Abstract
Coffee endophytes have been studied for almost 74 years, and several studies have demonstrated coffee-endophytic fungi with antibacterial and antifungal potential for human and plant pathogens. In this study, we isolated and identified a total of 235 strains of endophytic fungi from coffee leaf tissues collected in four coffee plantations in Pu’er city, Yunnan province, China. Molecular identification was carried out using maximum likelihood phylogenetic analysis of nuclear ribosomal internal transcribed spacer (ITS1-5.8S rDNA-ITS2) sequences, while the colonization rate and the isolation frequency were also calculated. Two pathogenic fungi (Alternaria alternata and Penicillium digitatum) and two pathogenic bacteria (Pseudomonas syringae and Salmonella enterica subsp. enterica) were used for screening the antagonistic activities of 61 strains of coffee-endophytic fungi by a dual-culture test assay while maximum likelihood phylogenetic analysis confirmed their natural classification. This is the first study of coffee-leaf-endophytic fungal diversity in China, and the results revealed that coffee-endophytic fungi from this study belong to the Ascomycota, distributed among two classes, 10 orders, and 17 families. Concurrently, endophytic fungi isolates distributed in Arthrinium, Biscogniauxia, Daldinia, Diaporthe, and Nigrospora showed strong antagonistic activities against the pathogens. For the pathogens Alternaria alternata and Pseudomonas syringae, Nigrospora XCE-7 showed the best inhibitory effects with inhibition rates of 71.76% and 61.11%, respectively. For the pathogen Penicillium digitatum, Daldinia ME-9 showed the best inhibitory effect with a 74.67% inhibition rate, while Biscogniauxia PTE-7 and Daldinia T5E-1-3 showed the best inhibitory effect with a rate of 60.42% against the pathogen Salmonella enterica subsp. enterica. Overall, our study shows the diversity of coffee endophytes in four coffee-growing areas in Pu’er city, Yunnan province, China, and their potential use as biological control agents against two fungal and two bacterial pathogens.
Keywords: biocontrol agents, Coffea, coffee-endophytic fungi, pathogenic fungi, pathogenic bacteria
1. Introduction
Coffee is one of the most important crops around the world that grow in tropical and subtropical areas, and it is the second-largest export commodity after crude oil [1,2]. Coffee is susceptible to diseases and pests, but most coffee diseases are caused by pathogenic fungi, which can infect various tissues of coffee before and after coffee harvest and affect the yield and quality of the fruit [3,4]. Cerda et al. [5] showed that pests and diseases cause up to 38% of coffee yield losses. In order to reduce the impact of diseases on coffee, cultural control, chemical control, and biological control strategies have been developed [6,7]. Cultural control can avoid and reduce some plant diseases and generally requires workers to implement it manually, which is time-consuming and labor-intensive [7]. Chemicals can control diseases and increase yields, such as the use of the fungicides cyproconazole and thiamethoxam in the soil to control rust disease [8], but the chemical methods can result in environmental and food contamination, which forces society to use nonchemical methods for plant disease control [9]. Carl von Tubeuf was the first person to use biological control to manage plant diseases in 1914 [10,11,12,13]. The biological control of plant diseases occurs with several distinct mechanisms, including competition for nutrients between a pathogen and a harmless species, parasitism, and production of antibiotics [14,15]. Biological controls have the advantages of being environmentally compatible, can exhibit broad or narrow targets depending on the organism, can be site-specific, less prone to resistance, safe, and inexpensive compared with chemical pesticides. Due to these advantages, biocontrol has become a popular method of plant disease management [16,17,18]. Currently, some fungi and bacteria as biocontrol agents are available as commercial products [17]. A total of 101 microbial biological control agents were registered for disease control in Australia, Brazil, Canada, Europe, Japan, New Zealand, and the United States in 2017 [19].
Endophytes are microorganisms such as actinomycetes, bacteria, or fungi that live in plant tissues and do not cause the host to produce any disease symptoms [20,21,22]. They is generally considered reciprocal and neutral, but can cause disease or become saprophytic when senescence occurs in the host, its vitality decreases, and other factors are weakened [23,24]. The host strongly restricts the growth of endophytes, but it can overcome the host’s defense by secreting biologically active metabolites or changing the balance of plant hormones in the host [24,25,26]. Endophytes spend all or part of their life cycle in the healthy tissues of plants [27,28]. The bioactive compounds synthesized by endophytes enhance the resistance of plants to pathogenic microorganisms and pests, protect the host from environmental stresses such as high temperature and drought, and help the host to more easily access nutrients to promote the growth necessary for a stable symbiosis [21,26,29]. These bioactive metabolites produced by endophytes can be used directly or indirectly as potential biological fertilizers to promote plant growth and biocontrol agents to control pathogenic-causing diseases [28,30,31,32,33]. Fadiji and Babalola [34] summarized the applications of endophytes into four categories. These are: (i) plant growth promotion, including nutrient uptake, photostimulation, and phosphate solubilization; (ii) plant health and protection (antimicrobial activities and pathogen displacement); (iii) pollution control and phytoremediation; and (iv) medical and industrial applications (anticancer and antiviral). Since endophytes can produce large amounts of biologically active substances, the use of endophytes to control plant diseases and to increase crop yields has become the focus of the agricultural industry [17,35,36,37].
Endophytic fungi are one of the groups of microorganisms that colonize plant tissues and interact with plants, which can be used for biological control and resistance induction, and these fungi are one of the most interesting groups with high utilization value and high diversity [38]. Endophytic fungi are known to produce important secondary metabolites such as anticancer, antifungal, antidiabetic, and immunosuppressant compounds, which can be used as a source of some important secondary metabolites [39,40]. Numerous attempts have been made to control wilt diseases through biocontrol agents using endophytic fungi, and this way holds great promise for improving crop productivity while serving as an alternative to chemical fungicides and synthetic fertilizers [41,42,43,44]. A huge number of studies have shown that fungal endophytes have potential research values (e.g., endophytic fungi secrete hormones such as indoleacetic acid and gibberellins that aid in plant developmental processes and improve plant growth and crop productivity) [45,46]. The endophytic fungus Penicillium indica can regulate the production of phytohormones and contributes to root growth in crops such as barley and tobacco [47,48]. The endophytic fungus Acremonium alternatum controls the damage caused by the moth Plutella xylostella in beans and induces resistance against Leveillula taurica (the powdery mildew pathogen) in tomatoes [49]. Ek-Ramos et al. [49] showed that the cotton fungal endophyte Phomopsis sp. reduces caterpillar herbivory activity in cotton plants. The endophytic fungi Aspergillus terreus and Penicillium citrinum reduce the pathogenic Sclerotium rolfsii and induce increases in the biomass yield of sunflower plants [50]. Many studies have revealed that endophytic fungi, such as Ampelomyces quisqualis, Aureobasidium pullulans, Beauveria bassiana, Botryosphaeria sp., Candida oleophila, Coniothyrium minitans, Gliocladium catenulatum, Metarhizium anisopliae, Paecilomyces lilacinus, Pestalotiopsis microspora, Phlebiopsis gigantea, Purpureocillium lilacinum, Trichoderma asperellum, T. atroviride, T. gamsii, T. polysporum, and Verticillium lecanii, can be used as biocontrol agents in commercial products [51,52,53,54,55,56]. Therefore, endophytic fungi represent an effective agent in managing diseases and promoting plant growth; they also provide great opportunities for researching new products.
Studies of endophytic fungi in coffee were pioneered by Rayner in 1948 [57]. Since 1948, many studies have been published on coffee endophytes but no research has been carried out on the antagonistic effects of coffee endophytes as evaluated by a dual-culture assay. Beauveria bassiana can be established in coffee seedlings through different inoculation methods and the plants can resist coffee berry borers [58]. Fernandes et al. [59] showed that the fungal endophytes isolated from the healthy leaves of Coffea arabica could produce bacteriostatic, antifungal, and antitumor bioactive substances. Trichoderma sp. isolated from healthy coffee roots by Mulaw et al. [60] was demonstrated to have an antagonistic effect on the fungus Fusarium sp., which causes vascular wilt diseases. Monteiro et al. [61] isolated endophytes from Coffea arabica, and the results showed that Acremonium, Muscodor, and Simplicillium have antibacterial activities against Aspergillus ochraceus, while six different endophytic fungal strains can inhibit the growth of Fusarium verticillioides. The endophyte Simplicillium coffeanum isolated from coffee branches by Gomes et al. [62] has antimicrobial activities against Aspergillus niger, A. ochraceus, A. sydowii, and A. tubingensis. Trichoderma harzianum (CRF1) and T. viride (CRF-2), isolated from coffee rhizosphere soil by Ranjini et al. [63], can control Myrothecium roridum in vivo, a fungus that causes stem necrosis and leaf spot of coffee seedlings.
Both bacteria and fungi cause diseases in coffee [7]. Alternaria alternata was identified in Brazil in 1999 as the causative agent of necrotic spots on coffee leaves [64]. This pathogen is more serious when present in other fruits and vegetables, and the current control methods include fungicides, synthetic chemical fungicides, and biological control [65]. Penicillium digitatum is a destructive postharvest pathogen causing green mold in fruit, especially citrus fruits [66]. Fungicides are still the main control agent of postharvest green mold, and it is necessary to develop novel and safer strategies for effectively controlling plant diseases [67]. Bacterial halo blight disease caused by Pseudomonas syringae has affected coffee plantations in Brazil and Ethiopia. Due to only a few efficient commercial products being available on the market to control this disease and the low efficiency of the chemical control on field conditions, biological control and disease-resistant variety development should be the most promising management methods [68,69,70]. Salmonella enterica subsp. enterica is a common pathogen of animals, humans, or plants [71]. Salmonella can adhere to plant surfaces before actively infecting the interior of different plants, leading to the colonization of plant organs [72,73] and suppression of the plant immune system [74]. In addition, Salmonella originating from plants reduces its virulence toward animals [75]. Therefore, plants act as an alternative host for Salmonella pathogens and have a role in transmitting back to animals. Moreover, Salmonella can adapt to survive in different environments outside the host organism, such as low pH or high temperatures [76].
The microfungal diversity of the Greater Mekong Subregion (GMS) has been relatively well-studied, but their secondary metabolites have not been well-studied [77]. Since endophytic fungi have become the source of new antibacterial and antifungal agents, with some examples producing valuable secondary metabolites, this study was planned to identify and examine the antagonistic abilities against two fungal and two bacterial pathogens of endophytic fungi, which were isolated from healthy coffee leaves from four coffee-growing areas in Pu’er city, Yunnan province, China.
2. Materials and Methods
2.1. Sampling Location and Leaf Collection
Pu’er city has the largest coffee planting area, with the highest yield, and the best-quality coffee production in Yunnan province, China [78]. This study was conducted on mature and healthy leaf samples collected from four coffee-planting areas in Pu’er city, Yunnan province, China (Figure 1, Table 1). All sites were >30 km and <210 km apart. More than 50 healthy leaves were collected from each coffee plant host, placed in clean plastic bags, kept in iceboxes, brought to the laboratory, and refrigerated at 4 °C until further processing.
Figure 1.
(a) Aerial view of a coffee plantation in Pu’er city; (b) Arabica coffee trees; (c) Catimor coffee trees; (d) Yellow Bourbon coffee trees.
Table 1.
