Abstract
The effect of selenite on the growth rate and protein synthesis has been investigated in Rhodobacter sphaeroides. This photosynthetic bacterium efficiently reduces selenite with intracellular accumulation under both dark aerobic and anaerobic photosynthetic conditions. Addition of 1 mM selenite under these two growth conditions does not affect the final cell density, although a marked slowdown in growth rate is observed under aerobic growth. The proteome analysis of selenite response by two-dimensional gel electrophoresis shows an enhanced synthesis of some chaperones, an elongation factor, and enzymes associated to oxidative stress. The induction of these antioxidant proteins confirms that the major toxic effect of selenite is the formation of reactive oxygen species during its metabolism. In addition, we show that one mutant unable to precipitate selenite, selected from a transposon library, is affected in the smoK gene. This encodes a constituent of a putative ABC transporter implicated in the uptake of polyols. This mutant is less sensitive to selenite and does not express stress proteins identified in the wild type in response to selenite. This suggests that the entry of selenite into the cytoplasm is mediated by a polyol transporter in R. sphaeroides.
Selenium, a naturally occurring element, is essential for biological systems at low concentrations but toxic at higher levels. In aerobic conditions, selenium is present predominantly in the high valence toxic and soluble forms selenate (SeO42−, +VI) and selenite (SeO32−, +IV), while the dominant species in anaerobic sediments is the elemental selenium (Se0). In the environment, the reduction of these oxyanions occurs principally by biotic processes. The reduction of selenate or selenite into selenide is required, for example, for the synthesis of selenocysteine, an essential residue involved in the active site of various enzymes (12, 38). For a few species of bacteria, selenate or selenite acts as electron acceptors in the first steps of an anaerobic respiratory process similar to denitrification (35). To date, only four species (Thauera selenatis, Sulfospirillum barnesii SES-3, Bacillus arsicoselenatis, and Bacillus selenitireducens) that present such a potential have been isolated (22, 29, 36). Reduction of selenate and selenite into elemental selenium, which is insoluble and nontoxic, is also used by various species of bacteria to overcome the toxic character of the oxyanions. Detoxification of the selenium oxyanions can also be achieved by methylation of these compounds. Both reduction and/or methylation of selenate and selenite have been demonstrated in the case of purple nonsulfur photosynthetic bacteria (25, 39). Intracellular sequestration of the metal after reduction has been demonstrated for Rhodobacter sphaeroides cells in the case of tellurite (26). On the other hand, an extracellular reduction of selenite occurs for bacteria such as Rhodospirillum rubrum (17) or a marine photosynthetic bacterium (41). In addition to their tolerance to high concentrations to various toxic metals, these photosynthetic bacteria display an extraordinary metabolic versatility. Indeed, they are able to grow using a variety of bioenergetic processes such as anaerobic photosynthesis and aerobic and anaerobic respiration.
Although the exact mechanisms of toxicity of selenate and selenite is not known, there is increasing evidence that the toxic character of these compounds is related to their oxidant capacity. The high reactivity of selenite with thiols may explain its toxic character. Selenite reacts in particular with glutathione to form selenodiglutathione (9), producing the highly toxic compounds H2O2 and O2− (18). An important effect of the addition of selenite on the bacterial growth and resistance is therefore expected depending upon the presence or absence of oxygen.
In the present study, we combined biochemical and genetic approaches to better characterize the mechanisms of selenite reduction and toxicity in the photosynthetic bacterium R. sphaeroides depending on the growth conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
R. sphaeroides forma sp. denitrificans IL-106 was grown under anaerobic photosynthetic conditions at 30°C, in the light (10 W m−2) in 100-ml screw-cap bottles, in Hutner medium (7) or Siström's minimal medium A (32) containing one carbon source (succinate, citrate, malate, ethanol, or butyrate) or in Siström's minimal medium A lacking glutamate and succinate supplemented with d-mannitol. Dark aerobic cultures were grown in 250-ml Erlenmeyer flasks containing 100 ml of medium by shaking (150 rpm, 30°C). Kanamycin (10 μg/ml) was added when required. Escherichia coli strains were grown in liquid Luria-Bertani (LB) medium under aerobic conditions at 37°C. Plasmid pAK30, a generous gift from S. Kaplan, is a derivative of pRK415 (16) with an 8.0-kb EcoRI chromosomal insertion from R. sphaeroides Si4 genomic DNA containing the complete coding sequences of smoK, smoS, and mtlK (30). Plasmid was mobilizated into R. sphaeroides IL-106 by diparental mating with E. coli S17–1 as a donor. Cultures were exposed to various stress conditions at mid-exponential growth phase (i.e., an optical density at 660 nm [OD660] of 0.6 to 0.7). Heat shock was performed by incubation of the cells at 42°C for 12 h.
Library construction and screening
An R. sphaeroides transposon library was constructed by mixing E. coli S17–1 (harboring the suicide plasmid PSUP2021::Tn5) cells and R. sphaeroides IL-106 cells in a 1:10 ratio. Conjugal mating was performed by spotting the mixture onto LB solid medium and aerobic incubation in the dark at 30°C for 16 h. Tn5 insertion mutants were subsequently selected on minimal Siström medium plates containing kanamycin (10 μg/ml) and incubated for several days at 30°C. Screening for mutants unable to reduce selenite was performed by plating this library on minimal Siström medium plates containing selenite (200 μM).
MIC determination
Determination of the MIC was performed as described previously but at 30°C (1, 37).
