Abstract
Take-all, caused by Gaeumannomyces graminis var. tritici, is one of the most important fungal diseases of wheat worldwide. Knowing that microbe-based suppression of the disease occurs in monoculture wheat fields following severe outbreaks of take-all, we analyzed the changes in rhizosphere bacterial communities following infection by the take-all pathogen. Several bacterial populations were more abundant on diseased plants than on healthy plants, as indicated by higher counts on a Pseudomonas-selective medium and a higher fluorescence signal in terminal restriction fragment length polymorphism analyses of amplified 16S ribosomal DNA (rDNA). Amplified rDNA restriction analysis (ARDRA) of the most abundant cultured populations showed a shift in dominance from Pseudomonas to Chryseobacterium species in the rhizosphere of diseased plants. Fluorescence-tagged ARDRA of uncultured rhizosphere washes revealed an increase in ribotypes corresponding to several bacterial genera, including those subsequently identified by partial 16S sequencing as belonging to species of alpha-, beta-, and gamma-proteobacteria, sphingobacteria, and flavobacteria. The functional significance of some of these populations was investigated in vitro. Of those isolated, only a small subset of the most abundant Pseudomonas spp. and a phlD+ Pseudomonas sp. showed any significant ability to inhibit G.graminis var. tritici directly. When cultured strains were mixed with the inhibitory phlD+ Pseudomonas strain, the Chryseobacterium isolates showed the least capacity to inhibit this antagonist of the pathogen, indicating that increases in Chryseobacterium populations may facilitate the suppression of take-all by 2,4-diacetylphloroglucinol-producing phlD+ pseudomonads.
Take-all is a root disease of wheat and barley caused by the fungus Gaeumannomyces graminis var. tritici (2). Several practices can be used to limit the disease on susceptible crops, including tillage, rotation, choice of variety and N fertilizer, and chemical and biological seed treatments (7, 10, 30, 38). However, in regions where monoculturing of small grains is climatically and economically favored, continuous crops of wheat or barley can be productively maintained despite the presence of the pathogen. Unlike most root diseases, a severe outbreak of take-all followed by 4 to 6 years of monoculturing of wheat or barley will induce a natural suppression of the disease called take-all decline (TAD) (13). The suppressive factor in TAD soils is known to be a heat-labile fraction of soil microbial communities (2, 13). The occurrence of TAD in different wheat-growing regions of the world is remarkable because soil microbial communities are affected by both plant and soil factors (12, 17). In the state of Washington, studies have shown that antibiotic-producing fluorescent Pseudomonas spp. are present in TAD soils and can suppress take-all (5, 8, 24, 32, 34, 36, 42, 44, 47, 48).
Recently, work has focused on understanding the contributions of fluorescent Pseudomonas spp. that synthesize 2,4diacetylphloroglucinol (2,4-DAPG) to take-all suppression. Pseudomonads that produce this broad-spectrum antibiotic can suppress a variety of fungal root pathogens when applied as seed and soil inoculants (14, 35, 39, 44). Indigenous populations of these bacteria are abundant in TAD soils, and take-all suppression has been correlated with their presence in soils(34, 36). The diversity of these functionally important pseudomonad populations has been investigated (22, 29), and the capacity of different genotypes to control root diseases is the subject of ongoing study.
Other bacterial populations, including pseudomonads that do not produce 2,4-DAPG, are known to increase in abundance in the rhizosphere of diseased roots (23, 37, 38, 46); however, their involvement in the induction of TAD and their impact on populations of 2,4-DAPG-producing Pseudomonas spp. are unknown. To better understand the roles of different microbial populations in the development of take-all and its subsequent decline, it is necessary to compare bacterial communities inhabiting the rhizospheres of both healthy and diseased wheat plants. Past approaches for analyzing microbial community structure in take-all pathosystems have focused on isolating specific bacteria on selective media and identifying shifts in responses to pathogen infections (23, 37, 38, 46). The limitations of culture-based approaches are well known, and it has been suggested that complementary methods should be used to properly assess microbial communities in soil (18). In this study, we used two high-throughput methods: a culture-dependent method for enumerating specific populations of pseudomonads (28) and a culture-independent method for analyzing terminal restriction fragment length polymorphisms (T-RFLPs) of amplified 16S ribosomal DNA (rDNA) (19), called fluorescence-tagged amplified rDNA restriction analysis (FT-ARDRA) (25, 26).
The purpose of this study was to assess the changes in rhizosphere bacterial communities that occur when wheat roots go from a healthy to a diseased state. Our objectives were to (i) determine the association between different populations of Pseudomonas spp. and take-all disease on wheat roots, (ii) identify new bacterial taxa not previously associated with the ecology of take-all or TAD, and (iii) determine the capacity of the identified and cultured bacterial populations to inhibit the take-all pathogen and 2,4-DAPG-producing biocontrol bacteria.
MATERIALS AND METHODS
Bacterial and fungal culturing.
All chemicals were obtained from Sigma Chemical Co., St. Louis, Mo., unless noted otherwise. All bacterial and fungal cultures were incubated at room temperature (23 ± 2oC) in the dark. Bacteria were maintained on a Pseudomonas-selective medium, 1/3× KMB+++ (28), based on Simon-Ridge medium (40). Stock cultures of all strains were stored in 1/3× KMB (no antibiotics added)–18% glycerol at −80°C. G. graminis var. tritici strain R3-111a-1 (32) was used to infest soil and for in vitro inhibition assays. Fungi were maintained on fresh 1/5× PDA (containing, per liter, 4 g of dextrose, infusion from 40 g of freshly boiled potatoes [pH 6.3], and 18 g of agar). Oat kernel inoculum of G. graminis var. tritici was prepared by adding sliced agar cultures of G. graminis var. tritici to sterilized oat kernels in 1-liter flasks and incubating the mixtures at room temperature for 3 to 4 weeks. Colonized oat seeds were then air dried in a laminar flow hood and stored at room temperature for up to 6 months prior to use. Virulent stock cultures of G. graminis var. tritici were stored at 5°C.
Soils used for comparisons.
Soils at the Washington State University Mount Vernon Research and Extension Unit, Mount Vernon, are conducive to take-all and develop suppressiveness for take-all with several years of wheat monoculturing. On 3 November 1997, winter wheat (Triticum aestivum L. cv. Madsen) was mechanically sown at a rate of 90 lb of seed per acre. Part of the field was infested with G. graminis var. tritici-infested oat kernels (20% [wt/wt] of seed, or 0.6 g of oat kernels per row meter) (32). Infested and noninfested plots were established side by side across the field. Soil from the noninfested portions of the field was designated soil A, and soil from the infested portions was designated soil B. In October 1998 and October 1999, the field was again seeded with wheat, but no G. graminis var. tritici inoculum was added. During the 1997–1998 growing season, plants growing in soil A were significantly more healthy than those growing in soil B (P < 0.05), as indicated by lower root disease ratings, higher head counts, and yield (data not shown). During the following growing season, disease was observed across the entire field, and no significant differences in crop health were noted between the two soils (data not shown). Soils A and B were collected in the fall seasons of 1998 and 1999 from field plots following harvest for use in growth chamber assays.
Growth chamber assays.
Tapered plastic tubes (SC-10 Supercell containers; Hummert International, Earth City, Mo.) were filled with 0.1 kg of fresh soil into which two seeds of wheat (T. aestivum L. cv. Penewawa) were planted 1 cm below the surface. To half of the tubes, five to eight G. graminis var. tritici-infested oat kernels were added to the soil in a single layer 3 cm below the seed. The tubes were watered to saturation, covered with clear plastic wrap to maintain humidity, and placed in a growth chamber (15°C, 12-h light-dark cycle). After 5 days, the plastic wrap was removed, and the tubes were watered biweekly as needed. At 10 to 14 days after planting, plants were thinned to one per container and returned to the chamber. At 5 weeks after planting, the plants were harvested for microbiological analyses. Each tube served as a replicate, and a total of 15 to 18 replicates of each treatment were used in each experiment.
Processing of rhizosphere samples.
