ABSTRACT
Burkholderia vietnamiensis LMG10929 and Paraburkholderia kururiensis M130 are bacterial rice growth-promoting models. Besides this common ecological niche, species of the Burkholderia genus are also found as opportunistic human pathogens, while Paraburkholderia species are mostly environmental and plant associated. In this study, we compared the genetic strategies used by B. vietnamiensis and P. kururiensis to colonize two subspecies of their common host, Oryza sativa subsp. japonica (cv. Nipponbare) and O. sativa subsp. indica (cv. IR64). We used high-throughput screening of transposon insertional mutant libraries (Tn-seq) to infer which genetic elements have the highest fitness contribution during root surface colonization at 7 days postinoculation. Overall, we detected twice more genes in B. vietnamiensis involved in rice root colonization than in P. kururiensis, including genes contributing to the tolerance of plant defenses, which suggests a stronger adverse reaction of rice toward B. vietnamiensis than toward P. kururiensis. For both strains, the bacterial fitness depends on a higher number of genes when colonizing indica rice compared to japonica. These divergences in host pressure on bacterial adaptation could be partly linked to the cultivars’ differences in nitrogen assimilation. We detected several functions commonly enhancing root colonization in both bacterial strains, e.g., Entner-Doudoroff (ED) glycolysis. Less frequently and more strain specifically, we detected functions limiting root colonization such as biofilm production in B. vietnamiensis and quorum sensing in P. kururiensis. The involvement of genes identified through the Tn-seq procedure as contributing to root colonization, i.e., ED pathway, c-di-GMP cycling, and cobalamin synthesis, was validated by directed mutagenesis and competition with wild-type (WT) strains in rice root colonization assays.
IMPORTANCE Burkholderiaceae are frequent and abundant colonizers of the rice rhizosphere and interesting candidates to investigate for growth promotion. Species of Paraburkholderia have repeatedly been described to stimulate plant growth. However, the closely related Burkholderia genus includes both beneficial and phytopathogenic species, as well as species able to colonize animal hosts and cause disease in humans. We need to understand to what extent the bacterial strategies used for the different biotic interactions differ depending on the host and if strains with agricultural potential could also pose a threat toward other plant hosts or humans. To start answering these questions, we used in this study transposon sequencing to identify genetic traits in Burkholderia vietnamiensis and Paraburkholderia kururiensis that contribute to the colonization of two different rice varieties. Our results revealed large differences in the fitness gene sets between the two strains and between the host plants, suggesting a strong specificity in each bacterium-plant interaction.
KEYWORDS: Paraburkholderia kururiensis, Burkholderia vietnamiensis, root colonization, Tn-seq, PGPR, rice
INTRODUCTION
Species of the Burkholderia and closely related Paraburkholderia genera are highly prolific rhizosphere colonizers (1, 2). Their persistence and competitiveness in the rhizosphere environment can be explained by strong secondary-metabolite production as well as efficient nitrogen cycling, mineral solubilization, and phytohormone biosynthesis (3–5). Furthermore, several Paraburkholderia and at least one Burkholderia species can fix atmospheric nitrogen. Two well-studied models, Paraburkholderia kururiensis strain M130 and Burkholderia vietnamiensis strain LMG10929, demonstrate strong rice root colonization, endophytic lifestyles, and significant plant growth promotion through transfer of fixed nitrogen (6–9). Despite this convergence in their plant-beneficial features, these bacteria belong to distinct genetic backgrounds. While Paraburkholderia species are often found in beneficial relationships and symbioses with plants (10–12), Burkholderia members comprise human pathogens and opportunists as well as fungal and plant pathogens (13).
Bacterial genes used for plant colonization have been screened in several model bacteria but also on a broader scale using microbiome approaches to reveal plant-associated functions (1). A few studies have compared plant-adapted Burkholderia (the former genus that is now regrouping the newly defined Burkholderia, Paraburkholderia, Caballeronia, and others) at the genomic level (4, 11) or profiled the transcriptome of bacteria stimulated by root exudates (14, 15). However, there is no record of a comparison between the strategies used by plant-adapted Burkholderia and Paraburkholderia species. The host plant genotype’s impact on bacterial colonization strategies also remains poorly explored for these bacterial genera. Rice is an interesting model to assess Burkholderia sensu largo adaptation to the plant environment, as it hosts plant growth-promoting model strains from both the Burkholderia and Paraburkholderia genera. It was repeatedly demonstrated that rice genotypes influence the composition of their microbiome at the rhizosphere and rhizoplane levels (16, 17). In particular, a study on 95 different Oryza sativa subsp. indica and O. sativa subsp. japonica varieties showed significant differences in microbiome compositions between these subspecies, which were related to their nitrogen use efficiency and the presence of a particular nitrate transporter in the indica variety (18).
Transposon mutagenesis sequencing (Tn-seq) is a high-throughput screening method that combines transposon insertional mutagenesis followed by sequencing of the insertion sites (19). It is a powerful tool that leads to immediate identification of genes of interest improving or reducing the bacterium’s fitness in a tested condition. This methodology has been successfully used to unravel important genes and functions in plant-pathogenic or plant-symbiotic interactions (20–22). Commonly between pathogens and mutualists, genes functioning in amino acid and purine metabolism as well as in cell motility were detected to be required for root colonization (21, 23, 24).
In the present study, we used Tn-seq to perform a genome-wide identification of genes involved in rice root colonization in B. vietnamiensis and P. kururiensis. In detecting which genes influence the fitness of B. vietnamiensis and P. kururiensis, we aim to unravel their commonalities and differences in root colonization strategies. We also analyzed the association strategies of B. vietnamiensis and P. kururiensis with the two rice genotypes Oryza sativa subsp. japonica (cv. Nipponbare) and O. sativa subsp. indica (cv. IR64) to understand how the host factor can influence colonization strategies. Overall, we identified totals of 1,404 and 540 genes that influence the fitness of B. vietnamiensis and P. kururiensis, respectively, during rice root colonization. Our results underline the importance of motility, amino acid and vitamin metabolism, and stress response as well as biofilms for the efficient association of these bacteria with rice roots.
RESULTS
Quality of Tn-Seq libraries and essential genomes in P. kururiensis and B. vietnamiensis.
To generate a genome-wide library of insertion mutants for P. kururiensis and B. vietnamiensis strains we used a mariner transposon that targets genomic thymine-adenosine (TA) sites (see Materials and Methods). The genome of B. vietnamiensis contains 95,075 TA sites, and we estimated the mutant population at 1.6 × 108 CFU, which represents a 1,683× coverage of the total TA sites. The saturation level of the P. kururiensis library was lower, although still significant, at a 38× coverage given the 4.0 × 106 CFU obtained after transposon mutagenesis, for a total of 106,136 total TA sites contained in the genome.
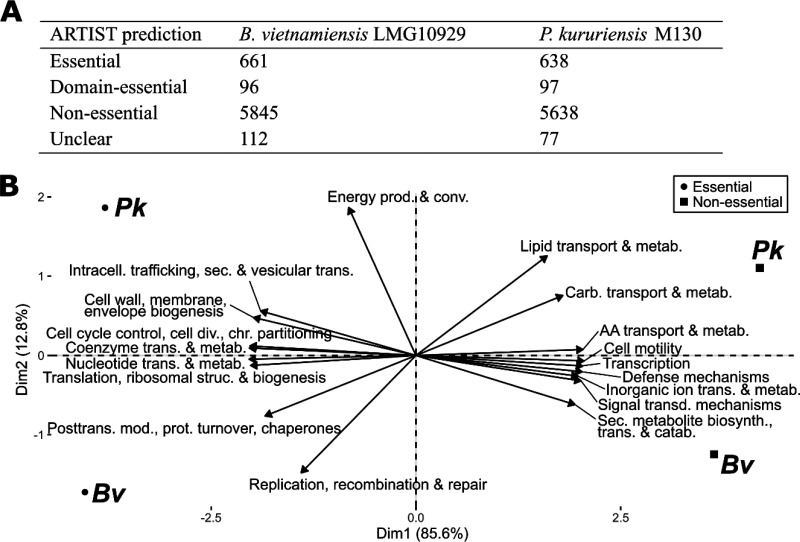
To further assess the quality of the Tn-seq libraries, we determined the essential genomes required by both bacteria for optimal development in a rich liquid growth medium. The two bacteria have similar genome sizes (6,820 and 6,436 genes for B. vietnamiensis and P. kururiensis, respectively) and comparable proportions of genes (661 [9.7%] and 638 [9.9%], respectively) that are required for optimal growth in a rich liquid medium (Fig. 1A; see also Data Set S1 in the supplemental material). The sizes of the essential genomes determined in a controlled liquid medium are in the order of magnitude of what has been observed for other prokaryotes, including Burkholderia spp. (25–29). We used the Minimal Gene Set tool implemented in the MicroScope platform (see Materials and Methods) to extract a core list of the predicted minimal bacterial gene sets from B. vietnamiensis and P. kururiensis. On a total of 206 core bacterial genes defined by their conservation among multiple bacterial genomes (30), 151 and 150 were identified as essential by our approach in B. vietnamiensis and P. kururiensis, respectively. In 10 and 9 cases of genes belonging to the minimal gene set but predicted as nonessential by our approach in B. vietnamiensis and P. kururiensis, respectively, there are duplicates present in the genome.
FIG 1.
Distribution of genes significantly impacting bacterial fitness in a rich liquid medium. (A) Number of genes predicted to be essential, domain essential (only insertions in part of the gene lead to abundance decrease), or nonessential by the EL-ARTIST pipeline. (B) Principal-component analysis (PCA) of essential and nonessential genes of B. vietnamiensis (Bv) and P. kururiensis (Pk) in a rich liquid medium. The genes were classified according to their COG categories and the PCA was built in regard to the abundance of these categories.
According to their distribution in clusters of orthologous groups (COG) categories, little variance differentiates the essential genomes of P. kururiensis and B. vietnamiensis (Fig. 1B). As expected, in a liquid rich medium, genes involved in motility, defense and nutrition are largely nonessential. Conversely, structural components of the cell and the general replicative machinery are predictably essential (Data Set S1).
