Abstract
Filamentous bacteria containing bacteriochlorophylls c and a were enriched from hypersaline microbial mats. Based on phylogenetic analyses of 16S rRNA gene sequences, these organisms form a previously undescribed lineage distantly related to Chloroflexus spp. We developed and tested a set of PCR primers for the specific amplification of 16S rRNA genes from filamentous phototrophic bacteria within the kingdom of “green nonsulfur bacteria.” PCR products recovered from microbial mats in a saltern in Guerrero Negro, Mexico, were subjected to cloning or denaturing gradient gel electrophoresis and then sequenced. We found evidence of a high diversity of bacteria related to Chloroflexus which exhibit different distributions along a gradient of salinity from 5.5 to 16%.
Chloroflexus aurantiacus is a filamentous, bacteriochlorophyll-containing, metabolically versatile bacterium that is capable of anoxygenic photoautotrophy, photoheterotrophy, and aerobic chemoorganotrophy. A great interest in this organism from evolutionary, biochemical, and biogeochemical perspectives has been fueled by its deep branching in the bacterial phylogenetic tree, indicating its evolutionary antiquity (10, 27, 38), and by its unique pathway of carbon dioxide fixation (32, 33).
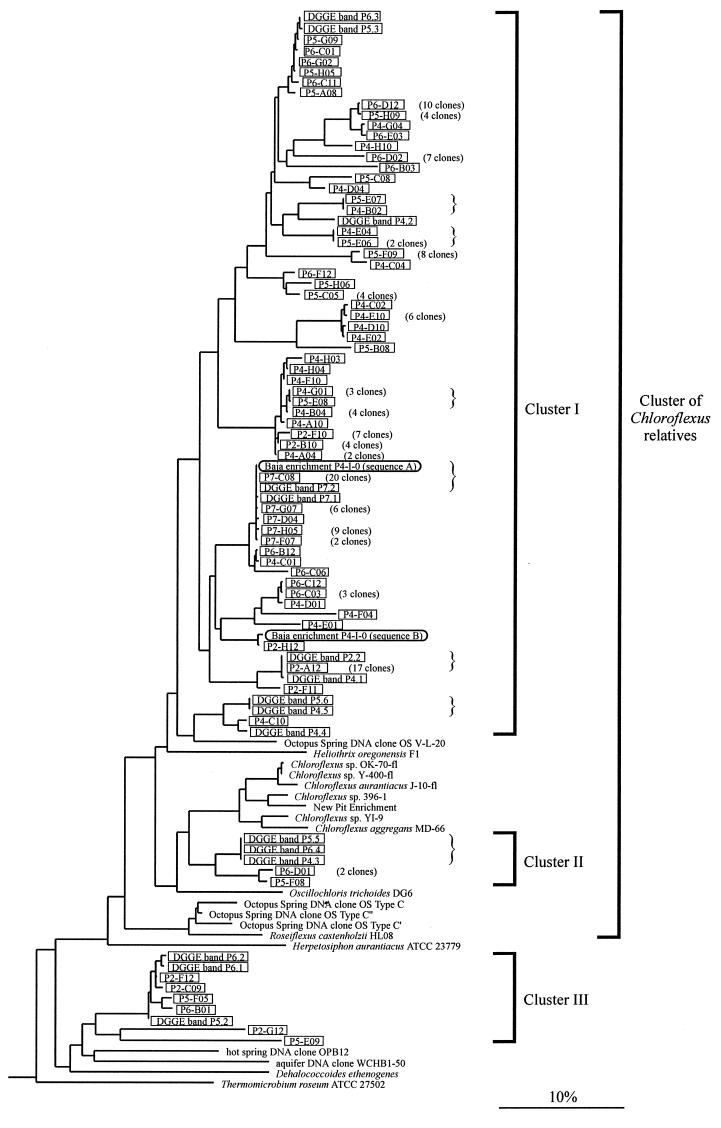
Additional filamentous phototrophs phylogenetically related to C. aurantiacus have been cultivated from hot spring habitats (Chloroflexus aggregans [(11)] and Heliothrix oregonensis [(29)]) and a freshwater spring (Oscillochloris trichoides [(14)]). Molecular analyses have revealed the existence of bacteria, termed “type C,” that inhabit microbial mats in neutral to alkaline hot springs and branch just outside the cluster containing Chloroflexus, Heliothrix, and Oscillochloris (35, 37) (Fig. 1). While the phenotype of type C organisms cannot be reliably inferred merely from their phylogenetic relationships to these other phototrophs, the recent cultivation of a thermophilic, filamentous phototrophic bacterium related to type C, Roseiflexus castenholzii, suggests that the members of this lineage may indeed be phototrophic (Fig. 1) (11a). Here, we use the term “Chloroflexus relatives” to refer to the phylogenetic cluster that contains all known filamentous photosynthetic bacteria in the “green nonsulfur bacteria” kingdom (Fig. 1).