Rhizospheres from individual plants were processed separately as described previously (28). Briefly, samples from different treatments were processed in parallel. The intact portion of each root system was recovered from the soil, separated from the shoot, and placed in 7.5 ml of sterile distilled water. Bacteria and adhering soil were dislodged from the roots by vortexing and subsequent incubation in a sonication bath. One-hundred microliters of each sample was serially diluted 1:3 in a 96-well microtiter plate (Costar, Corning, N.Y.), each well of which was prefilled with sterile distilled water. Aliquots from these “rhizosphere-wash” plates were used to inoculate dilution plates for culturing bacteria and as templates for PCR amplification of rDNA sequences (see below).
Enumeration of cultured bacterial populations.
Growth chamber assays were used to determine the abundance and diversity of cultured rhizosphere bacterial populations. From rhizosphere-wash plates, 50 μl of each dilution was transferred to other 96-well plates containing 200 μl of 1/3× KMB+++ per well. These culture plates were incubated at room temperature in the dark for 48 ± 4 h, and bacterial growth was assayed spectrophotometrically (an optical density at 600 nm of ≥0.05 was scored as positive). Replica plates were made by transferring half of each culture into an equal volume of 35% glycerol, and these plates were stored at −80°C. The original culture plates were frozen at −80°C for a minimum of 1 h and then transferred to a −20°C freezer for storage.
The abundance and diversity of 2,4-DAPG-producing Pseudomonas spp. in the dilution cultures were determined using a previously described PCR-based assay (28). Briefly, portions of the phlD gene (4) were amplified using gene-specific primers B2BF (ACC CAC CGC AGC ATC GTT TAT GAG C) and BPR4 (CCG CCG GTA TGG AAG ATG AAA AAG TC). To determine the genotype of phlD+ populations, amplification products were digested with HaeIII or TaqI (New England Biolabs, Beverly, Mass.). DNA fragments were separated on agarose gels in 0.5× Tris-borate-EDTA and visualized by ethidium bromide staining. Gel images were processed using a Kodak (Rochester, N.Y.) EDAS120 or EDAS290 digital imaging system.
Characterization of cultured bacterial populations.
The most abundant cultured bacterial populations were recovered from the highest dilution scored positive for growth (i.e., the terminal dilution culture [TDC]) and stored in glycerol. In most instances, a single dominant bacterial morphotype was isolated from each TDC by streaking on 1/3× KMB+++ agar. In instances when two morphotypes appeared to be equally dominant, both were purified and used in subsequent analyses. The distributions of morphotypes recovered from fresh and frozen TDCs matched exactly (data not shown). Sixty-nine isolates were obtained from the 1998 samples, and 98 more were obtained from the 1999 samples. All isolates were classified initially by morphotype and genomic fingerprinting using the primer BOXAIR (BOX=PCR) as described previously (29) (data not shown), and 53 representative isolates from independent replicates of each treatment in each year were chosen for further analyses. The genotypes of these representative isolates were determined using amplified rDNA restriction analysis (ARDRA) of 16S rDNA, and partial sequences of multiple representative ribotypes were subsequently determined as described below. In addition, two phlD+ isolates, MtV1 and MtV2, present in Mount Vernon soils A and B, respectively, were obtained on glycerol replica plates from the terminal dilution in which they were detected as described previously (28).
Characterizations of amplified 16S rDNA sequences.
Nearly full-length portions of 16S rDNA were PCR amplified from eubacterial templates with high-pressure liquid chromatography-purified oligonucleotide primers 8F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-ACG GCT ACC TTG TTA CGA CTT-3′), based on those described by Weisburg et al. (45) (Operon Technologies, Alameda, Calif.). For FT-ARDRA, a fluorescence-labeled primer, 8F-HEX, was used instead of 8F. Templates for PCR amplification were obtained from freeze-thaw lysates of either (i) whole cells of isolated bacteria, (ii) the third and fourth dilutions of the rhizosphere-wash plates, or (iii) isolated plasmids containing cloned 16S sequences. Amplification was carried out with a 25-μl reaction mixture containing 2.5 μl of 10× buffer (Promega Corp., Madison, Wis.), 1.8 μl of 25 mM MgCl2, 2.5 μl of 2 mM deoxynucleoside triphosphates, 14.4 μl of sterile distilled water, 1 μl of each primer (50 pmol per μl), 0.2 μl of RNase A (2 mg/ml), 0.33 μl of Taq DNA polymerase (5 U/μl) (Promega), and 5 μl of template. Amplification was performed with a PTC-200 Thermocycler (MJ Research Inc., Watertown, Mass.). The cycling program included a 5-min initial denaturation step at 95°C; 30 cycles of 94°C for 60 s, 54°C for 45 s, and 70°C for 60 s; and an 8-min final extension step at 70°C. Amplification products were separated on agarose gels and visualized as described above.
Restriction digestions were performed with 96-well plates. Each digestion included 7 μl of a PCR reaction mixture and 10 U of either RsaI or MspI (New England Biolabs) in a total volume of 30 μl. Reaction mixtures were incubated at 37°C for 2 to 4 h and then stored at −20°C. For standard ARDRA, restriction fragments were separated by electrophoresis on 1.6% agarose gels and visualized as described above. For FT-ARDRA, digests were submitted to the Laboratory for Biotechnology and Bioanalysis, Washington State University, Pullman, for electrophoresis and imaging. There, 2 μl of each digest was mixed with 2 μl of loading buffer containing a fluorescence-labeled DNA size standard (G2500-ROX; Applied Biosystems, Foster City, Calif.) and loaded onto 6% urea-containing polyacrylamide gels. Electrophoresis was performed with an ABI373 sequencer (Applied Biosystems) such that DNA fragments of ≥1,100 bp could be sized. Data were collected automatically using the B filter and analyzed with GeneScan 2.1 software and Genotyper 2.1 (Applied Biosystems).
To identify the bacteria corresponding to different ribotypes in T-RFLP profiles, we initially analyzed our results using web-based TRFLP-TAP software (21) available through the Ribosomal Database Project (http://www.cme.msu.edu/RDP/html/analyses.html; Center for Microbial Ecology, Michigan State University, East Lansing) (20). Using this software, we assembled lists of bacteria whose sequences were present in the 16S ribosomal database (release 7.0) and predicted to have terminal restriction fragments (TRFs) corresponding to three possible situations: (i) one terminal restriction fragment (TRF) increasing in each profile with similar peak areas (matched pairs), (ii) one TRF increasing in each profile but with dissimilar peak areas (unmatched pairs), and (iii) one TRF increasing in one profile and a predicted matching signal that was visible but not actually increasing in the second profile (lone peak). Given the extensiveness of the lists, only matched and unmatched pairs were considered useful for predicting the bacterial genera corresponding to specific ribotypes given the two restriction enzymes used in our analyses.
Additional analyses relied on partial sequencing of amplified16S rDNA. For identification of the cultured isolates, direct sequencing was performed using primer 8F. To supplement FT-ARDRA, a clone bank was prepared from rDNA amplified from three different diseased plants. To do so, amplification products were separated on an 0.7% agarose gel, extracted using a QIAEX II gel extraction kit (Qiagen, Valencia, Calif.), and ligated into the pGEM-T Easy vector (Promega) according to the manufacturer's protocol. Plasmids were introduced into Escherichia coli cells and isolated from transformants using standard methods (3). Plasmids containing inserts representing distinct ribotypes were identified from a collection of over 70 transformants by ARDRA of whole-cell templates. The nucleotide sequences of cloned inserts and PCR products from individual isolates were determined by using an ABI Prism dye terminator cycle sequencing kit (Perkin-Elmer) according to the manufacturer's instructions. Sequence data were analyzed with BLAST using the web server housed at the National Center for Biotechnology Information (1). Taxonomic designations were made following the terminology of Bergey's Manual Trust (http://www.cme.msu.edu/bergeys/taxonomyinfo.html), Michigan State University, East Lansing.
In vitro inhibition assays.
