Abstract
The mechanisms by which cellulolytic enzymes and enzyme complexes in Ruminococcus spp. bind to cellulose are not fully understood. The product of the newly isolated cellulase gene endB from Ruminococcus flavefaciens 17 was purified as a His-tagged product after expression in Escherichia coli and found to be able to bind directly to crystalline cellulose. The ability to bind cellulose is shown to be associated with a novel cellulose-binding module (CBM) located within a region of 200 amino acids that is unrelated to known protein sequences. EndB (808 amino acids) also contains a catalytic domain belonging to glycoside hydrolase family 44 and a C-terminal dockerin-like domain. Purified EndB is also shown to bind specifically via its dockerin domain to a polypeptide of ca. 130 kDa present among supernatant proteins from Avicel-grown R. flavefaciens that attach to cellulose. The protein to which EndB attaches is a strong candidate for the scaffolding component of a cellulosome-like multienzyme complex recently identified in this species (S.-Y. Ding et al., J. Bacteriol. 183:1945–1953, 2001). It is concluded that binding of EndB to cellulose may occur both through its own CBM and potentially also through its involvement in a cellulosome complex.
Cellulolytic Ruminococcus spp. play an important role in the degradation of plant cell wall polysaccharides in the rumen and hindgut of mammals (5, 15, 20). Early biochemical and microscopic evidence indicated that their plant cell wall-degrading enzymes are organized into high-molecular-weight complexes on the cell surface (22, 24, 43). Analysis of cloned polysaccharidase genes has, however, produced somewhat conflicting evidence concerning the molecular organization of these enzymes, and the mechanisms by which they might attach to their substrate and to the cell surface have thus remained unclear. Many of the cellulase genes first isolated from R. flavefaciens and R. albus (7, 18, 30, 33, 40, 41) were reported to encode single domain enzymes smaller than 50 kDa that carry no obvious substrate binding domains, and no regions that might be responsible for the types of protein-protein interactions that are found, for example, in the assembly of cellulosome complexes from Clostridium spp. (3, 4, 9). On the other hand, it has been known for some time that xylanases from R. flavefaciens display complex multidomain organization (11, 45). Furthermore, the endoglucanase EndA from R. flavefaciens 17 was shown to be a multidomain enzyme carrying an 80-amino-acid dockerin-like region that is also present in three xylanases and an esterase from the same strain (2, 19). Dockerin-like regions have also been reported recently in multidomain endoglucanases from R. albus F40 (29, 30). Since dockerins are responsible for the assembly of cellulosome complexes via specific dockerin-cohesin interactions in Clostridium species (3, 4, 17, 31, 34), this provides a strong indication that complexes resembling cellulosomes may be involved in the organization of many plant cell wall-degrading enzymes in ruminococci. In support of this, two linked genes that encode structural proteins containing cohesin domains have recently been identified in R. flavefaciens 17 (8). Cellulosome organization provides one potential mechanism for binding of cellulolytic enzymes to their substrate since cellulosomal scaffolding proteins from clostridia carry cellulose-binding modules (CBMs) (4, 39). On the other hand, recent evidence from R. albus has also demonstrated a role for binding modules associated with pilus proteins in attachment to cellulose (27, 32).
We show here that the product of a newly isolated cellulase gene from R. flavefaciens 17, EndB, is a multidomain enzyme that carries its own CBM, which is unrelated to previously described CBMs. In addition, however, EndB possesses a dockerin region that is shown to be involved in a specific interaction with a putative scaffolding protein of 130 kDa present among R. flavefaciens 17 proteins that bind to crystalline cellulose (Avicel).
MATERIALS AND METHODS
Growth conditions.
R. flavefaciens 17 was grown anaerobically as described previously (10) at 39°C in M2GSC medium (26) or, for large-scale culture with cellulose as energy source, in Hungate Stack medium (16) containing 0.2% Avicel PH101. Escherichia coli strains XL1-Blue, used for cloning, and BL21(DE3)pLys Gold, used for expression of pET28a His6-tagged constructs, were obtained from Stratagene. pET28a was from Novagen (Madison, Wis.).
Gene isolation.
The clone pCMCP3 was isolated from a pUC13 plasmid library as described previously (7). Carboxymethyl cellulase (CMCase) activity was visualized on plates by using an overlay of 0.1% carboxymethyl cellulose (CMC)–0.4% (wt/vol) agarose in 25 mM sodium phosphate buffer (pH 6.8). The plates were incubated for 4 h at 37°C, followed by staining with 0.1% (wt/vol) Congo red and destaining with 1 M NaCl. Plasmid DNA isolation and other molecular biology techniques were done according to standard procedures (35).
DNA sequence analyses.
DNA sequences were determined on both strands by using an ABI377 automated sequencer and appropriate oligonucleotide primers. Computer analysis was carried out by using the UWGCG software available through the HGMP facility (Cambridge, United Kingdom). Multiple alignment was done by using CLUSTALW. Database screening made use of the National Library of Medicine retrieval system (http://www.ncbi.nlm.nih.gov/) and the BLAST-P program.
Overexpression of EndB.
