.Y.; formal analysis, X.-D.Y.; resources, X.-D.Y.; data curation, X.-D.Y. and S.-N.Z.; writing—original draft preparation, X.-D.Y.; writing—review and editing, X.-D.Y., S.-N.Z. and J.-K.L.; supervision, J.-K.L.; project administration, J.-K.L.; funding acquisition, J.-K.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences data were submitted to GenBank.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study is supported by the Joint Fund of the National Natural Science Foundation of China and the Karst Science Research Center of Guizhou province (Grant No. U1812401).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kirk P., Cannon P., Minter D., Stalpers J. Ainsworth and Bisby’s Dictionary of the Fungi. 10th ed. CABI Publishing; Wallingford, UK: 2008. [Google Scholar]
- 2.Mapook A., Boonmee S., Liu J.K., Jones E.B.G., Bahkali A.H., Hyde K.D. Taxonomic and phylogenetic placement of Phaeodimeriella (Pseudoperisporiaceae, Pleosporales) Cryptogam. Mycol. 2016;37:157–176. doi: 10.7872/crym/v37.iss2.2016.157. [DOI] [Google Scholar]
- 3.Ferdinandez H.S., Manamgoda D.S., Udayanga D., Deshappriya N., Munasinghe M.S., Castlebury L.A. Molecular phylogeny and morphology reveal three novel species of Curvularia (Pleosporales, Pleosporaceae) associated with cereal crops and weedy grass hosts. Mycol. Prog. 2021;20:431–451. doi: 10.1007/s11557-021-01681-0. [DOI] [Google Scholar]
- 4.Matsumura M., Kato W., Hashimoto A., Takahashi Y.S., Shirouzu T., Tanaka K. Crassiperidium (Pleosporales, Dothideomycetes), a new ascomycetous genus parasitic on Fagus crenata in Japan. Mycosphere. 2018;9:1256–1267. doi: 10.5943/mycosphere/9/6/13. [DOI] [Google Scholar]
- 5.Liu Z.P., Zhang S.N., Cheewangkoon R., Zhao Q., Liu J.K. Crassoascoma gen. nov. (Lentitheciaceae, Pleosporales): Unrevealing Microfungi from the Qinghai-Tibet Plateau in China. Diversity. 2022;14:15. doi: 10.3390/d14010015. [DOI] [Google Scholar]
- 6.Li W.L., Ba D.F., Liu N.G., Hyde K.D., Liu J.K. Aquatisphaeria thailandica gen. et sp. nov. (Tetraplosphaeriaceae, Pleosporales) from freshwater habitat in Thailand. Phytotaxa. 2021;513:118–128. doi: 10.11646/phytotaxa.513.2.3. [DOI] [Google Scholar]
- 7.Jones E.B.G., Devadatha B., Abdel-Wahab M.A., Dayarathne M.C., Zhang S.N., Hyde K.D., Liu J.K., Bahkali A.H., Sarma V.V., Tibell S., et al. Phylogeny of new marine Dothideomycetes and Sordariomycetes from mangroves and deep-sea sediments. Bot. Mar. 2020;63:155–181. doi: 10.1515/bot-2019-0014. [DOI] [Google Scholar]
- 8.Rupcic Z., Chepkirui C., Hernández-Restrepo M., Crous P.W., Luangsa-Ard J.J., Stadler M. New nematicidal and antimicrobial secondary metabolites from a new species in the new genus, Pseudobambusicola thailandica. MycoKeys. 2018;33:1–23. doi: 10.3897/mycokeys.33.23341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luttrell E.S. The Ascostromatic Ascomycetes. Mycologia. 1955;47:511–532. doi: 10.1080/00275514.1955.12024473. [DOI] [Google Scholar]
- 10.Barr M.E. Prodromus to Class Loculoascomycetes. University of Massachusetts; Amherst, MA, USA: 1987. [Google Scholar]
- 11.Barr M.E. New taxa and combinations in the Loculoascomycetes. Mycotaxon. 1987;29:501–505. [Google Scholar]