Agar plugs (7-mm diameter) of G. graminis var. tritici were transferred to the center of a fresh plate of 1/5× PDA and incubated at room temperature in the dark. On the following day, a bacterial inoculum was prepared by resuspending isolated colonies in 150 μl of 1/3× KMB to an optical density at 600 nm of between 0.2 and 0.4. Ten microliters of the bacterial inoculum (106 cells) from four different bacterial strains was inoculated at four equidistant positions along the perimeter of the assay plate. For each assay, the positions of the inoculated strains were randomized, each strain appeared on four different assay plates inoculated on two separate occasions, and three uninoculated control plates were used. Plates were incubated for up to 8 days at room temperature in the dark. Growth of the fungus was assayed at two different time points in each assay, when the growth on the uninoculated control plates was approximately 0.7 times and 1.0 times the radius of the plate. Two measurements were made: the distance from the edge of the plug to the growing edge of the fungus (x) and the distance from the edge of the bacterial growth to the growing edge of the fungus (y). For each replicate, an inhibition index (i) was calculated as follows: i = y/(x+y). The impact of each bacterial strain on the activity of 2,4-DAPG-producing fluorescent Pseudomonas spp. was assessed in similar assays in which a mixed bacterial inoculum was used, with each individual strain and strain MtV1 being present at a 3:1 ratio.
Statistical analyses.
Statistical analyses were performed with the assistance of the JMP statistical software package (SAS Inc., Cary, N.C.) and the data analysis package bundled with Microsoft Excel 97 (Microsoft Corp., Redmond, Wash.) and with appropriate parametric and nonparametric procedures (27). Disease ratings, median population counts, and TRF peak areas were compared using the two-sample Mann-Whitney test, and inhibition indices were compared using the Tukey-Kramer multiple-range test.
Nucleotide sequence accession numbers.
Cloned sequences were deposited into GenBank under accession numbers AF375827 through AF375850.
RESULTS
Changes in cultured bacterial populations.
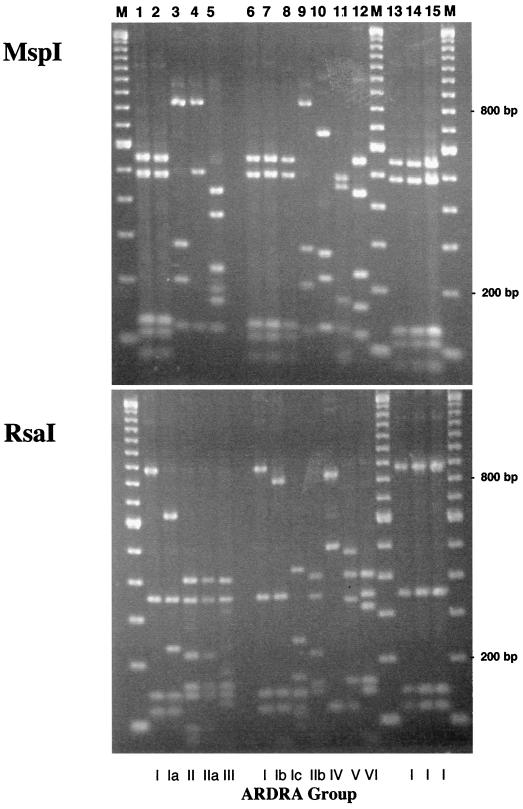
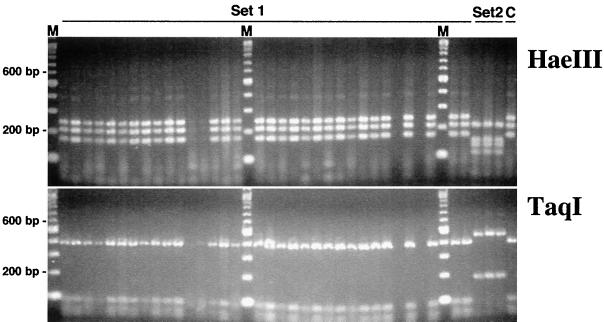
Using a Pseudomonas-selective liquid medium (i.e., 1/3× KMB+++), we observed differences in the size and diversity of bacterial populations cultured from healthy and diseased roots of wheat plants grown in a growth chamber. Changes in specific bacterial populations were associated with increased levels of disease in four different comparisons (Table 1). In each of the comparisons, the abundance of cultured bacteria was higher in diseased than in healthy rhizospheres (P < 0.05). Amplified rDNA restriction analyses of 16S sequences obtained from the dominant bacterial populations (i.e., those recovered from TDCs) revealed several different genotypes (Fig. 1). In these experiments, two groups of bacteria were found in the majority of TDCs, ARDRA groups I and II. These two ARDRA-defined genotypes belong to Pseudomonas and Chryseobacterium, respectively, based on >500 bp of16S rDNA sequence obtained from several isolates of each group. The ARDRA group II bacteria were isolated more frequently from the TDCs obtained from diseased plants than any other ARDRA-defined genotype. In contrast, ARDRA group I isolates were obtained less frequently from the rhizospheres of diseased roots than from those of healthy roots. Populations of phlD+ Pseudomonas spp. were more abundant in the rhizospheres of diseased plants only when they were grown in the previously noninfested field soil (soil A). These differences were observed as both counts of and frequency of rhizosphere colonization by indigenous phlD+ bacteria. In all instances, the RFLP pattern of the detected phlD sequences matched that of known BOX D genotypes of phlD-containing pseudomonads (28), and phlD-containing pseudomonads corresponding to that genotype were isolated (Fig. 2).
TABLE 1.
Changes in cultured bacterial populations associated with take-all disease on wheat grown in two Mount Vernon soils in growth chambers
| Yr | Soila | Diseaseb
|
Most abundant bacteriac
|
phlD+ bacteriad
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of samples | DRe | Frequency | Counte | ARDRA groupf
|
Count7 | Frequency | ||||
| I | II | Other | ||||||||
| 1998 | A + GGT | 15 | 4 a | 1.00 | 7.9 a | 0.18 | 0.64 | 0.18 | 4.6 a | 0.80 |
| A | 16 | 0 b | 0.06 | 6.7 b | 0.33 | 0.42 | 0.25 | <3.1 b | 0.31 | |
| B + GGT | 18 | 3 a | 0.56 | 7.4 a | 0.17 | 0.33 | 0.50 | 3.4 a | 0.50 | |
| B | 18 | 0 b | 0.00 | 6.9 b | 0.35 | 0.48 | 0.17 | 4.1 a | 0.67 | |
| 1999 | A + GGT | 18 | 2.5 a | 0.89 | 6.5 a | 0.25 | 0.42 | 0.33 | 4.1 a | 0.78 |
| A | 18 | 0 b | 0.00 | 5.7 b | 0.53 | 0.11 | 0.37 | <3.1 b | 0.22 | |
| B + GGT | 18 | 2 a | 0.83 | 6.5 a | 0.33 | 0.40 | 0.27 | <3.1 b | 0.28 | |
| B | 18 | 0 b | 0.00 | 5.0 b | 0.52 | 0.05 | 0.43 | <3.1 b | 0.06 | |
Two soils, A and B, were taken from a field in Mount Vernon, Wash., following harvest in 1998 and 1999 for these assays. Soil B had been infested with a G. graminis var. tritici (GGT) inoculum at planting in the fall of 1997. Each soil was tested with and without the added GGT inoculum.
No. of samples, number of individually grown plants sampled. Take-all disease ratings (DR) ranged from 0 (no disease) to 8 (dead plants). The frequency of significant disease was tabulated as the proportion of plants with a DR greater than or equal to the combined median DR in each year (i.e., a DR of 3 in 1998 and a DR of 2 in 1999).
The median log count of cells growing on 1/3× KMB+++ is given on a per-rhizosphere basis (count) and as the proportions of rhizosphere samples dominated by bacteria belonging to different groups of strains isolated from the TDCs obtained from each sample (ARDRA groups).
The median log count of phlD+ bacteria detected in each sample is given on a per-rhizosphere basis (count) and as the proportion of rhizosphere samples with detectable populations above the limit of detection (i.e., ≥3.1 log units per rhizosphere) (frequency).
Values followed by different letters are significantly different, as determined by the Mann-Whitney U test (P < 0.05).
Genotypes of isolates based on ARDRA of 16S sequences with MspI and RsaI.
FIG. 1.
