Abstract
Mitogen-activated protein kinase (MAPK) pathways constitute key regulatory elements linking extracellular stimuli to nuclear gene expression. Immediate-early responsive genes (IEGs) of the activator protein 1 (AP-1) family, such as fos, achieve peak expression levels shortly after cells are stimulated with growth factors and sharply decrease thereafter. Several AU-rich binding proteins (AUBPs), including HuR (Hu-antigen R, Elav-like protein 1, ELAVL1) and KSRP (far upstream element-binding protein 2, KHSRP) bind to a fos AU-rich element (ARE) present in the 3′-UTR (untranslated region) of fos mRNA regulating its stability by a still poorly defined mechanism. We show in the present study that, whereas HuR binds and stabilizes transcribed reporter mRNAs bearing the fos 3′-UTR, KSRP counteracts this effect. Furthermore, we found that fos mRNA stability and HuR phosphorylation status are dependent on the activity of p38 MAPK in both epithelial cells and fibroblasts upon proliferative stimulation. Analysing PPI (protein–protein interaction) networks, we performed a thorough query of interacting proteins for p38 MAPKs, HuR and other AUBPs upon growth factor stimulation. This revealed novel HuR interactors including inhibitors of protein phosphatase 2 (PP2A) activity. Over-expression of two of these interactors, pp32 and APRIL (acidic leucine-rich nuclear phosphoprotein 32 family member B, ANP32B) and pharmacological inhibition of PP2A stabilized a fos reporter mRNA. Our results indicate that p38 MAPK regulates fos mRNA decay by affecting the state of phosphorylation of HuR while controlling yet to be fully elucidated PP regulatory networks.
Keywords: stability of mRNAs, c-fos, p38 MAPKs, AUBPs
INTRODUCTION
Immediate-early responsive genes (IEGs) are rapidly and transiently induced in response to a variety of stimuli in living cells. Members of the AP-1 (activator protein 1) family of transcription factors (composed mainly of the c-jun subfamily and c-fos-related genes) are among its most conspicuous representatives [1]. Particularly for the c-fos gene (referred to as fos herein), its mRNA level peaks shortly after 30 min of stimulation and decreases rapidly afterwards [2,3].
MAPKs (mitogen-activated protein kinases) are key components of important signalling pathways that transduce extracellular signals and ultimately regulate gene expression. Regulation of fos mRNA levels and protein activity are regulated by mechanisms involving MAPK signalling pathways [1,3,4]. Whereas the fos promoter exhibits a complex array of regulatory elements targeted by different transcription factors, the c-Fos (Fos hereinafter) protein contains a number of serine and threonine residues that represent targets for phosphorylation and concomitant transcriptional regulation [1,4–6].
Besides regulation of promoter and protein activity, post-transcriptional decay of fos mRNA provides one more regulatory instance for the fos gene family. AU-rich elements (AREs) are present in the mRNA of fos (and many other IEGs), which are essential for the regulation of mRNA decay processes [7,8]. ARE sequences are believed to be the main docking regions for most proteins involved in regulating mRNA stability [7]. There are several types of ARE regions, mostly characterized by the presence and repetition of the consensus sequence AUUUA; specifically, fos has a well characterized class I ARE in its 3’-UTR (3’-untranslated region) [9,10].
Trans-acting factors associated to the AREs are known as AU-rich binding proteins (AUBPs). Some of the better studied AUBPs are ELAVL1 or HuR (Hu-antigen 1, Elav-like protein 1 and Hu family members HuB, HuC, HuD) [11], HNRNPD or AUF1 (heterogenous nuclear ribonucleoprotein D0) or [12], KHSRP or KSRP (far upstream element-binding protein 2) [13], ZFP36 (tristetraprolin, TTP) [14] and ZFP36L1 (zinc finger protein 36, C3H1 type-like 1, BRF1) [15] among others. Some of them are involved in mRNA stabilization (HuR), whereas others stimulate its degradation (TTP, KSRP, BRF1 and AUF1). KSRP has been shown to bind to AREs of unstable mRNAs, associate with TTP and together recruit the exosome complex, leading to mRNA degradation [16]. KSRP and TTP were reported to be phosphorylated by p38 MAPK family members (p38β and p38α respectively) and by the MAPK-activated protein 2 (MK2) [17–19]. Phosphorylated forms are unable to bind to their target sequence in mRNAs, provoking a delay in the degradation process, reversed by phosphatase reactivation. On the other hand, HuR binds to AREs and, when overexpressed, stabilizes mRNAs preventing their degradation [20]. HuR is also reported to be targeted by different kinases, such as Chk2 (serine/threonine-protein kinase Chk2, CHEK2), CDK1 (cyclin-dependent kinase 1), PKCδ (protein kinase C delta type) and p38α [21–24]. Phosphorylation of different residues within the sequence of HuR (Ser88, Thr118, Ser202, Ser221 and Ser318) is involved in increased nucleo-cytoplasmic shuttling and RNA binding [21–24].
AUBPs reported to associate to the fos ARE in different cellular contexts include HuR, HuB, HuD, AUF1 and KSRP [8]. Although there are several studies of signalling pathways regulating the activity of these proteins and the fos mRNA decay process, details regarding the regulatory mechanisms in which they participate remain elusive.