The region of endB coding for residues 20 to 808 (i.e., omitting the N-terminal signal peptide) was amplified by using the forward primer aattccatggCGCCCGTCAACGGTCTG and the reverse primer cacgctcgagTTCGGGAAGCTTGTCTAT (lowercase letters indicating additional 5′ residues that carry XhoI or NcoI sites) (see Fig. 2). The product was cloned in an NcoI/XhoI-cut pET28a(+) vector such that His6 residues were fused at the C terminus of EndB. The C-terminal truncated EndB (retaining amino acid residues 20 to 702) was produced by using another reverse primer, tatactcgagAGTTACCTTCGGAGCCTCTCC. The putative CBM (residues 499 to 702) was amplified by using this reverse primer with the forward primer atatccatggTGCCTGCCTTCTCTGCTGCA. Successful constructs were transformed into E. coli BL21(DE3)pLysS, and the cloned product was expressed as follows. A single colony was transferred to 50 ml of Luria-Bertani (LB) medium containing 30 μg of kanamycin/ml and grown overnight at 37°C. Then, 1 liter of LB medium containing 1.2% (vol/vol) glycerol was inoculated with the 50-ml overnight culture and allowed to grow to an optical density at 600 nm between 0.8 and 1.0. The culture was then cooled in ice for 1 h, and 1 mM of IPTG was added, followed by incubation at 16°C without shaking for 1 h and then overnight shaking at 200 rpm. The culture was centrifuged at 5,000 × g at 4°C for 10 min, and the cells were washed twice in 100 ml of lysis buffer (50 mM sodium phosphate buffer [pH 8.0], 0.3 M NaCl, and 10 mM imidazole). The cells were resuspended in 10 ml of lysis buffer containing protease and lysozyme (8), incubated at 37°C for 30 min, and then sonicated at full speed (MSE Soniprep) for three cycles of 1 min with 2 min cooling on ice. The sonicated cell suspension was centrifuged at 15,000 × g at 4°C for 10 min, and the supernatant was collected for further purification.
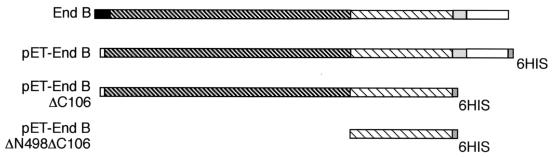
FIG. 2.
Diagram showing domain structures of EndB and of the His-tagged derivatives used here. The N-terminal signal peptide is shown in black, the C-terminal dockerin is shown in white, the T-rich region is shown in light gray, the family 44 catalytic domain is indicated by dark cross-hatching, and the unknown region (containing a new CBM) is indicated by light cross-hatching. The primers used for construction of the pET28 clones in order to overexpress the full-length and truncated polypeptides as His6-tagged products are described in the Materials and Methods.
Purification of His-tagged proteins.
His-tagged proteins were purified from sonicated E. coli cells by binding to Ni-NTA resin, as described previously (36). Reducing sugar assays were performed at pH 6.5 and 37°C, unless otherwise stated, by the method of Lever (23) as described previously (10).
Polysaccharide binding assay for the cloned EndB product.
The purified His-tagged EndB products (20 μg) were incubated at either 37 or 4°C for a specified time with 5 mg of prewashed Avicel, phosphoric acid-swollen Avicel, or oat spelt xylan in 8 μl of 50 mM potassium phosphate buffer (pH 6.8) containing 2 mM dithiothreitol (DTT) (14). After binding, the Avicel was washed four times with 200 μl phosphate buffer at ambient temperature. Proteins remaining attached were then eluted with sodium dodecyl sulfate (SDS) sample buffer at 100°C for 5 min (21) and separated by SDS-polyacrylamide gel electrophoresis (PAGE). After Western blotting onto Immobilon P membranes (Millipore, Mass.) as described previously (8), His-tagged proteins were detected by using specific antibodies (Invitrogen) and chemiluminescent detection kits (Super Signal West Pico, Pierce, Ill.) according to the manufacturers' instructions. Size markers were detected by Coomassie blue staining.
Binding of R. flavefaciens proteins to Avicel.
R. flavefaciens 17 was grown in Hungate Stack medium containing 0.2% Avicel for 150 h. Cultures were still growing at this stage, as judged by the continued production of acetate, which was measured by gas chromatography. Next, 1,600 ml of culture was harvested by centrifugation at 13,000 × g, and the supernatant was retained. Culture supernatants were freeze-dried, redissolved, and desalted by using Vivascience 10 KDa concentrators to give a final 50-fold concentration. The pellet containing cells and substrate was resuspended in 50 mM sodium phosphate (pH 6.5)–1 mM DTT and left at room temperature for 30 min (43). Cell debris was then spun at 2,600 × g and washed three times in 5 ml of 50 mM Tris-HCl (pH 7.5) at room temperature. Supernatants from these washes were combined with those from the initial sodium phosphate wash, and fresh undegraded Avicel was added (1% final concentration). Similarly, fresh Avicel was added to the original culture supernatant. In both cases, after incubation at 37°C for 15 min, the added Avicel was spun down and washed three times in 50 mM sodium phosphate buffer (pH 6.5) containing 2 mM EDTA (by centrifugation at 13,000 × g), and both the buffer washes and the final pellet were analyzed by SDS-PAGE to detect eluted or attached proteins.