ARDRA of distinct strains of the most abundant bacterial populations isolated from wheat rhizospheres in this study. All bacteria were cultured on 1/3× KMB+++. ARDRA of 16S sequences was performed using MspI and RsaI. Restriction digests of 16S sequences amplified from the most abundant cultured bacteria obtained from the 1998 (lanes 1 through 5) and 1999 (lanes 6 through 12) growth chamber experiments are shown, as are digests of sequences obtained from bacteria with the capacity to inhibit G. graminis var. tritici in vitro (lanes 13 through 15). The ARDRA group designation for each strain is indicated at the bottom of the figure. The five ARDRA groups identified in the1998 samples are represented by isolates U5 (lane 1), dI14 (lane 2), U3 (lane 3), dU1 (lane 4), and U1 (lane 5). The seven ARDRA groups identified in the 1999 samples are represented by strains C201A (lane 6), C201 (lane 7), C2+6 (lane 8), C2+4 (lane 9), E1+1 (lane 10), E102 (lane 11), and C204B (lane 12). In addition, representative strains with the ability to inhibit the take-all pathogen are also displayed: strain dI1 (lane 13), strain E206 (lane 14), and phlD+ strain MtV1 (lane 15). The sizes of individual fragments were determined based on the 100-bp ladder shown in lanes M.
FIG. 2.
Genotyping of phlD-containing bacteria present in the soils of Mount Vernon, Wash. Representative results for the growth chamber assays are shown. The phlD sequences were amplified using gene-specific primers B2BF and BPR4 and subsequently digested with either HaeIII or TaqI. Set 1 includes samples of several terminal phlD+ dilutions and isolates obtained from rhizospheres of wheat grown in soils A and B. Set 2 includes three isolates of a different genotype obtained from a Lind, Wash., soil for contrast. A known 2,4-DAPG-producing, BOX D genotype strain of Pseudomonas fluorescens (Q8r1–96) was used as a positive control for comparison (lane C). The sizes of individual fragments were determined based on the 100-bp ladder shown in lanes M.
Changes in uncultured bacterial populations.
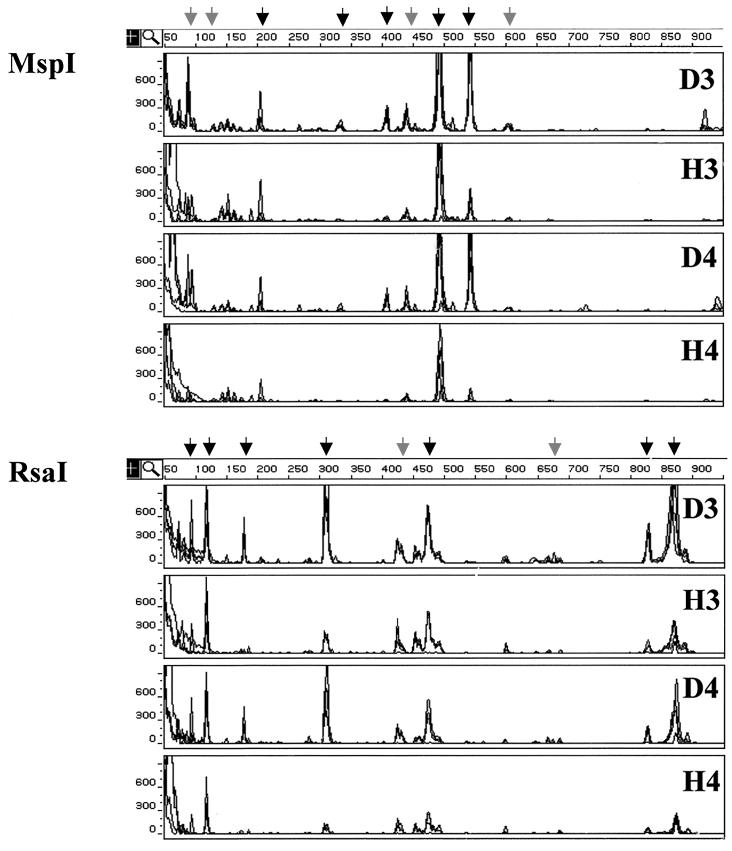
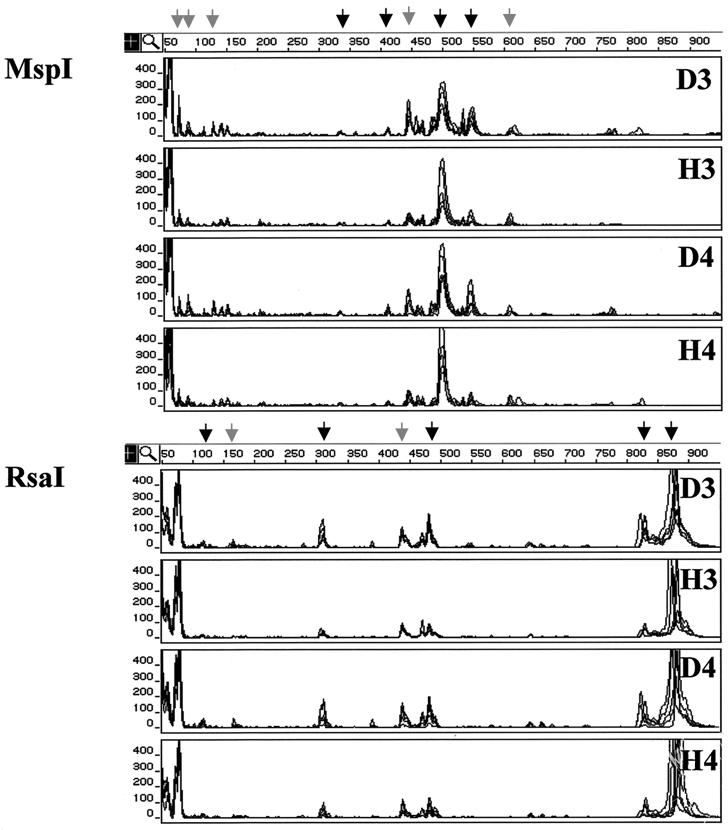
To identify differences in other bacterial populations, we examined rhizosphere bacterial communities using FT-ARDRA. Analyses were performed with fluorescence-tagged sequences amplified from washes of healthy and diseased roots grown in growth chambers (n = 8) and in field conditions (n = 6). Four comparisons of the bacterial communities inhabiting diseased and healthy roots were made using the same growth chamber-grown plants as those used for the culture-based assays (see above). The T-RFLP profiles generated for one of these comparisons (1998 soil A) are shown in Fig. 3. The results of the other three comparisons were similar (data not shown). Additionally, a fifth comparison was made of the bacterial communities inhabiting healthy and diseased roots of mature wheat grown in the field in 1998 (Fig. 4) because differences in take-all disease were readily apparent that year. In all five comparisons, the total fluorescence signal (i.e., total PCR product) was greater in amplifications of diseased samples than in those of healthy ones (P < 0.05). The threefold serial dilution of the wash template resulted in a significant reduction of the amplified signal (P < 0.01 for each experiment), but the overall topology of the T-RFLP profiles (i.e., the rank order of TRF peak areas) was maintained (Fig. 3 and 4). In each profile, approximately 24 TRFs were clearly visible above the background noise level, and approximately half of those contributed the large majority of the fluorescence signal in each T-RFLP profile.
FIG. 3.
FT-ARDRA of bacterial communities inhabiting the rhizospheres of healthy and diseased wheat grown in growth chambers. The analysis shown was perfomed on rhizosphere washes of diseased (D) and healthy (H) plants grown in soil A which had been collected from Mount Vernon, Wash., following harvest in 1998. The T-RFLP profiles were generated from amplified 16S sequences digested with either MspI or RsaI. In each panel, overlaid chromatographic traces from four independent samples are displayed. The data for two serial dilutions (3 and 4) of rhizosphere washes from each condition (D and H) are shown. Signals corresponding to TRFs that increased significantly in the rhizospheres of diseased plants in all of the 1998 and 1999 growth chamber experiments are indicated by black arrows, while those specific to this 1998 experiment are indicated by grey arrows. The size of each TRF in base pairs is indicated by the horizontal scale at the top the GeneScan results display, and the abundance of each is correlated with the peak area given in arbitrary fluorescence units on the vertical scale.