Information on the host species and sample collection sites.
| Host (Varieties) | Coffee Plantation Site in Pu’er City | Location (Latitude; Longitude; Altitude) | Abbreviation | Collection Date |
|---|---|---|---|---|
| Coffea arabica (Catimor) | Xiao Ao Zi coffee plantation (小凹子咖啡庄园) | 22.6618° N; 100.9584° E; 1066 m | XCE | 21 December 2020 |
| Qi Xiang coffee plantation (奇象咖啡庄园) | 22.7040° N; 101.3462° E; 900 m | QCE | 22 December 2020 | |
| Mo Jiang Jing Gong coffee plantation (墨江晶工咖啡庄园) | 23.2543° N; 101.7390° E; 1100 m | ME | 23 December 2020 | |
| Coffea arabica (Aika) | Xiang Yuan Shu He coffee plantation (香橼树河咖啡庄园) | 22.7541° N; 101.2869° E; 1006 m | AKE | 22 December 2020 |
| Coffea arabica (P4) | P4E | 22 December 2020 | ||
| Coffea arabica (PT) | PTE | 22 December 2020 | ||
| Coffea arabica (T5175) | T5E | 22 December 2020 | ||
| Coffea arabica (T8667) | T8E | 22 December 2020 | ||
| Coffea arabica (Yellow Bourbon) | Xiao Ao Zi coffee plantation (小凹子咖啡庄园) | 22.6618° N; 100.9584° E; 1066 m | XYE | 21 December 2020 |
2.2. Coffee Leaves Processing and Fungal Endophytes Isolation
The isolation process of endophytic fungi refers to the slightly modified method of Tibpromma et al. [79] and Du et al. [80]. Sterile water, 70% alcohol, 1% sodium hypochlorite, a sterile scalpel, sterile tissue paper, sterile fine-tip forceps, and potato dextrose agar (PDA) plates (supplemented with amoxicillin) were prepared. Ten healthy leaves were randomly selected for each host and each leaf was washed under running tap water and then air-dried. Working under an aseptic laminar hood, a hole puncher was used to randomly punch the leaves to obtain 5 mm size pieces (10 pieces/leaf) and these were washed in sterile water for one minute, dried on sterile tissue paper, soaked in 70% alcohol for 30 s, transferred to 1% sodium hypochlorite and soaked for one minute, then soaked in 70% alcohol for 30 s, and finally transferred to sterile water for one minute to remove chemicals. Afterwards, they were dried on sterile tissue paper; then, five pieces were placed on 90 mm-diameter PDA plates. A total of 30 leaf pieces were placed on six PDA plates for each host and each location. All PDA plates were incubated at 28 °C and observation of the culture plates was performed every day to check emerging hypha from the edge of the leaves. After colonies had grown, a small piece of mycelium with agar was cut and transferred to new PDA plates to obtain a pure culture [26,81,82]. There was a total of 270 coffee leaf pieces, including Arabica coffee (150 pieces × 5 plants × 1 place), Catimor (90 pieces × 1 plant × 3 places), and Yellow Bourbon (30 pieces × 1 plant × 1 places). A total of 235 pure cultures were isolated in this study (Table 2).
Table 2.
Records of coffee leaf fungal endophytes.
| Class | Genus | AKE | ME | P4E | PTE | QCE | T5E | T8E | XCE | XYE | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dothideomycetes | Cladosporium | - | 1 | - | - | - | - | - | 1 | - | 2 |
| Phyllosticta | 2 | 2 | 1 | 1 | 2 | 2 | - | 1 | 5 | 16 | |
| Pyrenochaetopsis | - | - | - | - | - | 1 | - | - | - | 1 | |
| Shiraia | - | - | - | - | 1 | - | - | - | - | 1 | |
| Sordariomycetes | Annulohypoxylon | 1 | - | 1 | 1 | - | - | 1 | - | - | 4 |
| Arthrinium | - | - | - | - | - | - | - | 2 | 3 | 5 | |
| Biscogniauxia | 1 | - | - | 1 | - | - | - | - | - | 2 | |
| Cladorrhinum | 1 | - | 1 | 1 | - | - | 2 | - | - | 5 | |
| Colletotrichum | 8 | 15 | 7 | 5 | 20 | 6 | 8 | 17 | 14 | 100 | |
| Daldinia | 5 | 1 | 4 | 3 | 4 | 3 | 4 | - | 1 | 25 | |
| Diaporthe | - | 2 | 1 | - | - | - | - | - | - | 3 | |
| Fusarium | - | - | - | - | 1 | - | - | - | - | 1 | |
| Hypoxylon | - | - | - | 1 | - | - | 2 | 1 | 1 | 5 | |
| Kretzschmaria | - | - | - | 1 | - | - | - | - | - | 1 | |
| Naviculispora | - | - | - | - | - | 1 | - | - | - | 1 | |
| Nemania | - | - | - | - | - | - | - | - | 2 | 2 | |
| Neurospora | - | - | 1 | - | - | - | - | - | - | 1 | |
| Nigrospora | - | - | 1 | - | - | 3 | 1 | 2 | - | 7 | |
| Nodulisporium | - | - | - | - | - | - | 2 | - | - | 2 | |
| Pestalotiopsis | - | - | - | - | - | 1 | - | - | - | 1 | |
| Xylaria | 6 | - | 5 | 4 | 1 | 3 | 6 | 1 | 3 | 29 | |
| - | Unknown | 2 | - | 1 | 5 | 2 | 4 | 6 | 1 | - | 21 |
| Total | 21 | 26 | 21 | 23 | 23 | 31 | 24 | 32 | 26 | 29 | 235 |
2.3. DNA Extraction, PCR Amplification, and ITS Gene Sequencing
The Biospin Fungus Genomic DNA Extraction Kit-BSC14S1 (BioFlux, Beijing, China) was used for DNA extraction from pure cultures grown on PDA for 7–10 days. The extraction process followed the protocol of the manufacturer. Internal transcribed spacer (ITS) regions 1 and 2 of the nuclear ribosomal DNA operon, including the 5.8S region, were amplified by the primers ITS5 and ITS4 [83]. The methods of Lu et al. [84] were followed for the polymerase chain reaction (PCR). Amplification reactions were performed in a 25 μL reaction volume which contained 2 μL DNA, 1 μL of each reverse and forward primers and 8.5 μL ddH2O, 12.5 μL 2 × FastTaq Premix (mixture of FastTaq TM DNA Polymerase, buffer, dNTP Mixture, and stabilizer) (Beijing Qingke Biological Technology Co., Ltd., Beijing, China). The amplified PCR products were sent to Bioer Technology Co., Ltd., Hangzhou, and Beijing Kinco Biotechnology Co., Ltd. Kunming Branch, China for sequencing.
2.4. Molecular Identification and Phylogenetic Analyses
A total of 235 sequences were obtained, and the newly generated sequences were merged forward and reversed by Geneious 9.1.2 (https://www.geneious.com/ (Auckland, New Zealand), accessed on 30 June 2022). The BLAST search of the sequences was carried out and the top five high-similarity results were recorded. Among our isolates, resistant genera based on published papers and less frequently isolated genera were selected (61 isolates) for the dual-culture assay. The closest match of sequences for the 61 isolates that were used for dual-culture assay were retrieved from GenBank. Multiple alignments were prepared using the online tool MAFFT version 7 (https://mafft.cbrc.jp/alignment/server/, accessed on 1 June 2022) [85], manually edited in BioEdit v. 7.0.5.2 [86], and then their format converted from FASTA to PHYLIP by ALTER online tool (https://www.sing-group.org/ALTER/, accessed on 1 April 2022) [87]. Phylogenetic analyses of the aligned sequences were conducted using the maximum likelihood method [88]. Maximum likelihood trees were generated via RAxML-HPC version 8 on XSEDE (8.2.12) [89,90] in the CIPRES Science Gateway platform [91] using the GTR+I+G model of evolution. The resulting trees were visualized with FigTree version 1.4.0 and annotated in Microsoft 365 PowerPoint. Sixty-one generated sequences were deposited in GenBank and accession numbers were obtained.
2.5. Diversity Analysis
The doughnut chart in Excel 365 was used to show the family, order, and class distribution of endophytic fungi. The colonization rate (CR) and the isolation frequency (IF) of the calculated endophytic fungi: CR = NS/NL × 100%, IF = NS/NT × 100%, NS = number of strains isolated from the location, NL = number of leaf pieces prepared from the location, NT = all number of isolates from all location. Then, the taxonomic diversity based on the species/genera (S/G) ratio was calculated to evaluate the endophyte diversity of each host. In addition, the relative abundances of the species were calculated and the similarity between the fungal communities was estimated using Sorensen’s index [92]. The species diversity of different hosts in each sampling site was calculated by the Shannon diversity (H’), Margalef diversity (d), and the Pielou evenness indices (J’), H’ = -∑ (Pi ln [Pi]), Pi = ni/N, ni = number of individuals of the species i, and N = total number of individuals of all species; d = S-1/Log N, where S is the number of species and N is the total number of specimens in the sample; J’ = H’/Log (S), where H’ is the value obtained by the Shannon index and S is species richness [26,93,94].
2.6. Tests for Antagonism: Endophytic Fungi vs. Pathogenic Fungi/Bacteria
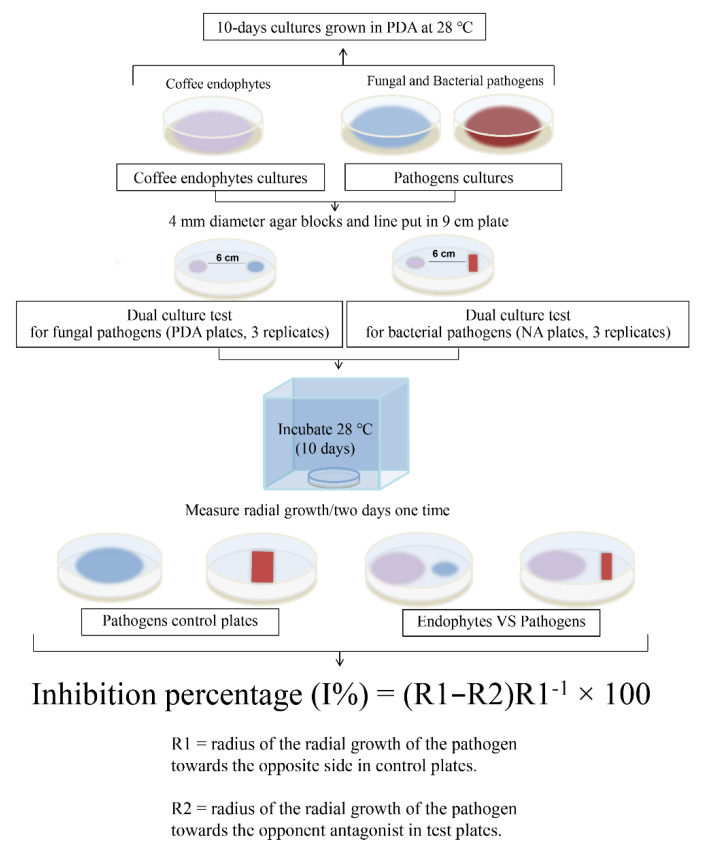
Endophytes were selected for dual-culture testing in vitro based on the information reported in the literature [95,96] and the less frequent strains from our results. The living cultures of the 61 endophytic fungi were deposited in the Kunming Institute of Botany Culture Collection (KUMCC). The 61 endophytic fungi were tested for dual-culture assays in 9 cm Petri dishes with two fungal and bacterial pathogens (i.e., Alternaria alternata, Penicillium digitatum, Pseudomonas syringae, and Salmonella enterica subsp. enterica) (Table 3) from China General Microbiological Culture Collection Center (CGMCC). These pathogens were selected for our experiments because they are common and cause different diseases on numerous hosts (Table 3) and have been used in biological control tests [97,98,99,100,101,102,103]. The PDA was supplemented with amoxicillin antibiotic and nutrient agar (NA) was prepared without amoxicillin antibiotic. The steps used are given in Figure 2.
Table 3.
Information of the pathogens obtained from CGMCC.
| Pathogen Group | Species Name | Strain Number | Major Disease |
|---|---|---|---|
| Bacteria | Pseudomonas syringae | CGMCC 1.3333 | Canker, halo blight of bean [70] |
| Salmonella enterica subsp. enterica | CGMCC 1.10603 | Food-borne illness [72] | |
| Fungi | Alternaria alternata | CGMCC 3.15535 | Black spots [64] |
| Penicillium digitatum | CGMCC 3.15410 | Fruit post-harvest diseases [66] |
Figure 2.
Dual-culture assay steps and the percentage inhibition formula.