Given the strong saturation level of the TA insertion sites and the coherent essentiality results observed in the control setting, we can safely assume that both mutant libraries allow a reliable analysis of the impact of genes on the bacterial fitness.
Colonization of the two rice genotypes by P. kururiensis and B. vietnamiensis.
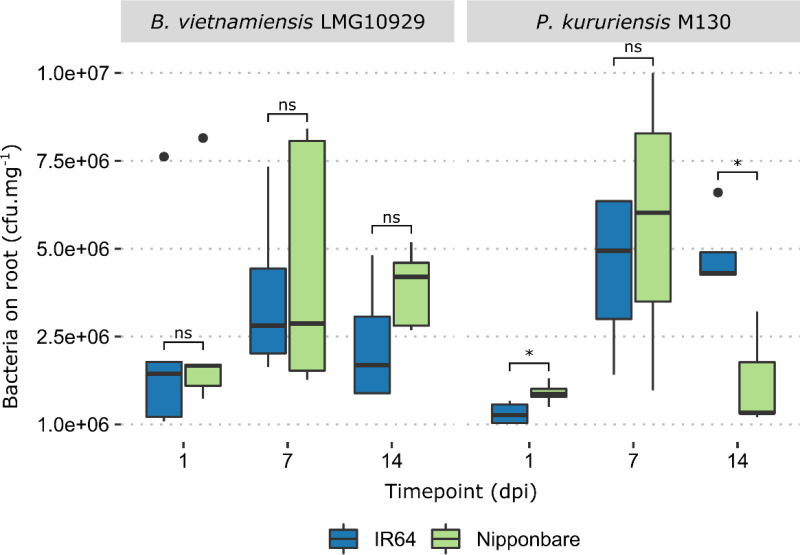
Prior to genetic analyses, we assessed the colonization efficiencies of B. vietnamiensis and P. kururiensis on the Nipponbare (japonica) and IR64 (indica) rice genotypes (Fig. 2). Overall, the two bacteria displayed similar colonization dynamics, with an increasing population density during the first week and a decrease in the second week. A host genotype effect was observed in the colonization phenotype displayed by P. kururiensis, as significantly different root colonizing populations were observed on IR64 and Nipponbare at 3 days postinoculation (dpi) and 14 dpi (Fig. 2). B. vietnamiensis, on the other hand displayed a steady colonization pattern between both plant genotypes at all measured time points. These observations confirm that the rice-colonizing populations of B. vietnamiensis and P. kururiensis can be compared, especially at 7 dpi, which was selected for further analyses. In our following analyses, we consider that the bacterial adaptations observed result purely from root surface colonization, as the endophytic populations at 7 dpi are inferior by several log scales to the surface colonizing population (6).
FIG 2.
Rice colonization efficiencies for B. vietnamiensis and P. kururiensis. Nipponbare and IR64 rice seedlings were inoculated at 5 days postgermination with 1 × 107 bacterial cells, total root systems were harvested at different time points, and the associated bacterial population was estimated (P < 0.05, Wilcoxon test). The boxplots were generated using 5 replicates for each condition. Outliers are represented as black dots.
Identification of rice colonization genes.
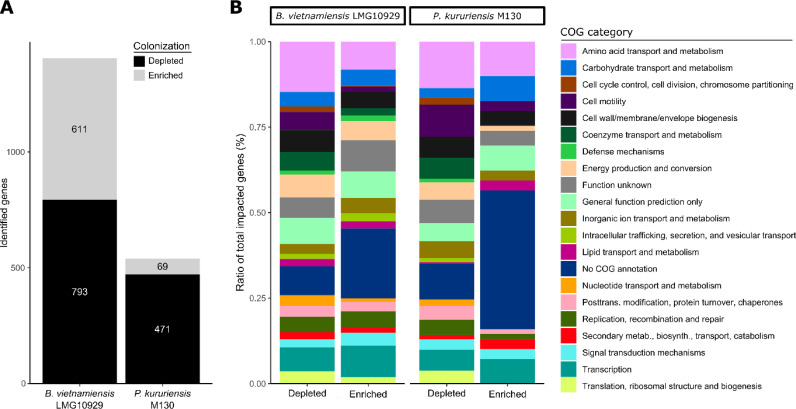
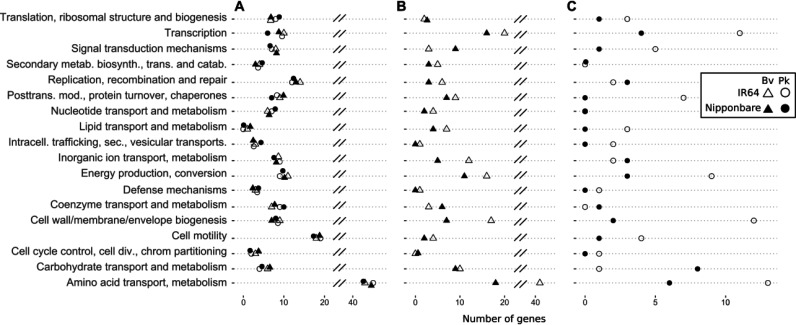
To assess gene fitness for rice root colonization, and its host-dependent variation in the two model bacteria, we inoculated the Tn-Seq mutant libraries on the Nipponbare and IR64 rice genotypes. Seven days after inoculation, we harvested and pooled five root systems per replicate, representing a total of 1.2 × 107 colonization events (Fig. 2) and a >100-fold coverage of the available mutant diversity. We performed a first Tn-seq analysis by pooling the reads of IR64 and Nipponbare isolates together to infer the genes globally required for the association with rice. The read frequencies were compared to a control condition grown in a rich medium with limited growth generations (see Materials and Methods) to establish a root fitness score for each gene. In this manner, we identified 1,404 and 540 genes as significantly impacted (enriched or depleted) after root colonization by B. vietnamiensis and P. kururiensis, respectively (Fig. 3A). Colonization-depleted genes are our major focus, as they are positively associated with bacterial fitness. Inversely, colonization-enriched genes diminish the bacterial fitness during root colonization. We organized the identified colonization genes according to their general function based on their COG annotation (Fig. 3B). Colonization-enriched and -depleted genes share similar distributions, with amino acid metabolism, cell wall/membrane/envelope biogenesis, transcription, and cell motility being among the most frequent categories, consistent with the expected implication of nutrition, motility, and morphological adaptations involved in plant colonization.
FIG 3.
Distribution of genes significantly impacting bacterial fitness during rice colonization. Colonization events on IR64 and Nipponbare rice cultivars were pooled for this analysis. (A) Transposon insertions in genes leading to a 1.5-fold or higher decrease in abundance on roots at a Padj value of <0.05 were identified to be colonization depleted. Those leading to a 1.5-fold or higher abundance were identified to be colonization enriched. (B) Distribution of genes significantly depleted or enriched during rice colonization along COG categories in ratio of total impacted genes.
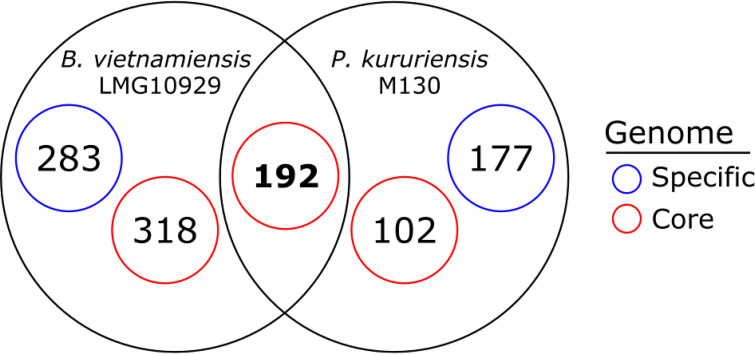
We identified a total of 2,071 core gene families for B. vietnamiensis and P. kururiensis, sharing homologues in both strains (Data Set S2). In B. vietnamiensis and P. kururiensis, 192 genes, respectively 24% and 41% of the colonization-depleted genes, are part of the core genome and equally depleted in both strains (Fig. 4). B. vietnamiensis displays 68% more colonization-depleted genes than P. kururiensis (Fig. 3). Interestingly, a majority of these B. vietnamiensis-specific colonization-depleted genes (53%) are part of the core genome (Fig. 4). Similarly, a lower, although substantial, portion of the genome shared with B. vietnamiensis (37%) is required by P. kururiensis specifically for efficient root colonization (Fig. 4). Thus, although a large portion of the core genes contribute to root colonization in both strains, many core genes are required for colonization in only one or the other strain, indicating that the impact of these genes on root colonization is dependent on the genome context.
FIG 4.
Distribution of colonization-depleted genes along the core and specific genomes of B. vietnamiensis and P. kururiensis. There are a total of 2,072 genes that present significant homologies in the genomes of B. vietnamiensis and P. kururiensis and are thus considered part of the core genome (see Materials and Methods). A fraction of this core genome is colonization depleted in both species. Additionally, a substantial proportion of the genes that are found to be colonization depleted in a species or the other also belong to the core genome.
Rice cultivar-dependent colonization specialization.
Next, we performed a separate analysis of the reads for each rice cultivar and then compared the lists of significantly colonization-depleted and -enriched genes to infer cultivar specificities. For both strains, the majority (~65%) of identified genes impacted the bacterial fitness on both IR64 and Nipponbare rice (Table 1). However, both bacteria displayed a greater number of colonization-depleted genes (41% and 49% higher for B. vietnamiensis and P. kururiensis, respectively) on IR64 than on Nipponbare rice (Table 1).
TABLE 1.