FIG. 1.
Phylogenetic affiliations of Chloroflexus relatives detected in hypersaline microbial mats. Sequences from enrichments, DNA clones, and DGGE bands (compare with Fig. 3) are framed. Clone and band designations contain abbreviations (P2 to P7) indicating the sources of the respective sequences (evaporation ponds 2 to 7). For sequences detected several times in the same mat sample, only single representatives are shown, and the respective numbers of identical clones are in parentheses. Identical sequences that were detected in several different mat samples are indicated by braces. The maximum-likelihood tree was calculated by using the phylogeny software package ARB and is based on almost-complete 16S rRNA gene sequences. Thirty-five sequences from organisms with various phylogenetic affiliations (not shown) were used to root the tree. Phylogenetic affiliations of organisms represented by partial 16S rRNA gene sequences were reconstructed by applying parsimony criteria without changing the overall tree topology. The scale bar indicates 10% estimated sequence divergence.
In microbial mats in marine and hypersaline environments, filamentous organisms that resemble Chloroflexus in terms of morphology, ultrastructure, and pigmentation have repeatedly been observed (5, 8, 18, 20, 30, 31). In some mat systems, such as in the saltern in Guerrero Negro, Baja California, Mexico, the presence of phenotypically different filaments suggests that these organisms may be diverse (5, 30). While knowledge about these filaments is very limited, it has been speculated that they might be related to and occupy ecological niches analogous to those of their hot spring counterparts (30). However, no pure cultures of these organisms are available, and their phylogeny has not yet been resolved. We investigated microbial mats in hypersaline brines of the saltern in Guerrero Negro. Structurally coherent mat ecosystems flourish in evaporation ponds of this system at salinities from approximately 6 to 16%. The mats are almost exclusively composed of microorganisms and have been the subject of considerable scientific interest in the past (for reviews and detailed descriptions of these sites, see references (6 and 12). Dense populations of halotolerant filamentous phototrophs have been reported to form macroscopically visible layers in anoxic zones of some of these microbial mats (8, 13, 30). In the present study, we investigated the diversity and phylogeny of these organisms and surveyed the distribution of rRNA-defined populations along a salinity gradient.
This work is part of a comparative study of microbial mats in geothermal and hypersaline environments in the framework of the NASA Ames Astrobiology Institute Program. By investigating generality versus site specificity of particular mat features, we attempt to gain insights into ecological and geochemical patterns that might be common among or unique to mat communities or their fossil equivalents, stromatolites, found in different geographic locations and environmental settings.
Diversity of cultivated Chloroflexus spp.
To evaluate diversity among Chloroflexus strains currently available, we sequenced their 16S rRNA genes. Results of the phylogenetic analysis are shown in Fig. 1. Strains OK-70-fl and Y-400-fl are closely related to the type strain of C. aurantiacus, J-10-fl (maximum sequence divergence within this cluster, 1.6%). Strain YI-9 is related to the type strain of C. aggregans, MD-66 (divergence between the two sequences, 4.1%). Strain 396-1, together with organisms enriched from the sulfidic “New Pit” hot spring (36), forms a lineage more distantly related to C. aurantiacus and C. aggregans (divergence between sequence from 396-1 and sequences from J-10-fl and MD-66, 5.6 and 6.7%, respectively).
Enrichments.