FIG. 4.
FT-ARDRA of bacterial communities inhabiting the rhizospheres of healthy and diseased wheat grown in the field. The analysis shown was performed on rhizosphere washes of diseased (D) and healthy (H) plants grown at Mount Vernon, Wash., in 1998. The T-RFLP profiles were generated from amplified 16S sequences digested with either MspI or RsaI. In each panel, overlaid chromatographic traces from six independent samples are displayed. The data for two serial dilutions (3 and 4) of rhizosphere washes from each condition (D and H) are shown. Signals corresponding to TRFs that increased significantly in the rhizospheres of diseased plants both in the field and in the growth chamber experiments are indicated by black arrows, while those specific to the field samples are indicated by the grey arrows. The size of each TRF in base pairs is indicated by the horizontal scale at the top the GeneScan results display, and the abundance of each is correlated with the peak area given in arbitrary fluorescence units on the vertical scale.
Comparisons of the T-RFLP profiles from diseased and healthy plants revealed differences in several fluorescence signals, each comprising >1% of the total peak area. Most of the significant differences were observed in all four growth chamber comparisons, but some were soil and/or year specific (Fig. 3). In these assays, the peak areas of fluorescence-labeled TRFs observed at 204, 334, 404, 490, and 542 bp in the MspI-generated profiles and at 94, 118, 179, 310, 474, 829, 884 bp in the RsaI-generated profiles were greater in the profiles of G. graminis var. tritici-infected root samples (P < 0.05 for each signal, all experiments). These differences indicated that the bacterial genomes that gave rise to each of these signals were more abundant in the rhizospheres of take-all-infected plants. The relative abundance, i.e., the proportion of an individual peak area relative to the total peak area, was also greater for some of these TRFs: M404, M542, R179, and R310 (P < 0.10 for each signal, all comparisons). Similar results were obtained in comparisons of the T-RFLP profiles of the more mature healthy and diseased plants grown in the field in 1998 (Fig. 4). In these rhizospheres, the peak areas of most of the fluorescence-labeled TRFs observed to increase in the growth chamber experiments were also observed to increase in the field (P < 0.05), including M334, M404, M490, M542, R118, R310, R474, R829, and R884 (Fig. 4).
In the course of doing these experiments, we observed soil-to-soil and year-to-year variations in the T-RFLP profiles. For example, the profiles of the younger, growth chamber-grown plants did not completely match those obtained from the mature, field-grown plants (compare Fig. 3 and Fig. 4). Some of these differences included signals that were identified as significant in comparisons made for individual growth chamber assays (Fig. 3) or the single field assay (Fig. 4). One example is the 440-bp TRF in the MspI-generated profile that was increased in all of the1998 experiments but not in the 1999 growth chamber experiments. There were also changes in the relative abundances of some signals. For example, TRF R118 represented a much smaller fraction of the total signal in the profiles obtained from the field samples than in those obtained from the growth chamber samples (compare Fig. 4 to Fig. 3).
Identification of significant ribotypes.
Having determined the sizes of the major MspI- and RsaI-generated TRFs, especially those associated with take-all infection, we attempted to identify the bacterial species giving rise to these ribotypes using two different methods. The first used the web-based TRFLP-TAP tool to generate a list of bacterial genera that could have given rise to those TRFs. The second approach was to sequence 16S rDNA sequences directly from rhizosphere washes and cultured isolates and compare their individual T-RFLP profiles to those generated from the whole-rhizosphere samples.
In total, over 24, primarily gram-negative, prokaryotic genera belonging to 10 different bacterial divisions were predicted to be potentially present in our profiles by the TRFLP-TAP tool. These included the well-known soil- and root-inhabiting genera Pseudomonas, Aeromonas, Caulobacter, Sphingomonas, Azospirillum, Zooglea, Burkholderia, Klebsiella, Serratia, Enterobacter, Erwinia, Pantoea, Rhizobium, Agrobacterium, Rhodopseudomonas, Cytophaga, Flavobacterium, and Flexibacter. Gram-positive genera that were predicted to be present in the profiles, included Achromobacter, Acetobacter, Bacillus, and Actinobacillus, all well known as soil inhabitants. Arthrobacter-like signals were conspicuously absent in this study. Additionally, mycoplasma-like and chloroplast-like signals were present in the profiles.
We next tried to identify bacterial ribotypes corresponding to matched pairs of TRFs that were more abundant in the profiles of take-all-infected roots. The first of these, M542 and R310, corresponded to the greatest single difference in terms of total peak area in comparisons of healthy and diseased plants (Fig. 3 and 4). Four possible genera contained strains that could be responsible for these signals: Flexibacter, Cytophaga, Pedobacter, and Psychrospora. (It is important to note here that not all known species of these genera were predicted to give TRFs M542 and R310). The second matched pair was M204 and R94 (Fig. 3), and the bacteria that could give rise to these signals were predicted to belong to Chryseobacterium or Riemerella. Some signals that were more prominent in the diseased samples than in the healthy samples were putatively identified using the TRFLP-TAP tool as unmatched pairs. In this way, the M204 signal might also be ascribed to two other lineages known only by their 16S sequences, “str. DCM-like” (R878) and “clone 1-60-like” (R309) bacteria. This is because the corresponding MspI-generated TRFs predicted by the TRFLP-TAP tool were observed to be much more intense (i.e., much larger peak area) than the observed MspI-generated peaks. Similar observations were made for M334, which was matched only to an uncultured lineage similar to clone JW32 (R490). In contrast, most of the other unmatched pairs were similar to previously cultured genera listed above. For example, the M404 signal might have originated from a strain(s) of Sphingomonas or Zymomonas (R118), Rhizobium (R826), or Bartonella (R828 and R870). However, little could be inferred from most of these predictions because so many of them consisted of very different genera. Furthermore, without having additional digests, it was impossible to deduce which combinations of peaks could be reasonably combined to give rise to the observed profiles.
To further elucidate which bacterial taxa gave rise to specific TRFs, we analyzed individual 16S rDNA sequences amplified from (i) the dominant bacterial isolates growing on the Pseudomonas-selective media and (ii) a clone bank of sequences amplified from the rhizospheres of diseased roots. In the first situation, we partially sequenced the 16S rDNAs amplified from 18 different isolates, a set which represented all of the distinct ARDRA groups obtained from both the 1998 and the 1999 growth chamber experiments (Fig. 2). Direct sequencing of these amplified sequences followed by BLAST comparisons to GenBank revealed a high degree of sequence similarity to known species of Pseudomonas (ARDRA group I), Chryseobacterium (ARDRA group II), Sphingobacterium (ARDRA group III), Agrobacterium (ARDRA group IV), Stenotrophomonas (ARDRA group V), and Flavobacterium (ARDRA group VI). We also sequenced 31 clones representing distinct ARDRA genotypes present in a clone bank prepared from washes of diseased rhizospheres and found that they had significant similarity to a number of known soil-inhabiting bacterial genera (data not shown). Of these 49 sequences, 17 contained MspI and RsaI restriction sites that could give rise to matched pairs of TRFs that increased in abundance in the rhizospheres of diseased roots (Table 2). Based on partial sequencing, the TRFLP-TAP tool predictions, and the TRFs of the isolated sequences, we can reasonably assert that the bacterial taxa responsible for particular TRFs identified can be ascribed to strains belonging to the genera listed in Table 2. Interestingly, more than one unique sequence and/or isolate could account for each of the six different matched signals. For example, clones A13 and A20 had identical patterns of TRFs (i.e., M490 and R119) but differed somewhat in their actual sequences. Likewise, isolates dI1, I5B, C101A, U5, E206, and E201 all gave rise to TRFs M490 and R880 but differed in their colony morphologies (data not shown) and capacity to inhibit the take-all pathogen (see below) as well as their 16S rDNA sequences. These results indicate that some degree of diversity is present in these signals at the subgenus level.
TABLE 2.