Electron microscopy and X-ray analysis
Cells were fixed in 2.5% glutaraldehyde and 0.1 M cacodylate buffer (pH 7.1) for 30 min. After two washes with the same medium, the cells were postfixed in 1% OsO4 in 0.02 M cacodylate buffer (pH 7.1) for 1 h and subsequently dehydrated with a graded ethanol-water series and embedded in low-viscosity epoxy resin (Epon). Microtome-cut thin sections were contrasted with uranyl acetate and lead citrate as described by Hess (13) and observed with a Philips CM 120 transmission electron microscope. For energy-dispersive X-ray (EDX) analysis, thin sections were applied to carbon-coated transmission electron microscopy grids and dried at room temperature. The EDX analysis was performed with a Jeol model 2010 F electron microscope operating at 200 kV equipped with an EDAX-KEVEX microanalysis system.
Determination of metal accumulation
Overnight cultures were used to inoculate fresh 100-ml Hutner cultures under aerobic or anaerobic conditions to an initial OD660 of ca. 0.1. Na2SeO4 or Na2SeO3 were added to a final concentration of 1 mM. Control cultures were grown under identical conditions without any added oxyanions. Aliquots of bacteria culture were sampled at different time intervals during the cell growth. Cell yield was determined by the measurement of the OD660 or with a Thoma counting chamber. A good correlation between the two methods was obtained. After centrifugation of the aliquots at low speed, cell pellets were washed with fresh medium and resuspended in concentrated HNO3 before transfer in an acid digestion cell (Parr Instrument Company). The cells were then heated at 150°C for 3 h. The solutions were analyzed for their selenium content using a Perkin-Elmer AAnalyst 100 atomic absorption spectrophotometer. Standard solutions of selenium were prepared immediately before use by the solubilization of selenium powder (Interchim) in HNO3.
Inverse PCR
Chromosomal DNA from mutant strains was extracted and digested by NotI in the presence of RNase A, followed by enzyme inactivation (65°C, 15 min). An intramolecular ligation was carried out in a total volume of 0.05 ml and incubated at 14°C for 16 h. The ligated DNA was then used directly as a template for PCR amplification by using the oligonucleotide primers designed from the Tn5 sequence: TR1 (5′-CCGCCGAAGAGAACACAGATTTA-3′) and TR2 (5′-ACCCTGCCGATGCGGATGAAAA-3′). Varying the PCR hybridation conditions leads to a single PCR product demonstrating the absence of multiple Tn5 insertions. The PCR product was purified (QIAquick; Qiagen) and used directly for automated sequencing (ABI Prism 310; Applied Biosystems) in the presence of one of the oligonucleotide primers described above to obtain the sequence flanking the Tn5.
Preparation of cell extracts
Cells of IL-106 were harvested by centrifugation for 15 min at 5,000 × g (4°C) and were resuspended in ice-cold 50 mM Tris-HCl (pH 8)–1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF). The cells were disrupted by two passages through a French press (1.4 107 Pa). Unbroken cells were removed by centrifugation at 18,000 × g (4°C) for 20 min. The soluble and membrane fractions of cell extracts were separated by ultracentrifugation for 1 h at 150,000 × g (4°C). Bradford reagents (4) were used to determine protein concentrations, with bovine serum albumin as the standard.
2D gel electrophoresis and protein microsequencing.
High-resolution two-dimensional (2D) gel electrophoresis was performed according to the method of O'Farrell (28) in a Bio-Rad Investigator apparatus. Samples prepared from untreated cells or from cells exposed to 1 mM Na2SeO4 or Na2SeO3 were precipitated in acetone and resuspended in a loading buffer containing 9.5 M urea, 6% Triton X-100, 0.04% 3/10 and 0.01% 4/6 carrier ampholytes (Bio-Rad), and 0.5% dithiothreitol. Proteins were first applied onto an isoelectric focusing gel. The capillary gel was applied to a second-dimension electrophoretic gel containing 12% polyacrylamide. Gels were stained with silver nitrate (3). For quantitative densitometry, 35S-labeled proteins were extracted and submitted to the separation method described by Maillet et al. (23). The radioactive gels were recorded by using PhosphoImager technology (Molecular Dynamics) and analyzed with 2D gel analysis software (Melanie II; Bio-Rad). For N-terminal sequences, proteins were transferred to polyvinyidene difluoride membranes in a 10 mM 3-[cyclohexylamino]1-propanesulfonic acid (CAPS)–20% methanol buffer (pH 11) using a Bio-Rad Transblotter. The membranes were stained with Coomassie brilliant blue R-250. Proteins of interest were excised and identified by Edman degradation (10 to 15 cycles) with an Applied Biosystems sequencer (model 477A) equipped with a phenylthiohydantoin derivative analyzer (model 120A). Peptide sequences were matched against proteins of the Swiss-Prot database.
Immunoblot assays.
Proteins separated on a 10 to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) minigels (20) were transferred onto nitrocellulose membranes (Schleicher & Schuell) by a semidry transfer system, and the membranes were blocked for 1 h at room temperature in 5% bovine serum albumin in in 10 mM Tris buffer (pH 7.5), 100 mM NaCl, and 0.1% (vol/vol) Tween (TBST). The membranes were incubated overnight at 4°C with the appropriate antisera diluted into blocking buffer. After an extensive washing in TBST, membranes were incubated for at least 1 h at room temperature with alkaline phosphatase-coupled secondary antiserum (Bio-Rad) diluted in 5% lowfat milk-TBST. After further washing, the immunocomplexes were revealed by using BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium (Sigma) as a substrate.