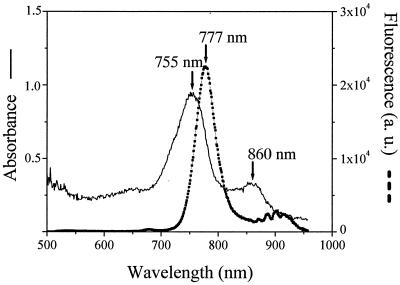
Microbial mats were sampled in June 1999 from evaporation ponds 4 to 7 of the saltern in Guerrero Negro (6). For enrichments of anoxygenic phototrophic bacteria, a medium for cyanobacteria (9) containing 9% (wt/vol) commercial sea salt was modified by adding NH4Cl (200 mg liter−1), yeast extract (250 mg liter−1), Na2S2O4 (70 mg liter−1), Na2S · 9H2O (150 mg liter−1), 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU; 1.2 mg liter−1), and resazurin (0.0001%, wt/vol). The medium was boiled to drive out dissolved oxygen, and 10 ml was aliquoted into 20-ml screw-cap tubes under a stream of nitrogen. Tubes were immediately closed with butyl rubber septa and autoclaved. Aliquots from a series of mat samples that had been homogenized and diluted to extinction in growth medium were injected through the rubber septa. Tubes were incubated at room temperature (20 to 25°C) and received natural daylight (approximately 10 μmol of photons m−2 s−1 at midday). After 5 weeks of incubation, one enrichment tube, inoculated with an undiluted sample from the mat in pond 4 and labeled P4-I-0, contained a green pellicle consisting of gliding filaments with a diameter of 1 μm and indeterminate length. These enriched filaments resembled the “marine Chloroflexus-like organisms” described previously by Pierson et al. (30), except that we did not observe any sheaths. Spectral absorption and fluorescence characteristics of filaments air dried onto a microscope slide were measured by using an epifluorescence microscope connected to a Hamamatsu PMA-11 spectrometer (250–950 nm; Hamamatsu Photonics) as described previously (17). Absorption maxima at 755 and 860 nm suggested that these filaments contained bacteriochlorophylls c and a, respectively (15) (Fig. 2). A fluorescence maximum at 777 nm (Fig. 2) further confirmed the presence of bacteriochlorophyll c (2). Following PCR amplification of DNA from these filaments (26), molecular cloning (applying the TOPO TA cloning kit; Invitrogen), and sequencing (applying the ABI Big Dye terminator kit and an ABI Prism 310 capillary sequencer; Applied Biosystems), two almost complete 16S rRNA gene sequences (designated A and B in Fig. 1), differing from each other by 8.4%, were recovered from enrichment P4-I-0. Phylogenetic analyses indicated that these filamentous organisms form a separate, previously undescribed branch within the lineage containing Chloroflexus relatives (Fig. 1). However, even though the described procedure enriched bacteriochlorophyll-containing filamentous bacteria, as judged on the basis of microscopic observation, spectrometry measurements, and molecular cloning results, it was not effective for their continued cultivation. Subsequent anaerobic transfers to fresh medium did not grow, and, consequently, all enrichments were lost.
FIG. 2.
Absorption and fluorescence spectra obtained by microscope spectrometry on airdried filaments from enrichment P4-I-0. Wavelengths of absorbance and fluorescence maxima are indicated. a. u., arbitrary units.
PCR primers to amplify 16S rRNA genes from Chloroflexus relatives.
Based on the 16S rRNA gene sequences newly determined in this study, together with sequences available from public databases, we developed a PCR protocol for the specific amplification of rRNA gene segments from Chloroflexus relatives. PCR primers CCR-344-F (ACGGGAGGCAGCAGCAAG) and CCR-1338-R (ACGCGGTTACTAGCAACT) were designed by using the PROBE DESIGN and PROBE MATCH options of the phylogeny software package ARB (available at http://www.mikro.biologie.tu-muenchen.de) and the BLAST program (1) at the National Center for Biotechnology Information (Washington, D.C.). Target regions within the 16S rRNA gene are 344 to 361 and 1338 to 1355 (Escherichia coli numbering [3]). These primers match all gene sequences within the Chloroflexus relatives cluster described herein, encompassing organisms from both temperate and hot spring freshwater environments as well as those enriched from hypersaline brines. Outside this phylogenetic cluster no nucleotide sequence currently deposited in public databases (as of June 2001) has fewer than two mismatches total to primers CCR-344-F and CCR-1338-R. Therefore, the combined use of both primers should result in a PCR specific for Chloroflexus relatives. PCR mixtures (50 μl) contained Taq polymerase, nucleotides, and buffer according to the manufacturer's recommendations (Fisher Scientific) and 50 pmol of each primer. Temperature cycling (cycler PTC-100; MJ Research) included an initial denaturation step (2 min at 94°C), 30 incubation cycles (each consisting of 45 s at 94°C, 45 s at 60°C, and 45 s at 72°C), and a final elongation step (7 min at 72°C). Under these conditions, amplification products were obtained from all Chloroflexus relatives tested, whereas other bacteria were discriminated against (Table 1).