- 12.Zhang Y., Crous P.W., Schoch C.L., Hyde K.D. Pleosporales. Fungal Divers. 2012;53:1–221. doi: 10.1007/s13225-011-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyde K.D., Jones E.B.G., Liu J.K., Ariyawansa H., Boehm E., Boonmee S., Braun U., Chomnunti P., Crous P.W., Dai D.Q., et al. Families of Dothideomycetes. Fungal Divers. 2013;63:1–313. doi: 10.1007/s13225-013-0263-4. [DOI] [Google Scholar]
- 14.Sun Y.R., Liu N.G., Hyde K.D., Jayawardena R.S., Wang Y. Pleocatenata chiangraiensis gen. et. sp. nov. (Pleosporales, Dothideomycetes) from medicinal plants in northern Thailand. MycoKeys. 2022;87:77–98. doi: 10.3897/mycokeys.87.79433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hongsanan S., Hyde K.D., Phookamsak R., Wanasinghe D.N., McKenzie E.H.C., Sarma V.V., Boonmee S., Luecking R., Bhat D.J., Liu N.G., et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere. 2020;11:1553–2107. doi: 10.5943/mycosphere/11/1/13. [DOI] [Google Scholar]
- 16.Wijayawardene N., Hyde K., Dai D.Q., Sánchez G., Tomio Goto B., Saxena R., Erdoğdu M., Selcuk F., Rajeshkumar K.C., Aptroot A., et al. Outline of Fungi and fungus-like taxa—2021. Mycosphere. 2022;13:53–453. doi: 10.5943/mycosphere/13/1/2. [DOI] [Google Scholar]
- 17.Antonelli A., Smith R., Fry C., Simmonds M.S., Kersey P.J., Pritchard H., Abbo M., Acedo C., Adams J., Ainsworth A. Ph.D. Thesis. Sfumato Foundation; Richmond, UK: 2020. State of the World’s Plants and Fungi. Royal Botanic Gardens (Kew) [Google Scholar]
- 18.Wanasinghe D.N., Mortimer P.E., Xu J. Insight into the Systematics of Microfungi Colonizing Dead Woody Twigs of Dodonaea viscosa in Honghe (China) J. Fungi. 2021;7:180. doi: 10.3390/jof7030180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheek M., Nic Lughadha E., Kirk P., Lindon H., Carretero J., Looney B., Douglas B., Haelewaters D., Gaya E., Llewellyn T. New scientific discoveries: Plants and fungi. Plants People Planet. 2020;2:371–388. doi: 10.1002/ppp3.10148. [DOI] [Google Scholar]
- 20.Wang S., Li H., Niu S. Empirical Research on Climate Warming Risks for Forest Fires: A Case Study of Grade I Forest Fire Danger Zone, Sichuan Province, China. Sustainability. 2021;13:7773. doi: 10.3390/su13147773. [DOI] [Google Scholar]
- 21.Wanasinghe D.N., Wijayawardene N.N., Xu J.C., Cheewangkoon R., Mortimer P.E. Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China) PLoS ONE. 2020;15:e0235855. doi: 10.1371/journal.pone.0235855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Pujol J., Zhang F.M., Ge S. Plant biodiversity in China: Richly varied, endangered, and in need of conservation. Biodivers. Conserv. 2006;15:3983–4026. doi: 10.1007/s10531-005-3015-2. [DOI] [Google Scholar]
- 23.Senanayake I., Rathnayake A., Marasinghe D., Calabon M., Gentekaki E., Lee H. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere. 2020;11:2678–2754. doi: 10.5943/mycosphere/11/1/20. [DOI] [Google Scholar]
- 24.Liu J.K., Chomnunti P., Cai L., Phookamsak R., Chukeatirote E., Jones E.B.G., Moslem M., Hyde K.D. Phylogeny and morphology of Neodeightonia palmicola sp. nov. from palms. Sydowia. 2010;62:261–276. [Google Scholar]
- 25.Crous P.W., Gams W., Stalpers J.A., Robert V., Stegehuis G. MycoBank: An online initiative to launch mycology into the 21st century. Stud. Mycol. 2004;50:19–22. [Google Scholar]