Detection of EndB binding to R. flavefaciens proteins.
R. flavefaciens 17 cultures were grown as described above, and proteins from concentrated culture supernatant and from pelleted cells plus substrate were separated by SDS-PAGE. Proteins were transferred by Western blotting onto Immobilon-P membranes and probed with His-tagged EndB. After extensive washing, bound EndB was detected by chemiluminescence as described previously (8).
SDS-PAGE zymograms.
Proteins were eluted after attachment to insoluble polysaccharide substrates by heating at 60°C for 20 min in SDS sample buffer (21). To detect CMCase activity after SDS-PAGE, 0.2% CMC was included in the polyacrylamide solution before the gels were cast. After electrophoresis, the gel was washed and the enzymes were allowed to renature overnight at 4°C before staining with Congo red to reveal activity bands according to the method of Saul et al. (36).
RESULTS
Binding of R. flavefaciens endoglucanases to cellulose.
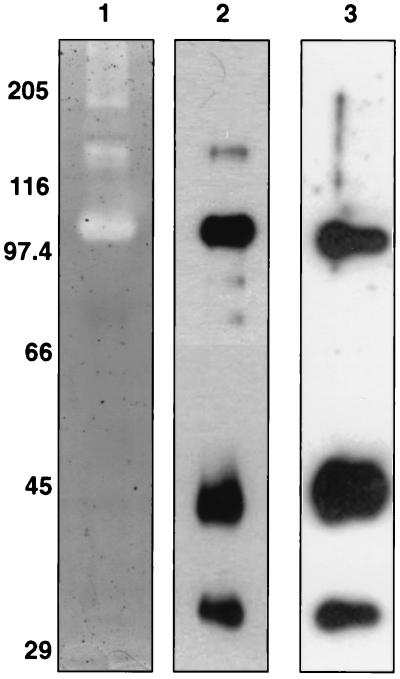
R. flavefaciens 17 cultures were grown for 150 h with crystalline cellulose (Avicel) as an energy source. Culture supernatant and supernatants obtained from washing harvested cells in buffer were allowed to bind to fresh, undegraded Avicel (see Materials and Methods). Polypeptides that remained bound after three washes of the added Avicel in phosphate buffer were eluted with 2% SDS (in sample buffer [21]) and analyzed in SDS-PAGE zymograms (Fig. 1). These showed that at least five major endoglucanases ranging in molecular mass from 60 to 125 kDa were among the extracellular proteins that bound to Avicel. These observations demonstrate substrate binding but do not reveal whether binding occurs directly through binding domains present in the endoglucanases or indirectly through the mediation of other components.
FIG. 1.
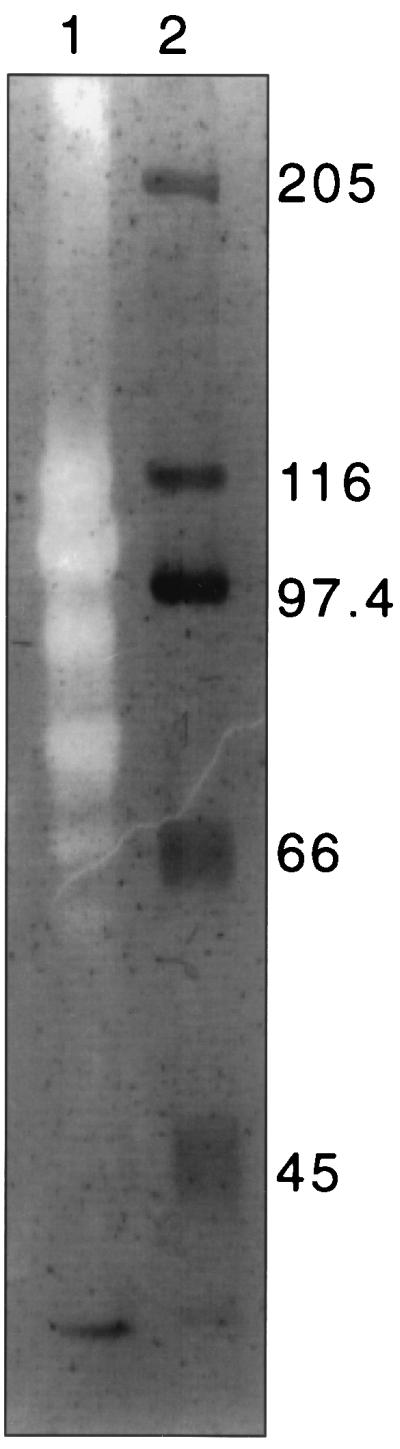
Binding of R. flavefaciens 17 endoglucanases to crystalline cellulose (Avicel). Lane 1 shows an SDS-PAGE CMC zymogram of extracellular proteins from an Avicel-grown (150-h) culture of R. flavefaciens 17 that bound to Avicel. Clear zones are the result of CMCase activity, as revealed by Congo red staining. In this case the proteins were from the supernatants obtained from washing harvested cells in buffer (see Materials and Methods), but similar results were obtained with the original culture supernatant (not shown). Lane 2 shows molecular size markers (in kilodaltons), stained with Coomassie blue.
Multidomain organization of the endoglucanase EndB from R. flavefaciens 17.