TABLE 1.
Bacterial cultures tested for amplification by PCR specific for Chloroflexus relatives
| Strain | Phylogeny | Sourcea | Amplificationb |
|---|---|---|---|
| Chloroflexus aurantiacus J-10-fl (DSM 635) | Chloroflexus relatives | DSMZ | + |
| Chloroflexus aurantiacus OK-70-fl (DSM 636) | Chloroflexus relatives | DSMZ | + |
| Chloroflexus aurantiacus Y-400-fl (DSM 637) | Chloroflexus relatives | DSMZ | + |
| Chloroflexus aggregans MD-66 (DSM 9485) | Chloroflexus relatives | DSMZ | + |
| Chloroflexus aggregans YI-9 (DSM 9486) | Chloroflexus relatives | DSMZ | + |
| Chloroflexus sp. strain 396-1 | Chloroflexus relatives | This study | + |
| Baja enrichment P4-I-0 | Chloroflexus relatives | This study | + |
| Herpetosiphon aurantiacus DSM 785 | Green nonsulfur bacteria | DSMZ | − |
| Microcoleus chthonoplastes PCC 7420 | Cyanobacteria | PCC | − |
| Spirulina sp. strain PCC 6313 | Cyanobacteria | PCC | − |
| Bacillus subtilis DSM 402 | Gram positive, low G+C | DSMZ | − |
| Desulfobacterium vacuolatum DSM 3385 | Proteobacteria, δ subdivision | DSMZ | − |
| Escherichia coli DSM 489 | Proteobacteria, γ subdivision | DSMZ | − |
| Thiomicrospira thyasirae | Proteobacteria, γ subdivision | TB | − |
| Cytophaga johnsonae DSM 2064 | Cytophaga group | DSMZ | − |
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; PCC, Pasteur Culture Collection, Paris, France; TB, T. Brinkhoff, Universität Oldenburg, Oldenburg, Germany.
Amplification products obtained with primers CCR-344-F and CCR-1338-R.
Diversity in hypersaline microbial mats.
For molecular biological analyses, microbial mat samples were collected from evaporation ponds 2, 4 (sampled in April 1996), 5, 6 (April 1997), and 7 (June 1999). Salinities of brines overlying microbial mats were measured refractometrically to be 5.5, 9, 11, 14, and 16% (in ponds 2 to 7) at times of collection (24, 25). Mat samples were frozen on site, transported to the laboratory in liquid nitrogen, and stored at −80°C until processed. Subsequent analyses were restricted to the upper 5 to 8 mm of mats, including the illuminated zone, where anoxygenic phototrophic filaments have been observed. To analyze the phylogenetic diversity of Chloroflexus relatives in hypersaline mats, community DNAs were extracted (by applying a previously published protocol [21] modified by omission of the DNA purification step) and segments of 16S rRNA genes were amplified by applying the newly developed PCR protocol. PCR products were cloned and sequenced. Nucleotide sequences (length, 674 to 909 nucleotides) of 166 plasmid inserts were determined, including 30 to 39 sequences from each of five evaporation ponds investigated. In total, 62 different clone sequences were found, indicating that sequence diversity was high (Fig. 1). Independent analyses of 300 nucleotide sequence segments from 5′ and 3′ ends did not reveal any evidence for chimeric DNA molecules composed of 16S rRNA genes from different organisms (16). Most segments of 16S rRNA genes were amplified from organisms affiliated with the cluster containing Chloroflexus relatives (clusters I and II in Fig. 1), though a few fell just outside (cluster III in Fig. 1). Numbers of different sequences detected in ponds 2, 4, 5, 6, and 7 were 8, 22, 16, 14, and 5, respectively. Rarefaction analysis (19) (performed by using the freeware program aRarefactWin, available at http: //www.uga.edu/∼strata/AnRareReadme.html) indicated that the investigation of additional plasmid inserts from microbial mats in ponds 4, 5, and 6 very likely would have led to the discovery of a significant number of additional 16S rRNA gene sequences (data not shown). Based on extrapolations of the rarefaction curves (25), the total numbers of different 16S rRNA gene sequences from Chloroflexus relatives probably present in samples from these mats were estimated to be 52, 25, and 15, respectively. In contrast, the sequence diversity in ponds 2 and 7 seems well represented by the actual sequences determined (eight and five different sequences estimated to be present).