- 26.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990;18:315–322. doi: 10.1016/b978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- 28.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 29.Rehner S.A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 30.Kazutaka K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall T.A. Nucleic Acids Symposium Series. Volume 41. Information Retrieval Ltd.; London, UK: 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; pp. 95–98. [Google Scholar]
- 32.Dissanayake A., Bhunjun C., Maharachchikumbura S., Liu J. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere. 2020;11:2652–2676. doi: 10.5943/mycosphere/11/1/18. [DOI] [Google Scholar]
- 33.Silvestro D., Michalak I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2011;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 34.Swofford D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sinauer Associates; Sunderland, UK: 2003. [Google Scholar]
- 35.Nylander J. MrModeltest v2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
- 36.Rannala B., Yang Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 37.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 38.Thambugala K.M., Hyde K.D., Tanaka K., Tian Q., Wanasinghe D.N., Ariyawansa H.A., Jayasiri S.C., Boonmee S., Camporesi E., Hashimoto A., et al. Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers. 2015;74:199–266. doi: 10.1007/s13225-015-0348-3. [DOI] [Google Scholar]
- 39.Mantle P.G., Hawksworth D.L., Pazoutova S., Collinson L.M., Rassing B.R. Amorosia littoralis gen. sp. nov., a new genus and species name for the scorpinone and caffeine-producing hyphomycete from the littoral zone in The Bahamas. Mycol. Res. 2006;110:1371–1378. doi: 10.1016/j.mycres.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 40.Crous P.W., Carnegie A.J., Wingfield M.J., Sharma R., Mughini G., Noordeloos M.E., Santini A., Shouche Y.S., Bezerra J.D.P., Dima B., et al. Fungal Planet description sheets: 868–950. Persoonia. 2019;42:291–473. doi: 10.3767/persoonia.2019.42.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jayasiri S.C., Hyde K.D., Jones E.B.G., McKenzie E.H.C., Jeewon R., Phillips A.J.L., Bhat D.J., Wanasinghe D.N., Liu J.K., Lu Y.Z., et al. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere. 2019;10:1–186. doi: 10.5943/mycosphere/10/1/1. [DOI] [Google Scholar]
- 42.Crous P.W., Wingfield M.J., Lombard L., Roets F., Swart W.J., Alvarado P., Carnegie A.J., Moreno G., Luangsa-ard J., Thangavel R., et al. Fungal Planet description sheets: 951–1041. Persoonia. 2019;43:223–425. doi: 10.3767/persoonia.2019.43.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai D.Q., Bhat D.J., Liu J.K., Zhao R.L., Hyde K.D. Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogam. Mycol. 2012;33:363–379. doi: 10.7872/crym.v33.iss3.2012.363. [DOI] [Google Scholar]
- 44.Wijesinghe S.N., Wang Y., Camporesi E., Wanasinghe D.N., Boonmee S., Hyde K.D. A new genus of Bambusicolaceae (Pleosporales) on Corylus avellana (Fagales) from Italy. Biodivers. Data J. 2020;8:e55957. doi: 10.3897/BDJ.8.e55957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J.K., Hyde K.D., Jones E.B.G., Ariyawansa H.A., Bhat D.J., Boonmee S., Maharachchikumbura S.S.N., Mckenzie E.H.C., Phookamsak R., Phukhamsakda C. Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- 46.Dai D.Q., Bahkali A.H., Li W.J., Bhat D.J., Zhao R.L., Hyde K.D. Bambusicola loculata sp. nov.(Bambusicolaceae) from bamboo. Phytotaxa. 2015;213:122–130. doi: 10.11646/phytotaxa.213.2.5. [DOI] [Google Scholar]