16S rDNA sequence-based identification of bacterial isolates and cloned sequences corresponding to matched pairs of T-RFLP signals that increased following take-all infection
| Matched-pair TRFsa | Sequence identificationb | Sequence matchc
|
TRF sizes (bp) observed with:
|
|||
|---|---|---|---|---|---|---|
| Order | Genus | % | MspI | RsaI | ||
| M204-R94 | U3, dU3, I19, C2+4 | Flavobacteria I | Chryseobacterium | 94–97 | 204 | 94 |
| M404-R118 | A6 | Alpha Proteobacteria IV | Sphingomonas | 95 | 406 | 116 |
| M490-R118 | A13 | Beta Proteobacteria I | Herbaspirillum | 93 | 490 | 119 |
| M490-R118 | A20 | Beta Proteobacteria VI | Zooglea | 94 | 490 | 119 |
| M490-R118 | F8 | Beta Proteobacteria VI | Zooglea | 96 | 490 | 116 |
| M490-R880 | dU, I5B, C101A, U5, E206 | Gamma Proteobacteria VIII | Pseudomonas | 98–100 | 490 | 880 |
| M490-R94 | dU1 | Flavobacteria I | Chryseobacterium | 94 | 490 | 94 |
| M542-R310 | F1 | Sphingobacteria I | Cytophaga | 95 | 536 | 304 |
| M542-R310 | C4 | Sphingobacteria I | Flexibacter | 91 | 537 | 308 |
| M542-R310 | C204B | Flavobacteria I | Flavobacterium | 97 | 542 | 311 |
The sizes of the TRFs observed in the bacterial community profiles are given in base pairs preceded by an indicator of the restriction enzyme used to generate the profile, either MspI (M) or RsaI (R).
Identifiers for GenBank accession numbers (AF375827 through AF375850) of partial 16S sequences obtained in this study.
Partial sequences of the 16S rDNA genes obtained from bacterial isolates and clones were compared to GenBank data using BLAST 2.0 (1). The phylogenetic groups with the highest BLAST score are given along with the percent sequence identity to the samples corresponding to each matched-pair TRF.
In vitro interactions among dominant cultured populations, G. graminis var. tritici, and 2,4-DAPG producers.
Having identified differences in specific rhizosphere populations, we investigated the capacity of some of these bacteria to inhibit G. graminis var. tritici in the presence and absence of phlD+ bacteria in vitro (Table 3). The phlD+ strains had the greatest capacity to inhibit the take-all pathogen. Of the 53 phlD-negative isolates tested, only five (dI1, dI1B, I1, I5B, and E206) displayed any significant inhibition of the take-all pathogen. These five isolates represented approximately 10% of the dominant cultured populations in terms of the random sample of 53 isolates and the full set of 167 isolates obtained from the TDCs and analyzed by BOX-PCR (data not shown). Notably, all five belonged to ARDRA group I, the same group as that of phlD+ strain MtV1 (Fig. 2). The 22 other ARDRA group I isolates did not show any significant capacity to inhibit G. graminis var. tritici (Table 3). When each of the 53 isolates was mixed 3:1 with the phlD+ strain, several of them appeared to reduce the capacity of the phlD+ strain to inhibit the pathogen in vitro; however, none of the observed differences was statistically significant (P > 0.10). Notably, though, the ARDRA group II isolates showed the least capacity to interfere with the inhibition of G. graminis var. tritici by the phlD+ strain (Table 3). In most instances, the morphology of the mixed cultures differed from that of the individual isolates on the inhibition plates. However, mixtures containing I1 or E206 developed a morphology indistinguishable from that of the isolates themselves, indicating that these two isolates prevented the growth of the phlD+ strain.
TABLE 3.
Inhibition of G. graminis var. tritici and phlD+ strain MtV1 by representative isolates of the most abundant bacterial strains cultured from the rhizospheres of healthy and diseased wheat
| Isolatea | phlDb | ARDRA groupc | Inhibition indexd for the following treatment at the indicated time:
|
|||
|---|---|---|---|---|---|---|
| Isolate alone
|
Isolate plus MtV1e
|
|||||
| T1 | T2 | T1 | T2 | |||
| Q8r1 | + | I | 0.57 a | 0.48 ab | 0.58 | 0.50 |
| MtVB | + | I | 0.60 a | 0.54 a | 0.64 | 0.53 |
| dI1 | − | I | 0.32 bc | 0.26 c | 0.43 | 0.36 |
| dI1B | − | I | 0.41 ab | 0.29 bc | 0.35 | 0.26 |
| I1 | − | I | 0.35 abc | 0.33 bc | 0.34 | 0.29 |
| I5B | − | I | 0.35 abc | 0.23 c | 0.49 | 0.41 |
| E206 | − | I | 0.34 ab | 0.26 c | 0.34 | 0.30 |
| Other ARDRA group I, min | − | I | 0.05 c | 0.00 e | 0.34 | 0.27 |
| Other ARDRA group I, max | − | I | 0.30 bc | 0.08 e | 0.59 | 0.55 |
| ARDRA group II, min | − | II | 0.07 bc | 0.00 e | 0.51 | 0.44 |
| ARDRA group II, max | − | II | 0.23 bc | 0.07 e | 0.63 | 0.58 |
| All other, min | − | Other | 0.00 c | 0.00 e | 0.46 | 0.39 |
| All other, max | − | Other | 0.29 bc | 0.00 e | 0.62 | 0.56 |
Isolates with significantly different inhibition phenotypes are grouped together for brevity, with the minimum (min) and maximum (max) inhibition indices for each genotype displayed. The total sample sizes were 22, 9, and 17 for other ARDRA group I, ARDRA group II, and all other isolates, respectively.
Presence (+) or absence (−) of the phlD gene, a marker for 2,4-DAPG biosynthesis.
Genotypes of isolates based on ARDRA of 16S sequences with MspI and RsaI.
Inhibition indices range from 0 (no inhibition) to 1 (total inhibition of fungal growth). Measurements were taken at two different time points, T1 (where uninhibited fungal growth extended 50 to 75% of the radius of the assay plate) and T2 (where uninhibited fungal growth extended the full radius of the assay plate). Values followed by different letters are significantly different, as determined by the Tukey-Kramer multiple-range test (P < 0.10).
Tested strains were mixed 3:1 with strain MtV1 prior to inoculation on the assay plates.
DISCUSSION
Because no single method can give an absolutely comprehensive view of soil microbial communities (18), we used two independent measures of bacterial population structure to obtain a more complete picture of the monotonic changes in rhizosphere bacterial populations that occurred following take-all infection. Culturing on Pseudomonas-selective media and FT-ARDRA of rhizosphere washes revealed that the diversity of rhizosphere bacterial populations changed following the development of take-all disease (Table 1 and Fig. 3 and 4). The two types of assays also allowed us to independently detect significant increases in two different bacterial populations associated with take-all infection. These populations were subsequently identified as species of Pseudomonas and Chryseobacterium (Table 2). Other differences in bacterial populations were observed only in the culture-independent T-RFLP analyses of amplified16S rDNA sequences. Web-based analyses can be performed to identify the phlyogenetic group(s) giving rise to specific TRFs in these types of analyses (21), but the limits to this approach are only now becoming fully recognized (9, 21). In this study, the genera of the cultured and uncultured ribotypes could not be accurately determined, except by partial sequencing of amplified 16S rDNA (Table 2), but sequence-based identification using rDNA amplified from environmental samples is not without its limitations as well (33, 41). Nonetheless, there was a correspondence between these two approaches at the level of bacterial genera. Indeed, the genus for the highest-scoring match from GenBank always corresponded to one of the bacterial genera identified by the TRFLP-TAP tool as a potential source of the ribotype of that sequence. The use of primers specific to a subset of bacterial taxa below the superkingdom level of eubacteria might alleviate the need for partial sequencing of isolated 16S sequences, but such a choice would also limit the breadth of bacterial taxa examined. Using our combined approach, we identified a number of changes in rhizosphere bacterial communities following take-all infection.