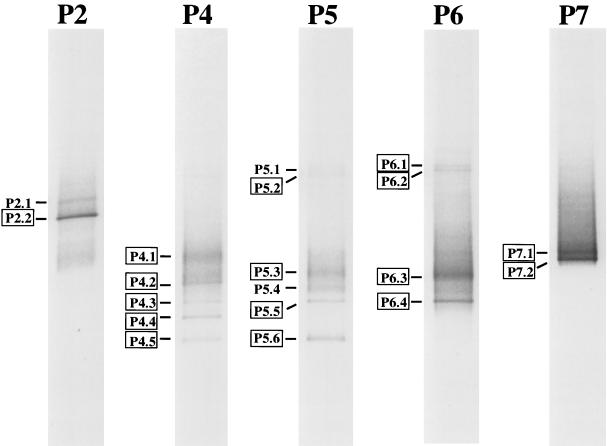
In parallel, PCR products with GC clamps (40 bp, synthesized onto the 5′ end of the reverse primer [(22)]) were analyzed by denaturing gradient gel electrophoresis (DGGE) [22]) using a Hoefer SE 600 gel electrophoresis unit in a 40-liter aquarium (7). This technique separates DNA molecules of equal size but different sequences by their differential melting behavior in a gradient of denaturants. When analyzed by DGGE, PCR products derived from the different mats appeared in characteristic patterns of two to six bands. Nucleotide sequences could be determined for 16 out of 19 bands total (Fig. 3). In cases where rarefaction analyses had indicated clone libraries to sufficiently represent extant sequence diversity (ponds 2 and 7), the most frequently found clone sequences (P2-A12 [17 plasmids out of 33] and P7-C08 [20 plasmids out of 39]) (Fig. 1) were identical to sequences from the respective most intense DGGE bands (P2.2 and P7.2) (Fig. 3). However, when rarefaction analyses indicated insufficient sampling for the cloning approach (ponds 4, 5, and 6), clone sequences did not match any DGGE band sequences.
FIG. 3.
Negative image of Sybr green-stained DGGE separation patterns of segments of 16S rRNA genes amplified by a Chloroflexus relative-specific PCR protocol. The denaturing gradient was from 30 to 80% (100% denaturant is 40% [vol/vol] formamide–7 M urea), and electrophoresis was run for 20 h at 60°C and 72 V. Results are shown for microbial mats from evaporation ponds 2 to 7 (salinities of brines overlying microbial mats were 5.5, 9, 11, 14, and 16%). Framed band designations indicate bands from which nucleotide sequences could be determined.
Phylogeny of uncultivated Chloroflexus relatives.
Phylogenetic analysis arranged cloned PCR products and DGGE bands into three clusters, termed I, II, and III in Fig. 1. Cluster I branches off within the lineage of Chloroflexus relatives. It encompasses 167 clones and DGGE bands from microbial mats from all five evaporation ponds. Some of these sequences were found to be identical or highly similar to those from enrichment P4-I-0 (Fig. 1). It is interesting, however, that such sequences were not detected in the mat from pond 4, from which P4-I-0 was enriched. The sequence divergence within cluster I is large. Sequences differ by up to 14% in the positions analyzed (nucleotide positions 400 to 1,300, E. coli numbering), and differences among the respective full gene sequences would likely be even larger. Cluster II is composed of six clones and DGGE bands from ponds 4, 5, and 6 and also branches off within the lineage of Chloroflexus relatives (Fig. 1). Sequences in cluster III are affiliated with the kingdom of green nonsulfur bacteria but branch outside the lineage of Chloroflexus relatives. The most similar sequences available from public databases are derived from uncultivated bacteria previously detected in a hot spring and in an aquifer (Fig. 1).