Detection of SOD activity.
Superoxide dismutase (SOD) activity was measured according to an in situ staining procedure described previously (2), after electrophoresis of the total soluble extracts in nondenaturing 8% polyacrylamide gels.
Reagents
All chemicals used were analytical grade. Sodium selenate- and selenite-specific GroES, GroEL, thioredoxin antisera were purchased from Sigma-Aldrich.
RESULTS
Effect of selenite on the growth of R. sphaeroides
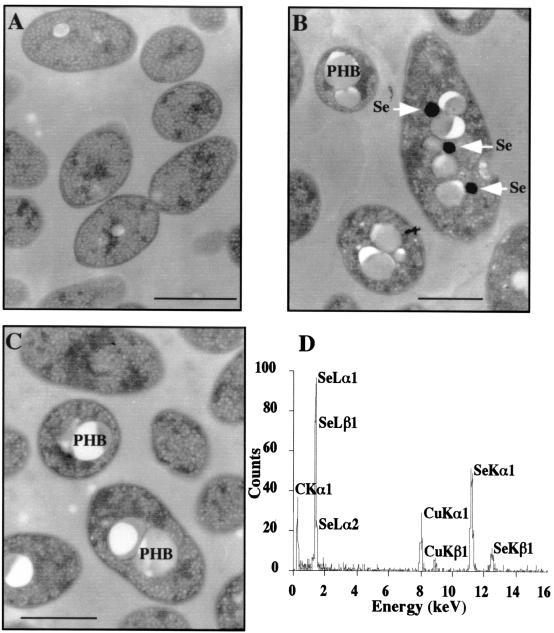
R. sphaeroides cells, grown in liquid medium containing 1 mM (175 ppm) SeO32− became bright red in color as described by Moore and Kaplan (25). This coloration results from the reduction of selenite into elemental selenium. The intracellular accumulation of metallic selenium in the cytoplasmic compartment is clearly demonstrated by the presence of high-electron-density particles in electron micrographs for cells grown under both anaerobic photosynthetic (Fig. 1B) and dark aerobic (data not shown) conditions in the presence of selenite. These electron-dense particles presented an energy-dispersive X-ray spectrum with characteristic peaks of selenium at 1.37, 11.22, and 12.49 keV (Fig. 1D). Metallic selenium particles were occasionally found outside the cells probably due to the lysis of some of them. On the other hand, the appearance of metallic selenium was only barely or not detectable (Fig. 1C) when cells were grown in the presence of selenate whatever the growth medium, in particular the carbon source (citrate, malate, succinate, ethanol, or butyrate). However, growth in the presence of 1 mM SeO42− or SeO32− induced the appearance of white granules of polyhydroxybutyrate usually found under stress conditions (Fig. 1B and C) and caused a slight increase (1.2 factor in the average) of the length of the bacteria.
FIG. 1.
Thin-section micrographs of R. sphaeroides IL-106 grown under anaerobic photosynthetic conditions in the absence (A) or in the presence of either 1 mM SeO32− (B) or 1 mM SeO42− (C). Arrows indicate the presence of electron dense particles of selenium (Se). White particles correspond to polyhydroxybutyrate (PHB) granules. Bars, 1 μm. (D) Energy-dispersive X-ray spectrum of electron-dense particles indicated by arrows in the panel C.
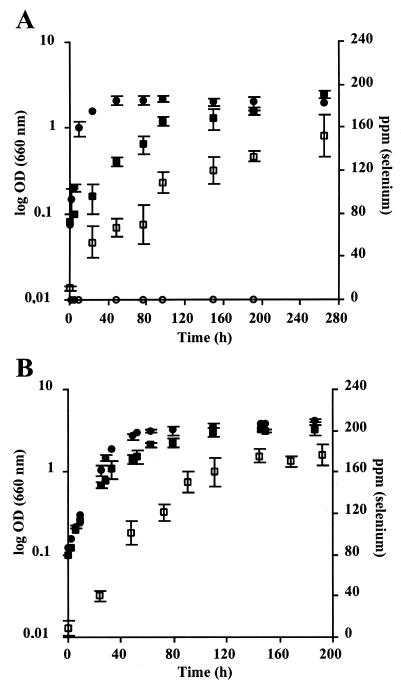
Reduction of selenite into metallic selenium occurred during the exponential growth phase under both dark aerobic and anaerobic photosynthetic conditions (Fig. 2). The addition of 1 mM SeO32− did not affect the cells density reached at the end of the growth phase for culture grown under both conditions. However, the growth rate was significantly affected depending on growth conditions. An important decrease in growth rate was observed for cells grown in the presence of selenite under dark aerobic conditions (Fig. 2A), while no significant decrease was measured under anaerobic photosynthetic conditions (Fig. 2B). In agreement with the important effect on the growth rate observed under aerobic condition, we found a lower level of resistance to selenite under these conditions than under anoxic photosynthetic conditions with MIC equal to 225 μg/ml (1.3 mM) and 800 μg/ml (4.6 mM) for oxic and anoxic conditions, respectively.
FIG. 2.
Growth of R. sphaeroides IL-106 under dark aerobic (A) or anaerobic photosynthetic (B) conditions in the absence (●) or presence (▪) of 1 mM SeO32−. □, Selenium concentration accumulated by the cells. Selenite was added at the beginning of the growth. Each curve shows mean values based on the results of three experiments.
Protein induction.