The new gene endB was identified from sequencing the insert in the plasmid clone CMCP3 that had been isolated by the expression of CMCase activity (7). Sequence analysis (accession number AJ298117) predicted a gene product (EndB) of 808 amino acids. As for most other polysaccharidase genes studied from R. flavefaciens 17, the stop codon is followed by a region containing a perfect 14-base palindrome that represents a possible ρ-independent terminator. The N-terminal region of EndB consists of a family 44 endoglucanase catalytic domain following a putative signal peptide sequence of about 30 amino acids (Fig. 2). This portion of EndB shows 81% amino acid identity with CelB from R. flavefaciens FD1 (40). The C terminus of EndB consists of a threonine-rich linker of 26 amino acids, as was also found in the R. flavefaciens 17 enzymes XynB, XynD, and EndA (11, 19), followed by a dockerin-like region of 81 amino acids. The EndB dockerin shows 42% amino acid identity with that of EndA, and the conserved features noted previously between the dockerins of XynB, XynD, and EndA are also present in the EndB dockerin. These include two copies of a putative Ca2+ binding motif, which accounts for almost all of the similarity between the Ruminococcus dockerins and those of Clostridium spp. (Fig. 3). Between the family 44 catalytic domain of EndB and the T-rich region is a sequence of ca. 200 amino acids that shows no close similarity to other proteins in database searches.
FIG. 3.
Multiple alignment of dockerin domains from R. flavefaciens 17 XynD, XynB, EndB and EndA with dockerin from LicB from Clostridium thermocellum (37). Residues conserved in all five sequences are highlighted in black, while those conserved in 4 out of 5 sequences are highlighted in grey. Numbers on the right hand side refer to amino acid sequence positions within the relevant gene product.
The complete EndB coding region, excluding residues encoding the signal peptide, was amplified by PCR, and the product was expressed as a C-terminal His6-tagged fusion product in the vector pET28a (see Materials and Methods). The resulting construct was introduced into E. coli BL21, and the overexpressed 87-kDa tagged EndB product was purified by means of the His6 tag. Activity was confirmed for the purified enzyme by clear zone formation in polysaccharide-containing agar plates against CMC or lichenan (β1-3:1-4 glucan) but was not detected against mannan, phosphoric acid-swollen cellulose, or Avicel. The specific activity estimated by reducing sugar release was 2.5 × 10−3 μmol/min/mg of protein for CMC and 0.97 × 10−3 μmol/min/mg of protein for lichenan. No activity was detectable against p-nitrophenyl cellobioside. EndB showed a pH optimum of ca. 5.8, and the enzyme was quite temperature labile, its activity when assayed at 50°C being only 40% that observed at 37°C (results not shown).
Binding of EndB to cellulose.
The purified His-tagged EndB protein was found to bind strongly to Avicel, at both 4 and 37°C. Zymograms performed with CMC showed a major active band that migrated slightly slower than the predicted size of 87 kDa for the pET-EndB product (Fig. 4). In addition, smaller inactive bands of 30 and 40 kDa carrying C-terminal His tags were detected that retained the ability to bind to cellulose. The smallest of these bands (30 kDa) was recovered and shown by peptide sequencing to contain the N-terminal sequence XEFTDI (corresponding to residues 551 to 556 of EndB) and the internal sequence SYNLPLGS (corresponding to residues 617 to 624). This suggested that the cellulose-binding capacity of EndB resides in the C-terminal 258 amino acid residues of the protein. Apart from the dockerin and T-rich regions, this fragment contains only the unknown region that follows the catalytic domain. Residues 499 to 702, representing this unknown region, were therefore expressed as a His-tagged fusion product (pET-EndBΔN498ΔC106) after amplification of the relevant coding sequence (see Fig. 2). The purified product, whose predicted molecular size is 21.17 kDa, bound strongly to cellulose, as shown in Fig. 5. We concluded that cellulose binding is due to the region between residues 551 and 702 of EndB. The full-length EndB enzyme was shown to bind Avicel and acid-swollen Avicel and also showed some binding to birch wood xylan when tested at 37°C (results not shown).
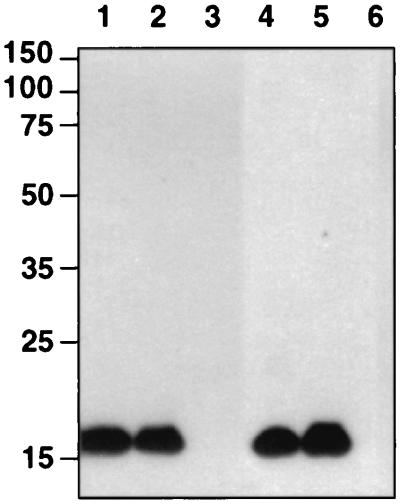
FIG. 4.
Binding of purified, His-tagged pET-EndB to Avicel. Binding was assayed by incubating the purified EndB protein with Avicel. The Avicel was then washed three times in phosphate buffer, and attached polypeptides were then eluted with SDS sample buffer (see Materials and Methods). CMCase activity was detected by a zymogram technique (36) by incorporating CMC into the gel (lane 1). His-tagged protein was detected by using specific antibodies (see Materials and Methods) (lanes 2 and 3). Incubation with Avicel was at 37°C for 5 min (lanes 1 and 2) or at 4°C for 20 h (lane 3).