There are many sets of closely related sequences, all of which include clones from different ponds and therefore different salinities. It cannot be excluded at this time that some of the sequence diversity detected might be due to PCR misincorporations or to heterogeneities of 16S rRNA genes within genomes of individual bacteria (23). However, numerous (28 out of 62) sequences were found at least twice and sometimes in independent PCRs based on different microbial mat samples (clones P4-B02/P5-E07 and P4-E04/P5-E06, DGGE bands P4.3/P5.5/P6.4 and P4.5/P5.6). DGGE bands showing unique distributions along the salinity gradient suggest that they likely represent genetically different microbial populations as opposed to different genes within single organisms. Such clusters of highly similar yet distinct 16S rRNA gene sequences have been observed in molecular studies of microbial diversity in numerous habitats, including hot springs (reviewed in reference 35). Unique distributions of closely related hot spring cyanobacterial 16S rRNA genes indicate the evolutionary divergence of closely related microorganisms into ecologically distinct populations (35). Following the evolution of halotolerance, ancestors of recent Chloroflexus relatives may have adapted to particular conditions in hypersaline habitats (e.g., with respect to carbon sources, light conditions, salinity-growth rate relationships, etc.) through evolutionary radiations that are reflected in rRNA gene sequences today.
Distribution along a salinity gradient.
Several DGGE bands were common among patterns derived from different microbial mats (P4.3/P5.5/P6.4 and P4.5/P5.6; sequence identity was confirmed by sequence analyses), indicating some overlap of mat composition. In contrast, all other DGGE bands were unique to mats from certain ponds. For ponds 2 and 7, DGGE results were strongly supported through clone library analyses. Presumably due to the more complex compositions of the microbial communities in ponds 4, 5, and 6, rarefaction analyses indicated that the numbers of clones analyzed from these ponds were too low to sufficiently represent the sequence diversity present in the mats and, consequently, may not enable reliable estimates of PCR product composition. Through DGGE analysis, in contrast, DNA molecules differing in sequence were first separated according to their electrophoretic behavior and then chosen for sequencing on the basis of band intensity. Thus, the sequences determined for DGGE bands should represent the most abundant amplification products (and, possibly, genes). However, both cloning and DGGE are based on PCR amplification of 16S rRNA gene segments. In part reflecting its wide application in the study of microbial diversity, various pitfalls and potential biases of this technique have been reported (reviewed in reference 34). Abundances of Chloroflexus related populations and their small-scale distribution within the microbial mats could be investigated in more detail through hybridization studies, applying probes directed against the various 16S rRNA gene sequences or phylogenetic clusters reported here.
The presence of euryhaline Chloroflexus relatives in the Guerrero Negro microbial mats would be consistent with the widespread distribution of some of the 16S rRNA gene sequences observed. Similarly, salinity was previously found to govern the distribution of cyanobacteria and diatoms in the same saltern system (4, 24). However, it must be cautioned that rRNA genes are more conserved in structure and function than most protein encoding genes, and, consequently, identical 16S rRNA gene sequences may conceal several physiologically and ecologically different microbial populations (e.g., see reference 28). Ultimately, cultivated strains will be necessary to investigate the ecophysiology of Chloroflexus relatives in hypersaline environments in more detail.
Conclusions.
Our data strongly suggest that the diversity of Chloroflexus relatives in hypersaline environments is considerably larger than had previously been imagined based on microscopic observations (5, 30). Even though the phenotypes of these organisms cannot be safely inferred from their phylogenetic relationships, it seems likely that a large variety of filamentous phototrophic bacteria in these habitats still await cultivation.
No close relatives of freshwater (including hot spring) filamentous anoxygenic phototrophs (e.g., Chloroflexus, Oscillochloris, Roseiflexus, and type C) were detected in the hypersaline habitats investigated. Very similarly, cyanobacteria in the same hypersaline microbial mats were recently found to form several phylogenetic lineages only distantly related to their freshwater morphological counterparts (9, 24, 26). In different habitats, analogous ecological niches may be occupied by different microorganisms, each adapted to the respective physicochemical conditions. Similarities with respect to easily observable phenotypic characters (for example, morphology and pigmentation) may therefore mask significant differences in ecophysiology and phylogeny.
Nucleotide sequence accession numbers.
EMBL database accession numbers for the sequences reported in this study are AJ308496 to AJ308502 and AJ309574 to AJ309653.
Acknowledgments
We are indebted to Dave Des Marais and coworkers for the coordination of microbial mat research within the NASA Astrobiology Institute, the skillful organization of a field trip to Baja California, and generous hospitality. We thank Exportadora de Sal, S. A. de C. V., BCS, Mexico, for support in our field collections, Satoshi Hanada for sharing unpublished data and Steven M. Holland for providing the freeware program aRarefactWin.