In the present article, we focus on fos mRNA decay regulation by MAPKs in cultured cells under proliferative stimulation. We found that activation of the p38 MAPK pathway promotes the decay of the fos mRNA through the regulation of regulatory sequences that require an intact ARE element in its 3′-UTR. We also observed that HuR binds to the fos ARE, stabilizing the fos mRNA. On the other hand, KSRP counteracts this effect, promoting fos mRNA degradation. Although we observed that p38 MAPK affects HuR mRNA binding and stabilizing properties, we did not detect a direct phosphorylation of HuR by p38 MAPK. We found that p38 MAPK activation affects HuR serine phosphorylation state in a negative fashion, diminishing its ability to bind to the fos ARE and promoting fos mRNA decay, possibly due to the activation of phosphatases acting on HuR.
MATERIALS AND METHODS
Culture and treatments of cell lines
Human embryonic kidney (HEK)293 and HeLa TetOff (Clontech) cells (cells stably expressing the transactivator tTa) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) high glucose (Invitrogen), with L-glutamine and sodium pyruvate supplemented with 10% FBS and penicillin/streptomycin/amphotericin B (Invitrogen). NIH3T3 mouse fibroblasts and NIH3T3 expressing the human M1 muscarinic acetylcholine receptor (m1.2 cells, described in [3]) were grown in DMEM high glucose, with L-glutamine, containing10%calfserumorbovinegrowthserumsupplemented calf (Thermo-Scientific) and the above anti-microbial mixture. Before the stimuli, cells were starved for 2, 12 or 24 h, as indicated, and then treated with human recombinant plateletderived growth factor (PDGF)-BB 100 ng/ml final concentration (Beta Laboratories) for the NIH3T3 cell line or epidermal growth factor (EGF) at 10 ng/ml final concentration, in the case of HEK293 cells. The MAPK inhibitors (PD98059 at final concentration of 20 μM, UO126 at 10 μM and SB203580 at 10 μM) and MAPKAPK2 inhibitor or MK2ai (also known as CMPD1, at 50 μM) (Calbiochem) were pre-incubated for 1 h before stimulation. Protein phosphatase 2 (PP2A) and PP1 inhibitor endothall (at final concentration of 50 μM) was added to the medium from 1 to 2 h before treatment. For reporter mRNA decay assays, cells were incubated with tetracycline (Sigma–Aldrich) at a final concentration of 1 μg/μl for a maximum of 5 h.
Transient transfections
All cell lines were plated in complete medium and allowed to grow overnight to 70–80% confluence. The cells were transfected using linear polyethyleneimine (PEI; Polysciences) with a protocol adjusted for each cell line. Transfection complexes were achieved using the appropriate volume of PEI (1 μg/μl, pH 7.2), in a 3:1 ratio (PEI–DNA) for HEK293 and HeLa TetOff cells or in a 7:1 ratio for NIH3T3 cells.
DNA constructs
The PCR fragment corresponding to the entire 3′-UTR of the murine fos mRNA (1283–2091 bp) was amplified from cDNA of PDGF-stimulated NIH3T3 cells with appropriate primers and subcloned into the pTRE2hygLuc (Luc represents luciferase) plasmid (Clontech) using the NheI and ClaI restriction sites (Luc–fos). The 3′-UTR ΔARE of fos was obtained by deletion of 69 bp of the described AU-rich region or ARE (Luc–fosΔARE). This deletion was achieved by amplification of both regions up- and down-stream of the ARE region of the fos 3′-UTR from Luc–fos and ligating them with an AgeI restriction site, replacing the 69 bp with 6 bp of this site. pTRE2hygLuc with the entire 3′-UTR of β-globin mRNA was used as control (Luc–β-globin). The ARE region, plus 100 bp downstream, was subcloned into the pBluescriptSK(+) using the restriction sites NheI and HindIII. A fragment of equivalent size from β-globin was subcloned in the same vector and used as a control. Labelled in vitro transcripts obtained from these vectors were used in the experiments of RNA gel mobility-shift assays. AUBP expression vectors were engineered as follows: cDNA of murine HuR, TTP and BRF1 were subcloned into the following plasmids pCEFL–HA (mHuR) and pGEX4T3 (GST–HuR, GST–TTP and GST–BRF1). The cDNA encoding human HuR in its wild-type (WT) version and mutant in three phosphorylation sites (T118A, S202A, S221A) were a gift from Angel Nebreda and were subcloned in the pCEFL–HA (hHuR WT and hHuR 3×M). For expression of Streptagged human HuR, the pcDNA3Zeo(+)Strep2HuR was used. Expression vectors for pCEFL–HA-tagged c-Jun N-terminal kinase 1 (JNK1), extracellular-signal-regulated kinase (ERK)2, ERK5, p38α, p38β, p38γ and p38δ MAPK and pGEX4T3–ATF2 have been described previously [1]. pET15b–KSRP (histidine–KSRP) and pcDNA4TOEGFP–KSRP [25] were gifts from Hartmut Kleinert. pcDNA3–FLAG–pp32 and pcDNA3–FLAG–APRIL were a gift from Joan Steitz [26].