FIG. 5.
Binding of a purified, His-tagged protein fragment (from pET-EndBΔ N498Δ C106) that carries residues 499 to 702 of EndB to insoluble cellulose (Avicel). The putative EndB CBM was incubated with Avicel for 20 h at 4°C (lanes 1, 2, and 3) or for 1 h at 37°C (lanes 4, 5, and 6), followed by four washes in buffer (see Materials and Methods). Lanes 1 and 4 were loaded with the unbound protein, lanes 2 and 5 were loaded with protein eluted with 2% SDS after binding to Avicel and washing, and lanes 3 and 6 were loaded with the 18-fold-concentrated final buffer wash.
Binding of EndB to a 130-kDa R. flavefaciens extracellular protein.
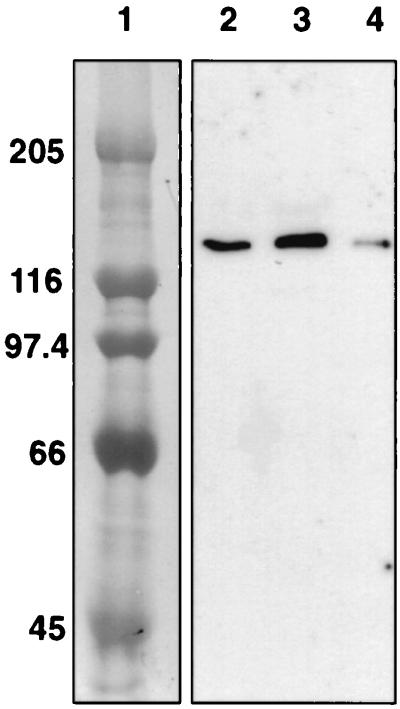
The His-tagged EndB protein was also used to probe native R. flavefaciens 17 proteins from Avicel-grown cultures after separation by SDS-PAGE (Fig. 6). Specific binding to a polypeptide of 130 kDa was detected in cell extracts, in culture supernatant, and in culture supernatant proteins recovered after binding to Avicel. A His-tagged truncated derivative of EndB (EndBΔ C106) was also constructed that lacks the C-terminal 106 amino acids representing the T-rich linker and dockerin regions (Fig. 2). Under the experimental conditions described in Fig. 6, we did not detect binding of the purified truncated EndBΔ C106 enzyme to any R. flavefaciens protein. This suggests strongly that the dockerin region is responsible for binding to the 130-kDa protein.
FIG. 6.
Binding of purified His-tagged pET-EndB to R. flavefaciens 17 proteins from an Avicel-grown culture. Lane 1, molecular size markers (in kilodaltons); lane 2, R. flavefaciens culture pellet; lane 3, R. flavefaciens culture supernatant; lane 4, R. flavefaciens supernatant proteins after absorption onto Avicel at 37°C, four washes in buffer, and subsequent elution with 2% SDS.
DISCUSSION
EndB is only the second cellulase to be characterized fully from R. flavefaciens 17, and it is the first protein from R. flavefaciens to be shown to carry a CBM. We show here that the CBM lies within a region of 152 amino acids and that it represents a novel binding module that is unrelated to other described protein sequences. The predicted secondary structure (12) of the 200 amino acids following the family 44 catalytic domain contains alpha-helices at each end flanking a central region of ca. 60 amino acids that is composed of beta sheets and that includes several aromatic residues. This central region appears to be the best candidate for the cellulose-binding activity based on structural analyses of other families of CBMs (38, 39). Further analysis of the binding specificity and structure-function relationships of this module will clearly be of interest. There have been several instances of novel bacterial CBMs that show unique sequences, suggesting that there is considerable sequence diversity among CBMs (6, 25, 44).
EndB is one of only five enzymes thus far reported to carry a catalytic domain belonging to glycoside hydrolase family 44 (http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html). CelJ from C. thermocellum carries both a family 9 and a family 44 catalytic domain; the purified family 44 domain of CelJ was reported to have some activity against the crystalline cellulose Avicel, and a key role in the cellulosome complex was proposed (1). While we did not detect activity against Avicellase for EndB, recovery of the active enzyme was relatively poor. Interestingly the CelB endoglucanase from R. flavefaciens FD1 (40) was also reported to encode a family 44 catalytic domain that shares 81% amino acid sequence identity with R. flavefaciens 17 EndB. CelB was reported to terminate at a point corresponding to 176 amino acids before the end of EndB, i.e., before the dockerin-like domain of EndB, and was considered to be a single domain enzyme lacking a dockerin domain (40). It is possible that CelB is a partially homologous, but shorter, enzyme than EndB that does not contain a dockerin-like domain. Alternatively, it is not ruled out that the celB clone isolated in E. coli might carry a mutation that results in the C-terminal coding region being out of frame. It may be noted that R. flavefaciens 17 EndB shares 45% identical amino acid residues with a translation of the sequence immediately downstream of the celB gene of R. flavefaciens FD1.