3. Results
3.1. Isolation and Identification of Endophytic Fungi
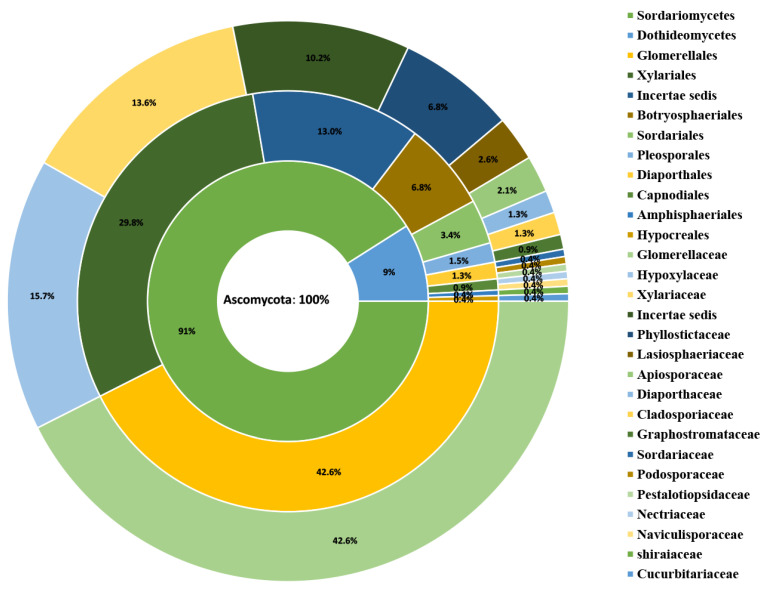
A total of 235 fungal endophytes were isolated from 270 coffee leaf pieces obtained from four coffee plantations in Yunnan province, China. All endophytes were used for the molecular identification, based on the blast result of ITS sequences, and the closest species information was obtained. Overall, 235 endophytes belong to Ascomycota (100%) and are distributed among two classes, 10 orders, and 17 families (Figure 3). At the class level, 91% are members of the Sordariomycetes, while 9% are members of the Dothideomycetes. At the order level, the Glomerellales (42.6%) is the largest group, while the Amphisphaeriales (0.4%) and the Hypocreales (0.4%) are reported as the least important. At the family level, the Glomerellaceae (42.6%) is the most common family, while the Cucurbitariaceae (0.4%), Naviculisporaceae (0.4%), Nectriaceae (0.4%), Podosporaceae (0.4%), Pestalotiopsidaceae (0.4%), Sordariaceae (0.4%), and Shiraiaceae (0.4%) are the less important families.
Figure 3.
Taxonomic distribution of the fungal taxa (n = 235) from the coffee.
Table 2 shows that 235 species are distributed in 21 genera. Colletotrichum is the most common genus, composed of 100 isolates, and found in each location and on each host. This is followed by Xylaria (composed of 29 isolates) and Daldinia (composed of 25 isolates). Seven genera (Fusarium, Kretzschmaria, Naviculispora, Neurospora, Pestalotiopsis, Pyrenochaetopsis, and Shiraia) have a low frequency of occurrence; all of these are represented by one isolate and are distributed in two coffee plantations. In addition, 21 genus names could not be determined from only the blast results of ITS, which indicated these as unidentified fungal endophytes, unidentified fungal species, or an undetermined species in the Sordariomycetes.
3.2. Phylogenetic Analyses
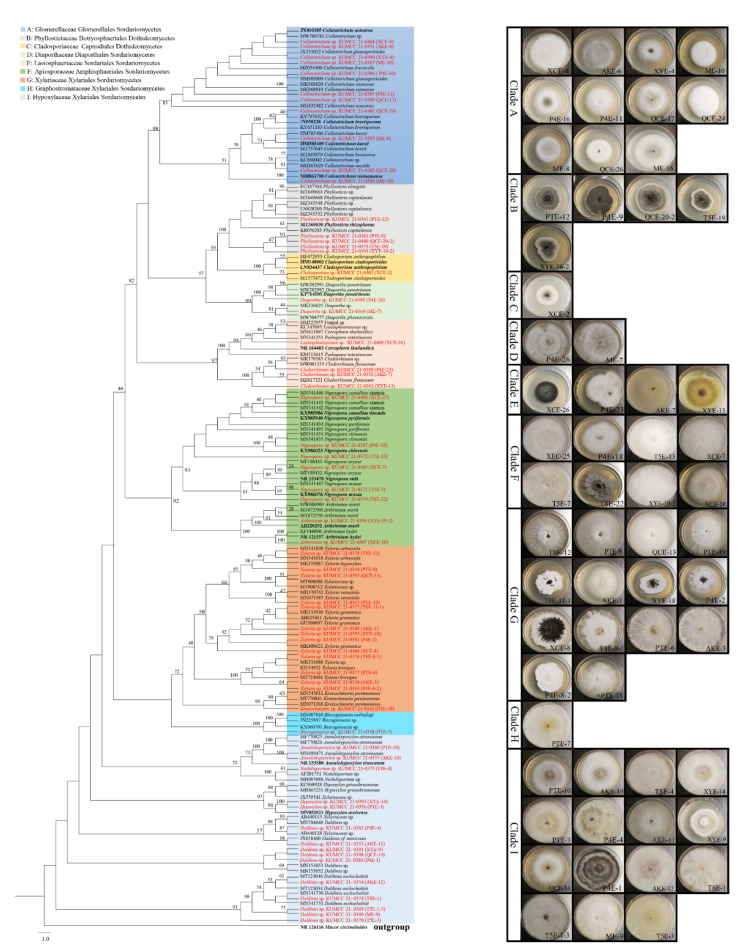
Phylogenetic analyses were carried out on the 61 endophytes that were used for the dual-culture testing. Our ITS sequences were combined with the sequences of close relatives in GenBank to construct an ITS phylogenetic tree that was composed of 167 sequences. The ML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of − 13283.937651. The alignment has 684 distinct alignment patterns, with 32.41% completely undetermined characters and gaps. Parameters for the GTR+I+G model: estimated base frequencies A = 0.246497, C = 0.253353, G = 0.238501, T = 0.261649; substitution rates AC = 0.867207, AG = 1.955827, AT = 1.257643, CG = 0.680575, CT = 2.861925, GT = 1.000000; and gamma distribution shape parameter α = 0.259865. The RAxML analysis resulted in a tree which is shown in Figure 4.
Figure 4.
A maximum likelihood analysis showing the phylogenetic relationships of coffee fungal endophytes. The tree has a total of 167 fungal sequences, of which 61 fungal sequences came from the present study and 106 reference sequences of close relatives came from the GenBank. Mucor circinelloides (NR126116; Mucoraceae; Mucorale; Mucoromycetes) was used as the outgroup taxon. The bootstrap support values (≥50%) are indicated at the nodes. The bold font indicates the type species from the GenBank and data presented in the red font are the isolate code and culture number of the fungal endophytes obtained from the coffee leaves. Fungal isolates are further highlighted in different colors according to the family classification. Images on the right side of the phylogeny tree are the morphotypes of each clade.
The phylogenetic tree includes two classes, seven orders, and nine families. Five orders (Amphisphaeriales, Diaporthales, Glomerellales, Sordariales and Xylariales) and seven families (Apiosporaceae, Diaporthaceae, Glomerellaceae, Graphostromataceae, Hypoxylaceae, Lasiosphaeriaceae and Xylariaceae) belong to the class Sordariomycetes. Another two orders (Botryosphaeriales and Capnodiales) and two families (Cladosporiaceae and Phyllostictaceae) belong to the class Dothideomycetes (Figure 4).
Since the blast result of XCE-26 has a high similarity with both Podospora intestinacea (MN341351, 99.40%) and Cercophora thailandica (NR_164483, 99.12%), we were not able to determine the genus name and it was treated as Lasiosphaeriaceae sp.
3.3. Diversity of Coffee Fungal Endophytes
In Table 4, the highest colonization rate (CR) and the isolation frequency (IF) were reported from Coffea arabica (T8667) from the Xiang Yuan Shu He coffee plantation (CR = 106.67%, IF = 13.62%), while Catimor (ME) from the Mo Jiang Jing Gong coffee plantation reported the lowest (CR = 70.00%, IF = 8.94%). By analyzing the taxonomic diversity through the computation of the (S/G) ratio, and the relative abundance of the species through the calculation of Margalef diversity (d), Shannon diversity (H’), and the Pielou evenness indices (J’), we found that Coffea arabica (P4) from the Xiang Yuan Shu He coffee plantation showed the highest species richness (d = 5.7407, H’ = 2.8944, J’ = 0.9830), followed by Coffea arabica (T5175) from the same coffee plantation (d = 4.7199, H’ = 2.6582, J’ = 0.9587). The number of species and the genera of these two hosts are P4 (18 species, nine genera, S/G = 2.00) and T5175 (15 species, eight genera, S/G = 1.88). Furthermore, the best S/G ratio was reported from Coffea arabica (PT) from the Xiang Yuan Shu He coffee plantation, which was reported as 14 species belonging to nine genera, S/G = 1.56.
Table 4.
Records and diversity of fungal endophytes associated with coffee hosts from different population sites. R = individuals/records, S = number of species, G = number of genera, d = Margalef diversity, H’ = Shannon diversity index, J’ = Pielou evenness index, CR = colonization rate, IF = isolation frequency.
| Isolate Name | Leaf Pieces | R | S | G | S/G Ratio | d | H’ | J’ | CR (%) | IF (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| AKE | 30 | 26 | 12 | 7 | 1.71 | 3.9901 | 2.4382 | 0.9506 | 86.67% | 11.06% |
| ME | 30 | 21 | 11 | 5 | 2.20 | 3.2846 | 2.2524 | 0.9393 | 70.00% | 8.94% |
| P4E | 30 | 23 | 18 | 9 | 2.00 | 5.7407 | 2.8944 | 0.9830 | 76.67% | 9.79% |
| PTE | 30 | 23 | 14 | 9 | 1.56 | 4.4650 | 2.5218 | 0.9312 | 76.67% | 9.79% |
| QCE | 30 | 31 | 16 | 6 | 2.67 | 4.3681 | 2.5246 | 0.9106 | 103.33% | 13.19% |
| T5E | 30 | 24 | 15 | 8 | 1.88 | 4.7199 | 2.6582 | 0.9587 | 80.00% | 10.21% |
| T8E | 30 | 32 | 15 | 9 | 1.67 | 4.3281 | 2.6205 | 0.9452 | 106.67% | 13.62% |
| XCE | 30 | 26 | 13 | 7 | 1.86 | 3.9901 | 2.2125 | 0.8384 | 86.67% | 11.06% |
| XYE | 30 | 29 | 13 | 7 | 1.86 | 3.5637 | 2.3303 | 0.9085 | 96.67% | 12.34% |
3.4. Dual-Culture Assay
To evaluate the antagonistic abilities of coffee endophytes, 61 coffee endophytes along with four pathogens were selected for a dual-culture assay in our study. The results are shown in Table 5. We had some dual-culture plates grown for 10 days with an inhibition rate greater or equal to 60%. For bacterial pathogens, only two endophytes exhibited significant values of growth inhibition against Pseudomonas syringae (1.3333) and Salmonella enterica subsp. enterica (1.10603) (Figure 5). In contrast, the effect of fungal pathogens (Figure 6) was appreciably greater than that of bacteria. For example, 13 endophytes exhibited significant values of growth inhibition against Alternaria alternata (3.15535) and 10 endophytes exhibited significant values of growth inhibition against Penicillium digitatum (3.15410).
Table 5.