Distribution of genes significantly impacting bacterial fitness on IR64 and Nipponbare ricea
| Strain | Colonization impact | No. of genes: |
Total | ||
|---|---|---|---|---|---|
| Common to both cultivars | Present on IR64 | Present on Nipponbare | |||
| B. vietnamiensis LMG10929 | Enriched | 507 | 83 | 130 | 720 |
| Depleted | 635 | 225 | 134 | 994 | |
| Total | 1,142 | 308 | 264 | 1,714 | |
| P. kururiensis M130 | Enriched | 48 | 57 | 36 | 141 |
| Depleted | 374 | 98 | 50 | 522 | |
| Total | 422 | 155 | 86 | 663 | |
Colonization events on IR64 and Nipponbare rice cultivars were analyzed separately.
Core genes of B. vietnamiensis and P. kururiensis that were colonization depleted on both cultivars belong primarily to housekeeping categories such as central metabolism, cell cycle control, and motility (Fig. 5A). The additional genes depleted during IR64 colonization are chiefly attributed to amino acid metabolism, transcription, cell wall/membrane/envelope biogenesis, and energy production in both bacterial strains (Fig. 5B and C). Interestingly, contrary to the global trend, in P. kururiensis the COG categories “replication, recombination & repair,” “coenzyme transport & metabolism,” “carbohydrate metabolism,” and “inorganic ion transport” are more strongly impacted on Nipponbare than on IR64 (Fig. 5C). The same trend was observed in B. vietnamiensis for the COG categories “signal transduction mechanisms” and “coenzyme transport and metabolism” (Fig. 5B).
FIG 5.
Distribution of colonization-depleted genes on IR64 and Nipponbare rice cultivars. Distribution is along COG categories of core genes of B. vietnamiensis and P. kururiensis that are colonization depleted on both rice cultivars (A), genes that are specifically colonization depleted in B. vietnamiensis (B), and genes that are specifically colonization depleted in P. kururiensis (C).
Tn-seq reveals bacterial functions involved in early rice colonization.
Many genes significantly impacting the colonization fitness of B. vietnamiensis and P. kururiensis are clustered together within operons, supporting the validity of our results. In these cases, there is furthermore a strong conservation of either gene enrichment or depletion within the same operon. In several cases explored here, the lack of detection of a complete operon can be explained by the presence of homologues for some of the genes resulting in functional complementation.
Functions required for bacterial fitness during rice root colonization.
As expected in this kind of colonization assay, mutants affected in motility and chemotaxis functions (flg, flh, fli, and mot) were depleted from the root-colonizing population (Table 2 and Data Set S3). For both bacteria, we further detected many genes involved in amino acids (arg, his, ilv, leu, lys, met, pro, ser, and trp) and nucleotide synthesis (pur and pyr) that, when mutated, negatively impacted the bacteria’s fitness on plants. Genes involved in the synthesis of enzymatic cofactors for amino acid metabolism, such as vitamin B1 (thiamine), were also colonization depleted in both strains. Multiple Tn-seq studies reported that auxotrophy for certain amino acids is disadvantageous for root colonization and can limit plant growth promotion and biocontrol potential (23, 31).
TABLE 2.
Selection of functions and number of corresponding genes impacting rice colonization efficiencies when mutateda
| Function | No. of genes |
|||
|---|---|---|---|---|
|
B. vietnamiensis
|
P. kururiensis
|
|||
| Depleted | Enriched | Depleted | Enriched | |
| Motility | 35 | 35 | ||
| Chemotaxis | 4 | 5 | 9 | 1 |
| Cobalamin synthesis (B12) | 1 | 8 | 15 | |
| Thiamine synthesis (B1) | 4 | 5 | ||
| Biotin synthesis (B7) | 5 | |||
| Potassium uptake | 5 | 5 | ||
| Ribose metabolism | 5 | |||
| Entner-Doudoroff pathway | 1 | 2 | ||
| Aromatic compound metabolism | 5 | |||
| c-di-GMP cycling | 1 | 7 | 1 | |
| Putrescine metabolism | 3 | 1 | ||
| Putrescine uptake | 5 | |||
| Cellulose synthase | 5 | |||
| Cell wall integrity | 5 | 10 | ||
| Peptidoglycan synthesis | 3 | |||
| Lytic transglycolases | 4 | 1 | ||
| Tol-Pal system | 5 | 3 | ||
| Hopanoid synthesis | 7 | 1 | ||
| O-antigen synthesis | 2 | |||
| Osmoprotection | 2 | 1 | ||
| DNA repair | 9 | 9 | ||
| Flp/Tad pilus | 12 | |||
| Arginine biosynthesis | 5 | 4 | ||
| Histidine biosynthesis | 9 | 9 | ||
| Isoleucine biosynthesis | 6 | 4 | ||
| Leucine biosynthesis | 3 | 4 | ||
| Lysine biosynthesis | 1 | |||
| Methionine biosynthesis | 3 | 5 | ||
| Proline biosynthesis | 2 | 2 | ||
| Serine biosynthesis | 2 | 2 | ||
| Tryptophan biosynthesis | 6 | 6 | ||
| Purine metabolism | 9 | 1 | 1 | |
| Pyrimidine metabolism | 5 | 6 | ||
| Queuosine synthesis | 6 | 1 | ||
| Quorum sensing | 2 | |||
| T2SS | 9 | 8 | ||
| Autotransporter assistance | 2 | |||
| Prophage | 22 | |||
A detailed list of genes is provided in Table S3. Pathways in bold are the three pathways for which new insertion mutants were produced to confirm implication in root colonization.
Additionally, genes involved in potassium nutrition (kdpA-F) were similarly depleted. Mutants of both B. vietnamiensis and P. kururiensis affected in central elements of the Entner-Doudoroff glycolysis (ED) pathway (edd and zwf) suffered a significant fitness decrease on rice. This pathway is involved in the metabolism of gluconate, which is not a dominant sugar of rice exudates (32), suggesting an alternative role than sole sugar assimilation for this pathway. The activation of the ED pathway was suggested to be a tolerance strategy toward oxidative stress through the generation of NADPH as an essential cofactor for thioredoxins (33, 34).
Genes of the type 2 secretion system (T2SS; gspD-M) are the only ones belonging to a macromolecular secretory pathway found to be colonization depleted in both bacteria. While B. vietnamiensis possesses a single T2SS, P. kururiensis bears two copies (4), of which only one is colonization depleted, suggesting that these systems are not redundant but are used differently in specific conditions.
Several genes involved in DNA maintenance and reparation, i.e., ruvABC, xerCD, and recABCD, were colonization depleted in both B. vietnamiensis and P. kururiensis, while their absence was tolerated in the relatively stress-free control medium (Data Set S1). Thus, rice appears to inflict considerable genotoxic stress during the process of colonization. Furthermore, the presence of osmotic stress is exemplified by the depletion of B. vietnamiensis and P. kururiensis mutants involved in the synthesis of the osmoprotectant trehalose (otsAB).
B. vietnamiensis seems to suffer additional stress as multiple functions maintaining cell wall and membrane integrity are colonization depleted. We found several genes involved in hopanoid synthesis (hpnDEFHJKN), peptidoglycan synthesis (murAI), and maintenance (tolAQR and pal) as well as lytic murein transglycosylation (mltA, rlpA, and mtgA) to be depleted specifically in B. vietnamiensis mutant populations. This indicates that B. vietnamiensis has an increased requirement to maintain its cellular integrity compared to P. kururiensis. Consistent with an increased need in membrane maintenance, the loss of vitamin B7 (biotin) synthesis genes (bioABCDF) had a negative impact on the colonization of B. vietnamiensis. Biotin is a cofactor for many enzymes, especially those involved in fatty acid biosynthesis and amino acid metabolism (35, 36). Roots are known to secrete aromatic phenolic compounds which are toxic to various soil microbes. Accordingly, B. vietnamiensis mutants were depleted during colonization when impacted in genes of the β-ketoadipate pathway (pcaBCDK), which allow metabolism of 4-hydroxybenzoate and protocatechuate. A final sign of the stress B. vietnamiensis is exposed to during colonization is found in the depletion of mutants for the queuosine synthesis pathway (queACEF and tgt). This hypermodified nucleoside improves translation accuracy, a need that only arose in B. vietnamiensis during rice colonization.
While no genes annotated as coding for autotransporter proteins (T5SS) were identified through our approach, the genes tamAB were colonization depleted in P. kururiensis. TamA and TamB can be involved in outer membrane assembly, allowing surface structuration, which is essential for adhesion and host invasion by bacteria in several pathogenic models (37). Genes for vitamin B12 (cobalamine) synthesis are present in both B. vietnamiensis and P. kururiensis but were colonization depleted only in P. kururiensis (cobBDHIKLMNQSTUW, cbiB, and btur). Intriguingly, several genes of this synthesis pathway were actually colonization enriched in B. vietnamiensis, suggesting an adverse role in colonization for this cofactor involved in a multitude of enzymatic reactions.
Functions detrimental for bacterial fitness during rice root colonization.
Only two genes were identified to be colonization enriched in B. vietnamiensis and P. kururiensis simultaneously, one of which (AK36_1927/ANSKv1_30041) has an unknown function. The other (pdeR) is involved in cyclic di-GMP (c-di-GMP) cycling. In B. vietnamiensis, four other genes having a homologous role in c-di-GMP degradation (phosphodiesterases [Data Set S3]) and two involved in c-di-GMP production (diguanylate cyclases [Data Set S3]) were also colonization enriched. c-di-GMP levels can play a role in colonization, as they were demonstrated to influence biofilm production and motility in Burkholderia species (38). Interestingly, while several genes of the putrescine catabolism (puuBD and speG) were colonization depleted in B. vietnamiensis, the entire cluster responsible for putrescine uptake (potFGHI and puuP) was colonization enriched. Together, these findings suggest that putrescine accumulation has a negative impact on root colonization. An accumulation of intracellular putrescine could have an impact on biofilm production as recently suggested (23). Among additional biofilm-related colonization-enriched genes in B. vietnamiensis, we identified a cluster involved in cellulose synthesis (bcsABCE). Another B. vietnamiensis surface element that increases colonization efficiency when mutated is a Tad pilus (tadABCEVZ, rcpAB, and flp), suggesting that this attachment or motility mechanism is suitable for an alternative condition than rice colonization. While many genes involved in carbohydrate metabolism in both bacteria were colonization depleted, B. vietnamiensis mutants were colonization enriched when impacted in ribose import and catabolism genes (rbsABCK).