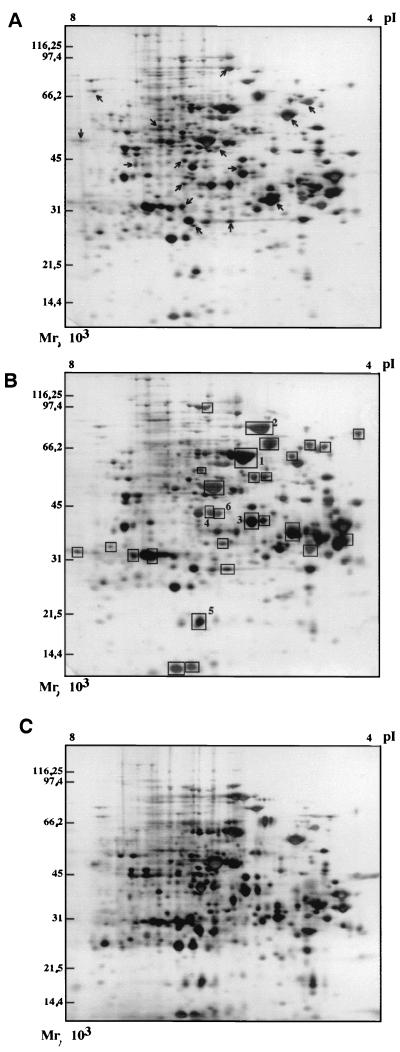
We have investigated the global stress response generated by selenite to identify potentially important proteins involved in selenite resistance and metabolism. Exponentially growing cultures under anaerobic photosynthetic conditions were treated with 1 mM SeO32− for 12 h. The soluble protein contents from control untreated (Fig. 3A) and selenite-treated cells (Fig. 3B) were then subjected to comparative 2D gel electrophoresis and visualized by silver nitrate staining (see Materials and Methods). Exposure to selenite results in a strong change in gene expression since up to 25 proteins are specifically induced and more than 20 are repressed. The addition of 1 mM SeO42− led to smaller alterations of the protein pattern of R. sphaeroides (data not shown). To quantify the modifications in protein expression, cells were treated with 1 mM SeO42− or SeO32− for 30 to 120 min and pulse-labeled with [35S]methionine. Changes in the intensity of protein spots, relative to the initial signal (at time = 0 min), were quantified by use of PhosphoImager technology and software analysis (see Materials and Methods for details). Exposure to selenite for 120 min enhanced the synthesis of 16% of the proteins by a factor of 2 to 10, whereas the synthesis was repressed in 21% (data not shown). Selenate treatment had a lesser effect on the protein synthesis since only 10 and 14% of the total proteins were enhanced or repressed, respectively, by a factor >2 (data not shown).
FIG. 3.
Comparative 2D gel electrophoresis analysis of total R. sphaeroides IL-106 proteins expressed in response to selenite treatment. Equal amounts of protein (about 50 μg) were loaded onto each gel. (A) Extracts prepared from control untreated cells. Some proteins whose synthesis is repressed are indicated by arrows. (B) Extracts prepared from cells exposed to 1 mM SeO32− for 12 h. Newly induced proteins are indicated by squares. Proteins 1 to 6 were analyzed by N-terminal sequencing. (C) Extracts prepared from ΔsmoK1 cells exposed to 1 mM SeO32−.
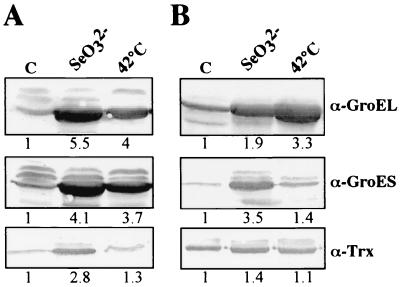
In order to characterize the major proteins induced by selenite treatment, the larger spots (numbered 1 to 6 in Fig. 3B) were excised from the gels and their N-terminal amino acid sequence were determined (Table 1). The spots numbered 1 and 2 correspond to the heat shock proteins HSP60 (99% identity with GroEL of R. sphaeroides) and HSP70 (92% identity with DnaK of R. capsulatus), respectively. Spot 3 corresponds to an elongation factor (82% identity with EF-Ts of Bacillus subtilis). Spot 4 presents 66 and 55% identities with a xenobiotic reductase and a morphinone reductase of Pseudomonas aeruginosa, respectively. These two NAD(P)H-dependent reductases reduce aliphatic nitroester compounds and other electrophilic xenobiotics, including 2-cyclohexen-1-one, N-ethylmaleimide, morphinone and codeinone, and TNT. No significant match has been found between the N-terminal amino acid sequences of spots 5 and 6 and sequences deposited in the Swiss-Prot data bank. The implication of chaperones synthesis in the response to selenite stress under both aerobic and anaerobic culture conditions was further demonstrated by Western blots analysis and comparison with cells heated at 42°C. The synthesis of GroEL, GroES (Fig. 4), and HSP70 (data not shown) occurred irrespective of the growth conditions.
TABLE 1.