Results of the dual-culture assay of 61 selected endophytes against the four pathogens. The type of fungal interaction for each isolate against each pathogen and the percentage of growth inhibition (I%) ± standard deviation (SD) is indicated.
| Isolate Code | Species Name | Strain Number | GenBank Accession Number (ITS) | Percent Inhibition (PI) of 10 Days ± SD | |||
|---|---|---|---|---|---|---|---|
| Penicillium digitatum CGMCC 3.15410 | Alternaria alternata CGMCC 3.15535 |
Pseudomonas syringae CGMCC 1.3333 |
Salmonella enterica subsp. enterica CGMCC 1.10603 |
||||
| AKE-1 | Xylaria sp. | KUMCC 21-0349 | ON100618 | 52.00 ± 3.56 | 43.98 ± 5.57 | 28.89 ± 9.88 | 41.67 ± 8.68 |
| AKE-3 | Xylaria sp. | KUMCC 21-0350 | ON100619 | 52.00 ± 14.22 | 44.44 ± 5.14 | 0.00 ± 0.00 | 39.58 ± 8.68 |
| AKE-6 | Colletotrichum sp. | KUMCC 21-0351 | ON100620 | 59.11 ± 1.58 | 44.44 ± 1.29 | 24.44 ± 2.47 | 43.75 ± 26.04 |
| AKE-7 | Cladorrhinum sp. | KUMCC 21-0352 | ON100621 | 52.00 ± 15.41 | 43.52 ± 0.43 | 22.22 ± 9.88 | 52.08 ± 8.68 |
| AKE-11 | Daldinia sp. | KUMCC 21-0353 | ON100622 | 67.56 ± 5.14 | 60.00 ± 2.04 | 31.11 ± 9.88 | 39.58 ± 8.68 |
| AKE-12 | Daldinia sp. | KUMCC 21-0354 | ON100623 | 64.00 ± 4.74 | 60.19 ± 0.43 | 53.33 ± 29.63 | 54.17 ± 8.68 |
| AKE-19 | Annulohypoxylon sp. | KUMCC 21-0355 | ON100624 | 53.33 ± 1.19 | 43.98 ± 3.00 | 20.00 ± 29.63 | 43.75 ± 26.04 |
| ME-7 | Diaporthe sp. | KUMCC 21-0364 | ON100633 | 64.89 ± 7.51 | 63.43 ± 0.43 | 37.78 ± 9.88 | 40.63 ± 8.68 |
| ME-8 | Colletotrichum sp. | KUMCC 21-0365 | ON100634 | 50.22 ± 0.40 | 45.83 ± 1.29 | 28.89 ± 39.51 | 41.67 ± 8.68 |
| ME-9 | Daldinia sp. | KUMCC 21-0366 | ON100635 | 74.67 ± 8.30 | 71.30 ± 0.43 | 60.00 ± 7.41 | 50.00 ± 6.51 |
| ME-10 | Colletotrichum sp. | KUMCC 21-0367 | ON100636 | 51.56 ± 5.14 | 46.30 ± 3.00 | 31.11 ± 2.47 | 41.67 ± 8.68 |
| ME-16 | Colletotrichum sp. | KUMCC 21-0368 | ON100637 | 57.78 ± 0.40 | 44.44 ± 1.29 | 26.67 ± 29.63 | 29.17 ± 8.68 |
| PTE-3 | Hypoxylon sp. | KUMCC 21-0356 | ON100625 | 58.67 ± 3.56 | 56.94 ± 5.14 | 4.44 ± 9.88 | 39.58 ± 8.68 |
| PTE-6 | Xylaria sp. | KUMCC 21-0357 | ON100626 | 50.22 ± 0.40 | 37.50 ± 9.00 | 0.00 ± 0.00 | 45.83 ± 34.72 |
| PTE-7 | Biscogniauxia sp. | KUMCC 21-0358 | ON100627 | 67.56 ± 0.40 | 61.11 ± 1.29 | 53.33 ± 0.00 | 60.42 ±8.68 |
| PTE-9 | Xylaria sp. | KUMCC 21-0359 | ON100628 | 50.22 ± 0.40 | 43.06 ± 1.29 | 35.56 ± 9.88 | 25.00 ± 26.04 |
| PTE-10 | Annulohypoxylon sp. | KUMCC 21-0360 | ON100629 | 57.78 ± 2.77 | 58.33 ± 0.00 | 31.11 ± 17.28 | 39.58 ± 34.72 |
| PTE-12 | Phyllosticta sp. | KUMCC 21-0361 | ON100630 | 46.67 ± 18.96 | 44.44 ± 1.29 | 28.89 ± 17.28 | 37.50 ± 6.51 |
| PTE-18 | Kretzschmaria sp. | KUMCC 21-0362 | ON100631 | 47.56 ± 0.40 | 37.96 ± 3.00 | 0.00 ± 0.00 | 39.58 ± 8.68 |
| PTE-19 | Xylaria sp. | KUMCC 21-0363 | ON100632 | 47.56 ± 0.40 | 41.20 ± 5.57 | 0.00 ± 0.00 | 39.58 ± 8.68 |
| P4E-1 | Daldinia sp. | KUMCC 21-0380 | ON072525 | 64.89 ± 6.32 | 54.63 ± 1.71 | 46.67 ± 29.63 | 41.67 ± 8.68 |
| P4E-2 | Xylaria sp. | KUMCC 21-0381 | ON072526 | 46.67 ± 1.19 | 29.63 ± 3.00 | 33.33 ± 0.00 | 37.50 ± 0.00 |
| P4E-4 | Daldinia sp. | KUMCC 21-0382 | ON072527 | 47.56 ± 0.40 | 53.70 ± 3.00 | 4.44 ± 39.51 | 47.92 ± 8.68 |
| P4E-8-2 | Xylaria sp. | KUMCC 21-0383 | ON072528 | 49.78 ± 0.40 | 30.56 ± 1.29 | 8.89 ± 69.14 | 39.58 ± 8.68 |
| P4E-9 | Phyllosticta sp. | KUMCC 21-0384 | ON072529 | 44.89 ± 2.77 | 30.56 ± 1.29 | 33.33 ± 0.00 | 41.67 ± 8.68 |
| P4E-11 | Colletotrichum sp. | KUMCC 21-0385 | ON072530 | 47.56 ± 2.77 | 26.39 ± 1.29 | 22.22 ± 39.51 | 37.50 ± 0.00 |
| P4E-16 | Colletotrichum sp. | KUMCC 21-0386 | ON072531 | 40.89 ± 0.40 | 32.87 ± 3.00 | 33.33 ± 29.63 | 39.58 ± 8.68 |
| P4E-18 | Nigrospora sp. | KUMCC 21-0387 | ON072532 | 43.56 ± 0.40 | 29.17 ± 1.29 | 28.89 ± 69.14 | 45.83 ± 8.68 |
| P4E-23 | Cladorrhinum sp. | KUMCC 21-0388 | ON072533 | 43.56 ± 0.40 | 28.24 ± 3.00 | 31.11 ± 32.10 | 43.75 ± 0.00 |
| P4E-26 | Diaporthe sp. | KUMCC 21-0389 | ON072534 | 46.22 ± 0.40 | 34.72 ± 1.29 | 31.11 ± 9.88 | 41.67 ±8.68 |
| QCE-13 | Xylaria sp. | KUMCC 21-0397 | ON072542 | 37.33 ± 1.19 | 24.07 ± 3.00 | 2.22 ± 9.88 | 25.00 ± 26.04 |
| QCE-14 | Daldinia sp. | KUMCC 21-0398 | ON072543 | 44.89 ± 0.40 | 53.70 ± 1.71 | 24.44 ± 9.88 | 41.67 ± 8.68 |
| QCE-17 | Colletotrichum sp. | KUMCC 21-0399 | ON072544 | 44.89 ± 1.58 | 34.26 ± 3.00 | 6.67 ± 29.63 | 45.83 ± 34.72 |
| QCE-20-2 | Phyllosticta sp. | KUMCC 21-0400 | ON072545 | 44.44 ± 1.58 | 30.56 ± 1.29 | 0.00 ± 0.00 | 41.67 ± 8.68 |
| QCE-24 | Colletotrichum sp. | KUMCC 21-0401 | ON072546 | 55.56 ± 2.77 | 44.44 ± 1.29 | 22.22 ± 69.14 | 47.92 ± 8.68 |
| QCE-26 | Colletotrichum sp. | KUMCC 21-0402 | ON072547 | 46.67 ± 1.19 | 34.72 ± 1.29 | 0.00 ± 0.00 | 39.58 ± 8.68 |
| T5E-1-3 | Daldinia sp. | KUMCC 21-0369 | ON100638 | 70.67 ± 4.74 | 63.43 ± 3.00 | 55.56 ± 9.88 | 60.42 ± 8.68 |
| T5E-3 | Daldinia sp. | KUMCC 21-0370 | ON100639 | 68.00 ± 1.19 | 68.52 ± 0.43 | 21.11 ± 17.28 | 39.58 ± 8.68 |
| T5E-7 | Nigrospora sp. | KUMCC 21-0371 | ON100640 | 72.00 ± 1.19 | 65.74 ± 3.00 | 13.33 ± 29.63 | 41.67 ± 6.51 |
| T5E-13 | Nigrospora sp. | KUMCC 21-0372 | ON100641 | 65.78 ± 2.77 | 48.61 ± 1.29 | 27.78 ± 17.28 | 52.08 ± 8.68 |
| T5E-19 | Phyllosticta sp. | KUMCC 21-0373 | ON100642 | 51.56 ± 2.77 | 36.11 ± 5.14 | 24.44 ± 9.88 | 35.42 ± 8.68 |
| T8E-1 | Daldinia sp. | KUMCC 21-0374 | ON100643 | 52.89 ± 11.06 | 55.56 ± 5.14 | 46.67 ± 29.63 | 5.00 ± 0.00 |
| T8E-4 | Nodulisporium sp. | KUMCC 21-0375 | ON100644 | 58.22 ± 2.77 | 45.37 ± 3.00 | 35.56 ± 9.88 | 43.75 ± 26.4 |
| T8E-8-1 | Xylaria sp. | KUMCC 21-0376 | ON100645 | 45.78 ± 2.77 | 43.98 ± 3.00 | 13.33 ± 29.63 | 41.67 ± 8.68 |
| T8E-11-1 | Xylaria sp. | KUMCC 21-0377 | ON100646 | 43.56 ± 2.77 | 45.37 ± 0.43 | 35.56 ± 9.88 | 35.42 ± 8.68 |
| T8E-12 | Xylaria sp. | KUMCC 21-0378 | ON100647 | 44.44 ± 5.14 | 43.06 ± 1.29 | 28.89 ± 39.51 | 41.67 ± 15.19 |
| T8E-22 | Nigrospora sp. | KUMCC 21-0379 | ON100648 | 53.33 ± 1.19 | 43.98 ± 0.43 | 2.22 ± 9.88 | 43.75 ± 26.04 |
| XCE-2 | Cladosporium sp. | KUMCC 21-0403 | ON072548 | 42.67 ± 1.19 | 32.41 ± 0.43 | 8.89 ± 39.51 | 39.58 ± 8.68 |
| XCE-4 | Colletotrichum sp. | KUMCC 21-0404 | ON072549 | 47.56 ± 2.77 | 37.50 ± 1.29 | 4.44 ± 9.88 | 37.50 ± 26.04 |
| XCE-7 | Nigrospora sp. | KUMCC 21-0405 | ON072550 | 68.00 ± 1.19 | 71.76 ± 0.43 | 61.11± 32.10 | 56.25 ± 0.00 |
| XCE-8 | Xylaria sp. | KUMCC 21-0406 | ON072551 | 46.67 ± 1.19 | 30.56 ± 1.29 | 40.00 ± 29.63 | 29.17 ± 8.68 |
| XCE-10 | Arthrinium sp. | KUMCC 21-0407 | ON072552 | 64.90 ± 2.77 | 57.87 ± 3.00 | 28.89 ± 39.51 | 35.42 ± 15.19 |
| XCE-25 | Nigrospora sp. | KUMCC 21-0408 | ON072553 | 67.56 ± 6.32 | 62.50 ± 1.29 | 15.56 ± 9.88 | 45.83 ± 34.82 |
| XCE-26 | Lasiosphaeriaceae sp. | KUMCC 21-0409 | ON072554 | 48.44 ± 0.40 | 34.26 ± 3.00 | 26.67 ± 0.00 | 33.33 ± 8.68 |
| XYE-4 | Colletotrichum sp. | KUMCC 21-0390 | ON072535 | 42.22 ± 1.58 | 32.87 ± 1.71 | 15.56 ± 39.51 | 41.67 ± 8.68 |
| XYE-9 | Daldinia sp. | KUMCC 21-0391 | ON072536 | 47.56 ± 0.40 | 31.48 ± 0.43 | 22.22 ± 9.88 | 41.67 ± 8.68 |
| XYE-13 | Cladorrhinum sp. | KUMCC 21-0392 | ON072537 | 44.44 ± 0.40 | 29.17 ± 1.29 | 46.67 ± 29.63 | 56.25 ± 0.00 |
| XYE-14 | Hypoxylon sp. | KUMCC 21-0393 | ON072538 | 44.44 ± 2.77 | 34.72 ± 1.29 | 33.33 ± 0.00 | 35.42 ± 8.68 |
| XYE-16-2 | Phyllosticta sp. | KUMCC 21-0394 | ON072539 | 46.22 ± 0.40 | 31.02 ± 5.57 | 2.22 ± 9.88 | 39.58 ± 8.68 |
| XYE-18 | Xylaria sp. | KUMCC 21-0395 | ON072540 | 38.22 ± 5.14 | 31.94 ± 1.29 | 33.33 ± 0.00 | 39.58 ± 8.68 |
| XYE-19-2 | Arthrinium sp. | KUMCC 21-0396 | ON072541 | 44.44 ± 2.77 | 34.72 ± 1.29 | 15.56 ± 9.88 | 27.08 ± 8.68 |
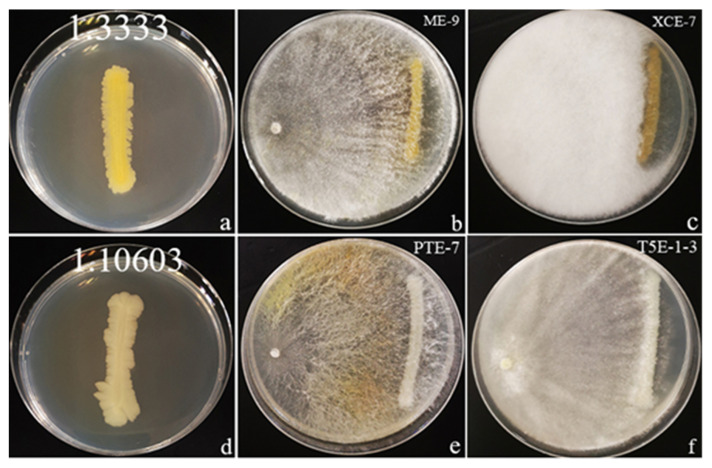
Figure 5.