Reporter mRNA decay assays
The quantitative PCRs (qPCRs) to measure luciferase mRNA were performed as in [27]. The primer set used was the following: forward, 5′-ccgccgttgttgttttg-3′; reverse, 5′-acacaactcctccgcgc-3′. The amount of luciferase mRNA was normalized using primers for the hygromycin gene, present in the reporter plasmid: forward, 5-ggaatccccgaacatcg-3′; reverse, 5′-gcagacgcgctacttcg-3′.
Northern blots and real-time qPCR assays
Total cellular RNA was isolated using TRIzol (Invitrogen) or TRIReagent (Genbiotech) using the manufacturer’s protocols. mRNAs were reverse-transcribed using oligo-dT primers (Biodynamics). Real-time qPCR was performed using a Stratagene MX 3000P QPCR with SYBR Green detection (Agilent). For real-time qPCR, 2 μg of total RNA was subjected to reverse transcription. A dilution of all samples was then used to measure the expression of the endogenous fos, using a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) product as normalizer. For Northern blots, 30 μg of RNA extracted were ran in a 1% agarose gel in TAE (Tris base, acetic acid and EDTA buffer) 1×, transferred on to a positively charged nylon membrane (HyBond, GE Healthcare) and incubated with a fos-specific probe 32P-labelled by PCR.
RNA electrophoretic mobility-shift assays (EMSAs)
RNA probes were synthesized with a T7 RNA transcription kit (Promega) from the pBSfosARE as well as the β-globin control. For radioactive probes [α−32P]rUTP (NEN PerkinElmer) was added. Total extracts were obtained from NIH3T3 cells plated in 6-cm plates and grown to 90% confluence, starved overnight and then treated with PDGF (10 ng/ml) and pretreated (or not) with SB 203580 and other inhibitors as indicated. The protocol for protein extraction and probe incubation has been previously described [28].
Solid-phase kinase assays
NIH3T3 cell were incubated in 6-cm plates to 80% confluence, starved for 12 h and stimulated with PDGF for different times as described in Results and the corresponding figure legends. The plates were washed with ice-cold PBS and lysed at 4°C in a buffer containing 25 mM HEPES, pH 7.5, 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1% Triton X-100, 0.1% SDS, 10 mM β-glycerophosphate, 1 mM sodium vanadate and 1 mM PMSF. A mix of glutathione–Sepharose beads (GE Healthcare) with bound GST–HuR was added corresponding to 4 μg of protein to each tube and incubated at 4°C overnight. The following day, the beads were collected and washed and developed as described for the kinase assay protocol in vitro.
Western blot analysis
The protocol is described elsewhere [27]. The primary antibodies used were the following: anti-KSRP (Cell Signaling Technology), anti-HuR (Santa Cruz Biotechnology), anti-c-Fos (Santa Cruz Biotechnology), anti-actin (Santa Cruz Biotechnology), anti-β-tubulin (Santa Cruz Biotechnology), anti-α-tubulin (Cell Signaling Technology), anti-haemgglutinin (HA) (Santa Cruz Biotechnology), anti-GFP (Santa Cruz Biotechnology) anti-FLAG (Sigma–Aldrich) and anti-phosphoserine (Millipore), anti-ERK2 (Santa Cruz Biotechnology), anti-ERK5 (Cell Signaling Technology), anti-p38α (Cell Signaling Technology), anti-p38β (Santa Cruz Biotechnology) and anti-p38γ (Santa Cruz Biotechnology) and anti-p38δ (Santa Cruz Biotechnology).
Biotin–RNA pull-down assays
5′-End biotin-labelled RNA oligonucleotides (Bi–RNA) were used to isolate proteins that associate with the ARE of fos. The following probes were used: (1) ARE–WT oligonucleotide, 40-bp-long that includes the three AUUUA consensus sequences (Dharmacon Thermo-Scientific); (2) MUT3x, also 40-bp-long and has the three uracils of AUUUA mutated to guanidine (Dharmacon Thermo-Scientific); (3) negative control, non-ARE RNA sequence from the EDI (ectodysplasin-A) gene 47-bp-long (Dharmacon Thermo-Scientific). Lysates from HEK293 and NIH3T3 cells were quantified, precleared with streptavidin–agarose beads (Thermo-Scientific) and 100 μg of protein was incubated in the presence of 0.5 nmol of each probe for 10 min at room temperature and 10 min at 4°C. After several washes, SDS sample buffer was added and a SDS/PAGE was performed. The gels were either stained or transferred and subjected to Western blot analysis.