P. kururiensis mutants impacted in the ability to sense the bacterial community through quorum sensing (braR and rsaL) were enriched during colonization, as can be expected for organisms that have lost the ability to sense population density and reduce growth rates accordingly. Genes belonging to a region enriched in elements of phage origin (ANSKV1_30083-120) had the same effect when mutated, indicating that the prophage might be induced by the plant colonization process and that its inactivation through mutation benefits the bacteria’s fitness.
Rice cultivar-specific adaptations.
Genes which are colonization depleted or enriched specifically on one cultivar can be more rarely grouped in metabolic pathways and operon structures (Data Set S4). Still, single genes can have determining impacts on a strain’s ecology and metabolism. There are two genes that were colonization depleted for both B. vietnamiensis and P. kururiensis on Nipponbare rice. One encodes an outer membrane porin of the OmpC family (AK36_1494/ANSKv1_11218) and the other, amtB, is involved in ammonium uptake (Table 3 and Data Set S4). B. vietnamiensis mutants for genes involved in phosphate transport (pstAC) and glycerol metabolism (ugpABQ) were depleted on IR64 rice, further suggesting that the metabolic requirements of the plant force adaptations on rhizosphere microbes.
TABLE 3.
Selection of functions and number of corresponding colonization-depleted genes on IR64 and Nipponbare rice
| Function | No. of genes |
|||
|---|---|---|---|---|
|
B. vietnamiensis
|
P. kururiensis
|
|||
| IR64 | Nipponbare | IR64 | Nipponbare | |
| T6SS | 10 | |||
| Glycerol metabolism | 3 | 4 | ||
| Phosphate import | 3 | |||
| Ammonium transport (AmtB) | 1 | 1 | ||
| Oxoprolinase | 3 | |||
| O-antigen biosynthesis | 1 | 3 | ||
Several B. vietnamiensis genes involved in type 6 secretion (tssGHIJK) were colonization depleted on IR64. For P. kururiensis, mutants impacted in O-antigen biosynthesis (rfbABD) were depleted on IR64. In both cases we can hypothesize that surface elements have significant and diverging repercussions depending on the type of colonized rice host.
Validation of three candidate genes.
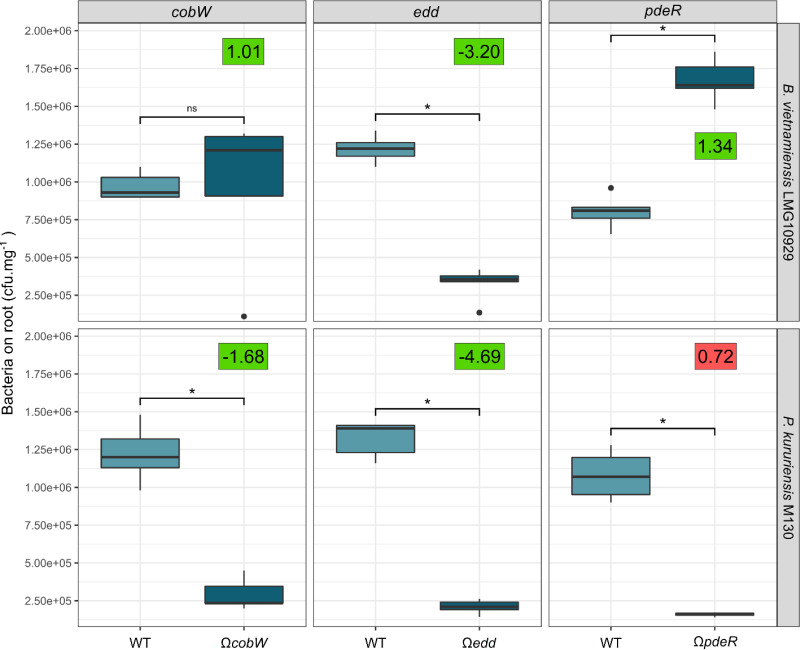
We underlined the importance of various genes involved in the ED pathway, c-di-GMP cycling, and cobalamin synthesis, among others. To validate their involvement in the colonization of rice roots, we used an insertional mutagenesis approach in B. vietnamiensis and P. kururiensis by targeting single-copy genes that are key in the respective processes. The 6-phosphogluconate dehydratase Edd is essential for the first step of the ED pathway (39). The metal delivery protein CobW is essential for vitamin B12 synthesis (40). Finally, we chose PdeR, an enzyme with domains predicted to be involved in both c-di-GMP production and degradation. The genomic context of each of these genes, including their fold change between root and rich-medium conditions, is presented in Fig. S1.
Mutants and wild-type (WT) strains were used jointly in a competition assay, and colonization efficiencies were surveyed at 7 dpi. In B. vietnamiensis, each mutation had the observable impact that had been predicted by the Tn-seq analysis (Fig. 6). Disruption of the colonization-depleted gene edd reduced B. vietnamiensis’s colonization capacity, while deletion of the colonization-enriched gene pdeR had the opposite effect. The disruption of cobW had been identified in Tn-seq data as having a deleterious impact on P. kururiensis but did not significantly alter the root colonization capacity of B. vietnamiensis. The mutagenesis approach confirms that the disruption of cobW has no negative impact on B. vietnamiensis but is required by P. kururiensis for efficient root colonization. The only inconsistent observation occurred for the pdeR mutant in P. kururiensis, which was predicted as slightly colonization enriched by the Tn-seq approach, but its disruption resulted in a colonization deficiency compared to the case with the WT strain.
FIG 6.
Colonization capacity of B. vietnamiensis and P. kururiensis insertion mutants in competition assays. Three genes, cobW, edd, and pdeR, predicted by the Tn-seq approach to be involved in rice root colonization were selected for targeted disruption in B. vietnamiensis and P. kururiensis. Mutants and WT strains were inoculated simultaneously on Nipponbare rice roots and enumerated at 7 dpi. Significance levels of pairwise comparisons were estimated using a Wilcoxon test (P < 0.05). For each mutant, the log2 fold change (FC) value observed in the Tn-seq on Nipponbare is displayed. Positive and negative correlations with the mutagenesis approach are expressed with green and red squares, respectively.
DISCUSSION
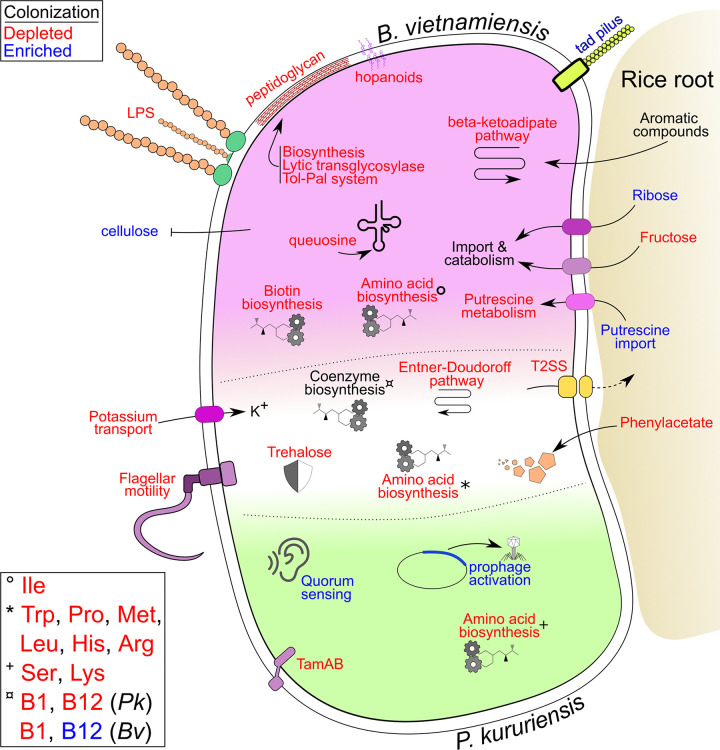
Understanding which genetic bases are involved in plant growth promoting rhizobacteria (PGPR)-cereal interaction is pivotal for a controlled and informed selection of beneficial organisms and a gateway to efficient and directed strain improvement. In rice, the microbiome composition was demonstrated to be significantly modulated by the plant genotype (16). This suggests a variance in selective pressure forced by the plant onto colonizing microbes. The present Tn-seq analysis demonstrated which genes have the strongest contribution to bacterial fitness in the early steps of rhizosphere colonization of two rice cultivars, Nipponbare and IR64, by two bacteria, P. kururiensis and B. vietnamiensis (Fig. 7). At this stage, we cannot exclude that additional genes might be required to colonize more mature plants as the bacteria progress and potentially reach different plant compartments. It is also known that Tn-seq approaches are insensitive to genes whose function is complemented by functional homology either within the same bacterium (gene duplicates and paralogues) or by the surrounding bacterial community, e.g., secretion systems and secreted molecules.
FIG 7.
Summary diagram of colonization-impacted bacterial functions involved in efficient rice root colonization. Pathways, functions, and genes described throughout this work are synthetized on this integrative representation. Elements specific to B. vietnamiensis are placed in the top part of the schematic bacteria (pink), and P. kururiensis-specific elements are at the bottom (green). Elements that are commonly found in both bacteria are placed at the interface (white). Function and molecular systems which were found to be colonization depleted are written in red font, whereas colonization-enriched genes are written in blue font.
The present study compared insertion mutant libraries that were grown on plant roots for 7 days to libraries grown in a rich liquid medium for a brief period. Thus, caution needs to be taken when interpreting the role of the detected genes in plant interaction. Especially detected nutritional function could be general prerequisites for a sound bacterial development in a poor medium and not specifically involved in a plant colonization task. Still, we found several examples of genes involved in plant interaction that are supported by independent approaches, as illustrated here.