Amino acid sequences of the N terminus of the soluble proteins induced by the addition of 1 mM SeO32−a
| Protein | Sequence | Match characteristics |
|---|---|---|
| 1 | AAKDVKFDTDARDRMLRGVNILADAVKVTLGPKGRNVVID | HSP60, 99% identity with GroEL of R. sphaeroides |
| 2 | AKVIGIDLGTTNSXVAIMDGAQPRVI | HSP70, 92% identity with DnaK of R. capsulatus |
| 3 | AITAQMVKELRESTGAGMMDAKKALTETDGDMXAAVDWL | Elongation factor, 82% identity with EF-Ts of B. subtilis |
| 4 | TEKLFTLIAFGDLTLKNRVVMAPLTRNRAEP | 66% identity with a xenobiotic reductase of P. aeruginosa |
| 5 | KDLAVPAXSXG | No significant match |
| 6 | ADLMPRAAXXRSE | No significant match |
Proteins are numbered according to Fig. 3B.
FIG. 4.
Western blots analysis upon SeO32− and heat shock induction. A 30-μg portion of the soluble fraction of R. sphaeroides IL-106 grown under dark aerobic (A) or anaerobic photosynthetic (B) conditions was separated by SDS-PAGE on minigels and transferred to nitrocellulose, and proteins were immunodetected using antisera specific for thioredoxin (Trx) and for two heat shock proteins (GroEL and GroES). Cells were either treated with SeO32− as described in Fig. 3 or heated at 42°C for 12 h. C, extracts from control untreated cells. Signals from the Western blot analyses were quantified, and the degrees of induction were calculated relative to the signal at time zero.
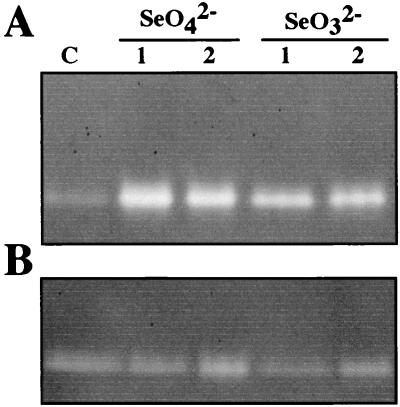
Since the reaction of selenite with glutathion generates in vitro H2O2 and O2− species (17), we looked for the effect of selenite addition on SOD activity. An enzyme staining for SOD activity was rapidly enhanced in the presence of either selenate or selenite after 30 min (Fig. 5A) to 12 h of incubation under aerobic condition but at a low level under anaerobic condition (Fig. 5B). To determine the SOD metal, the gels were incubated with 5 mM H2O2 or 1 mM KCN before staining for SOD activity was done (2). The dismutase activity was inhibited by H2O2 but remained unaffected by KCN treatment, indicating that the enzyme is an iron-containing superoxide dismutase (FeSOD). Glutathione reductase, an enzyme with antioxidant properties, is also induced upon exposure to selenite (data not shown). Additionally, an important induction of the synthesis of thioredoxin, another thiol-containing protein, is observed under aerobic conditions but not under anaerobic conditions (Fig. 4).
FIG. 5.
Effect of SeO42− and SeO32− on the activation of SOD of R. sphaeroides IL-106. For soluble extracts, 30 μl (ca. 80 μg) was loaded onto a nondenaturing 8% polyacrylamide gel, run under a 25-mA constant current, and then tested for SOD activity as described elsewhere (3). (A) Cells grown under dark aerobic conditions and treated with 1 mM SeO42− or SeO32− for 30 min (lane 1) or 12 h (lane 2). (B) Same as in panel A for cells grown under anaerobic photosynthetic conditions. Lane C refers to the control experiment in the absence of selenate or selenite.
Mutants unable to reduce selenite.
The formation of red amorphous Se0 as product of selenite reduction was used to select mutants affected in this reduction process from an R. sphaeroides transposon Tn5 library (see Materials and Methods). Indeed, clones unable to reduce selenite remained green after growth on petri dishes supplemented with 200 μM SeO32−, whereas clones unaffected in the assimilation and reduction of selenite turned bright red. On the basis of this screening, 10 mutants have been isolated out of 4,000 clones of the transposon Tn5 library. Amplification of the gene affected in each of these mutants was obtained by inverse PCR as described in Materials and Methods. Sequencing of the PCR products revealed that the Tn5 transposon is inserted, for the mutant denoted ΔsmoK1, in a gene presenting 94% identity with the smoK gene of the closely related strain R. sphaeroides Si4. The smoK gene is part of a polyol operon. This gene is located 1 nucleotide downstream the smoS gene, coding for sorbitol dehydrogenase, itself located 55 nucleotides upstream the mannitol dehydrogenase gene (mltK) (34). The smoK gene encodes a protein of 332 amino acid residues. The amino acid sequence presents a high similarity with various ATP-binding proteins of bacterial ABC transporters or traffic ATPases. It is therefore supposed that SmoK is a constituent of an ABC transporter involved in the uptake of sugar alcohols (34). To verify that the observed phenotype (no reduction of selenite) of ΔsmoK1 is linked to the disruption of the smoK gene, two experiments were performed. We have first looked for the capacity of the ΔsmoK1 mutant to grow on d-mannitol as sole carbon source. As expected, this mutant was unable to grow under these conditions contrary to the parental strain. Second, after transfer of pAK30, a fragment containing the smoK gene, to R. sphaeroides ΔsmoK1 by biparental mating, the transconjugant strain was able to reduce selenite, indicating that this fragment was able to complement the transposon-induced mutation in R. sphaeroides IL-106. In addition to the inability to reduce selenite into elemental selenium, the ΔsmoK1 mutant presents other specific features upon addition of selenite compared to the wild type (WT). The MIC for selenite of this mutant is increased more than 10-fold compared to the WT, reaching 2,500 μg/ml (14.4 mM) under aerobic conditions. In contrast to the WT, the addition of selenite under aerobic conditions does not slow down the rate of the exponential phase in the case of the ΔsmoK1 mutant (data not shown). Another important difference between the WT and the ΔsmoK1 mutant is the absence of the induction of the major stress proteins in response to 1 mM SeO32− added to growth medium (Fig. 3C). These data support the view that the polyol ABC transporter is involved in selenite transport to the cytoplasm.