EndB was able to bind specifically to a polypeptide of ca. 130 kDa in R.flavefaciens 17 proteins eluted after binding to Avicel. This indicates a specific protein-protein interaction that could be involved in the positioning of the EndB protein within a multienzyme complex. Since binding was shown to be dependent on the dockerin domain present in EndB, the 130-kDa protein to which EndB attaches is a possible candidate for the scaffolding protein component of a cellulosome complex, by analogy with the model proposed for cellulolytic Clostridium spp. (3). Recent work identified two adjacent genes, scaA and scaB, that encode likely cellulosomal structural components in R. flavefaciens 17 (8). The products of both genes carry multiple cohesin domains which were shown by immunoblotting experiments to recognize other R. flavefaciens proteins. ScaB cohesins 4 and 5 were shown to interact specifically with a dockerin at the C terminus of the putative scaffolding protein ScaA and recognized an R. flavefaciens protein of 130 kDa, assumed to be ScaA, in immunoblotting experiments. Furthermore, the purified xylanase–β-glucanase enzyme XynD, whose dockerin is structurally similar to that of EndB, was shown to bind the isolated ScaA cohesin 2 (8). It appears very likely, therefore, that EndB is one of many R. flavefaciens enzymes that interacts with cohesins in ScaA and represents a cellulosome-associated enzyme that carries its own CBM. This supposition is supported by the finding that a peptide sequence from the 130-kDa protein matches a region within the recently completed N-terminal domain of the ScaA protein (M. T. Rincón et al., unpublished data). There is recent evidence that several cellulosomal cellulases in Clostridium spp. carry CBMs (see, for example, references 13 and 46).
The only two cellulases thus far studied from R. flavefaciens 17 (EndA [19] and EndB), together with several cellulases from R. albus F40 (28, 29), have proved to be multidomain enzymes that carry dockerin sequences. On the other hand, there are many reports of single-domain endoglucanases from R. flavefaciens and R. albus that are smaller than 50 kDa (30, 33, 40, 41, 42). This may suggest that not all cellulases are cellulosome associated in ruminococci. It now appears likely that the genetic instability that is often encountered with cellulase genes from ruminococci when cloned in E. coli has resulted in a bias against the recovery of genes encoding the larger cellulases. For example, EndA from R. flavefaciens 17 was first reported to be a single-domain enzyme (7), but further investigation, and sequencing from chromosomal DNA by PCR walking, established it to be a multidomain enzyme that includes a dockerin (7, 19). The reasons for such instability remain unclear but might be attributable in part to toxic effects of certain protein domains, in particular the dockerin, in E. coli.
In R. flavefaciens 17 cultures grown on Avicel, we found that CMCases ranging in molecular size from 60 to 120 kDa attached to cellulose. Based on the present work with EndB, we can speculate that the attachment of R. flavefaciens cellulases to cellulose may occur through several mechanisms. These means include the direct binding via CBMs present in individual enzymes, as observed for EndB, and the indirect binding resulting from CBMs present in other components of a cellulosome-like complex. Indirect binding might involve CBMs present in a scaffolding protein, as are found in all cellulolytic clostridial species studied to date (4), in catalytic subunits such as EndB, or in other noncatalytic subunits yet to be defined. The recent discovery of pilus-like adhesins that bind to cellulose in R. albus (27, 32) suggests that there might be additional attachment mechanisms either for whole cells, or conceivably for the enzyme complexes, if such pilus proteins were in some way associated with the complexes themselves. Such pili have not yet been demonstrated in R. flavefaciens.
In conclusion, the present study provides the first report of a CBM in R. flavefaciens and also the first evidence for specific dockerin-mediated binding of an R. flavefaciens cellulase to another extracellular protein. More information on the distribution of CBMs in cellulosomal and noncellulosomal proteins is clearly still needed in order to fully elucidate the mechanisms of substrate attachment and hydrolysis in this important cellulolytic species.
ACKNOWLEDGMENTS
This work was supported by the Scottish Executive Rural Affairs Department. M.T.R. is supported by a CONICIT-Venezuela studentship.
We thank Mark Wilkinson (University of Liverpool) for peptide sequencing and Ed Bayer for valuable advice and discussion.