B. vietnamiensis and P. kururiensis have different strategies for rice root colonization.
As was previously suggested by microscopy observations and host transcriptomics, B. vietnamiensis and P. kururiensis have different approaches to rice colonization (6). The present analysis highlights the respective genetic requirements of B. vietnamiensis and P. kururiensis during their rice colonization process. Strikingly, a considerable higher number of genes is necessary for a successful colonization by B. vietnamiensis than by P. kururiensis, despite their similar genome sizes. This requirement could result from increased plant defenses that B. vietnamiensis is exposed to compared to P. kururiensis (6). Supposedly, the pathogenic genetic background and potential of B. vietnamiensis could be responsible for this adverse plant response and necessitate a higher number of genes to act against. Moreover, as B. vietnamiensis is known to colonize multiple eukaryotic hosts, both plants and animals (41–44), we can also hypothesize that B. vietnamiensis expresses many genes which are not of use for plant colonization; thus, a mutation in these genes will increase the bacterium’s fitness as the ensuing metabolic cost can be spared. This could explain the fact that plant-detrimental genes were found in relatively similar amounts to plant-essential genes in B. vietnamiensis, while they were considerably less frequent in P. kururiensis (Fig. 3A). One striking example is the case of vitamin B12 production. This cofactor is among the most complex in bacteria and requires at least 19 enzymatic steps to produce, while many enzymes further depend on B12 for their activity (45). Here, the synthesis of B12 is beneficial for the fitness of P. kururiensis during colonization, while it has an adverse effect on B. vietnamiensis. This observation is supported both by the Tn-seq and the insertional mutagenesis approaches. We can hypothesize that the cofactor is involved in enzymatic activities that play a crucial role for colonization in P. kururiensis but not in B. vietnamiensis. As the production of B12 is associated with a high metabolic cost, sparing its expense could explain the fitness advantage displayed by B. vietnamiensis when the B12 synthesis pathway is mutated.
Defects in secretory activities are readily complemented in bacterial communities, making the responsible secretion systems opaque for detection by Tn-seq approaches. Indeed, secretion systems are not detected by most studies relying on Tn-seq except when focusing on a more sparsely colonized environment, such as the apoplast (31). In the present study, however, a T2SS was colonization depleted in B. vietnamiensis and P. kururiensis and the type 6 secretion system (T6SS) was colonization depleted in B. vietnamiensis during IR64 colonization. While the T6SS can be used for cell adhesion (46), a role beyond complementable secretion remains elusive for the T2SS. Still, as only one T2SS is supposedly involved in colonization, it suggests a separate role from the second one present in P. kururiensis.
The only gene with an identified function that is predicted by the Tn-seq approach to be negatively involved in root colonization in both B. vietnamiensis and P. kururiensis is pdeR. Site-directed mutagenesis and a competition assay revealed that its mutation was highly beneficial for the root colonization activity of B. vietnamiensis, as expected from the Tn-seq results, but detrimental for P. kururiensis, which was inconsistent with the Tn-seq analysis. PdeR is involved in the turnover of the secondary messenger c-di-GMP. In Xanthomonas oryzae, the deletion of pdeR results in a decreased virulence on rice (47). Through its association in two-component signal transduction systems, PdeR could have a variety of indirect roles beside c-di-GMP cycling (48). The divergence observed between Tn-seq and directed mutagenesis for P. kururiensis could also be linked to the inoculum concentration, which is likely to have been superior in the latter approach and might have triggered a different plant response with the observed detrimental effects on the P. kururiensis mutant population (Fig. 6).
Tn-seq identifies common plant colonization traits.
To date, there are no transcriptome data available for B. vietnamiensis that would allow a comparison with our data. However, the transcriptomic response of P. kururiensis to rice root macerates has been assessed before (14). Out of the 471 colonization-depleted genes identified by the present approach, 267 are differentially expressed in P. kururiensis when stimulated with root macerate (Data Set S5). Dominantly, these genes are involved in amino acid metabolism, cell motility, and cell wall/membrane biogenesis. The prevalence of these functions has further been revealed by Tn-seq approaches in other root colonization models such as Dickeya dadantii and Pseudomonas spp. (21, 23, 24). Our results are congruent with those of most plant-microbe colonization studies, underlining the importance of genes involved in the production of surface components for cellular attachment to the host (49). Our analysis also presents substantial overlap of colonization-depleted genes with what has been found in the plant growth-promoting species Azoarcus olearius and Herbaspirillum seropedicae (50). Notably, genes involved in peptidoglycan (ampD) and cell wall (murAI) formation, chemotaxis (cheARW), iron uptake (ferredoxins), and cobalamin synthesis (cbiA and cobIO) were detected.
Disruption of the ED pathway was demonstrated to reduce root colonization capacity in Pseudomonas chlororaphis, with a subsequent loss of induced systemic resistance (ISR) stimulation (51). Later works on Pseudomonas fluorescens observed that the expression of edd was enhanced in the rhizosphere of Arabidopsis compared to liquid growing medium (52). Mutants for edd in the latter study also failed to stimulate ISR but without the colonization defect observed in P. chlororaphis. For pseudomonads, the importance of the ED pathway might be linked to its direct product, pyruvate, which is required for the synthesis of the ISR-eliciting butanediol (51). Other advantages might arise from the ED pathway, such as the production of NAD(P)H, which is not generated by the Embden-Meyerhof-Parnas (EMP) glycolysis pathway. This cofactor is used by thioredoxins and could be involved in plant colonization and associated oxidative stress tolerance (33, 34).
IR64 rice has more stringent requirements for colonization by B. vietnamiensis and P. kururiensis than Nipponbare rice.
For both B. vietnamiensis and P. kururiensis, a stronger host genotype effect on the number of colonization-impacted genes was observed during the association with IR64 than with Nipponbare rice. For P. kururiensis, this genotype effect was also observed on the root colonization profiles of the two cultivars (Fig. 2). One specificity of indica rice (IR64) over japonica (Nipponbare) is its improved nitrate uptake and assimilation capacity (53), which is linked with the presence of the nitrate transporter NRT1.1B in the former and proved to impact its microbiota (18). Nipponbare rice preferably imports the alternative nitrogen source ammonium, which can be correlated with the depletion of bacterial mutants impacted in the import of this nutrient (amtB) in the Nipponbare environment in our experimental setup, as bacteria would compete with rice for the same nitrogen source (Table 3).
It was interesting to observe that the T6SS, a major macromolecular system of B. vietnamiensis, was only colonization depleted during inoculation on IR64 roots. The T6SS can be employed by bacteria for competition with other prokaryotes but also interaction with eukaryotes (54). As secretory function should be complemented by the bacterial community, the T6SS of B. vietnamiensis appears to be rather involved in adherence to eukaryotic cells, in a host-dependent manner.
Still, most colonization-impacted genes are conserved between rice cultivars, and several functions, such as motility, amino acid metabolism, and biofilm production, have been repeatedly described for their role in the general association of bacteria with plants (Fig. 7). We have found evidence of an increased stress to which bacteria are exposed in the near vicinity of plants through the depletion of mutants involved in osmoprotection, toxic compound degradation, and DNA reparation. The identification of several functions that are part of the core genome shared by both bacteria but are colonization impacted in only one strain reinforces the validity of these observations and the different adaptations that both bacteria must undergo during colonization of rice. This is especially true in the present system, as B. vietnamiensis and P. kururiensis induce different levels of plant defenses (6).
An increasing amount of plant microbiome studies rely on Tn-seq to reveal the nature of genes underlying root colonization (20, 21, 23, 24, 31, 50). Tn-seq further offers the benefit over more common methods, such as transcriptome sequencing (RNA-seq), to inform on the genes obstructing colonization (mutants with higher colonization fitness), thus presenting a more complete catalogue of factors driving host-bacterium compatibility. Consistent with other trending methods, such as microfluidic visualization technics and synthetic bacterial communities (49), Tn-seq proves here to be a powerful tool for a better understanding of the genetic bases underlying colonization of different hosts.
MATERIALS AND METHODS
Bacterial and plant culture conditions.
The strains used in this study were Burkholderia vietnamiensis LMG 10929 and Paraburkholderia kururiensis M130, either wild type or modified by insertion of the pIN29 plasmid conferring chloramphenicol (Cm) resistance and DsRed fluorescence, spontaneous rifampicin (Rif)- and spectinomycin (Spt)-resistant strains, or insertional mutants of a nonreplicative plasmid in several candidate genes in each strain (Table 4) (6, 55). Spontaneous antibiotic-resistant strains for LMG 10929 and M130 were obtained by plating 100 μL of bacterial liquid culture at an optical density at 600 nm (OD600) of 1.0 on Luria’s low-salt LB (LBm; Sigma-Aldrich) with either rifampicin (30 μg·mL−1) or spectinomycin (50 μg·mL−1). After 48 h of incubation at 28°C, single colonies were selected and streaked on fresh LBm plates with rifampicin or spectinomycin, then grown in broth LBm with antibiotics, and stored in 20% glycerol at −80°C. From glycerol stocks, bacterial strains were cultured as follows. Bacterial cells conserved at −80°C were plated on LBm agar plates (with antibiotic for mutants) and incubated for 72 h at 28°C. Single colonies were used to inoculate 10 mL of LBm broth (with antibiotic for mutants) in 50-mL Falcon tubes and incubated for various amounts of time, allowing the different strains to reach an OD600 of 1.0. For inoculation purposes, cultures were adjusted to 5 × 107 CFU·mL−1.