Illustration of an antagonist test by the dual-culture technique with two bacterial pathogens and an inhibition rate greater than or equal to 60%. The two pathogens were cocultivated with the coffee fungal endophytes on NA plates and incubated for 10 days at 28 °C. (a) Pseudomonas syringae control plate; (b,c) Coffee endophytes inhibit Pse. syringae; (d) Salmonella enterica subsp. enterica control plate; (e,f) Coffee endophytes inhibit S. enterica subsp. enterica.
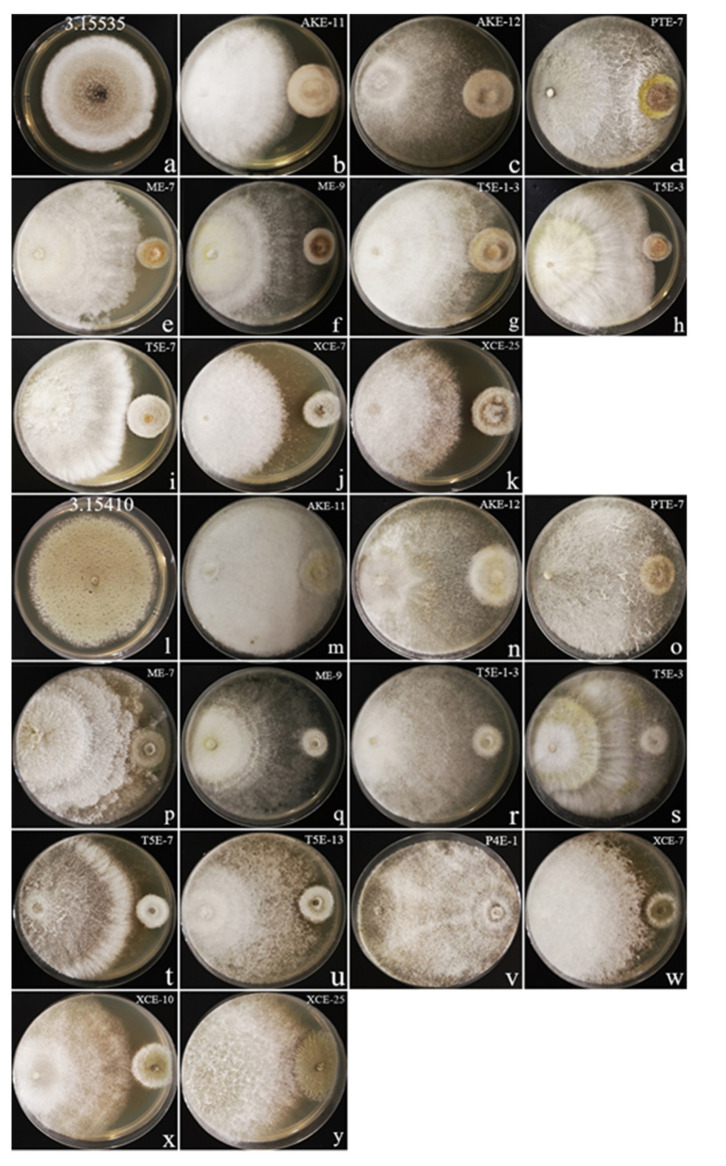
Figure 6.
Illustration of an antagonist test using the dual-culture technique with two fungal pathogens and an inhibition rate greater than or equal to 60%. The two pathogens were cocultivated with the coffee fungal endophytes on PDA plates and incubated for ten days at 28 °C. (a) Alternaria alternata control plate; (b–k) Coffee endophytes inhibit A. alternata; (l) Penicillium digitatum control plate; (m–y) Coffee endophytes inhibit Pen. digitatum.
3.4.1. Effect of Coffee Endophytes on the Growth of Pseudomonas syringae (1.3333)
Nigrospora sp. XCE-7 (Figure 5b) was able to inhibit the growth of Pse. syringae with an inhibition rate of 61.11%, and Daldinia sp. ME-9 (Figure 5c) showed an inhibition rate of 60.00%. The Pse. syringae control plate is shown in Figure 5a.
3.4.2. Effect of Coffee Endophytes on the Growth of Salmonella enterica subsp. enterica (1.10603)
Biscogniauxia sp. PTE-7 (Figure 5e) and Daldinia sp. T5E-1-3 (Figure 5f) showed the same inhibition rate of 60.42% against the growth of S. enterica subsp. enterica. PTE-7 produced a yellow mycelium after it was in contact with bacterial pathogens. The S. enterica subsp. enterica control plate is shown in Figure 5d.
3.4.3. Effect of Coffee Endophytes on the Growth of Alternaria alternata (3.15535)
Nigrospora sp. XCE-7 (Figure 6j) inhibited the growth of A. alternata with an interaction characterized by a clear zone around the pathogen and an inhibition rate of 71.76%. It was followed by Daldinia sp. ME-9 (Figure 6f) with an inhibition rate of 71.30%. Those demonstrating inhibition rates greater than or equal to 60 were Daldinia sp. T5E-3 (Figure 6h, 68.52%), Nigrospora sp. T5E-7 (Figure 6i, 65.74%), Daldinia sp. T5E-1-3 (Figure 6g, 63.43%), Diaporthe sp. ME-7 (Figure 6e, 63.43%), Nigrospora sp. XCE-25 (Figure 6k, 62.50%), Biscogniauxia sp. PTE-7 (Figure 6d, 61.11%), Daldinia sp. AKE-12 (Figure 6c, 60.19%), and Daldinia sp. AKE-11 (Figure 6b, 60.00%). The A. alternata control plate is shown in Figure 6a.
3.4.4. Effect of Coffee Endophytes on the Growth of Penicillium digitatum (3.15410)
Daldinia sp. ME-9 (Figure 6q) inhibited the growth of Pen. digitatum with an inhibition rate of 74.67%, and this was followed by Nigrospora sp. T5E-7 (Figure 6t) with an inhibition rate of 72.00%. Inhibition rates greater than or equal to 60 were found in Daldinia sp. T5E-1-3 (Figure 6r, 70.67%), Daldinia sp. T5E-3(Figure 6s, 68.00%), Nigrospora sp. XCE-7 (Figure 6w, 68.00%), Daldinia sp. AKE-11 (Figure 6m, 67.56%), Biscogniauxia sp. PTE-7 (Figure 6o, 67.56%), Nigrospora sp. XCE-25 (Figure 6y, 67.56%), Nigrospora sp. T5E-13 (Figure 6u, 65.78%), Arthrinium sp. XCE-10 (Figure 6x, 64.90%), Diaporthe sp. ME-7 (Figure 6p, 64.89%), Daldinia sp. P4E-1 (Figure 6v, 64.89%), and Daldinia sp. AKE-12 (Figure 6n, 64.00%). Among these, XCE-7 and T5E-13 inhibited the growth of Pen. digitatum by displaying a small zone around the pathogen. The Pen. digitatum control plate is shown in Figure 6l.
Overall, this dual-culture assay demonstrated that most coffee endophytes inhibit the growth of pathogens by competition for nutrients and space, while only two endophytes displayed the mechanism of antibiosis to inhibit the growth of pathogens by producing metabolites (antibiotics and enzymes). One of the Nigrospora sp. (XCE-7) isolates produced a clear zone of inhibition for Pseudomonas syringae, Alternaria alternata, and Penicillium digitatum, while Nigrospora sp. (T5E-13) had a zone of inhibition only for Pen. digitatum. In this study, Arthrinium sp., Biscogniauxia sp., Daldinia sp., Diaporthe sp. and Nigrospora sp. produced different levels of inhibitory effects on the growth of pathogens. Arthrinium sp. (XCE-10) inhibited only the growth of one fungal pathogen (Pen. digitatum), but Diaporthe sp. (ME-7) inhibited the growth of two fungal pathogens (Alternaria alternata and Pen. digitatum). Biscogniauxia sp. (PTE-7), Daldinia sp. (AKE-11, AKE-12, ME-9, P4E-1, T5E-1-3, and T5E-3), and Nigrospora sp. (T5E-13, T5E-7, XCE-7, and XCE-25) inhibited the growth of fungal pathogens as well as bacterial pathogens.
4. Discussion
The results of our study agree with the results of previous studies [59,81,93,104], which showed that Colletotrichum and Xylaria are the dominant endophytes in coffee leaves. However, the species richness and diversity of endophytes as determined in the present study are different and higher than those reported by Santamarıá and Baymanet [81] in Puerto Rico, Saucedo-García et al. [104] in Mexico, and Oliveira et al. [93] in Brazil for coffee leaf endophytes. Species of Colletotrichum are common saprobes, pathogens, and endophytes on a range of economically important plant hosts in tropical regions [105,106], and species of Xylaria are recognized as saprotrophic fungi and as endophytic fungi of many plants [107]. Other unique and less frequently encountered species were reported, most likely due to different climates, altitude, humidity, and sampling period. Herein, nine genera are reported for the first time to be isolated as coffee endophytes. These are Annulohypoxylon, Biscogniauxia, Kretzschmaria, Naviculispora, Neurospora, and Pyrenochaetopsis, and they were isolated from Coffea arabica in the Xiang Yuan Shu He coffee plantation. Fusarium sp. and Shiraia sp. were isolated from Catimor in the Qi Xiang coffee plantation, and Nemania sp. was isolated from Yellow Bourbon in the Xiao Ao Zi coffee plantation. In this study, only the PDA medium was used, and it is possible that some species were not capable of growing on PDA.