DISCUSSION
Response of R. sphaeroides to selenium oxyanions.
Nearly complete reduction of selenite is observed (Fig. 2) for R. sphaeroides cells grown under both dark aerobic and phototrophic anaerobic conditions. Our observation that R. sphaeroides cells reduce efficiently selenite into metallic selenium but do not significantly affect the redox state of selenate is in full agreement with the recent detailed analysis of Van Fleet-Stalder et al. (40). These authors reported that the bioconversion of selenite into metallic selenium reaches 94%, whereas only a small percentage of selenate is reduced by phototrophic cultures of R. sphaeroides. The addition of selenite induces a marked slowdown of the growth rate under aerobic but not anaerobic conditions, but it does not affect the final cell density. Different effects have been reported by Kessi et al. for the related photosynthetic species Rhodospirillum rubrum (17). For this species, the growth rate is not affected by the presence of 0.5 mM SeO32−, but the final cell density is reduced by more than a factor of 2 under anaerobic conditions whereas it is not affected under aerobic conditions. Moreover, only a small fraction (25%) of the selenite is reduced under aerobic conditions, whereas complete reduction is observed under photosynthetic anaerobic growth. Another difference between R. sphaeroides and Rhodospirillum rubrum is the observation that the reduction of selenite takes place during the exponential phase in R. sphaeroide, whereas it occurs during the transition from the exponential to the stationary phase in Rhodospirillum rubrum (17). R. sphaeroides and Rhodospirillum rubrum differ also in Se0 sequestration. While R. sphaeroides accumulates metallic selenium in the cytoplasmic compartment, Rhodospirillum rubrum appears to excrete the selenium granules across the plasma membrane and the cell wall after completion of the selenite reduction (17). Therefore, R. sphaeroides presents several advantages compared to other photosynthetic bacteria. These advantages, coupled with the metabolic diversity of R. sphaeroides, make this bacterium an excellent candidate in bioremediation processes (26).
Although R. sphaeroides efficiently reduces selenite under both dark aerobic and phototrophic anaerobic conditions, a marked slowdown of the growth rate is observed under the first condition but not under the latter. The higher toxic effect of selenite under aerobic condition compared to anaerobic condition is indicated by the lower level of resistance to selenite under this first condition. The oxidative stress generated by the addition of selenite (18) is partially overcome by an increase in the synthesis of proteins directly related to the cellular antioxidant defense (Fig. 3 to 5) in aerobic conditions. The bacteria detoxify the formation of reactive oxygen species, notably by inducing the expression of an iron-containing SOD (Fig. 5). In E. coli, the FeSOD, encoded by sodB, does not participate in oxidative stress response. Interestingly, the homologous sodB gene in R. capsulatus, a species closely related to R. sphaeroides, is regulated in response to oxidants in a way similar to the sodA (the MnSOD) of E. coli (8). The possible regulation of sodB in R. sphaeroides by selenite exposure or oxidative stress would illustrate a different adaptive response in the photosynthetic and enteric bacteria.
The addition of selenite under both dark aerobic and phototrophic anaerobic conditions induces an important increase in the expression of heat shock proteins and enzymes involved in protein synthesis. The induction of heat shock proteins is common with various stresses such as UV irradiation, H2O2, or heat stimuli (11). These are essential components of the cellular protection toward general stress (27). Since the SOD and thioredoxin are expressed at low level for cells of R. sphaeroides grown under phototrophic anaerobic conditions, the exact mechanisms of the selenite toxicity has to be elucidated under these conditions. Some proteins induced in response to selenite have yet to be identified, and this approach will benefit from the ongoing automated sequencing of the entire genome of R. sphaeroides.
Reduction and transport of selenite.
In addition to the induction of stress proteins synthesis, the proteome analysis shows that the synthesis of a protein, presenting a high identity with xenobiotic and morphinone reductases of P. aeruginosa, is clearly enhanced in the presence of selenite. This enzyme is therefore possibly involved in the reduction of selenite. There are, however, arguments against such a hypothesis. Analysis of one of the mutants unable to reduce selenite selected from our transposon library is affected in a moaA gene (98, 93, and 90% identities with the moaA gene of T. selenatis, R. capsulatus, and P. aeruginosa, respectively) (unpublished results). The moaA gene is part of the moa locus involved in the synthesis of molybdopterin and its dinucleotide derivatives, the organic component of the molybdenum cofactor (MoCo). This factor is found in various oxotransferases and hydroxylases (14, 15) and in particular in the highly specific selenate reductase purified from T. selenatis (31). Since the two reductases of P. aeruginosa do not contain a molybdenum cofactor, we cannot determine whether the xenobiotic-morphinone reductase is directly involved in one of the reduction steps of selenite or whether this enhanced synthesis is due to an indirect effect of the presence of selenite.