TABLE 4.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics and plasmid constructions | Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| MFDpir | Donor strain containing the pSAM-Ec vector and auxotroph for diaminopimelate, ΔdapA::(erm-pir) | 56 |
| B. vietnamiensis | ||
| LMG10929 | Wild-type strain | 42 |
| Rifr | Spontaneous rifampicin resistance clone of LMG10929 | This study |
| Sptr | Spontaneous spectinomycin resistance clone of LMG10929 | This study |
| DsRed | LMG1029 + pIN29 | 6 |
| ΩcobW | Insertion mutant of pSHAFT2 in AK36_2246 | This study |
| ΩpdeR | Insertion mutant of pSHAFT2 in AK36_4666 | This study |
| Ωedd | Insertion mutant of pSHAFT2 in AK36_178 | This study |
| P. kururiensis | ||
| M130 | Wild-type strain | 9 |
| Rifr | Spontaneous rifampicin resistance clone of M130 | This study |
| Sptr | Spontaneous spectinomycin resistance clone of M130 | This study |
| DsRed | M130 + pIN29 | 6 |
| ΩcobW | Insertion mutant of pSHAFT2 in ANSKv1_10390 | This study |
| ΩpdeR | Insertion mutant of pSHAFT2 in ANSKv1_51226 | This study |
| Ωedd | Insertion mutant of pSHAFT2 in ANSKv1_70910 | This study |
| Plasmids | ||
| pIN29 | Carries genes conferring DsRed fluorescence and chloramphenicol resistance. DsRed is under control of the highly active pTAC promoter. | 55 |
| pSAM-Ec | Contains Himar1C9 transposase and a mariner transposon with a kanamycin resistance cassette | 57 |
| pSHAFT2 | Vector used for insertional mutagenesis containing a chloramphenicol resistance gene (nonreplicative plasmid) | 62 |
Oryza sativa subsp. japonica cv. Nipponbare and Oryza sativa subsp. indica cv. IR64 were cultured as described by King et al. (6). Briefly, seeds were sterilized using successive 70% ethanol and 3.6% sodium hypochlorite treatments and germinated seedlings were transferred onto sterile perlite in an airtight hydroponic system. Five-day-old seedlings were inoculated with 1 mL of bacterial solution at 5 × 107 CFU·mL−1 and grown for up to 14 days at 28°C (16 h of light and 8 h of dark). For competition assays, five plants were inoculated with a mix at 1.107 CFU·mL−1 containing 0.5 mL of Sptr B. vietnamiensis or P. kururiensis strains (previously grown separately in broth LBm plus Spt at 50 μg·mL−1 and washed to remove antibiotic) and 0.5 mL of either one of the insertional mutants (ΩcobW, ΩpdeR, or Ωedd) of the corresponding strain (grown in LBm plus Cm at 100 μg·mL−1 and washed to remove antibiotic).
Estimation of root-colonizing bacterial population.
For the estimation of root-colonizing populations, the plants were prepared as described above and inoculated with WT strains at 1 × 107 CFU·mL−1. Three systems (15 plants) were prepared for each condition and grown for 1, 7, or 14 days (28°C; 16 h of light and 8 h of dark). At each time point, five plants were harvested for each condition. The entire root systems were sampled and separately transferred to screw-cap tubes containing 1 mL of sterile water and a sterile ceramic bead. Roots were weighed and then pulverized using a Fastprep-24 (MPbio) at 6 m·s−1 for 40 s. A serial dilution of the resulting solution was spotted out onto square LBm plates and incubated at 28°C for 48 h. The number of CFU was counted and adjusted to the weight of the root systems. The mean colonization values were compared between cultivars and between bacterial genotypes at each time point and for each bacterium separately using a Wilcoxon test. Results were considered significantly different at a P value of <0.05. For the competition assay between WT and mutant strains, plants were harvested at 7 dpi and processed as described above, except the bacterial dilutions were spotted onto LBm plates containing either streptomycin (30 μg·mL−1) or Cm (200 μg·mL−1) to select, respectively, the WT or the mutant strains.
Tn-seq library preparation.
A Himar1 mariner transposon carried by the pSAM_EC vector in Escherichia coli strain MFDpir (56, 57) was introduced into Paraburkholderia kururiensis M130 Rifr and Burkholderia vietnamiensis LMG 10929 Rifr through conjugation. Both donor and recipient were grown until they reached an OD600 of 1.0 in liquid LBm, and the medium of MFDpir was further supplemented with diaminopimelic acid (DAP; 300 μg·mL−1). Cells were spun down and washed once with LBm and concentrated to a final OD600 of 50. Donor and recipient strains were mixed 1:1 and 50-μL suspensions were spotted onto square petri dishes containing LBm and DAP. Growth rates were different for the two recipient species, and conjugation time was adapted accordingly. The mating mixtures were incubated for 6 h and 24 h at 28°C for LMG10929 and M130, respectively. The spots were then resuspended in 2 mL of LBm per plate. The mating mixtures were further spread on LBm petri dishes with rifampicin (30 μg·mL−1) and kanamycin (50 μg·mL−1) and incubated at 28°C. This positively selects for recipient strains having integrated the transposon and negatively selects for the DAP auxotroph E. coli donor. After growth, the colonies were resuspended in 1 mL of LBm per plate. The library was separated into 1-mL aliquots and stored in 20% glycerol at −80°C. Additionally, a serial dilution of the mating mixtures followed by spreading onto LBm with rifampicin (30 μg·mL−1) and kanamycin (50 μg·mL−1) was used to estimate the abundance of mutants in the library through CFU counting.
Tn-seq experimental setup.
Tn-seq libraries were thawed on ice, diluted in sterile water to an OD600 of 1, then inoculated into 50 mL of liquid LBm with rifampicin (30 μg·mL−1) and kanamycin (50 μg·mL−1) at a final OD600 of 0.1, and grown at 28°C and 150 rpm to an OD600 of 1.0. One part was conserved for plant inoculation, while the other was centrifuged at top speed for 10 min and flash-frozen for subsequent DNA extraction to serve as a control condition. Plants were grown in hydroponic systems as described above. After 5 days of growth, each plant was inoculated with 1 mL of Tn-seq library bacterial suspension at 5 × 107 CFU·mL−1. The experimentation was performed in triplicates, with each replicate consisting of five plants. At 7 dpi, the roots were harvested and placed in Tris-EDTA (TE) buffer.
DNA extraction and sequencing methods.
The bacterial genomic DNA isolation using the cetyltrimethylammonium bromide (CTAB) protocol (Joint Genome Institute [JGI]) was used for the extraction of bacterial DNA from the control and experimental conditions. For the latter, whole roots were used in the first stages of the protocol. Whole roots were immersed in the extraction solution and vortexed for 5 min at each step of the protocol to facilitate bacterial separation. Root grinding was avoided to prevent excessive DNA contamination from the plant material. Roots were removed from the extraction buffer after the lysis steps, before CTAB was added.
Ten micrograms of total DNA was digested with 1 μL of MmeI restriction enzyme (New England Biolabs) in a 250-μL total volume with 10 μL of S-adenosylmethionine (SAM; 1.5 mM) and 25 μL of CutSmart buffer for 1 h at 37°C. Then, 1 μL of FastAP thermosensitive alkaline phosphatase at 1 U·μL−1 (Thermo Scientific) was added and incubated for 1 h at 37°C. All enzymes were inactivated through a 5-min incubation at 75°C. Digested DNA was column purified using a NucleoSpin gel and PCR cleanup kit (Macherey-Nagel). Two micrograms of purified DNA was ligated to adapters containing specific barcode sequences. Adapters were obtained by annealing the primers 5′-TTCCCTACACGACGCTCTTCCGATCTXXXXXNN-3′ and 5′-YYYYYAGATCGGAAGAGCGTCGTGTAGGGAAAGAGT-3′ where NN are random nucleotides for annealing to the dinucleotide overhang generated by MmeI and XXXXX and YYYYY are complementary, barcode-specific sequences. Ligations were performed using 1 μL of 5 mM adapters, 1.5 μL of T4 DNA ligase at 1 Weiss U·μL−1 (Thermo Scientific), and 2.5 μL 10× ligase buffer in a total volume of 25 μL and incubated overnight at 16°C. One microliter of ligation product was amplified using PCR with a GO Taq DNA polymerase (Promega) and Illumina primers (P7 [5′-CAAGCAGAAGACGGCATACGAGATAGACCGGGGACTTATCATCCAACCTGT-3′] and P5 [5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′]). Twenty-two PCR cycles were run (30 s at 92°C, 30 s at 60°C, and 1 min at 72°C) with an initial step at 92°C for 2 min and a final step at 72°C for 10 min. PCR products were subjected to a final gel purification using the NucleoSpin gel and PCR cleanup kit (Macherey-Nagel).
Sequencing was performed at the high-throughput sequencing platform at I2BC (CNRS, Gif-sur-Yvette, France) using NextSeq500 (Illumina) technology and 80 sequencing cycles (single read). Preliminary data analysis (demultiplexing, trimming, and mapping) was performed by the sequencing platform. Sequences were trimmed from barcodes and mariner transposon sequences.
Tn-seq data analysis.
Gene essentiality for optimal growth in a liquid LBm broth under agitation was assessed through a hidden Markov model (HMM)-based analysis using the EL-ARTIST pipeline (window size of 7 TA sites; P value of 0.03) (58). The pipeline predicts whether a TA site is located in an essential or nonessential region. In some cases, the locus is predicted to be located in a region that contains both domains that are required and domains that are dispensable for growth. After the first refinement, the HMM will recalculate the transition probabilities based on the new data and repeat the run until the algorithm reaches convergence (see Data Set S6 for TA sites transposon insertion cover in each gene and condition).
For the identification of conditionally essential genetic regions, the sorted aligned sequences were first normalized using TRANSIT 3.1.0 (59) with the trimmed total reads method at default settings and an additional LOESS (locally estimated scatterplot smoothing) correction. The normalized data sets were then analyzed using the TnseqDiff function of the R package Tnseq (60). Adjusted P values (Padj) were calculated using the Benjamini and Hochberg correction. Genetic loci were considered colonization depleted when their occurrence under the experimental condition was at least 1.5-fold lower than under the control condition at a confidence level (Padj) of <0.05, with the inverse for colonization-enriched genes.
Comparative genomics procedures.