Endophytic fungi isolated from Coffea showed promising sources of bioactive compounds [61]. Fulthorpe et al. [108] reported the isolation of coffee root endophytes that have potential ecological roles with Coffea arabica based on next-generation sequencing, and Burkholderia, Enterobacter and Pantoea were reported as the dominant bacterial genera while Cladosporium, Exidiopsis, Mycena, Penicillium and Trechispora were reported as the dominant fungal genera [108]. Sette et al. [109] used a minimal inhibitory concentration test (MIC) to investigate the endophytic filamentous fungi against different human pathogenic bacteria from coffee plants (Coffea arabica and C. robusta) in Pedreira, Mococa, and Pindorama counties in Brazil. The study showed that 17 endophytic fungi were able to inhibit at least one of the bacterial pathogens while Guignardia sp. (CBMAI 69), Phomopsis sp. (CBMAI 164), and Trichoderma harzianum (CBMAI 43) were able to inhibit four to five bacterial species. Furthermore, the study showed that Aspergillus versicolor (CBMAI 46), Cladosporium sp. (CBMAI 64 and CBMAI 66), Fusarium oxysporum (CBMAI 53), and Glomerella sp. (CBMAI 63) can inhibit all pathogenic bacteria [109]. The previous studies show that coffee-endophytic fungi are diverse and different groups of fungi are able to inhibit different fungal and bacterial pathogens [58,59,60,61,62,63,108,109]. When comparing those results with our study, different endophytic fungi isolated in our study, such as Arthrinium sp., Biscogniauxia sp., Daldinia sp., Diaporthe sp. and Nigrospora sp., can also inhibit the growth of pathogens but they are not the same fungal genera that were reported in previous studies [58,59,60,61,62,63,108,109].
Arthrinium is known for its antifungal capacity [110]. The species A. aureum and A. phaeospermum have the potential to inhibit the growth of Fusarium oxysporum and F. niveum and thus can be applied in biological controls [111]. Our isolate Arthrinium sp. (XCE-10) showed an inhibition rate of 64.89% against the growth of Penicillium digitatum.
Biscogniauxia sp. (O-811) isolated from the fresh fruiting bodies of wild mushrooms acts against the rice blast disease Magnaporthe oryzae by producing an inhibitory compound [112], while our endophytic strain Biscogniauxia sp. (PTE-7) can inhibit the growth of the fungal pathogens Alternaria alternata and Penicillium digitatum while also inhibiting the bacterial pathogen Salmonella enterica subsp. enterica with inhibition rates of 61.11%, 67.56%, and 60.42%, respectively.
Daldinia cf. concentrica prevented the development of molds on organic dried fruits and eliminated Aspergillus niger infection in peanuts by emitting volatile organic compounds [113]. The endophytic fungus Daldinia eschscholtzii BPEF73 of black pepper showed nematocidal activity by producing metabolites [114]. Six Daldinia isolates (T5E-1-3, T5E-3, AKE-11, P4E-1, AKE-12, and ME-9) in our study showed good antagonistic abilities against all four pathogens tested. The isolate ME-9 inhibited Penicillium digitatum with the highest inhibition rate of 74.67%.
Studies have shown that some endophytic species belonging to the genus Diaporthe can produce metabolites that protect the host from infection or act as a biological control for disease. For example, the endophytic fungus Diaporthe phaseolorum isolated from tomatoes can produce 16 different compounds, and it has an inhibitory effect on the growth of Xanthomonas vesicatoria, which causes bacterial spot disease in tomatoes [115]. Dhakshinamoorthy et al. [116] showed that the endophytic fungus Diaporthe caatingaensis from Buchanania axillaris leaves can produce the bioactive metabolite Camptothecin (CPT). While our endophyte Diaporthe sp. ME-7 has inhibitory effects on the growth of the fungal pathogens Alternaria alternata (63.43%) and Penicillium digitatum (64.89%), it did not show any apparent inhibitory effects on bacterial pathogens.
Nigrospora oryzae was isolated as an endophytic fungus from the leaves of Coccinia grandis and its secondary metabolites can be used as drugs to control diabetes. They also exhibit strong antifungal activities against the plant pathogen Cladosporium cladosporioides [117]. Our four Nigrospora isolates T5E-13, T5E-7, XCE-7, and XCE-25 showed antagonistic abilities for all three pathogens. The best isolate among all of these was XCE-7 which showed the highest inhibition rate of 71.76% against Alternaria alternata.
Our study is the first report on coffee-leaf-endophytic fungal diversity in China. We also tested the antagonistic abilities of endophytic fungi present in coffee leaves against major bacterial and fungal pathogens. Two isolates (Nigrospora sp. XCE-7 and Daldinia sp. ME-9) showed antagonism against the growth of Alternaria alternata, with an inhibition rate of over 70%. Three isolates (Daldinia sp. ME-9, Nigrospora sp. T5E-7, and Daldinia sp. T5E1-3) displayed antagonism against the growth of Penicillium digitatum, with an inhibition rate of over 70%. It is necessary to carry out further research on secondary metabolites and identification of the four coffee endophytes ME-9, T5E-1-3, T5E-7, and XCE-7 to the species level.
Acknowledgments
Li Lu thanks Mae Fah Luang University for the award of a fee-less scholarship. Center for Mountain Futures, Kunming Institute of Botany, Chinese Academy of Science; Center for Yunnan Plateau Biological Resources Protection and Utilization, College of Biological Resource and Food Engineering, Qujing Normal University; and Center of Excellence in Fungal Research are thanked by Li Lu for the facilities provided for the research work. Li Lu is also very grateful to Xiong Yin-Ru and Dao Chen-Jiao for helping her with the data analyses.
Author Contributions
Conceptualization, S.C.K. and S.T.; Formal analysis, L.L.; Funding acquisition, S.C.K., A.M.E., S.A.-R., S.T. and N.S.; Methodology, L.L.; Software, L.L.; Supervision, S.C.K. and S.T.; Writing—Original draft, L.L.; Writing—Review and editing, S.C.K., K.D.H., N.S., A.M.E., S.L.S., S.A.-R., R.S.J. and S.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The authors extend their appreciation to the researchers supporting project number (RSP-2021/120) King Saud University, Riyadh, Saudi Arabia. This research work was partially supported by Chiang Mai University.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ghosh P., Venkatachalapathy N. Processing and drying of coffee–A review. Int. J. Eng. Res. Technol. 2014;3:784–794. [Google Scholar]
- 2.Asegid A. Impact of climate change on production and diversity of coffee (Coffea arabica L.) in Ethiopia. Int. J. Res. Stud. Sci. Eng. Technol. 2020;7:31–38. [Google Scholar]
- 3.Ribeyre F., Avelino J. Impact of Field Pests and Diseases on Coffee Quality. Specialty Coffee: Managing Quality. 2012. [(accessed on 15 May 2022)]. pp. 151–176. Available online: https://www.researchgate.net/profile/Fabienne-Ribeyre/publication/259574741_Impact_of_field_pests_and_diseases_on_coffee_quality/links/00b7d52cabcde3a0b5000000/Impact-of-field-pests-and-diseases-on-coffee-quality.pdf.
- 4.Lemessa F., Abera A., Adunga G., Garedew W. Association of mycoflora with coffee (Coffea arabica L.) beans at Limmu coffee plantation, Southwestern Ethiopia. Plant Pathol. J. 2015;14:136. doi: 10.3923/ppj.2015.136.141. [DOI] [Google Scholar]
- 5.Cerda R., Avelino J., Gary C., Tixier P., Lechevallier E., Allinne C. Primary and secondary yield losses caused by pests and diseases: Assessment and modeling in coffee. PLoS ONE. 2017;12:e0169133. doi: 10.1371/journal.pone.0169133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford M.A., Phiri N. Pests and Diseases of Coffee in Eastern Africa: A Technical and Advisory Manual. CAB International; Wallingford, UK: 2006. [Google Scholar]
- 7.Muller R.A., Berry D., Avelino J., Bieysse D. Coffee: Growing, Processing, Sustainable Production. A Guidebook for Growers, Processors, Traders and Researchers. Wiley-VCH; Weinheim, Germany: 2009. pp. 495–549. [Google Scholar]
- 8.Capucho A., Zambolim L., Lopes U., Milagres N. Chemical control of coffee leaf rust in Coffea canephora cv. conilon. Australas. Plant Pathol. 2013;42:667–673. doi: 10.1007/s13313-013-0242-y. [DOI] [Google Scholar]
- 9.Ghini R., Bettiol W., Hamada E. Diseases in tropical and plantation crops as affected by climate changes: Current knowledge and perspectives. Plant Pathol. 2011;60:122–132. doi: 10.1111/j.1365-3059.2010.02403.x. [DOI] [Google Scholar]
- 10.Baker K.F. Evolving concepts of biological control of plant pathogens. Annu. Rev. Phytopathol. 1987;25:67–85. doi: 10.1146/annurev.py.25.090187.000435. [DOI] [Google Scholar]
- 11.Cook R.J. Biological control and holistic plant-health care in agriculture. Am. J. Altern. Agric. 1988;3:51–62. doi: 10.1017/S0889189300002186. [DOI] [Google Scholar]
- 12.Khalil A.M.A., Abdelaziz A.M., Khaleil M.M., Hashem A.H. Fungal endophytes from leaves of Avicennia marina growing in semi-arid environment as a promising source for bioactive compounds. Lett. Appl. Microbiol. 2021;72:263–274. doi: 10.1111/lam.13414. [DOI] [PubMed] [Google Scholar]
- 13.Khalil A.M.A., Hassan S.E.D., Alsharif S.M., Eid A.M., Ewais E.E.D., Azab E., Gobouri A.A., Elkelish A., Fouda A. Isolation and characterization of fungal endophytes isolated from medicinal plant Ephedra pachyclada as plant growth-promoting. Biomolecules. 2021;11:140. doi: 10.3390/biom11020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faull J. Competitive antagonism of soilborne plant pathogens. Fungi Biol. Control. Syst. 1988;48:303–309. [Google Scholar]
- 15.Van Driesche R., Bellows T.S., Jr. Biological Control. New York (USA) Chapman & Hall; México, NA, USA: 2012. [Google Scholar]
- 16.Hokkanen H.M., Lynch J.M. Biological Control: Benefits and Risks. Cambridge University Press; Cambridge, UK: 2003. [Google Scholar]
- 17.De Silva N.I., Brooks S., Lumyong S., Hyde K.D. Use of endophytes as biocontrol agents. Fungal Biol. Rev. 2019;33:133–148. doi: 10.1016/j.fbr.2018.10.001. [DOI] [Google Scholar]
- 18.Elnahal A.S., El-Saadony M.T., Saad A.M., Desoky E.-S.M., El-Tahan A.M., Rady M.M., AbuQamar S.F., El-Tarabily K.A. The use of microbial inoculants for biological control, plant growth promotion, and sustainable agriculture: A review. Eur. J. Plant Pathol. 2022;162:759–792. doi: 10.1007/s10658-021-02393-7. [DOI] [Google Scholar]
- 19.Köhl J., Kolnaar R., Ravensberg W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019;10:845. doi: 10.3389/fpls.2019.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nair D.N., Padmavathy S. Impact of endophytic microorganisms on plants, environment and humans. Sci. World J. 2014;2014:250693. doi: 10.1155/2014/250693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan Y., Gao L., Chang P., Li Z. Endophytic fungal community in grape is correlated to foliar age and domestication. Ann. Microbiol. 2020;70:30. doi: 10.1186/s13213-020-01574-9. [DOI] [Google Scholar]
- 22.Hyde K.D., Soytong K. The fungal endophyte dilemma. Fungal Divers. 2008;33:e173. [Google Scholar]
- 23.Terhonen E., Blumenstein K., Kovalchuk A., Asiegbu F.O. Forest tree microbiomes and associated fungal endophytes: Functional roles and impact on forest health. Forests. 2019;10:42. doi: 10.3390/f10010042. [DOI] [Google Scholar]