Identification of proteins by LC–MS
To determine protein identity, after the isolation of Bi–RNA and bound protein complexes, the proteins were separated by SDS/PAGE. Each gel was fixed and stained using either a Silver Stain Kit (Pierce) or colloidal Commassie Blue. After the staining protocol, the bands were sliced. Stained gel pieces were first washed and then reduced-alkylated (dithioerithriol–iodoacetamide). Proteolytic digestion was performed overnight at 37°C using modified trypsin (Promega) in 50 mM ammonium bicarbonate (pH 8.3). Peptides were then extracted from gels and concentrated by Speed Vac prior to LC–MS measurements. Dried samples were resuspended in LC–MS loading solvent (2% acetonitrile, 0.1% formic acid) and separated by HPLC (nanoAcquity, Waters). Samples were first captured and washed at initial chromatographic conditions on a home-made capillary Pre-column [Magic C18; 3 μm, 200 Å (1 Å = 0.1 nm); 2 cm × 100 μm] prior to analytical separation. An 80 min biphasic gradient was run starting from 100% A solvent (2% acetonitrile, 0.1% formic acid) to 90% B solvent (100% acetonitrile, 0.1% formic acid) on a home-made capillary column (Magic C18; 3 μm-100 Å; 15 cm × 75 μm internal diameter at 250 nl/min). Online mass spectrometric detection was performed on an LTQ–Orbitrap XL (Thermo-Scientific) using data-dependent acquisition mode with dynamic exclusion. For each MS scan, the ten most-intense detected ions were fragmented and then excluded for the following 30 s. Experimentally generated data were submitted to protein database search through Proteome Discoverer 1.1 (Thermo-Scientific) and Mascot 2.3 (Matrix Science) search engine against the human restricted UniProt Protein database (UniProt release 2011_07, 31 May 2011 including its reversed format). Scaffold 3 Viewer (Proteome Software) was used to finally compile data results.
Bacterial expression of GST fusion proteins
The BL21 pLysS strain of Escherichia coli was transformed with the vector pGEX–4T3 encoding the fusion protein GST–mHuR WT and with pET15b–KSRP (histidine–tagged KSRP). The protocol used to purified recombinant proteins was described in [1].
siRNA
NIH3T3 were transfected with HiPerfect (Qiagen) with a siRNA oligonucleotide-targeting HuR sequence (Qiagen) and AllStars oligonucleotide as negative control (Qiagen). At 48 h post-transfection, cells were starved and stimulated with PDGF and lysed to analyse fos and HuR protein expression as indicated.
Luciferase reporter assays
Cells were seeded on six-well or 12-well dishes and transfected with different expression plasmids together with 0.1 μg of luciferase fos promoter reporter vector. The total amount of transfected DNA was normalized with a β-galactosidase assay. Cells were lysed in passive lysis buffer (Promega) 24 h post-transfection. Cell lysates were incubated with luciferin reagent of the luciferase reporter system (Promega).
Network-based data organization and discovery
Integration of experimental data in protein–protein interaction (PPI) networks to build explorable maps using Cytoscape (http://www.cytoscape.org) has been previously described [29,30]. Briefly, using the human-centred PPI database built by Echeverria et al. [31], it was possible to extract a network containing the PPIs between the query proteins and their interactors. Query proteins used in the process were: p38α (MAPK14), p38β (MAPK11), p38γ (MAPK12), p38δ (MAPK13), MK2 (MAPKAPK2), TTP (ZFP36), HuR (ELAVL1), BRF1 (ZFP36L1), KSRP (KHSRP) and AUF1 (HNRNPD). The set of PPIs was further enriched by manually curated literature mining. One ‘node’ in this network corresponds to a protein and the connection between them is called an ‘edge’ and it refers to the PPI between these two nodes. Information for the different members of the network was also loaded from different databases (UniProt, Gene Ontology) to include in the graph as metadata. These efforts were then complemented by literature mining to improve our understanding of the potential biological relevance of these connected modules.
TUNEL assay
The percentage of apoptotic cells was measured using the ApopTag Fluorescein in situ Kit (Millipore). Cells were plated on glass slides and, after treated as indicated, fixed in paraformaldehyde (1%) for 10 min. DNA fragmentation was detected according to the manufacturer’s instructions. The kit uses DNA terminal deoxynucleotidyltransferease (TdT) to catalyse the addition of digoxygenin–dNTP to 3′-ends of fragmented DNA. This tagged tail is finally detected with a specific fluoresceintagged antibody. The slides were counterstained with DAPI; several pictures were taken for each treated sample using a Zeiss Axioimager.Z1 with Apotome attachment. At least 1000 cells were quantified in each treatment. Positive cells were quantified using the ImageJ software (NIH) and the percentage of apoptotic cells was determined.
Statistical analysis
All experiments were repeated at least three times. The statistical significance was measured with one-way ANOVA test and multiple mean comparisons in GraphPad Prism 5 software. The symbols are informed as follows: ns: no significant difference; *: significant difference of the means when α = 0.05 is considered; **: α = 0.01; ***: α = 0.001.