Core genome compositions were calculated using the Phyloprofile exploration tool implemented in the MicroScope microbial genome annotation and analysis platform (61). Homology constraints were set at minLrap of ≥0.8 and identity of ≥50%. We used the COGNiTOR precomputed COG category classification available on MicroScope to infer COG category affiliations of B. vietnamiensis and P. kururiensis genes.
Construction of insertional mutants.
For the production of the cobW, pdeR, or edd insertional mutants of B. vietnamiensis LMG10929 and P. kururiensis M130, a gene fragment of either cobW, pdeR and edd of each strain was amplified using specific primers (Table S1). Genes fragments were inserted into the target vector pSHAFT2 (62) carrying a chloramphenicol resistance gene using XbaI/XhoI double digestion. The insert and plasmid were mixed at a 5:1 ratio with regard to their respective molecular size, ligated using a T4 DNA ligase (Promega), and cloned into heat shock-competent E. coli DH5α cells. Positive cells were multiplied and their plasmids were extracted using the Wizard Minipreps DNA purification system (Promega). Plasmids were electroporated into WT B. vietnamiensis LMG10929 and P. kururiensis M130 strains. Transformed cells were transferred in LBm for 3 h at 30°C and then spotted onto LBm plates containing Cm (200 μg·mL−1) for selection of mutants. Plasmid insertion in each targeted gene was checked by PCR. Mutants and WT strains showed identical growth rates in broth LBm or Hoagland medium supplemented with glucose (data not shown).
Data availability.
Sequences have been deposited in the European Nucleotide Archive under BioProject number PRJEB42565.
ACKNOWLEDGMENTS
We acknowledge the IRD itrop HPC (South Green Platform; https://bioinfo.ird.fr and http://www.southgreen.fr) at IRD Montpellier for providing HPC resources that have contributed to the research results reported here. We acknowledge the high-throughput sequencing facility of I2BC for its sequencing and bioinformatics expertise. The LABGeM (CEA/Genoscope & CNRS UMR8030), the France Génomique, and the French Bioinformatics Institute national infrastructures are acknowledged for support within the MicroScope annotation platform.
We gratefully acknowledge financial support from the CGIAR research program (CRP) RICE as well as from the French national research agency (ANR) funding the BURKADAPT project (ANR-19-CE20-0007). A.W., J.L., and L.G. were supported by Ph.D. fellowships from the French Ministry of Higher Education, Research and Innovation.
Footnotes
Supplemental material is available online only.
Contributor Information
Lionel Moulin, Email: lionel.moulin@ird.fr.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCES
- 1.Levy A, Salas Gonzalez I, Mittelviefhaus M, Clingenpeel S, Herrera Paredes S, Miao J, Wang K, Devescovi G, Stillman K, Monteiro F, Rangel Alvarez B, Lundberg DS, Lu T-Y, Lebeis S, Jin Z, McDonald M, Klein AP, Feltcher ME, Rio TG, Grant SR, Doty SL, Ley RE, Zhao B, Venturi V, Pelletier DA, Vorholt JA, Tringe SG, Woyke T, Dangl JL. 2017. Genomic features of bacterial adaptation to plants. Nat Genet 50:138–150. 10.1038/s41588-017-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Compant S, Nowak J, Coenye T, Clement C, Ait Barka E. 2008. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol Rev 32:607–626. 10.1111/j.1574-6976.2008.00113.x. [DOI] [PubMed] [Google Scholar]
- 3.Suárez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonça-Previato L, James EK, Venturi V. 2012. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb Ecol 63:249–266. 10.1007/s00248-011-9929-1. [DOI] [PubMed] [Google Scholar]
- 4.Dias GM, de Sousa Pires A, Grilo VS, Castro MR, de Figueiredo Vilela L, Neves BC. 2019. Comparative genomics of Paraburkholderia kururiensis and its potential in bioremediation, biofertilization, and biocontrol of plant pathogens. Microbiologyopen 8:e00801. 10.1002/mbo3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallner A, King E, Ngonkeu ELM, Moulin L, Béna G. 2019. Genomic analyses of Burkholderia cenocepacia reveal multiple species with differential host-adaptation to plants and humans. BMC Genomics 20:803. 10.1186/s12864-019-6186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King E, Wallner A, Rimbault I, Barrachina C, Klonowska A, Moulin L, Czernic P. 2019. Monitoring of rice transcriptional responses to contrasted colonizing patterns of phytobeneficial Burkholderia s.l. reveals a temporal shift in JA systemic response. Front Plant Sci 10:1141. 10.3389/fpls.2019.01141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trân Van V, Berge O, Ngô Kê S, Balandreau J, Heulin T. 2000. Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield components in low fertility sulphate acid soils of Vietnam. Plant Soil 218/2:273–284. 10.1023/A:1014986916913. [DOI] [Google Scholar]
- 8.Mattos KA, Pádua VLM, Romeiro A, Hallack LF, Neves BC, Ulisses TMU, Barros CF, Todeschini AR, Previato JO, Mendonça-Previato L. 2008. Endophytic colonization of rice (Oryza sativa L.) by the diazotrophic bacterium Burkholderia kururiensis and its ability to enhance plant growth. An Acad Bras Cienc 80:477–493. 10.1590/S0001-37652008000300009. [DOI] [PubMed] [Google Scholar]
- 9.Baldani V, Oliveira E, Balota E, Baldani J, Kirchhof G, Dobereiner J. 1997. Burkholderia brasilensis sp. nov., uma nova espécie de bactéria diazotrófica endofítica. An Acad Bras Cienc 69:116. [Google Scholar]
- 10.Bournaud C, de Faria SM, dos Santos JMF, Tisseyre P, Silva M, Chaintreuil C, Gross E, James EK, Prin Y, Moulin L. 2013. Burkholderia species are the most common and preferred nodulating symbionts of the Piptadenia group (tribe Mimoseae). PLoS One 8:e63478. 10.1371/journal.pone.0063478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angus AA, Agapakis CM, Fong S, Yerrapragada S, Estrada-de los Santos P, Yang P, Song N, Kano S, Caballero-Mellado J, de Faria SM, Dakora FD, Weinstock G, Hirsch AM. 2014. Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS One 9:e83779. 10.1371/journal.pone.0083779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estrada-de los Santos P, Palmer M, Chávez-Ramírez B, Beukes C, Steenkamp E, Briscoe L, Khan N, Maluk M, Lafos M, Humm E, Arrabit M, Crook M, Gross E, Simon M, dos Reis Junior F, Whitman W, Shapiro N, Poole P, Hirsch A, Venter S, James E. 2018. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes (Basel) 9:389. 10.3390/genes9080389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberl L, Vandamme P. 2016. Members of the genus Burkholderia: good and bad guys. F1000Res 5:1007. 10.12688/f1000research.8221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutinho BG, Licastro D, Mendonça-Previato L, Cámara M, Venturi V. 2015. Plant-influenced gene expression in the rice endophyte Burkholderia kururiensis M130. Mol Plant Microbe Interact 28:10–21. 10.1094/MPMI-07-14-0225-R. [DOI] [PubMed] [Google Scholar]
- 15.Klonowska A, Melkonian R, Miché L, Tisseyre P, Moulin L. 2018. Transcriptomic profiling of Burkholderia phymatum STM815, Cupriavidus taiwanensis LMG19424 and Rhizobium mesoamericanum STM3625 in response to Mimosa pudica root exudates illuminates the molecular basis of their nodulation competitiveness and symbiotic evolutionary history. BMC Genomics 19:105. 10.1186/s12864-018-4487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V, Jeffery LD. 2015. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA 112:E911–E920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shenton M, Iwamoto C, Kurata N, Ikeo K. 2016. Effect of wild and cultivated rice genotypes on rhizosphere bacterial community composition. Rice (N Y) 9:42. 10.1186/s12284-016-0111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Liu YX, Zhang N, Hu B, Jin T, Xu H, Qin Y, Yan P, Zhang X, Guo X, Hui J, Cao S, Wang X, Wang C, Wang H, Qu B, Fan G, Yuan L, Garrido-Oter R, Chu C, Bai Y. 2019. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol 37:676–684. 10.1038/s41587-019-0104-4. [DOI] [PubMed] [Google Scholar]
- 19.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole BJ, Feltcher ME, Waters RJ, Wetmore KM, Mucyn TS, Ryan EM, Wang G, Ul-Hasan S, McDonald M, Yoshikuni Y, Malmstrom RR, Deutschbauer AM, Dangl JL, Visel A. 2017. Genome-wide identification of bacterial plant colonization genes. PLoS Biol 15:e2002860. 10.1371/journal.pbio.2002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Royet K, Parisot N, Rodrigue A, Gueguen E, Condemine G. 2019. Identification by Tn-seq of Dickeya dadantii genes required for survival in chicory plants. Mol Plant Pathol 20:287–306. 10.1111/mpp.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheatley RM, Ford BL, Li L, Aroney STN, Knights HE, Ledermann R, East AK, Ramachandran VK, Poole PS. 2020. Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis. Proc Natl Acad Sci USA 117:23823–23834. 10.1073/pnas.2009094117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Beskrovnaya P, Melnyk RA, Hossain SS, Khorasani S, O’Sullivan LR, Wiesmann CL, Bush J, Richard JD, Haney CH. 2018. A genome-wide screen identifies genes in rhizosphere-associated Pseudomonas required to evade plant defenses. mBio 9:e00433-18. 10.1128/mBio.00433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sivakumar R, Ranjani J, Vishnu US, Jayashree S, Lozano GL, Miles J, Broderick NA, Guan C, Gunasekaran P, Handelsman J, Rajendhran J. 2019. Evaluation of inseq to identify genes essential for Pseudomonas aeruginosa PGPR2 corn root colonization. G3 (Bethesda) 9:651–661. 10.1534/g3.118.200928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moule MG, Spink N, Willcocks S, Lim J, Guerra-Assunção JA, Cia F, Champion OL, Senior NJ, Atkins HS, Clark T, Bancroft GJ, Cuccui J, Wren BW. 2015. Characterization of new virulence factors involved in the intracellular growth and survival of Burkholderia pseudomallei. Infect Immun 84:701–710. 