- 24.Langer G.J., Bußkamp J., Terhonen E., Blumenstein K. Forest Microbiology. Academic Press; Cambridge, MA, USA: 2021. Fungi inhabiting woody tree tissues; pp. 175–205. [Google Scholar]
- 25.Navarro-Meléndez A.L., Heil M. Symptomless endophytic fungi suppress endogenous levels of salicylic acid and interact with the jasmonate-dependent indirect defense traits of their host, lima bean (Phaseolus lunatus) J. Chem. Ecol. 2014;40:816–825. doi: 10.1007/s10886-014-0477-2. [DOI] [PubMed] [Google Scholar]
- 26.Fadiji A.E., Babalola O.O. Exploring the potentialities of beneficial endophytes for improved plant growth. Saudi J. Biol. Sci. 2020;27:3622–3633. doi: 10.1016/j.sjbs.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pecundo M.H., dela Cruz T.E.E., Chen T., Notarte K.I., Ren H., Li N. Diversity, phylogeny and antagonistic activity of fungal endophytes associated with endemic species of Cycas (Cycadales) in China. J. Fungi. 2021;7:572. doi: 10.3390/jof7070572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar V., Jain L., Kaushal P., Soni R. Fungi Bio-Prospects in Sustainable Agriculture, Environment and Nano-Technology. Elsevier, Academic Press; Cambridge, MA, USA: 2021. Fungal endophytes and their applications as growth promoters and biological control agents; pp. 315–337. [Google Scholar]
- 29.Azeem F., Rashid F., Shahzadi M., Abbas A., Batool R., Nadeem H., Moosa A., Siddique M.H., Hussain S., Ali M.A. Recent Developments in Microbial Technologies. Springer; Singapore: 2021. Endophytes as guardians of plants against diseases; pp. 221–242. [Google Scholar]
- 30.Saad M.M., Ghareeb R.Y., Saeed A.A. The potential of endophytic fungi as bio-control agents against the cotton leafworm, Spodoptera littoralis (Boisd.) (Lepidoptera: Noctuidae) Egypt. J. Biol. Pest Control. 2019;29:7. doi: 10.1186/s41938-019-0108-x. [DOI] [Google Scholar]
- 31.Ma X.Y., Kang J.C., Hyde K.D., Doilom M., Chomnunti P. Pezicula endophytica sp. nov., endophytic in Dendrobium in Thailand. Mycotaxon. 2021;136:563–577. doi: 10.5248/136.563. [DOI] [Google Scholar]
- 32.Monkai J., Tibpromma S., Manowong A., Mapook A., Norphanphoun C., Hyde K.D., Promputtha I. Discovery of three novel Cytospora species in Thailand and their antagonistic potential. Diversity. 2021;13:488. doi: 10.3390/d13100488. [DOI] [Google Scholar]
- 33.Saengket M., Hyde K.D., Kumar V., Doilom M., Brooks S. Endophytic fungi from Oncosperma sp. with promising in vitro plant growth promotion and antagonistic activities. Chiang Mai J. Sci. 2021;48:837–852. [Google Scholar]
- 34.Fadiji A.E., Babalola O.O. Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Front. Bioeng. Biotechnol. 2020;8:467. doi: 10.3389/fbioe.2020.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abo Nouh F.A. Endophytic fungi for sustainable agriculture. Microb. Biosyst. 2019;4:31–44. [Google Scholar]
- 36.Ortega H.E., Torres-Mendoza D., Cubilla-Rios L. Patents on endophytic fungi for agriculture and bio- and phytoremediation applications. Microorganisms. 2020;8:1237. doi: 10.3390/microorganisms8081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muñoz-Guerrero J., Guerra-Sierra B.E., Alvarez J.C. Fungal endophytes of tahiti lime (Citrus citrus × latifolia) and their potential for control of Colletotrichum acutatum J.H. simmonds causing anthracnose. Front. Bioeng. Biotechnol. 2021;9:255. doi: 10.3389/fbioe.2021.650351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontana D.C., de Paula S., Torres A.G., de Souza V.H.M., Pascholati S.F., Schmidt D., Dourado Neto D. Endophytic fungi: Biological control and induced resistance to phytopathogens and abiotic stresses. Pathogens. 2021;10:570. doi: 10.3390/pathogens10050570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulz B., Boyle C., Draeger S., Römmert A.K., Krohn K. Endophytic fungi: A source of novel biologically active secondary metabolites. Mycol. Res. 2002;106:996–1004. doi: 10.1017/S0953756202006342. [DOI] [Google Scholar]
- 40.Priti V., Ramesha B., Singh S., Ravikanth G., Ganeshaiah K., Suryanarayanan T., Uma Shaanker R. How promising are endophytic fungi as alternative sources of plant secondary metabolites? Curr. Sci. 2009;97:477–478. [Google Scholar]
- 41.Wang W., Zhai Y., Cao L., Tan H., Zhang R. Endophytic bacterial and fungal microbiota in sprouts, roots and stems of rice (Oryza sativa L.) Microbiol. Res. 2016;188:1–8. doi: 10.1016/j.micres.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 42.Wu G., Zhou H., Zhang P., Wang X., Li W., Zhang W., Liu X., Liu H.-W., Keller N.P., An Z. Polyketide production of pestaloficiols and macrodiolide ficiolides revealed by manipulations of epigenetic regulators in an endophytic fungus. Org. Lett. 2016;18:1832–1835. doi: 10.1021/acs.orglett.6b00562. [DOI] [PubMed] [Google Scholar]
- 43.Saravanakumar K., Yu C., Dou K., Wang M., Li Y., Chen J. Biodiversity of Trichoderma community in the tidal flats and wetland of Southeastern China. PLoS ONE. 2016;11:e0168020. doi: 10.1371/journal.pone.0168020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raza W., Ling N., Zhang R., Huang Q., Xu Y., Shen Q. Success evaluation of the biological control of Fusarium wilts of cucumber, banana, and tomato since 2000 and future research strategies. Crit. Rev. Biotechnol. 2017;37:202–212. doi: 10.3109/07388551.2015.1130683. [DOI] [PubMed] [Google Scholar]
- 45.Teale W.D., Paponov I.A., Palme K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 46.Fu X., Lei Z., Wang K., Yan J. Low cost carrier competition and route entry in an emerging but regulated aviation market–the case of China. Transp. Res. Part A Policy Pract. 2015;79:3–16. doi: 10.1016/j.tra.2015.03.020. [DOI] [Google Scholar]
- 47.Johnson L., Shields R., Clancy C. Epidemiology, clinical manifestations, and outcomes of Scedosporium infections among solid organ transplant recipients. Transpl. Infect. Dis. 2014;16:578–587. doi: 10.1111/tid.12244. [DOI] [PubMed] [Google Scholar]
- 48.Prasad R., Chhabra S., Gill S.S., Singh P.K., Tuteja N. Advancement in Crop Improvement Techniques. Elsevier; Amsterdam, The Netherlands: 2020. The microbial symbionts: Potential for crop improvement in changing environments; pp. 233–240. [Google Scholar]
- 49.Ek-Ramos M.J., Zhou W., Valencia C.U., Antwi J.B., Kalns L.L., Morgan G.D., Kerns D.L., Sword G.A. Spatial and temporal variation in fungal endophyte communities isolated from cultivated cotton (Gossypium hirsutum) PLoS ONE. 2013;8:e66049. doi: 10.1371/journal.pone.0066049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Potshangbam M., Devi S.I., Sahoo D., Strobel G.A. Functional characterization of endophytic fungal community associated with Oryza sativa L. and Zea mays L. Front. Microbiol. 2017;8:325. doi: 10.3389/fmicb.2017.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strobel G.A. Rainforest endophytes and bioactive products. Crit. Rev. Biotechnol. 2002;22:315–333. doi: 10.1080/07388550290789531. [DOI] [PubMed] [Google Scholar]
- 52.Phongpaichit S., Rungjindamai N., Rukachaisirikul V., Sakayaroj J. Antimicrobial activity in cultures of endophytic fungi isolated from Garcinia species. FEMS Immunol. Med. Microbiol. 2006;48:367–372. doi: 10.1111/j.1574-695X.2006.00155.x. [DOI] [PubMed] [Google Scholar]
- 53.Vinale F., Sivasithamparam K., Ghisalberti E.L., Marra R., Woo S.L., Lorito M. Trichoderma–plant–pathogen interactions. Soil Biol. Biochem. 2008;40:1–10. doi: 10.1016/j.soilbio.2007.07.002. [DOI] [Google Scholar]
- 54.Tranier M.-S., Pognant-Gros J., Quiroz R.D.l.C., González C.N.A., Mateille T., Roussos S. Commercial biological control agents targeted against plant-parasitic root-knot nematodes. Braz. Arch. Biol. Technol. 2014;57:831–841. doi: 10.1590/S1516-8913201402540. [DOI] [Google Scholar]
- 55.Larran S., Simon M.R., Moreno M.V., Siurana M.S., Perelló A. Endophytes from wheat as biocontrol agents against tan spot disease. Biol. Control. 2016;92:17–23. doi: 10.1016/j.biocontrol.2015.09.002. [DOI] [Google Scholar]
- 56.Herrera S.D., Grossi C., Zawoznik M., Groppa M.D. Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol. Res. 2016;186:37–43. doi: 10.1016/j.micres.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Rayner R. Latent Infection in Coffea arabica L. Nature. 1948;161:245–246. doi: 10.1038/161245a0. [DOI] [Google Scholar]
- 58.Posada F., Aime M.C., Peterson S.W., Rehner S.A., Vega F.E. Inoculation of coffee plants with the fungal entomopathogen Beauveria bassiana (Ascomycota: Hypocreales) Mycol. Res. 2007;111:748–757. doi: 10.1016/j.mycres.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 59.Fernandes M.d.R.V., Pfenning L.H., Costa-Neto C.M.d., Heinrich T.A., Alencar S.M.d., Lima M.A.d., Ikegaki M. Biological activities of the fermentation extract of the endophytic fungus Alternaria alternata isolated from Coffea arabica L. Braz. J. Pharm. Sci. 2009;45:677–685. doi: 10.1590/S1984-82502009000400010. [DOI] [Google Scholar]
- 60.Mulaw T.B., Druzhinina I.S., Kubicek C.P., Atanasova L. Novel endophytic Trichoderma spp. isolated from healthy Coffea arabica roots are capable of controlling coffee tracheomycosis. Diversity. 2013;5:750–766. doi: 10.3390/d5040750. [DOI] [Google Scholar]
- 61.Monteiro M.C.P., Alves N.M., Queiroz M.V.D., Pinho D.B., Pereira O.L., Souza S.M.C.D., Cardoso P.G. Antimicrobial activity of endophytic fungi from coffee plants. Biosci. J. 2017;33:381–389. doi: 10.14393/BJ-v33n2-34494. [DOI] [Google Scholar]
- 62.Gomes A., Pinho D.B., Cardeal Z., Menezes H.C., De Queiroz M.V., Pereira O.L. Simplicillium coffeanum, a new endophytic species from Brazilian coffee plants, emitting antimicrobial volatiles. Phytotaxa. 2018;333:188–198. doi: 10.11646/phytotaxa.333.2.2. [DOI] [Google Scholar]
- 63.Ranjini A., Naika R. Efficacy of biocontrol agents on Myrothecium roridum, the stem necrosis and leaf spot pathogen of coffee seedlings. J. Biopestic. 2019;12:109–113. [Google Scholar]
- 64.Amorim E.D.R., dos Santos P., Cruz M. Description of a new coffee tree-disease (Coffea arabica) caused by the fungus Alternaria alternata in Brazil Descricao de uma nova doenca do Cafeeiro (Coffea arabica) incitada pelo fungo Alternaria alternata no Brasil. Summa Phytopathol. 2002;28:280–281. [Google Scholar]
- 65.Troncoso-Rojas R., Tiznado-Hernández M.E. Postharvest Decay. Elsevier; London, UK: 2014. Alternaria alternata (black rot, black spot) pp. 147–187. [Google Scholar]
- 66.Julca I., Droby S., Sela N., Marcet-Houben M., Gabaldón T. Contrasting genomic diversity in two closely related postharvest pathogens: Penicillium digitatum and Penicillium expansum. Genome Biol. Evol. 2016;8:218–227. doi: 10.1093/gbe/evv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng Y., Lin Y., Cao H., Li Z. Citrus postharvest green mold: Recent advances in fungal pathogenicity and fruit resistance. Microorganisms. 2020;8:449. doi: 10.3390/microorganisms8030449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Destéfano S., Rodrigues L., Beriam L., Patrício F., Thomaziello R., Rodrigues Neto J. Bacterial leaf spot of coffee caused by Pseudomonas syringae pv. tabaci in Brazil. New Dis. Rep. 2010;22:106–970. doi: 10.1111/j.1365-3059.2010.02357.x. [DOI] [Google Scholar]