- 47.Dai D.Q., Phookamsak R., Wijayawardene N.N., Li W.J., Bhat D.J., Xu J.C., Taylor J.E., Hyde K.D., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2017;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 48.Thambugala K.M., Wanasinghe D.N., Phillips A.J.L., Camporesi E., Bulgakov T.S., Phukhamsakda C., Ariyawansa H.A., Goonasekara I.D., Phookamsak R., Dissanayake A., et al. Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere. 2017;8:697–796. doi: 10.5943/mycosphere/8/4/13. [DOI] [Google Scholar]
- 49.Yang C.L., Xu X.L., Liu Y.G. Two new species of Bambusicola (Bambusicolaceae, Pleosporales) on Phyllostachys heteroclada from Sichuan, China. Nova Hedwig. 2019;108:527–545. doi: 10.1127/nova_hedwigia/2019/0526. [DOI] [Google Scholar]
- 50.Monkai J., Wanasinghe D.N., Jeewon R., Promputtha I., Phookamsak R. Morphological and phylogenetic characterization of fungi within Bambusicolaceae: Introducing two new species from the Greater Mekong Subregion. Mycol. Prog. 2021;20:721–732. doi: 10.1007/s11557-021-01694-9. [DOI] [Google Scholar]
- 51.Brahmanage R.S., Dayarathne M.C., Wanasinghe D.N., Thambugala K.M., Jeewon R., Chethana K.W.T., Samarakoon M.C., Tennakoon D.S., De Silva N.I., Camporesi E., et al. Taxonomic novelties of saprobic Pleosporales from selected dicotyledons and grasses. Mycosphere. 2020;11:2481–2541. doi: 10.5943/mycosphere/11/1/15. [DOI] [Google Scholar]
- 52.Dong W., Wang B., Hyde K.D., McKenzie E.H.C., Raja H.A., Tanaka K., Abdel-Wahab M.A., Abdel-Aziz F.A., Doilom M., Phookamsak R., et al. Freshwater Dothideomycetes. Fungal Divers. 2020;105:319–575. doi: 10.1007/s13225-020-00463-5. [DOI] [Google Scholar]
- 53.Nitschke T. Grundlage eines systems der Pyrenomyceten. Verh. Des Nat. Ver. Der Preuss. Rheinl. Westfal. Und Des Regier. Osnabrück. 1869;26:70–77. [Google Scholar]
- 54.Saccardo P.A. Sylloge Fungorum Omnium Hucusque Cognitorum. Sylloge Fungorum. 1883;2:672. [Google Scholar]
- 55.Maharachchikumbura S.S., Wanasinghe D.N., Cheewangkoon R., Al-Sadi A.M. Uncovering the hidden taxonomic diversity of fungi in Oman. Fungal Divers. 2021;106:229–268. doi: 10.1007/s13225-020-00467-1. [DOI] [Google Scholar]
- 56.Hashimoto A., Hirayama K., Takahashi H., Matsumura M., Okada G., Chen C.Y., Huang J.W., Kakishima M., Ono T., Tanaka K. Resolving the Lophiostoma bipolare complex: Generic delimitations within Lophiostomataceae. Stud. Mycol. 2018;90:161–189. doi: 10.1016/j.simyco.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bao D.F., Su H.Y., Maharachchikumbura S.S.N., Liu J.K., Nalumpang S., Luo Z.L., Hyde K.D. Lignicolous freshwater fungi from China and Thailand: Multi-gene phylogeny reveals new species and new records in Lophiostomataceae. Mycosphere. 2019;10:1080–1099. doi: 10.5943/mycosphere/10/1/20. [DOI] [Google Scholar]
- 58.Doilom M., Dissanayake A.J., Wanasinghe D.N., Boonmee S., Liu J.K., Bhat D.J., Taylor J.E., Bahkali A.H., Mckenzie E.H.C., Hyde K.D. Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 2017;82:107–182. doi: 10.1007/s13225-016-0368-7. [DOI] [Google Scholar]