Selenate has been shown, in E. coli (21) and Saccharomyces cerevisiae (33), for example, to enter the cell through the sulfate permease system, in agreement with the similarities between the chemical properties of sulfur and selenium. Selenite transport appears to be carried out by an alternative transporter (19). Guzzo and Dubow (10) have recently presented evidence that, in this species, selenite may be translocated by a polypeptide of ca. 43 kDa. Secondary-structure analysis of this protein revealed 12 predicted transmembrane domains and a sugar transport protein signature motive. The conclusion that selenate and selenite are not incorporated through an identical pathway has been inferred for various other species, such as Clostridium pasteurianum (6) or Salmonella enterica serovar Typhimurium (5). In the case of R. sphaeroides, the markedly higher efficiency observed for the assimilation of selenite compared to selenate may also be due to the presence of two distinct transport systems (40).
In the case of the ΔsmoK1 mutant obtained in this study, this inability to reduce selenite is due to the knock out of the smoK gene, which encodes a constituent of a putative ABC transporter involved in the uptake of sugar alcohols (34). Moreover, the addition of selenite does not induce any changes in the major general stress proteins content for this mutant compared to the control cells (Fig. 3), in which the expression of several proteins is altered. These two observations strongly support the hypothesis that this polyol ABC transporter is also involved in the transport of selenite through the cytoplasmic membrane in the case of R. sphaeroides. Our results, together with those of Guzzo and Dubow (10), highlight the important role of polyol transporters in selenite transport. Future studies on different species able to assimilate and reduce selenite may provide evidence that this is a general mechanism in the bacterial kingdom.
In conclusion, R. sphaeroides efficiently reduces selenite with intracellular accumulation under both aerobic and anaerobic growth conditions. The combination of biochemical and genetic approaches emphasizes the oxidative properties of selenite under aerobic conditions and the involvement of a polyol transporter in the uptake of selenite in the photosynthetic bacterium R. sphaeroides.
ACKNOWLEDGMENTS
We thank our colleagues in the Laboratoire de Bioénergétique Cellulaire, particularly C. Berthomieu and J. Lavergne, as well as V. Méjean, for their support in improving the manuscript. We thank J. Labarre for helpful advice concerning 35S labeling and 2D gel electrophoresis. We thank S. Kaplan, who generously provided the plasmid pAK30 used in this work. We also gratefully acknowledge J. Gagnon for microsequencing of selenite-induced proteins.
REFERENCES
- 1.Avazéri C, Turner R J, Pommier J, Weiner J H, Giordano G, Verméglio A. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology. 1997;143:1181–1189. doi: 10.1099/00221287-143-4-1181. [DOI] [PubMed] [Google Scholar]
- 2.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 3.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Brown T A, Shrift A. Assimilation of selenate and selenite by Salmonella typhimurium. Can J Microbiol. 1980;26:671–675. doi: 10.1139/m80-117. [DOI] [PubMed] [Google Scholar]
- 6.Bryant R D, Laishley E J. Evidence for two transporters of sulfur and selenium oxyanions in Clostridium pasteurianum. Can J Microbiol. 1988;34:700–703. [Google Scholar]
- 7.Clayton R K. The induced synthesis of catalase in Rhodopseudomonas sphaeroides. Biochim Biophys Acta. 1960;37:503–512. doi: 10.1016/0006-3002(60)90507-2. [DOI] [PubMed] [Google Scholar]
- 8.Cortez N, Carrillo N, Pasternak C, Balzer A, Klug G. Molecular cloning and expression analysis of the Rhodobacter capsulatus sodB gene, encoding an iron superoxide dismutase. J Bacteriol. 1998;180:5413–5420. doi: 10.1128/jb.180.20.5413-5420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganther H E. Selenotrisulfides. Formation by the reaction of thiols with selenious acid. Biochemistry. 1968;8:2898–2905. doi: 10.1021/bi00848a029. [DOI] [PubMed] [Google Scholar]
- 10.Guzzo J, Dubow M S. A novel selenite- and tellurite-inducible gene in Escherichia coli. Appl Environ Microbiol. 2000;66:4972–4978. doi: 10.1128/aem.66.11.4972-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartke A, Bouche S, Laplace J M, Benachour A, Boutibonnes P, Auffray Y. UV-inducible proteins and UV-induced cross-protection against acid, ethanol, H2O2 or heat treatments in Lactococcus lactis subsp. lactis. Arch Microbiol. 1995;163:329–336. [Google Scholar]
- 12.Heider J, Bröck A. Selenium metabolism in microorganisms. Adv Microbiol Physiol. 1993;35:71–109. doi: 10.1016/s0065-2911(08)60097-1. [DOI] [PubMed] [Google Scholar]
- 13.Hess W M. Fixation and staining of fungus hyphae and host plant root tissues for electron microscopy. Stain Technol. 1966;41:27–35. doi: 10.3109/10520296609116276. [DOI] [PubMed] [Google Scholar]