- 69.Botrel D.A., Laborde M.C.F., Medeiros F.H.V.D., Resende M.L.V.D., Ribeiro Júnio P.M., Pascholati S.F., Gusmão L.F.P. Saprobic fungi as biocontrol agents of halo blight (Pseudomonas syringae pv. garcae) in coffee clones. Coffee Sci. 2018;13:283–291. doi: 10.25186/cs.v13i3.1438. [DOI] [Google Scholar]
- 70.Rodrigues L.M.R., Andrade V.T., Carneiro M.G., Destéfano S.A.L., Beriam L.O.S., Braghini M.T., Fazuoli L.C. The alarming vulnerability of Coffea canephora varieties and germplasm to a virulent strain of Pseudomonas syringae pv. garcae, the cause of bacterial halo blight of coffee in Brazil. J. Plant Pathol. 2021;103:45–49. doi: 10.1007/s42161-020-00715-y. [DOI] [Google Scholar]
- 71.Wiedemann A., Virlogeux-Payant I., Chaussé A.M., Schikora A., Velge P. Interactions of Salmonella with animals and plants. Front. Microbiol. 2015;5:791. doi: 10.3389/fmicb.2014.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klerks M.M., Franz E., van Gent-Pelzer M., Zijlstra C., Van Bruggen A.H. Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 2007;1:620–631. doi: 10.1038/ismej.2007.82. [DOI] [PubMed] [Google Scholar]
- 73.Gu G., Hu J., Cevallos-Cevallos J.M., Richardson S.M., Bartz J.A., Van Bruggen A.H. Internal colonization of Salmonella enterica serovar Typhimurium in tomato plants. PLoS ONE. 2011;6:e27340. doi: 10.1371/journal.pone.0027340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schikora A., Garcia A.V., Hirt H. Plants as alternative hosts for Salmonella. Trends Plant Sci. 2012;17:245–249. doi: 10.1016/j.tplants.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 75.Schikora A., Virlogeux-Payant I., Bueso E., Garcia A.V., Nilau T., Charrier A., Pelletier S., Menanteau P., Baccarini M., Velge P. Conservation of Salmonella infection mechanisms in plants and animals. PLoS ONE. 2011;6:e24112. doi: 10.1371/journal.pone.0024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Semenov A.V., Van Bruggen A.H., Van Overbeek L., Termorshuizen A.J., Semenov A.M. Influence of temperature fluctuations on Escherichia coli O157: H7 and Salmonella enterica serovar Typhimurium in cow manure. FEMS Microbiol. Ecol. 2007;60:419–428. doi: 10.1111/j.1574-6941.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 77.Chaiwan N., Gomdola D., Wang S., Monkai J., Tibpromma S., Doilom M., Wanasinghe D.N., Mortimer P.E., Lumyong S., Hyde K.D. https://gmsmicrofungi.org : An online database providing updated information of microfungi in the greater mekong subregion. Mycosphere. 2021;12:1409–1422. doi: 10.5943/mycosphere/12/1/19. [DOI] [Google Scholar]
- 78.Li Z.-W. Issues and suggestions on building an international brand of “Pu’er Coffee”. Sel. Pap. Rural. Agric. Reform Innov. Agric. Mod. 2014;2:719–721. [Google Scholar]
- 79.Tibpromma S., Hyde K.D., Bhat J.D., Mortimer P.E., Xu J., Promputtha I., Doilom M., Yang J.B., Tang A.M., Karunarathna S.C. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from Southern Thailand. MycoKeys. 2018;33:25–67. doi: 10.3897/mycokeys.33.23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Du T.Y., Karunarathna S.C., Hyde K.D., Mapook A., Wariss H.M., Aluthwattha S.T., Wang Y.H., Mortimer P.E., Xu J.C., Tibpromma S. The endophytic fungi of Aquilaria sinensis from southern China. Fungal Biotec. 2022;2:1–15. [Google Scholar]
- 81.Santamaría J., Bayman P. Fungal epiphytes and endophytes of coffee leaves (Coffea arabica) Microb. Ecol. 2005;50:1–8. doi: 10.1007/s00248-004-0002-1. [DOI] [PubMed] [Google Scholar]
- 82.Van Thiep N., Soytong K. Chaetomium spp. as biocontrol potential to control tea and coffee pathogens in Vietnam. J. Agric. Technol. 2015;11:1381–1392. [Google Scholar]
- 83.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 84.Lu L., Tibpromma S., Karunarathna S.C., Thiyagaraja V., Xu J., Jayawardena R.S., Lumyong S., Hyde K.D. Taxonomic and phylogenic appraisal of a novel species and a new record of Stictidaceae from coffee in Yunnan province, China. Phytotaxa. 2021;528:111–124. doi: 10.11646/phytotaxa.528.2.4. [DOI] [Google Scholar]
- 85.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hall T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 87.Glez-Peña D., Gómez-Blanco D., Reboiro-Jato M., Fdez-Riverola F., Posada D. ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010;38:W14–W18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dissanayake A., Bhunjun C., Maharachchikumbura S., Liu J. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere. 2020;11:2652–2676. doi: 10.5943/mycosphere/11/1/18. [DOI] [Google Scholar]
- 89.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 90.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 92.Brower J., Zar J. Field and Laboratory Methods for General Ecology. McGraw-Hil; Dubuque, IA, USA: 1984. Community similarity; pp. 161–164. [Google Scholar]
- 93.Oliveira R., Souza R., Lima T., Cavalcanti M. Endophytic fungal diversity in coffee leaves (Coffea arabica) cultivated using organic and conventional crop management systems. Mycosphere. 2014;5:523–530. doi: 10.5943/mycosphere/5/4/4. [DOI] [Google Scholar]
- 94.Kumar V., Prasher I. Seasonal variation and tissues specificity of endophytic fungi of Dillenia indica L. and their extracellular enzymatic ectivity. Arch. Microbiol. 2022;204:341. doi: 10.1007/s00203-022-02933-7. [DOI] [PubMed] [Google Scholar]
- 95.Grabka R., d’Entremont T.W., Adams S.J., Walker A.K., Tanney J.B., Abbasi P.A., Ali S. Fungal endophytes and their role in agricultural plant protection against pests and pathogens. Plants. 2022;11:384. doi: 10.3390/plants11030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lacava P.T., Bogas A.C., Moreira C.C. Endophytic Fungi. Nova Science Publishers, Inc.; New York, NY, USA: 2022. Diversity and antimicrobial activity of endophytic fungi from mangrove forests; p. 49. [Google Scholar]
- 97.Orole O., Adejumo T. Activity of fungal endophytes against four maize wilt pathogens. Afr. J. Microbiol. Res. 2009;3:969–973. [Google Scholar]
- 98.Berger C.N., Brown D.J., Shaw R.K., Minuzzi F., Feys B., Frankel G. Salmonella enterica strains belonging to O serogroup 1, 3, 19 induce chlorosis and wilting of Arabidopsis thaliana leaves. Environ. Microbiol. 2011;13:1299–1308. doi: 10.1111/j.1462-2920.2011.02429.x. [DOI] [PubMed] [Google Scholar]
- 99.Kustrzeba-Wójcicka I., Siwak E., Terlecki G., Wolańczyk-Mędrala A., Mędrala W. Alternaria alternata and its allergens: A comprehensive review. Clin. Rev. Allergy Immunol. 2014;47:354–365. doi: 10.1007/s12016-014-8447-6. [DOI] [PubMed] [Google Scholar]
- 100.Olmedo G.M., Cerioni L., González M.M., Cabrerizo F.M., Volentini S.I., Rapisarda V.A. UVA photoactivation of harmol enhances its antifungal activity against the phytopathogens Penicillium digitatum and Botrytis cinerea. Front. Microbiol. 2017;8:347. doi: 10.3389/fmicb.2017.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shen Z., Mustapha A., Lin M., Zheng G. Biocontrol of the internalization of Salmonella enterica and enterohaemorrhagic Escherichia coli in mung bean sprouts with an endophytic Bacillus subtilis. Int. J. Food Microbiol. 2017;250:37–44. doi: 10.1016/j.ijfoodmicro.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 102.Xin X.F., Kvitko B., He S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018;16:316–328. doi: 10.1038/nrmicro.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wicaksono W.A., Jones E.E., Casonato S., Monk J., Ridgway H.J. Biological control of Pseudomonas syringae pv. actinidiae (Psa), the causal agent of bacterial canker of kiwifruit, using endophytic bacteria recovered from a medicinal plant. Biol. Control. 2018;116:103–112. doi: 10.1016/j.biocontrol.2017.03.003. [DOI] [Google Scholar]
- 104.Saucedo-García A., Anaya A.L., Espinosa-García F.J., González M.C. Diversity and communities of foliar endophytic fungi from different agroecosystems of Coffea arabica L. in two regions of Veracruz, Mexico. PLoS ONE. 2014;9:e98454. doi: 10.1371/journal.pone.0098454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thurston H. Tropical Plant Diseases. 2nd ed. APS Press; St. Paul, MN, USA: 1998. Coffee rust; pp. 123–128. [Google Scholar]
- 106.Cao X., Xu X., Che H., West J., Luo D. Characteristics and distribution of Colletotrichum species in coffee plantations in Hainan, China. Plant Pathol. 2019;68:1146–1156. doi: 10.1111/ppa.13028. [DOI] [Google Scholar]
- 107.Davis E.C., Franklin J.B., Shaw A.J., Vilgalys R. Endophytic Xylaria (Xylariaceae) among liverworts and angiosperms: Phylogenetics, distribution, and symbiosis. Am. J. Bot. 2003;90:1661–1667. doi: 10.3732/ajb.90.11.1661. [DOI] [PubMed] [Google Scholar]
- 108.Fulthorpe R., Martin A.R., Isaac M.E. Root endophytes of coffee (Coffea arabica): Variation across climatic gradients and relationships with functional traits. Phytobiomes J. 2020;4:27–539. doi: 10.1094/PBIOMES-04-19-0021-R. [DOI] [Google Scholar]
- 109.Sette L.D., Passarini M.R.Z., Delarmelina C., Salati F., Duarte M. Molecular characterization and antimicrobial activity of endophytic fungi from coffee plants. World J. Microbiol. Biotechnol. 2006;22:1185–1195. doi: 10.1007/s11274-006-9160-2. [DOI] [Google Scholar]
- 110.Larrondo J., Calvo R., Agut M., Calvo M. Inhibitory activity of strains of the genus Arthrinium on Aspergillus and Penicillium species. Microbios. 1995;82:115–126. [PubMed] [Google Scholar]
- 111.Calvo M., Arosemena L., Agut M., Adelantado C., Aissaoui H. Biological control of bayoud in the palm trees by strains of Arthrinium. J. Biol. Sci. 2005;5:236–238. [Google Scholar]
- 112.Moriguchi Y., Kihara J., Ueno M. Suppression effects of a secondary metabolite of Biscogniauxia sp. strain O-811 obtained from mushrooms against the rice blast fungus Magnaporthe oryzae. Bull Fac. Life Env. Sci. Shimane Univ. 2019;24:14–18. [Google Scholar]
- 113.Liarzi O., Bar E., Lewinsohn E., Ezra D. Use of the endophytic fungus Daldinia cf. concentrica and its volatiles as bio-control agents. PLoS ONE. 2016;11:e0168242. doi: 10.1371/journal.pone.0168242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sreeja K., Anandaraj M., Bhai R. In vitro evaluation of fungal endophytes of black pepper against Phytophthora capsici and Radopholus similis. J. Spices Aromat. Crops. 2016;25:113–122. [Google Scholar]
- 115.Rashid T.S. Bioactive metabolites from tomato endophytic fungi with antibacterial activity against tomato bacterial spot disease. Rhizosphere. 2021;17:100292. doi: 10.1016/j.rhisph.2020.100292. [DOI] [Google Scholar]
- 116.Dhakshinamoorthy M., Packiam K.K., Kumar P.S., Saravanakumar T. Endophytic fungus Diaporthe caatingaensis MT192326 from Buchanania axillaris: An indicator to produce biocontrol agents in plant protection. Environ. Res. 2021;197:111147. doi: 10.1016/j.envres.2021.111147. [DOI] [PubMed] [Google Scholar]
- 117.Thanabalasingam D., Kumar N.S., Jayasinghe L., Fujimoto Y. Endophytic fungus Nigrospora oryzae from a medicinal plant Coccinia grandis, a high yielding new source of Phenazine-1-carboxamide. Nat. Prod. Commun. 2015;10:1659–1660. doi: 10.1177/1934578X1501001008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.