RESULTS
Regulation of fos mRNA expression by MAPKs
As an IEG, fos shows a pattern of rapid and transient expression of its mRNA upon the addition of proliferative stimuli to cultured cells. To begin exploring the mechanism of fos expression regulation, we incubated NIH3T3 cells with carbachol or PDGF. Whereas the latter acts on endogenously expressed tyrosine kinase growth factor receptors, the former acts on M1 G-protein-coupled receptors, which transduce potent mitogenic signals when ectopically expressed in NIH3T3 cells [3], thus enabling us to study the shared key elements activated by distinct growth promoting factors. Northern blot and real-time qPCR analysis showed that these proliferative stimuli enhance fos mRNA levels rapidly in NIH3T3 cells, which peaked around 30 min after treatment and quickly went down within 2 h post-stimulation (Figures 1A and 1B). In an attempt to identify signalling pathways involved, we first tested for kinases involved in the regulation of fos promoter activity. We performed luciferase reporter assays co-transfecting NIH3T3 cells with a reporter containing the luciferase gene downstream the fos promoter along with vectors that express MAPKs and its upstream activating MAPKKs (mitogen-activated protein kinase kinases; Figure 1C). All kinases were expressed at comparable levels (Supplementary Figures S1A and S1B) and we found that only activation of the ERK2 signalling pathway influences fos promoter activity. Similar results were obtained in a slightly different environment, when cells were co-transfected with the reporter and MAPK expression vectors and then treated with carbachol or PDGF. As shown in Figure 1(D), neither ERK5 nor JNK nor any of the p38 MAPKs-affected fos promoter activity to the extent induced by ERK2.
Figure 1. Analysis of fos expression as a response to mitogenic stimuli and MAPK involvement in fos promoter activity.
(A) NIH3T3 cells expressing the M1 muscarinic receptor (m1.2 cells) were stimulated with the agonist carbachol (Ch) to a final concentration of 100 mM following a time course. RNA was extracted in order to perform a Northern blot assay using a specific fos probe. (B) Real-time qPCR of fos mRNA in m1.2 stimulated with Ch (100 mM, left) and NIH3T3 incubated in the presence of PDGF (100 ng/ml, right), levels of GAPDH expression were used to normalize. (C) Luciferase assays. pc–fosLuc (pGL3–c-fos promoter–luciferase) was co-transfected with vectors expressing different MAPKs and their activating MAPKKs, in NIH3T3. (D) Similar to (C) stimulating MAPKs in m1.2 cells with indicated agonists (Ch, left, or PDGF, right).
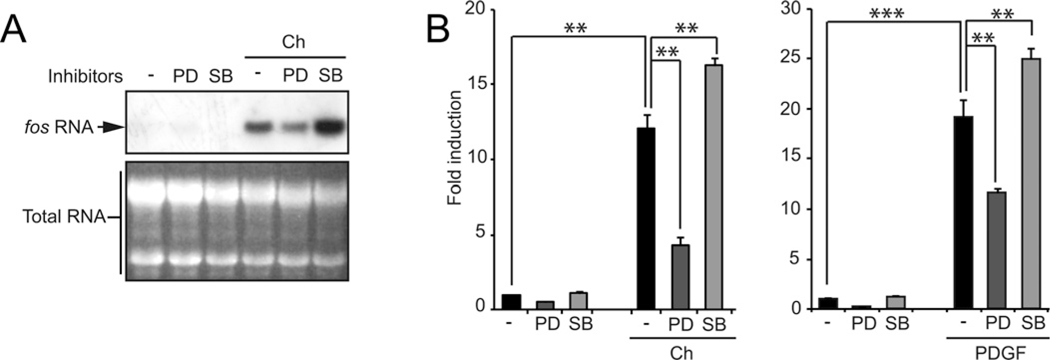
To study this using a complementary pharmacological approach, NIH3T3 cells were pretreated with MAPK inhibitors PD98059 (inhibits MEK activity, an ERK2 activator) and SB203580 (inhibits p38α MAPK activity) and stimulated with carbachol for 40 min, which stimulates all MAPKs in these cells (Supplementary Table S1). Figures 2(A) and 2(B) show that in Northern blots and qPCR assays, inhibition of the ERK2 pathway led to a decreased level of fos mRNA expression. However, after inhibiting p38 MAPK signalling, we found a significant increase in the amount of fos mRNA comparing with control conditions. Stimulating cells with PDGF rendered comparable results (Figure 2B, right panel). An alternative MEK inhibitor was tested (UO126) and rendered identical results (Supplementary Figure S2A). None of these kinase inhibitors triggered apoptosis to a significant level under the conditions used (Supplementary Figure S2B).
Figure 2. MAPK involvement on fos mRNA expression.
(A) Northern blot. Endogenous fos expression was analysed in mRNA extracts of m1.2 cells pretreated with MAPK inhibitors (MEK inhibitor, PD98059 20 μM, and p38 inhibitor, SB203580 10 μM) stimulated with carbachol (Ch) for 40 min. (B) Same experiment as in (A), but in this case fos mRNA was quantified by qPCR. m1.2 cells stimulated with Ch, left panel, and NIH3T3 cells stimulated with PDGF, right panel. * P < 0.05; **P < 0.01; ***P < 0.001.