Funding for this work was provided by the NASA Astrobiology Institute (CAN-97-01-OSS-004) through a cooperative agreement with the NASA Ames Research Center (NCC 2-1073) and through the Danish Natural Science Research Council (to M.K.). M.T.M. acknowledges support from the National Science Foundation (OPP 9809195).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amesz J, Vasmel H. Fluorescence properties of photosynthetic bacteria. In: Govindjee, Amesz, Fork D C, editors. Light emission by plants and bacteria. Orlando, Fla: Academic Press; 1986. pp. 423–450. [Google Scholar]
- 3.Brosius M, Dull T, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 4.Clavero E, Hernandez-Marine M, Grimalt O J, Garcia-Pichel F. Salinity tolerance of diatoms from thalassic hypersaline environments. J Phycol. 2000;36:1021–1034. [Google Scholar]
- 5.D'Amelio E D, Cohen Y, Des Marais D. Comparative functional ultrastructure of two hypersaline submerged cyanobacterial mats: Guerrero Negro, Baja California Sur, Mexico, and Solar Lake, Sinai, Egypt. In: Cohen Y, Rosenberg E, editors. Microbial mats—physiological ecology of benthic microbial communities. Washington, D.C.: American Society for Microbiology; 1989. pp. 97–113. [Google Scholar]
- 6.Des Marais D J. The biogeochemistry of hypersaline microbial mats. Adv Microb Ecol. 1995;14:251–274. doi: 10.1007/978-1-4684-7724-5_6. [DOI] [PubMed] [Google Scholar]
- 7.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Pichel F, Meshing M, Castenholz R W. Diel migrations of microorganisms within a benthic, hypersaline mat community. Appl Environ Microbiol. 1994;60:1500–1511. doi: 10.1128/aem.60.5.1500-1511.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Pichel F, Nübel U, Muyzer G. The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch Microbiol. 1998;169:469–482. doi: 10.1007/s002030050599. [DOI] [PubMed] [Google Scholar]
- 10.Gupta R S, Mukhtar T, Singh B. Evolutionary relationships among photosynthetic prokaryotes (Heliobacterium chlorum, Chloroflexus aurantiacus, cyanobacteria, Chlorobium tepidum and proteobacteria): implications regarding the origin of photosynthesis. Mol Microbiol. 1999;32:893–906. doi: 10.1046/j.1365-2958.1999.01417.x. [DOI] [PubMed] [Google Scholar]
- 11.Hanada S, Hiraishi A, Shimada K, Matsuura K. Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement. Int J Syst Bacteriol. 1995;45:676–681. doi: 10.1099/00207713-45-4-676. [DOI] [PubMed] [Google Scholar]
- 11a.Hanada, S., S. Takaichi, K. Matsuura, and K. Nakamura. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium which lacks chlorosomes. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 12.Javor B. Hypersaline environments. Berlin, Germany: Springer Verlag; 1989. [Google Scholar]
- 13.Jørgensen B B, Des Marais D J. Competition for sulfide among colorless and purple sulfur bacteria in cyanobacterial mats. FEMS Microbiol Ecol. 1986;38:179–186. doi: 10.1111/j.1574-6968.1986.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 14.Keppen O I, Baulina O I, Kondratieva E N. Oscillochloris trichoides neotype strain DG-6. Photosynth Res. 1994;41:29–33. doi: 10.1007/BF02184143. [DOI] [PubMed] [Google Scholar]
- 15.Kondratieva E N, Pfennig N, Trüper H G. The phototrophic prokaryotes. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. Berlin, Germany: Springer; 1992. pp. 313–330. [Google Scholar]
- 16.Kopczynski E D, Bateson M M, Ward D M. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kühl M, Fenchel T. Bio-optical characteristics and the vertical distribution of photosynthetic pigments and photosynthesis in an artificial cyanobacterial mat. Microb Ecol. 2000;40:94–103. doi: 10.1007/s002480000061. [DOI] [PubMed] [Google Scholar]
- 18.Kühl M, Jørgensen B B. Spectral light measurements in microbenthic phototrophic communities with a fiber-optic microprobe coupled to a sensitive diode array detector. Limnol Oceanogr. 1992;37:1813–1823. [Google Scholar]
- 19.Ludwig J A, Reynolds J F. Statistical ecology. New York, N.Y: John Wiley and Sons; 1988. [Google Scholar]