REFERENCES
- 1.Ahsan M M, Matsumoto M, Karita S, Kimura T, Sakka K, Ohmiya K. Purification of the family J catalytic domain derived from the Clostridium thermocellum endoglucanase CelJ. Biosci Biotech Biochem. 1997;61:427–431. doi: 10.1271/bbb.61.427. [DOI] [PubMed] [Google Scholar]
- 2.Aurilia V, Martin J C, McCrae S I, Scott K P, Rincon M T, Flint H J. Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology. 2000;146:1391–1397. doi: 10.1099/00221287-146-6-1391. [DOI] [PubMed] [Google Scholar]
- 3.Bayer E A, Morag E, Lamed R. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 1994;12:379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 4.Bayer E A, Shimon L J, Shoham Y, Lamed R. Cellulosomes: structure and ultrastructure. J Struct Biol. 1998;124:221–234. doi: 10.1006/jsbi.1998.4065. [DOI] [PubMed] [Google Scholar]
- 5.Bryant M P. Genus Ruminococcus Sijpestein, 1948. In: Holt J G, editor. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1093–1097. [Google Scholar]
- 6.Cann I K O, Kocherginskaya S, King M R, White B A, Mackie R I. Molecular cloning, sequencing, and expression of a novel multidomain mannanase gene from Thermoanaerobacterium polysaccharolyticum. J Bacteriol. 1999;181:1643–1651. doi: 10.1128/jb.181.5.1643-1651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham C, McPherson C A, Martin J, Harris W J, Flint H J. Sequence of a cellulase gene from the rumen anaerobe Ruminococcus flavefaciens 17. Mol Gen Genet. 1991;228:320–323. doi: 10.1007/BF00282484. [DOI] [PubMed] [Google Scholar]
- 8.Ding S-Y, Rincón M T, Lamed R, Martin J C, McCrae S I, Aurilia V, Shoham Y, Bayer E A, Flint H J. Cellulosomal proteins from Ruminococcus flavefaciens. J Bacteriol. 2001;183:1945–1953. doi: 10.1128/JB.183.6.1945-1953.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felix C R, Ljungdahl L G. The cellulosome: the exocellular organelle of Clostridium. Annu Rev Microbiol. 1993;47:791–819. doi: 10.1146/annurev.mi.47.100193.004043. [DOI] [PubMed] [Google Scholar]
- 10.Flint H J, McPherson C A, Martin J. Expression of two xylanase genes from the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 cloned in pUC13. Microbiology. 1991;137:123–129. doi: 10.1099/00221287-137-1-123. [DOI] [PubMed] [Google Scholar]
- 11.Flint H J, Martin J C, McPherson C A. A bifunctional enzyme, with separate xylanase and β(1-3:1-4) glucanase domains, encoded by the xynD gene of Ruminococcus flavefaciens. J Bacteriol. 1993;175:2943–2951. doi: 10.1128/jb.175.10.2943-2951.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 13.Gaudin C, Belaich A, Champ S, Belaich J P. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J Bacteriol. 2000;182:1910–1915. doi: 10.1128/jb.182.7.1910-1915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang L, Forsberg C W, Thomas D Y. Purification and characterization of a chloride-stimulated cellobiosidase from Bacteroides succinogenes S85. J Bacteriol. 1988;170:2923–2932. doi: 10.1128/jb.170.7.2923-2932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hungate R E. The anaerobic mesophilic cellulolytic bacteria. Bacteriol Rev. 1950;53:631–645. doi: 10.1128/br.14.1.1-49.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hungate R E, Stack R J. Phenylpropanoic acid: growth factor for Ruminococcus albus. Appl Environ Microbiol. 1982;44:79–83. doi: 10.1128/aem.44.1.79-83.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kagiuchi M, Isui A, Suzuki K, Fujino T, Fujino E, Kimura T, Karita S, Saki K, Ohmiya K. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J Bacteriol. 1998;180:4303–4308. doi: 10.1128/jb.180.16.4303-4308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karita S, Marioka K, Kajino T, Sakka K, Shimada K, Ohmiya K. Cloning and sequencing of a novel endo-1:4-β-glucanase gene from Ruminococcus albus. J Ferment Bioeng. 1993;76:439–444. [Google Scholar]
- 19.Kirby J, Martin J C, Daniel A S, Flint H J. Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol Lett. 1997;149:213–219. doi: 10.1111/j.1574-6968.1997.tb10331.x. [DOI] [PubMed] [Google Scholar]
- 20.Krause D O, Bunch R J, Smith W J M, McSweeney C S. Diversity of Ruminococcus strains: a survey of genetic polymorphisms and plant digestibility. J Appl Microbiol. 1999;86:487–495. [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lamed R, Naimark J, Morgenstern E, Bayer E A. Specialized cell surface structures in cellulolytic bacteria. J Bacteriol. 1987;169:3792–3800. doi: 10.1128/jb.169.8.3792-3800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lever M. Carbohydrate determination with 4-hydroxybenzoic acid hydrazide (PAHBAH): effect of bismuth on the reaction. Anal Biochem. 1977;81:21–27. doi: 10.1016/0003-2697(77)90594-2. [DOI] [PubMed] [Google Scholar]
- 24.Miron J, Yokoyama M T, Lamed R. Bacterial structures involved in Lucerne cell wall degradation by pure cultures of cellulolytic rumen bacteria. Appl Microbiol Biotechnol. 1989;32:218–222. [Google Scholar]
- 25.Mitsumori M, Minato H. Identification of the cellulose-binding domain of Fibrobacter succinogenes endoglucanase F. FEMS Microbiol Lett. 2000;183:99–103. doi: 10.1111/j.1574-6968.2000.tb08940.x. [DOI] [PubMed] [Google Scholar]