The p38 MAPK pathway regulates fos mRNA decay
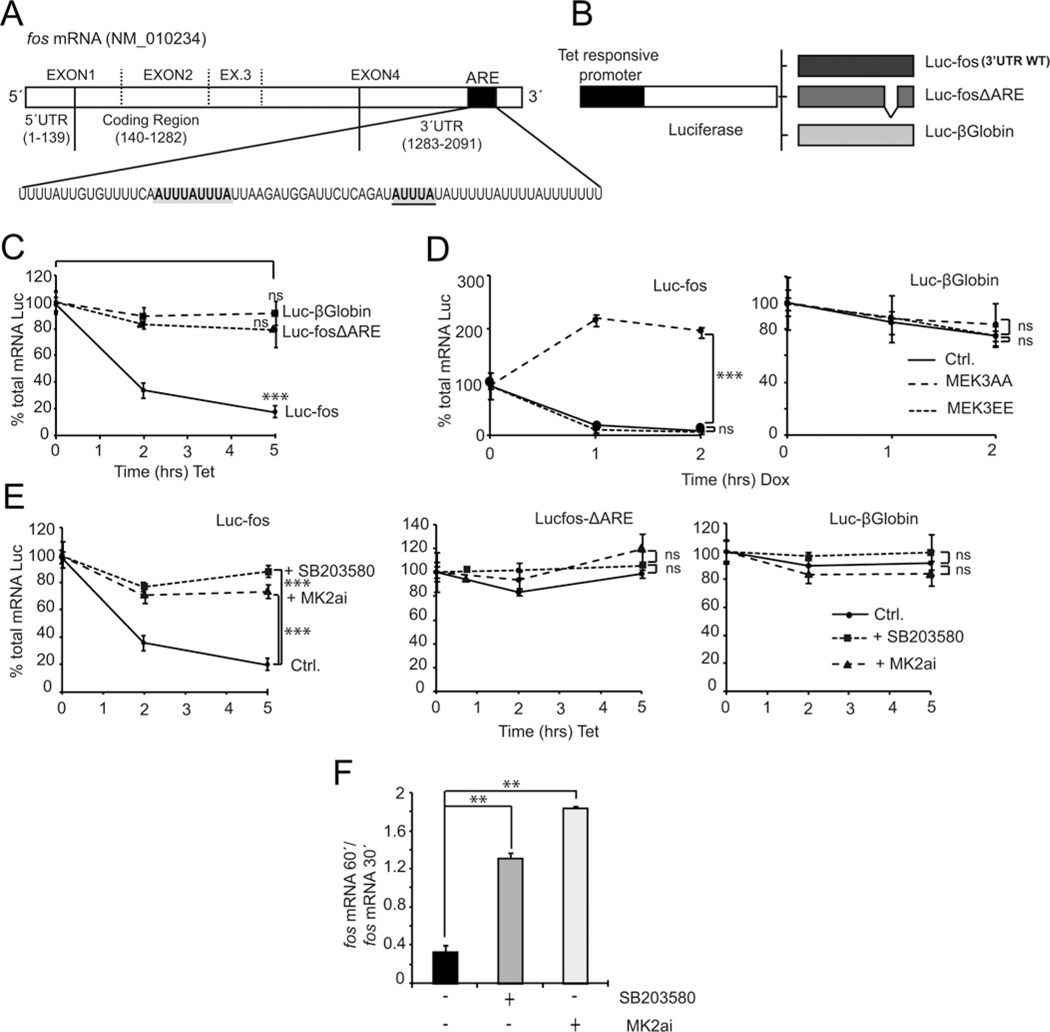
In order to study the effects of p38 signalling pathway on post-transcriptional events, we focused on elements present in the fos mRNA structure (Figure 3A) that may affect its stability or decay. Figure 3(A) shows a schematic representation of the murine fos mature mRNA structure, highlighting a 69 bp region (ARE) reported to be the main docking sequence for most proteins involved in regulating mRNA stability [9]. To address the regulatory mechanisms in the mRNA decay process and given the potential importance of this region, we cloned the entire fos 3′-UTR downstream luciferase to generate a different reporter construct. The new reporter plasmid has a promoter that is responsive to the addition of tetracycline, allowing the specific regulation of the reporter gene. We engineered two reporter constructs, Luc–fos containing the entire 3′-UTR of fos WT mRNA (Figure 3B) and Luc–fosΔARE containing the fos 3′-UTR with the ARE region deleted and replaced by an AgeI restriction site. An additional control is provided by Luc–β-globin, a reporter that expresses the luciferase gene fused to the 3′-UTR of the β-globin gene.
Figure 3. Role of the p38 MAPK pathway in regulating levels of fos mRNA expression.