- 59.Phookamsak R., Hyde K.D., Jeewon R., Bhat D.J., Jones E.B.G., Maharachchikumbura S.S.N., Raspé O., Karunarathna S.C., Wanasinghe D.N., Hongsanan S., et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019;95:1–273. doi: 10.1007/s13225-019-00421-w. [DOI] [Google Scholar]
- 60.Jiang H.B., Phookamsak R., Hyde K.D., Mortimer P.E., Xu J.C., Kakumyan P., Karunarathna S.C., Kumla J. A Taxonomic Appraisal of Bambusicolous Fungi in Occultibambusaceae (Pleosporales, Dothideomycetes) with New Collections from Yunnan Province, China. Life. 2021;11:932. doi: 10.3390/life11090932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu X.D., Zhang S.N., Cheewangkoon R., Liu J.K. Additions to Occultibambusaceae (Pleosporales, Dothideomycetes): Unrevealing Palmicolous Fungi in China. Diversity. 2021;13:516. doi: 10.3390/d13110516. [DOI] [Google Scholar]
- 62.Hyde K.D., Hongsanan S., Jeewon R., Bhat D.J., McKenzie E.H., Jones E.G., Phookamsak R., Ariyawansa H.A., Boonmee S., Zhao Q. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80:1–270. doi: 10.1007/s13225-016-0373-x. [DOI] [Google Scholar]
- 63.Zhang J.F., Liu J.K., Hyde K.D., Yang W., Liu Z.Y. Fungi from Asian Karst formations II. Two new species of Occultibambusa (Occultibambusaceae, Dothideomycetes) from karst landforms of China. Mycosphere. 2017;8:550–559. doi: 10.5943/mycosphere/8/4/4. [DOI] [Google Scholar]
- 64.Tanaka K., Hirayama K., Yonezawa H., Hatakeyama S., Harada Y., Sano T., Shirouzu T., Hosoya T. Molecular taxonomy of bambusicolous fungi: Tetraplosphaeriaceae, a new pleosporalean family with Tetraploa-like anamorphs. Stud. Mycol. 2009;64:175–209. doi: 10.3114/sim.2009.64.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hyde K.D., Dong Y., Phookamsak R., Jeewon R., Bhat D.J., Jones E.B.G., Liu N.G., Abeywickrama P.D., Mapook A., Wei D., et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020;100:5–277. doi: 10.1007/s13225-020-00439-5. [DOI] [Google Scholar]
- 66.Kelchner S.A. Higher level phylogenetic relationships within the bamboos (Poaceae: Bambusoideae) based on five plastid markers. Mol. Phylogenet. Evol. 2013;67:404–413. doi: 10.1016/j.ympev.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 67.Feng Y., Liu J.K., Lin C.G., Chen Y.Y., Xiang M.M., Liu Z.Y. Additions to the Genus Arthrinium (Apiosporaceae) From Bamboos in China. Front. Microbiol. 2021;720:12. doi: 10.3389/fmicb.2021.661281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dai D.Q., Tang L.Z., Wang H.B. A Review of Bambusicolous Ascomycetes. Bamboo Curr. Future Prospect. 2018;165:10. doi: 10.5772/intechopen.76463. [DOI] [Google Scholar]
- 69.Dlamini L.C., Fakudze S., Makombe G.G., Muse S., Zhu J. Bamboo as a Valuable Resource and its Utilization in Historical and Modern-day China. BioResources. 2022;17:1926–1938. doi: 10.15376/biores.17.1.Dlamini. [DOI] [Google Scholar]
- 70.Sun Y.R., Goonasekara I.D., Thambugala K.M., Jayawardena R.S., Wang Y., Hyde K.D. Distoseptispora bambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodivers. Data J. 2020;8:e53678. doi: 10.3897/BDJ.8.e53678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang H.B., Zhang S.J., Phookamsak R., Promputtha I., Kakumyan P., Xu J.C. Amphibambusa hongheensis sp. nov., a novel bambusicolous ascomycete from Yunnan, China. Phytotaxa. 2021;505:201–212. doi: 10.11646/phytotaxa.505.2.6. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences data were submitted to GenBank.