- 20.Mack E E, Pierson B K. Preliminary characterization of a temperate marine member of the Chloroflexaceae. In: Olson J M, Ormerod J G, Amesz J, Stackebrandt E, Trüper H G, editors. Green photosynthetic bacteria. New York, N.Y: Plenum Publishing Corp.; 1988. pp. 237–241. [Google Scholar]
- 21.Moré M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer G, Brinkhoff T, Nübel U, Santegoeds S, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. 1998. p. 3.4.4. /1-23. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 23.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nübel U, Garcia-Pichel F, Clavero E, Muyzer G. Matching molecular diversity and ecophysiology of benthic cyanobacteria and diatoms in communities along a salinity gradient. Environ Microbiol. 2000;2:217–227. doi: 10.1046/j.1462-2920.2000.00094.x. [DOI] [PubMed] [Google Scholar]
- 25.Nübel U, Garcia-Pichel F, Kühl M, Muyzer G. Spatial scale and the diversity of benthic cyanobacteria and diatoms in a salina. Hydrobiologia. 1999;401:199–206. [Google Scholar]
- 26.Nübel U, Garcia-Pichel F, Muyzer G. The halotolerance and phylogeny of cyanobacteria with helical, tightly coiled trichomes (Spirulina Turpin) and the description of Halospirulina tapeticola gen. nov., sp. nov. Int J Syst Evol Microbiol. 2000;50:1265–1277. doi: 10.1099/00207713-50-3-1265. [DOI] [PubMed] [Google Scholar]
- 27.Oyaizu H, Debrunner-Vossbrinck B, Mandelco L, Studier J A, Woese C R. The green non-sulfur bacteria: a deep branching in the eubacterial line of descent. Syst Appl Microbiol. 1987;9:47–53. doi: 10.1016/s0723-2020(87)80055-3. [DOI] [PubMed] [Google Scholar]
- 28.Palys T, Nakamura L K, Cohan F M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- 29.Pierson B K, Giovannoni S J, Stahl D A, Castenholz R W. Heliothrix oregonensis, gen. nov., sp. nov., a phototrophic filamentous gliding bacterium containing bacteriochlorophyll a. Arch Microbiol. 1985;142:164–167. doi: 10.1007/BF00447061. [DOI] [PubMed] [Google Scholar]
- 30.Pierson B K, Valdez D, Larsen M, Morgan E, Mack E E. Chloroflexus-like organisms from marine and hypersaline environments: distribution and diversity. Photosynth Res. 1994;41:35–52. doi: 10.1007/BF02184144. [DOI] [PubMed] [Google Scholar]
- 31.Stolz J F. Fine structure of the stratified microbial community at Laguna Figueroa, Baja California, Mexico. 1. Methods of in situ study of the laminated sediments. Precambrian Res. 1983;20:479–492. [Google Scholar]
- 32.Strauss G, Fuchs G. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur J Biochem. 1993;215:633–643. doi: 10.1111/j.1432-1033.1993.tb18074.x. [DOI] [PubMed] [Google Scholar]
- 33.van der Meer M T J, Schouten S, de Leeuw J W, Ward D M. Autotrophy of green non-sulphur bacteria in hot spring microbial mats: biological explanations for isotopically heavy organic carbon in the geological record. Environ Microbiol. 2000;2:428–435. doi: 10.1046/j.1462-2920.2000.00124.x. [DOI] [PubMed] [Google Scholar]
- 34.Von Wintzingerode F, Goebel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 35.Ward D M, Ferris M J, Nold S J, Bateson M M. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward D M, Santegoeds C M, Nold S C, Ramsing N B, Ferris M J, Bateson M M. Biodiversity within hot spring microbial mat communities: molecular monitoring of enrichment cultures. Antonie Leeuwenhoek. 1997;71:143–150. doi: 10.1023/a:1000131426164. [DOI] [PubMed] [Google Scholar]
- 37.Weller R, Bateson M M, Heimbuch B K, Kopczynski E D, Ward D M. Uncultivated cyanobacteria, Chloroflexus-like inhabitants, and Spirochete-like inhabitants of a hot spring microbial mat. Appl Environ Microbiol. 1992;58:3964–3969. doi: 10.1128/aem.58.12.3964-3969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong J, Fischer W M, Inoue K, Nakahara M, Bauer C E. Molecular evidence for the early evolution of photosynthesis. Science. 2000;289:1724–1730. doi: 10.1126/science.289.5485.1724. [DOI] [PubMed] [Google Scholar]