(A) Schematic representation of fos mRNA structure. An AU-rich region known as ARE is highlighted. (B) The entire murine 3′-UTR of fos in its WT version was inserted in the pTRE2hygLuc vector (Luc–fos); this plasmid has a tetracycline-responsive promoter (also responsive to analogues such as doxycycline) that drives transcription only in the absence of antibiotic. The Luc–fosΔARE reporter is identical with the WT except for a 69 bp deletion encompassing the ARE region. Luc–β-globin has the entire 3′-UTR of the β-globin gene cloned in pTRE2hygLuc and contains no ARE regulatory elements. (C) HeLa TetOff cells were transfected with the reporter constructs Luc–fos, Luc–fosΔARE and Luc–β-globin. At 24 h post-transfection cells were incubated with tetracycline (1μg/μl final concentration) for a maximum period of 5 h, RNA was extracted and a RT-qPCR (real-time qPCR) was performed to quantify luciferase total mRNA. (D) NIH3T3 TetOff cells were co-transfected with the reporters Luc–fos and Luc–β-globin and vectors expressing either the dominant-negative (MEK3AA) or constitutively activated (MEK3EE) versions of the MAPKK MEK3, a p38 regulator. At 24 h post-transfection cells were incubated with doxycycline and processed as in (C). (E) Same experiments as in (C) but pretreating cells with kinase inhibitors for p38 MAPK (SB203580, 10 μM final) and MK2 (MK2ai, 50 μM) before tetracycline addition. (F) Proliferating NIH3T3 cells were starved for 12 h and stimulated with PDGF following a time course, in the absence or presence of kinase inhibitors SB203580 and MK2ai. RT-qPCR of endogenous fos mRNA (normalized with GAPDH) was performed. The ratio of fos mRNA present at 60 min to mRNA present at 30 min is shown. *P < 0.05; **P < 0.01; ***P < 0.001.
Upon transfection of HeLa TetOff cells with the reporter vectors, tetracycline was added to the medium and cells were lysed following a time course. Reporter mRNA levels were then tested by qPCR. Results show that Luc–fos produces an mRNA that is unstable (Figure 3C). After 5 h of transcription inhibition, we could detect only about 20% of the original amount of luciferase mRNA. In contrast, mRNA expressed by control plasmids (both missing ARE elements) show no significant decay even 5 h after transcription was interrupted.
Co-transfection of the Luc–fos reporter construct with MEK3EE and MEK3AA (constitutively active and dominant negative forms of the dual p38 MAPKK, MEK3) revealed that the inhibition of the p38 MAPK signalling pathway prevents reporter mRNA decay (Figure 3D). Furthermore, treatment with the inhibitor SB203580 also prevents the decay of the WT reporter (Figure 3E, left panel). In order to test for a direct or indirect effect of p38 MAPK in the regulation of this post-transcriptional process by p38 MAPK, we tested MK2ai, an inhibitor targeting MAPKAPK2a (MK2α), a downstream target of p38 MAPK. MK2ai also prevented the decay of the Luc–fos reporter, suggesting a possible role for this kinase in the fos mRNA decay process (Figure3E,left panel).Neither inhibitor had any detectable effect upon the decay of reporter control mRNAs (Figure 3E, middle and right panels).
Following the time course expression profiles for endogenous fos mRNA in NIH3T3 cells stimulated with PDGF, we found that treatment with these two pharmacological inhibitors rendered a curve with delayed decay. We quantified levels of mRNA present at 30 and 60 min after the addition of PDGF in cells treated with vehicle or inhibitors for p38 MAPK and MK2. Figure 3(F) shows the ratio of the fos mRNA present at 60 min compared with at 30 min after stimulation for each treatment. Incubation with both kinase inhibitors resulted in a several fold increase in this ratio with respect to control conditions. These results indicate that fos mRNA decay may require active p38 MAPK signalling.
AUBPs and AUBP-interacting proteins are linked to the ARE region of fos mRNA
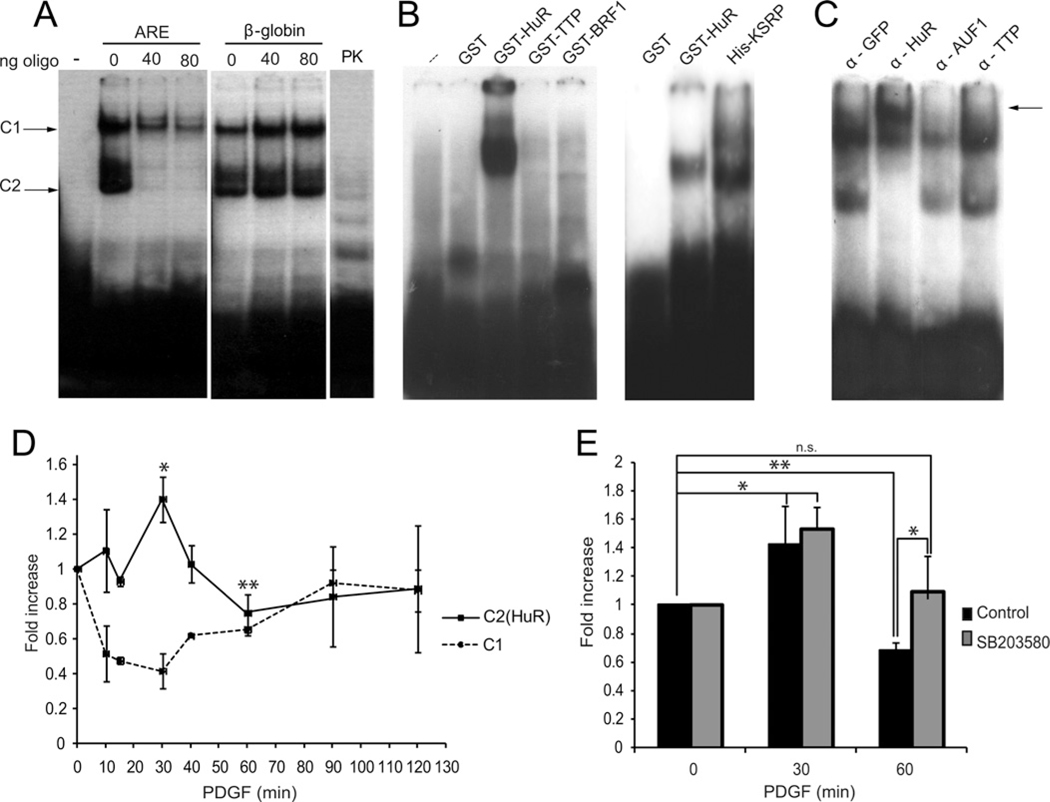