The rhizospheres of wheat plants infected with the take-all pathogen harbor larger populations of several bacterial species than healthy roots, as indicated by higher culturable counts (Table 1) and increased total fluorescence signal in the T-RFLP profiles (see Results). Previous work showed that the sizes of culturable bacterial populations, especially pseudomonads, were larger in the rhizospheres of wheat infected with G. graminis var. tritici (38, 46). In this study, we observed larger populations of cultured Pseudomonas spp. inhabiting G. graminis var. tritici-infected roots when plants were grown in growth chambers. While the data for these cultured populations were confounded by the occurrence of nonpseudomonads (especially Chryseobacterium spp.) growing on the semiselective media, the total pseudomonad populations were calculated to be larger on diseased roots, even when samples dominated by nonpseudomonads were excluded from the analysis (data not shown). Additional evidence for increased abundance of fluorescent pseudomonads in the rhizospheres of diseased plants came from the profiles generated by FT-ARDRA. The fluorescence signals corresponding to TRFs indicative of Pseudomonas spp. (e.g., M490 and R884) were more intense in the profiles of diseased plants than in those of healthy plants (P < 0.05) (Fig. 3 and 4), even though the difference was less dramatic in the comparison of field-grown plants because of a higher degree of plant-to-plant variation (Fig. 4).
Populations of phlD+ Pseudomonas spp. (i.e., 2,4-DAPG producers) were also significantly larger in the rhizospheres of diseased plants in three of four comparisons (Table 1). Interestingly, the one comparison in which no difference was observed used 1998 soil B, which had experienced severe take-all in the field (Table 1). In this instance, it is possible that the populations of phlD+ bacteria built up during the 1998 field season in this soil but not soil A, which saw little or no take-all. While the indigenous phlD+ pseudomonads had the capacity to inhibit G. graminis var. tritici, they represented only a small fraction of the total pseudomonad counts in our assays. Others have reported that indigenous populations of phlD+ pseudomonads inhabiting plant roots generally represent less than 10% of the total culturable Pseudomonas populations (31, 36), and in our assays they averaged less than 1% of the total (Table 1). While this is the first report of an increase in indigenous 2,4-DAPG-producing bacteria following take-all infection, a similar observation was made when the abundance of an inoculated 2,4-DAPG-producing strain on healthy and diseased roots was examined (23). In this study, a single genotype was identified as the dominant phlD+ strain in the soils obtained from Mount Vernon, Wash., as determined by RFLP analysis of the phlD gene (Fig. 2). Interestingly, this genotype was previously noted for its wide geographic distribution (29) and its unique ability to efficiently colonize wheat roots (35).
The abundance of two other bacterial populations also increased significantly in the rhizospheres of take-all-infected plants. The first population belongs to the genus Chryseobacterium, represented by both the ARDRA group II isolates (Table 1 and Fig. 1) and the M204 and R94 ribotypes detected in the T-RFLP profiles of rhizosphere washes (Fig. 3 and 4). The second population contains other members of the flavobacteria and the sphingobacteria, represented by the M542 and R310 ribotypes, one of which was isolated and determined to be a Flavobacterium sp. (ARDRA group VI; Fig. 1). Taxonomically similar bacteria were isolated from plant roots and composts, but their ability to inhibit the growth of plants and microorganisms in vitro varied tremendously from isolate to isolate (11, 15, 16, 43). This is the first report of these two bacterial populations being associated with take-all infection. The larger bacterial populations inhabiting the rhizospheres of diseased plants may be explained by the release of utilizable growth substrates from infected tissues. However, the partitioning of these substrates among detrimental (e.g., pathogenic), neutral (e.g., saprophytic), and beneficial (e.g., mutualistic) microbial populations is not well understood.
We investigated the ability of the most abundant cultured populations to interact with the take-all pathogen and a 2,4-DAPG-producing Pseudomonas sp. in vitro to determine which bacterial populations might contribute to the development of take-all suppression (Table 3). Only a few isolates of the dominant Pseudomonas spp. (ARDRA group I, M490 and R880) displayed significant in vitro inhibition (Table 3); however, they were not isolated any more frequently from the rhizospheres of diseased plants than from those of healthy plants. In contrast, the phlD+ pseudomonads (a subgroup of the ARDRA I strains; Fig. 2) had the greatest capacity of all the tested strains to inhibit the take-all pathogen (Table 3), and they were generally more abundant in the rhizospheres of G. graminis var. tritici-infected wheat roots (Table 1). These observations are consistent with other studies that indicated a key role for 2,4-DAPG-producing Pseudomonas spp. in TAD soils (34, 36). Isolates of the other dominant genera, including those belonging to Chryseobacterium (ARDRA group II, M204 and R94) and Flavobacterium (ARDRA group VI, M542 and R310), displayed no ability to inhibit the pathogen in vitro, counterindicating their direct involvement in pathogen suppression.
We also determined the potential for the dominant cultured bacteria to inhibit a known biocontrol population (i.e., 2,4-DAPG producers) in vitro. Several of the dominant Pseudomonas isolates reduced the ability of the indigenous phlD+ strain, MtV1, to inhibit the pathogen in vitro when coinoculated (Table 3). This result was not unexpected, because negative interactions between different strains of fluorescent pseudomonads have been observed in vitro and in the field when applied as seed treatments (32; L. S. Pierson III, personal communication). Such negative interactions could be mediated by signal molecules (6) as well as antibiotic activities (Pierson, personal communication). These results contrast with those for the other cultured isolates, especially the Chryseobacterium and Flavobacterium strains, which showed little or no capacity to reduce the inhibitory capacity of MtV1 (Table 3). Therefore, it is possible that shifts in rhizobacterial community structures that occur following take-all infection result in an environment more conducive to the biocontrol activity of 2,4-DAPG producers.
Nonetheless, much more work needs to be done to establish the functional role(s) of different rhizosphere bacterial populations in the ecology of take-all infection and suppression. We suspect that some of the observed changes in the rhizospheres of diseased plants will correspond to secondary colonization by saprophytes that have little direct impact on either the take-all pathogen or biological control bacteria. However, it is possible that some proportion of the ribotypes identified directly or indirectly contributes to the suppression of G. graminis var. tritici. In the future, studies aimed at isolating and characterizing multiple representatives of the ribotypes identified in this study will be useful in determining their role in the ecology of take-all and TAD.
ACKNOWLEDGMENTS
This research was supported by grants 97-35107-4804 and 01-35107-1011 from the U.S. Department of Agriculture, National Research Initiative, Competitive Grants Program.
We thank D. Pouchnik, K. Schroeder, D. Mavrodi, G. Philips, L. Morgan, E. Sachs, and E. Lutton for technical assistance in performing this study, L. Thomashow for many helpful discussions, and K. Nielsen for critically reading the manuscript prior to review.