- 26.Miyazaki K, Martin J C, Marinsek-Logar R, Flint H J. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- 27.Morrison M, Miron J. Adhesion to cellulase by Ruminococcus albus: a combination of cellulosomes and Pil-proteins? FEMS Microbiol Lett. 2000;185:109–115. doi: 10.1111/j.1574-6968.2000.tb09047.x. [DOI] [PubMed] [Google Scholar]
- 28.Ohara H, Karita S, Kimura T, Sakka K, Ohmiya K. Characterisation of the cellulolytic complex (cellulosome) from Ruminococcus albus. Biosci Biotech Biochem. 2000;64:254–260. doi: 10.1271/bbb.64.254. [DOI] [PubMed] [Google Scholar]
- 29.Ohara H, Noguchi J, Karita S, Kimura T, Sakka K, Ohmiya K. Sequence of egV and properties of EgV, a Ruminococcus albus endoglucanase containing a dockerin domain. Biosci Biotech Biochem. 2000;64:80–88. doi: 10.1271/bbb.64.80. [DOI] [PubMed] [Google Scholar]
- 30.Ohmiya K, Kajino T, Kato A, Shimizu S. Structure of a Ruminococcus albus endo-1:4-beta-glucanase gene. J Bacteriol. 1989;171:6771–6775. doi: 10.1128/jb.171.12.6771-6775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pages S, Belaich A, Tardif C, Reverbel-Leroy C, Gaudin C, Belaich J-P. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J Bacteriol. 1996;178:2279–2286. doi: 10.1128/jb.178.8.2279-2286.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pegden R S, Larson M A, Grant R J, Morrison M. Adherence of the gram-positive bacterium Ruminococcus albus to cellulose and identification of a novel form of cellulose-binding protein which belongs to the Pil family of proteins. J Bacteriol. 1998;180:5921–5927. doi: 10.1128/jb.180.22.5921-5927.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole D M, Hazlewood G P, Laurie J I, Barker P J, Gilbert H J. Nucleotide sequence of the Ruminococcus albus SY3 endoglucanase genes celA and celB. Mol Gen Genet. 1990;223:217–223. doi: 10.1007/BF00265057. [DOI] [PubMed] [Google Scholar]
- 34.Salamitou S, Raynaud O, Lemaire M, Coughlan M, Beguin P, Aubert J-P. Recognition specificity of the duplicated segments present in the Clostridium thermocellum endoglucanase gene celD and in the cellulosome integrating protein CipA. J Bacteriol. 1994;176:2822–2827. doi: 10.1128/jb.176.10.2822-2827.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Saul D J, Williams L C, Grayling R A, Chamley L W, Love D R, Bergquist P L. celB, a gene coding for a bifunctional cellulase from the extreme thermophile “Caldocellum saccharolyticum.”. Appl Environ Microbiol. 1990;56:3117–3124. doi: 10.1128/aem.56.10.3117-3124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schimming S, Schwarz W H, Staudenbauer W L. Structure of the Clostridium thermocellum gene licB and the encoded β-1:3-1,4-glucanase. Eur J Biochem. 1992;204:13–19. doi: 10.1111/j.1432-1033.1992.tb16600.x. [DOI] [PubMed] [Google Scholar]
- 38.Tomme P, Warren R A J, Miller R C, Kilburn D G, Gilkes N R. Cellulose-binding domains: classification and properties. In: Saddler J N, editor. Enzymatic degradation of insoluble carbohydrates. Washington, D.C.: American Chemical Society; 1995. pp. 142–163. [Google Scholar]
- 39.Tormo J, Lamed R, Chirino A J, Morag E, Bayer E A, Shoham Y, Steitz T A. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 40.Vercoe P E, Finks J L, White B A. DNA sequence and transcriptional characterization of a beta-glucanase gene (celB) from Ruminococcus flavefaciens FD-1. Can J Microbiol. 1995;41:869–876. doi: 10.1139/m95-120. [DOI] [PubMed] [Google Scholar]
- 41.Vercoe P E, Spight D H, White B A. Nucleotide sequence and transcriptional analysis of the celD beta-glucanase gene from Ruminococcus flavefaciens FD-1. Can J Microbiol. 1995;41:27–34. doi: 10.1139/m95-004. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Reid S J, Thomson J A. Transcriptional regulation of an endoglucanase and a cellodextrinase gene in Ruminococcus flavefaciens FD-1. J Gen Microbiol. 1993;139:1219–1226. doi: 10.1099/00221287-139-6-1219. [DOI] [PubMed] [Google Scholar]
- 43.Wood T M, Wilson C A, Stewart C S. Preparation of the cellulase from the cellulolytic anaerobic rumen bacterium Ruminococcus albus and its release from the bacterial cell wall. Biochem J. 1982;205:129–137. doi: 10.1042/bj2050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winterhalter C, Heinrich P, Candussio A, Wich G, Liebl W. Identification of a novel cellulose-binding domain within the multi-domain 120 kDa Xylanase XynA of the hyperthermophilic bacterium Thermotoga maritima. Mol Microbiol. 1995;15:431–444. doi: 10.1111/j.1365-2958.1995.tb02257.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J-X, Flint H J. A bifunctional xylanase encoded by the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 comprises two dissimilar domains linked by an asparagine/glutamine-rich sequence. Mol Microbiol. 1992;6:1013–1023. doi: 10.1111/j.1365-2958.1992.tb02167.x. [DOI] [PubMed] [Google Scholar]
- 46.Zverlov V V, Velikodvorskaya G V, Schwarz W H, Bronnenmeier K, Kellermann J, Staudenbauer W L. Multidomain structure and cellulosomal localization of the Clostridium thermocellum cellobiohydrolase CbhA. J Bacteriol. 1998;180:3091–3099. doi: 10.1128/jb.180.12.3091-3099.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]