10.1128/IAI.01102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong Y-C, Abd El Ghany M, Naeem R, Lee K-W, Tan Y-C, Pain A, Nathan S. 2016. Candidate essential genes in Burkholderia cenocepacia J2315 identified by genome-wide TraDIS. Front Microbiol 7:1288. 10.3389/fmicb.2016.01288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gislason AS, Turner K, Domaratzki M, Cardona ST. 2017. Comparative analysis of the Burkholderia cenocepacia K56-2 essential genome reveals cell envelope functions that are uniquely required for survival in species of the genus Burkholderia. Microb Genom 3:e000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grazziotin AL, Vidal NM, Venancio TM. 2015. Uncovering major genomic features of essential genes in Bacteria and a methanogenic Archaea. FEBS J 282:3395–3411. 10.1111/febs.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gil R, Silva FJ, Peretó J, Moya A. 2004. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev 68:518–537. 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutierrez MG, Yoder-Himes DR, Warawa JM. 2015. Comprehensive identification of virulence factors required for respiratory melioidosis using Tn-seq mutagenesis. Front Cell Infect Microbiol 5:78. 10.3389/fcimb.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helmann TC, Deutschbauer AM, Lindow SE. 2019. Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc Natl Acad Sci USA 116:18900–18910. 10.1073/pnas.1908858116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki K, Okazaki K, Tawaraya K, Osaki M, Shinano T. 2009. Gas chromatography–mass spectrometry associated global analysis of rice root exudates under aseptical conditions. Soil Sci Plant Nutr 55:505–513. 10.1111/j.1747-0765.2009.00390.x. [DOI] [Google Scholar]
- 33.Chavarría M, Nikel PI, Pérez-Pantoja D, De Lorenzo V. 2013. The Entner-Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ Microbiol 15:1772–1785. 10.1111/1462-2920.12069. [DOI] [PubMed] [Google Scholar]
- 34.Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D. 2004. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem 279:21749–21758. 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 35.Streit WR, Entcheva P. 2003. Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl Microbiol Biotechnol 61:21–31. 10.1007/s00253-002-1186-2. [DOI] [PubMed] [Google Scholar]
- 36.Salaemae W, Booker GW, Polyak SW. 2016. The role of biotin in bacterial physiology and virulence: a novel antibiotic target for Mycobacterium tuberculosis. Microbiol Spectr 4:VMBF-0008-2015. 10.1128/microbiolspec.VMBF-0008-2015. [DOI] [PubMed] [Google Scholar]
- 37.Selkrig J, Belousoff MJ, Headey SJ, Heinz E, Shiota T, Shen HH, Beckham SA, Bamert RS, Phan MD, Schembri MA, Wilce MCJ, Scanlon MJ, Strugnell RA, Lithgow T. 2015. Conserved features in TamA enable interaction with TamB to drive the activity of the translocation and assembly module. Sci Rep 5:12905–12912. 10.1038/srep12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter AM, Fazli M, Schmid N, Shilling R, Suppiger A, Givskov M, Eberl L, Tolker-Nielsen T. 2019. Key players and individualists of cyclic-di-GMP signaling in Burkholderia cenocepacia. Front Microbiol 10:3286. 10.3389/fmicb.2018.03286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peekhaus N, Conway T. 1998. What’s for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol 180:3495–3502. 10.1128/JB.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crouzet J, Levy-Schil S, Cameron B, Cauchois L, Rigault S, Rouyez MC, Blanche F, Debussche L, Thibaut D. 1991. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. J Bacteriol 173:6074–6087. 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Govindarajan M, Balandreau J, Muthukumarasamy R, Revathi G, Lakshminarasimhan C. 2006. Improved yield of micropropagated sugarcane following inoculation by endophytic Burkholderia vietnamiensis. Plant Soil 280:239–252. 10.1007/s11104-005-3223-2. [DOI] [Google Scholar]
- 42.Gillis M, Van Van T, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez MP. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol 45:274–289. 10.1099/00207713-45-2-274. [DOI] [Google Scholar]
- 43.Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, Vandamme P. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol 38:3165–3173. 10.1128/JCM.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang S-Y, Hara S, Melling L, Goh K-J, Hashidoko Y. 2010. Burkholderia vietnamiensis isolated from root tissues of nipa palm (Nypa fruticans) in Sarawak, Malaysia, proved to be its major endophytic nitrogen-fixing bacterium. Biosci Biotechnol Biochem 74:1972–1975. 10.1271/bbb.100397. [DOI] [PubMed] [Google Scholar]
- 45.Raux E, Schubert HL, Warren MJ. 2000. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell Mol Life Sci C 57:1880–1893. 10.1007/PL00000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lertpiriyapong K, Gamazon ER, Feng Y, Park DS, Pang J, Botka G, Graffam ME, Ge Z, Fox JG. 2012. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One 7:e42842. 10.1371/journal.pone.0042842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang F, Xue D, Tian F, Hutchins W, Yang CH, He C. 2019. Identification of c-di-GMP signaling components in Xanthomonas oryzae and their orthologs in xanthomonads involved in regulation of bacterial virulence expression. Front Microbiol 10:1402. 10.3389/fmicb.2019.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang F, Tian F, Sun L, Chen H, Wu M, Yang C-H, He C. 2012. A novel two-component system PdeK/PdeR regulates c-di-GMP turnover and virulence of Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact 25:1361–1369. 10.1094/MPMI-01-12-0014-R. [DOI] [PubMed] [Google Scholar]
- 49.Knights HE, Jorrin B, Haskett TL, Poole PS. 2021. Deciphering bacterial mechanisms of root colonization. Environ Microbiol Rep 13:428–444. 10.1111/1758-2229.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.do Amaral FP, Tuleski TR, Pankievicz VCS, Melnyk RA, Arkin AP, Griffitts J, Tadra-Sfeir MZ, Maltempi de Souza E, Deutschbauer A, Monteiro RA, Stacey G. 2020. Diverse bacterial genes modulate plant root association by beneficial bacteria. mBio 11:e03078-20. 10.1128/mBio.03078-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim HJ, Nam HS, Anderson AJ, Yang KY, Cho BH, Kim YC. 2007. Mutation in the edd gene encoding the 6-phosphogluconate dehydratase of Pseudomonas chlororaphis O6 impairs root colonization and is correlated with reduced induction of systemic resistance. Lett Appl Microbiol 44:56–61. 10.1111/j.1472-765X.2006.02029.x. [DOI] [PubMed] [Google Scholar]
- 52.Cheng X, Etalo DW, van de Mortel JE, Dekkers E, Nguyen L, Medema MH, Raaijmakers JM. 2017. Genome-wide analysis of bacterial determinants of plant growth promotion and induced systemic resistance by Pseudomonas fluorescens. Environ Microbiol 19:4638–4656. 10.1111/1462-2920.13927. [DOI] [PubMed] [Google Scholar]
- 53.Chanch T, Ohira K. 1982. Comparison of the uptake and assimilation of ammonium and nitrate in indica and japonica rice plants using the tracer 15N method. Soil Sci Plant Nutr 28:79–90. 10.1080/00380768.1982.10432373. [DOI] [Google Scholar]
- 54.Cianfanelli FR, Monlezun L, Coulthurst SJ. 2016. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24:51–62. 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 55.Vergunst AC, Meijer AH, Renshaw SA, O'Callaghan D. 2010. Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect Immun 78:1495–1508. 10.1128/IAI.00743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferrières L, Hémery G, Nham T, Guérout AM, Mazel D, Beloin C, Ghigo JM. 2010. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192:6418–6427. 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiles TJ, Norton JP, Russell CW, Dalley BK, Fischer KF, Mulvey MA. 2013. Combining quantitative genetic footprinting and trait enrichment analysis to identify fitness determinants of a bacterial pathogen. PLoS Genet 9:e1003716. 10.1371/journal.pgen.1003716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, Zhang YJ, Rubin EJ, Waldor MK. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet 10:e1004782. 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. 2015. TRANSIT—a software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11:e1004401. 10.1371/journal.pcbi.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao L, Anderson MT, Wu W, Harry HL, Bachman MA. 2017. TnseqDiff: identification of conditionally essential genes in transposon sequencing studies. BMC Bioinformatics 18:326. 10.1186/s12859-017-1745-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Médigue C, Calteau A, Cruveiller S, Gachet M, Gautreau G, Josso A, Lajus A, Langlois J, Pereira H, Planel R, Roche D, Rollin J, Rouy Z, Vallenet D. 2019. MicroScope—an integrated resource for community expertise of gene functions and comparative analysis of microbial genomic and metabolic data. Brief Bioinform 20:1071–1084. 10.1093/bib/bbx113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shastri S, Spiewak HL, Sofoluwe A, Eidsvaag VA, Asghar AH, Pereira T, Bull EH, Butt AT, Thomas MS. 2017. An efficient system for the generation of marked genetic mutants in members of the genus Burkholderia. Plasmid 89:49–56. 10.1016/j.plasmid.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Fig. S1. Download aem.00642-22-s0001.pdf, PDF file, 1.2 MB (1.2MB, pdf)
Data Set S1. Download aem.00642-22-s0002.xlsx, XLSX file, 0.4 MB (425.2KB, xlsx)
Data Set S2. Download aem.00642-22-s0003.xlsx, XLSX file, 0.1 MB (136.4KB, xlsx)
Data Set S3. Download aem.00642-22-s0004.xlsx, XLSX file, 0.9 MB (895.7KB, xlsx)
Data Set S4. Download aem.00642-22-s0005.xlsx, XLSX file, 0.06 MB (60.9KB, xlsx)
Data Set S5. Download aem.00642-22-s0006.xlsx, XLSX file, 0.2 MB (227.9KB, xlsx)
Data Set S6. Download aem.00642-22-s0007.xlsx, XLSX file, 12.9 MB (12.9MB, xlsx)
Data Availability Statement
Sequences have been deposited in the European Nucleotide Archive under BioProject number PRJEB42565.