REFERENCES
- 1.Altschul S F, Madden T L, Schiffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asher M J C, Shipton P J. Biology and control of take-all. New York, N.Y: Academic Press, Inc.; 1981. [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Bangera M G, Thomashow L S. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens 82-87. J Bacteriol. 1999;181:3155–3163. doi: 10.1128/jb.181.10.3155-3163.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull C T, Weller D M, Thomashow L S. Relationship between root colonization and suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens strain 2-79. Phytopathology. 1991;81:954–959. [Google Scholar]
- 6.Chancey S T, Wood D W, Pierson L S., III Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl Environ Microbiol. 1999;65:2294–2299. doi: 10.1128/aem.65.6.2294-2299.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook R J, Veseth R J. Wheat health management. St. Paul, Minn: APS Press; 1991. [Google Scholar]
- 8.Cook R J, Thomashow L S, Weller D M, Fujimoto D, Mazzola M, Bangera G, Kim D-S. Molecular mechanisms of defense by rhizobacteria against root disease. Proc Natl Acad Sci USA. 1995;92:4197–4201. doi: 10.1073/pnas.92.10.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbar J, Ticknor L O, Kuske C R. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol. 2001;67:190–197. doi: 10.1128/AEM.67.1.190-197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutteridge R J, Hornby D, Hollins T W, Prew R D. Take-all in autumn-sown wheat, barley, triticale and rye grown with high and low inputs. Plant Pathol. 1993;42:425–431. [Google Scholar]
- 11.Hebbar K P, Davey A G, Dart P J. Rhizobacteria of maize antagonistic to Fusarium moniliforme, a soil-borne fungal pathogen: isolation and identification. Soil Biol Biochem. 1992;24:979–987. [Google Scholar]
- 12.Hoitink H A J, Boehm M J. Biocontrol within the context of soil microbial communities: a substrate dependent phenomenon. Annu Rev Phytopathol. 1999;37:427–446. doi: 10.1146/annurev.phyto.37.1.427. [DOI] [PubMed] [Google Scholar]
- 13.Hornby D. Suppressive soils. Annu Rev Phytopathol. 1983;21:65–85. [Google Scholar]
- 14.Keel C, Schnider U, Maurhofer M, Voisard C, Laville J, Burger U, Wirthner, Haas P D, Defago G. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol Plant-Microbe Interact. 1992;5:4–13. [Google Scholar]
- 15.Kremer R J, Begonia M F T, Stanley L, Lanham E T. Characterization of rhizobacteria associated with weed seedlings. Appl Environ Microbiol. 1990;56:1649–1655. doi: 10.1128/aem.56.6.1649-1655.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwok O C H, Fahy P C, Hoitink H A J, Kuter G A. Interactions between bacteria and Trichoderma hamatum in suppression of rhizoctonia damping-off in bark compost media. Phytopathology. 1987;77:1206–1212. [Google Scholar]
- 17.Latour X, Corberand T, Laguerre G, Allard F, Lemanceau P. The composition of fluorescent pseudomonad populations associated with roots is influenced by plant and soil type. Appl Environ Microbiol. 1996;62:2449–2456. doi: 10.1128/aem.62.7.2449-2456.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liesack W, Janssen P H, Rainey F A, Ward-Rainey N L, Stackebrandt E. Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques. In: van Elsas J D, Trevors J, Wexler M, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc; 1997. pp. 375–440. [Google Scholar]
- 19.Liu W-T, Marsh T L, Cheng H, Forney L. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 1999;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh T L, Saxman P, Cole J, Tiedje J M. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl Environ Microbiol. 2000;66:3616–3620. doi: 10.1128/aem.66.8.3616-3620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavrodi O V, McSpadden Gardener B, Mavrodi D V, Bonsall R F, Weller D M, Thomashow L. Genetic diversity of phlD from 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. Phytopathology. 2001;91:35–43. doi: 10.1094/PHYTO.2001.91.1.35. [DOI] [PubMed] [Google Scholar]
- 23.Mazzola M, Cook R J. Effects of fungal root pathogens on the population dynamics of biocontrol strains of fluorescent pseudomonads in the wheat rhizosphere. Appl Environ Microbiol. 1991;57:2171–2178. doi: 10.1128/aem.57.8.2171-2178.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazzola M, Fujimoto D K, Thomashow L S, Cook R J. Variation in sensitivity of Gaeumannomyces graminis to antibiotics produced by fluorescent Pseudomonas spp. and effect on biological control of take-all of wheat. Appl Environ Microbiol. 1995;61:2554–2559. doi: 10.1128/aem.61.7.2554-2559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McSpadden Gardener B. Assessing the potential of creating biased rhizospheres. PhD thesis. East Lansing: Michigan State University; 1998. [Google Scholar]
- 26.McSpadden Gardener B, de Bruijn F J. FT-ARDRA of soil and rhizosphere bacterial communities. Phytopathology. 1998;88:S61. [Google Scholar]
- 27.McSpadden Gardener B, Lilley A K. Application of common statistical tools. In: van Elsas J D, Trevors J, Wexler M, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc; 1997. pp. 501–524. [Google Scholar]
- 28.McSpadden Gardener B, Mavrodi D V, Thomashow L S, Weller D M. A rapid polymerase chain reaction-based assay characterizing rhizosphere populations of 2,4-diacetylphloroglucinol-producing bacteria. Phytopathology. 2001;91:44–54. doi: 10.1094/PHYTO.2001.91.1.44. [DOI] [PubMed] [Google Scholar]
- 29.McSpadden Gardener B, Schroeder K, Kalloger S, Raaijmakers J, Thomashow L S, Weller D M. Genotypic and phenotypic diversity of phlD-containing Pseudomonas isolated from the rhizosphere of wheat. Appl Environ Microbiol. 2000;66:1939–1946. doi: 10.1128/aem.66.5.1939-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penrose L D J, Neate S M. Resistance to Gaeumannomyces graminis in wheat genotypes grown in field environments and sand culture. Soil Biol Biochem. 1994;26:719–726. [Google Scholar]
- 31.Picard C, Di Cello F, Ventura M, Fani R, Guckert A. Frequency and biodiversity of 2,4-diacetylphloroglucinol-producing bacteria isolated from the maize rhizosphere at different stages of plant growth. Appl Environ Microbiol. 2000;66:948–955. doi: 10.1128/aem.66.3.948-955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierson E A, Weller D M. Use of mixtures of fluorescent pseudomonads to suppress take-all and improve the growth of wheat. Phytopathology. 1994;84:940–947. [Google Scholar]
- 33.Qiu X, Wu L, Huang H, McDonel P E, Palumbo A V, Tiedje J M, Zhou J. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl Environ Microbiol. 2001;67:880–887. doi: 10.1128/AEM.67.2.880-887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raaijmakers J, Weller D M. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant-Microbe Interact. 1998;11:144–152. [Google Scholar]
- 35.Raaijmakers J, Bonsall R F, Weller D M. Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology. 1999;89:470–475. doi: 10.1094/PHYTO.1999.89.6.470. [DOI] [PubMed] [Google Scholar]
- 36.Raaijmakers J, Weller D M, Thomashow L S. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl Environ Microbiol. 1997;63:881–887. doi: 10.1128/aem.63.3.881-887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarniguet A, Lucas P. Evaluation of populations of fluorescent pseudomonads related to the decline of take-all patch of turfgrass. Plant Soil. 1992;145:11–15. [Google Scholar]
- 38.Sarniguet A, Lucas P, Lucas M. Relationship between take-all, soil conduciveness to the disease, populations of fluorescent pseudomonads and nitrogen fertilizers. Plant Soil. 1992;145:17–27. [Google Scholar]
- 39.Sharifi-Tehrani A, Zala M, Natsch A, Moenne-Loccoz Y, Defago G. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur J Plant Pathol. 1998;104:631–643. [Google Scholar]
- 40.Simon A, Ridge E H. The use of ampicillin in a simplified selective medium for the isolation of fluorescent pseudomonads. J Appl Bacteriol. 1974;37:459–460. doi: 10.1111/j.1365-2672.1974.tb00464.x. [DOI] [PubMed] [Google Scholar]
- 41.Specksnijder A G C L, Kowalchuk G A, de Jong S, Kline E, Stephen J R, Laanbroek H J. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl Environ Microbiol. 2001;67:469–472. doi: 10.1128/AEM.67.1.469-472.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomashow L S, Weller D M, Bonsall R F, Pierson L S., III Production of the antibiotic phenazine 1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol. 1990;56:908–912. doi: 10.1128/aem.56.4.908-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tunlid A, Hoitink H A J, Low C, White D C. Characterization of bacteria that suppress Rhizoctonia solani damping-off in bark compost media by analysis of fatty acid biomarkers. Appl Environ Microbiol. 1989;55:1368–1374. doi: 10.1128/aem.55.6.1368-1374.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincent M N, Harrison L A, Brackin J, Kovacevich P, Mukerji P, Weller D M, Pierson E A. Genetic analysis of the antifungal activity of a soilborne Pseudomonas aureofaciens strain. Appl Environ Microbiol. 1991;57:2928–2934. doi: 10.1128/aem.57.10.2928-2934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisburg W G, Barnes S M, Pelletier D A, Lane D J. 16S rDNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weller D M. Colonization of wheat roots by a fluorescent pseudomonad suppressive to take-all. Phytopathology. 1983;73:1548–1553. [Google Scholar]
- 47.Weller D M, Cook R J. Suppression of take-all on wheat by seed treatment with fluorescent pseudomonads. Phytopathology. 1983;73:463–469. [Google Scholar]
- 48.Weller D M, Howie W J, Cook R J. Relationship between in vitro inhibition of Gaeumannomyces graminis var. tritici and suppression of take-all of wheat by fluorescent pseudomonads. Phytopathology. 1988;78:1094–1100. [Google Scholar]