- 14.Hille R. The mononuclear molybdenum enzymes. Chem Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 15.Johnson J L, Rajagopalan K V. Involvement of chlA, chlE, chlM, and chlN loci in Escherichia coli molydopterin biosynthesis. J Bacteriol. 1987;169:117–125. doi: 10.1128/jb.169.1.117-125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 17.Kessi J, Ramuz M, Wehrli E, Spycher M, Bachofen R. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl Environ Microbiol. 1999;65:4735–4740. doi: 10.1128/aem.65.11.4734-4740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer G F, Ames B S. Mechanisms of mutagenicity and toxicity of sodium selenite in Salmonella typhimurium. Mutat Res. 1988;201:169–180. doi: 10.1016/0027-5107(88)90123-6. [DOI] [PubMed] [Google Scholar]
- 19.Kredich N M. Synthesis of cysteine. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American. Society for Microbiology Press; 1996. pp. 514–527. [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Linblow-Kull C, Kull F J, Shrift A. Single transporter of sulfate, selenate, and selenite in Escherichia coli K-12. J Bacteriol. 1985;163:1267–1269. doi: 10.1128/jb.163.3.1267-1269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macy J M, Rech S, Auling G, Dorsch M, Stackebrandt E, Sly L I. Thauera selenatis gen.nov., sp. nov., a member of the beta subclass of Proteobacteria with a novel type of anaerobic respiration. Intern J Syst Bacteriol. 1993;43:135–142. doi: 10.1099/00207713-43-1-135. [DOI] [PubMed] [Google Scholar]
- 23.Maillet I, Lagniel G, Perrot M, Boucherie H, Labarre J. Rapid identification of yeast proteins on two-dimensional gels. J Biol Chem. 1996;271:10263–10270. doi: 10.1074/jbc.271.17.10263. [DOI] [PubMed] [Google Scholar]
- 24.McEwan A G, Jackson J B, Fergusson S J. Rationalization of the properties of nitrate reductase from Rhodopseudomonas capsulata. Arch Microbiol. 1984;137:344–349. [Google Scholar]
- 25.Moore M D, Kaplan S. Identification of intrinsic high-level of resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J Bacteriol. 1992;174:1510–1514. doi: 10.1128/jb.174.5.1505-1514.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore M D, Kaplan S. Members of the family Rhodospirillaceae reduce heavy-metal oxyanions to maintain redox poise during photosynthetic growth. ASM News. 1994;60:17–23. [Google Scholar]
- 27.Nepple B B, Bachofen R. Induction of stress proteins in the phototrophic bacterium Rhodobacter sphaeroides. FEMS Microbiol Lett. 1997;153:173–180. [Google Scholar]
- 28.O'Farrell P H. High-resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 29.Oremland R S. Biogeochemical transformations of selenium in anoxic environments, p 389–420. In: Frankenberger J R, Benson S, editors. Selenium in the environment. New York, N.Y: Marcel Dekker, Inc; 1994. [Google Scholar]
- 30.Schneider K H, Giffhorn F, Kaplan S. Cloning, nucleotide sequence, and characterization of the mannitol dehydrogenase gene from Rhodobacter sphaeroides. J Gen Microbiol. 1993;139:2475–2484. doi: 10.1099/00221287-139-10-2475. [DOI] [PubMed] [Google Scholar]
- 31.Schröder I, Rech S, Krafft T, Macy J M. Purification and characterization of the selenate reductase from Thauera selenatis. J Biol Chem. 1997;272:23765–23768. doi: 10.1074/jbc.272.38.23765. [DOI] [PubMed] [Google Scholar]
- 32.Siström W R. Transfer of chromosomal genes mediated by plasmid r68.45 in Rhodopseudomonas sphaeroides. J Bacteriol. 1977;131:526–532. doi: 10.1128/jb.131.2.526-532.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith F W, Hawkesford M J, Prosser I M, Clarkson D T. Isolation of cDNA from Saccharomyces cerevisiae that encodes a high-affinity sulfate transporter at the plasma membrane. Mol Gen Genet. 1995;247:709–715. doi: 10.1007/BF00290402. [DOI] [PubMed] [Google Scholar]
- 34.Stein M A, Schäfer A, Giffhorn F. Cloning, nucleotide sequence, and overexpression of smoS, a component of a novel operon encoding an ABC transporter and polyol dehydrogenases of Rhodobacter sphaeroides Si4. J Bacteriol. 1997;179:6335–6340. doi: 10.1128/jb.179.20.6335-6340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stolz J F, Oremland R S. Bacterial respiration of arsenic and selenium. FEMS Microbiol Rev. 1999;23:615–627. doi: 10.1111/j.1574-6976.1999.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 36.Switzer-Blum J, Bindi A B, Buzzelli J, Stolz J F, Oremland R S. Bacillus arsenicoselenatis sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California, which respire oxyanions of selenium and arsenic. Arch Microbiol. 1998;171:19–30. doi: 10.1007/s002030050673. [DOI] [PubMed] [Google Scholar]
- 37.Turner R J, Weiner J H, Taylor D E. The tellurite-resistance determinants tehAtehB and klaAklaBtelB have different biochemical requirements. Microbiology. 1995;141:3133–3140. doi: 10.1099/13500872-141-12-3133. [DOI] [PubMed] [Google Scholar]
- 38.Turner R J, Weiner J H, Taylor D E. Selenium metabolism in Escherichia coli. Biometals. 1998;11:223–227. doi: 10.1023/a:1009290213301. [DOI] [PubMed] [Google Scholar]
- 39.Van Fleet-Stalder V, Gürleyük H, Bachofen R, Chasteen T G. Effects of growth conditions on production of methyl selenides in cultures of Rhodobacter sphaeroides. Ind Microbiol Biotechnol. 1997;19:98–103. [Google Scholar]
- 40.Van Fleet-Stalder V, Chasteen T G, Pickering I J, George G N, Prince R C. Fate of selenate and selenite metabolized by Rhodobacter sphaeroides. Appl Environ Microbiol. 2000;66:4849–4853. doi: 10.1128/aem.66.11.4849-4853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada A, Miyashita M, Inoue K, Matsunaga T. Extracellular reduction of selenite by a novel marine photosynthetic bacterium. Appl Microbiol Biotechnol. 1997;48:367–372. doi: 10.1007/s002530051064. [DOI] [PubMed] [Google Scholar]