In an attempt to identify the proteins that may link the p38 MAPK pathway to the decay fos mRNA, we performed Bi–RNA pull-down assays. A 40-bp-long 5′-biotinylated RNA probe with nucleotide sequence corresponding to the WT fos ARE including all three consensus AUUUA pentamer sequences was commercially synthesized. In addition, a control mutant probe with each core AUUUA sequence replaced by AGGGA was prepared. Proteins obtained from cell lysates were incubated with the probes, unbound material washed and retained proteins were run on SDS/PAGE gels (Figure 4A). Proteins present in the recovered protein–RNA complexes were subsequently identified by high-resolution LC–MS. This analysis rendered a list of putative ARE-associated factors as shown in Supplementary Table S2. Of interest, most of these proteins exhibit RNA-binding domains and four of them are known AUBPs: HuR, HuB, AUF1 and KSRP. Comparing the information obtained with the alternative probes, these AUBPs preferentially associated to the WT rather than the mutant probes (Figure 4A and Supplementary Table S2). We validated this information by performing Western blots using material pulled down with both of the fos probes and an unrelated sequence. HuR and KSRP bind only to the ARE sequences and mostly to the WT probe, with limited affinity for the mutant version (Figure 4A, lower panels).
Figure 4. AUBP binding to the fos ARE sequence.
(A) Biotinylated probes of fos mRNA ARE region (either WT or mutated as described in the Materials and methods section) were incubated with HEK293 cell lysates stimulated with EGF (10 ng/ml) and then purified with streptavidin–agarose. Silver-stained SDS/PAGE gel (upper panel) is shown. Western blot of these purified complexes (lower panel) identified HuR and KSRP binding differentially to the WT probe. (B) Network-based analysis of p38 MAPK and AUBP interactors. PPIs for the query proteins were retrieved and organized in maps together with literature curation data and functional metadata using the software Cytoscape [30]. Nodes (proteins) are connected by edges (lines) representing their known physical interactions. In grey are shown those proteins also found in the LC–MS sequencing experiments.
We next used systems biology approaches to provide additional predictive information that may help us to understand the potential relationship between p38 MAPKs, AUBPs binding to the fos ARE and putative additional interacting molecules. Organizing and integrating PPI data from public sources in biological maps allowed us to explore new features in signalling systems using network-based discovery methodologies [31], using a previously reported approach [29–31]. These proteins included all four isoforms of p38 MAPK (α, β, γ and δ), the MAPKAPK2 or MK2 and all AUBPs reported in the literature to bind to fos ARE: HuR, KSRP, AUF1 and TTP [8] and a non-associating AUBP, BRF1. Figure 4(B) shows one possible result of this analysis. Only KSRP was reported to interact physically with one isoform of p38 MAPK (β) [32]. The network prediction analysis revealed that KSRP and AUF1 relate to many mRNA degradation proteins such as exosome complex members (EXOCS2, EXOCS3, PARN, DCP2 and EXOCS4), regulators of the deadenylation and the translationally coupled mRNA decay process (PABPC1, PAIP1, IGF2BP2, CSDE1, SYNCRIP and YBX1) or other decay-related AUBPs (TTP). It is also interesting that these two AUBPs could be part of the same type of complex as they are connected by two hnRNP proteins (HNRNPA1 and HNRNPH1). On the other hand, interactors of HuR seem to be related to proteins regulating phosphatase activity, as shown in the non-characterized predicted interaction with a regulatory subunit of the phosphatase PP2A (PPP2R2B) and the interactions with SET, pp32 and APRIL, three PP2A inhibitors [33]. In grey, we highlight those interactors that were found experimentally in the pull-down studies of biotinylated-RNA probe. Proteins pinpointed by both the pull-down assay using a fos ARE probe as bait and the bioinformatics approach represent putative candidates as part of a regulatory mechanism linking protein kinase signalling to fos mRNA stability.
We next co-transfected a plasmid that expresses murine HA-tagged HuR (mHuR) with each luciferase–3′-UTR reporter. Figure 5(A) shows that overexpression of HuR prevents decay of the Luc–fos reporter (left panel) whereas no effect is observed over the decay pattern of control reporters (middle and right panel, inset shows effective expression of HuR). In addition, knockdown of HuR expression has a clear detrimental effect on endogenous fos protein expression (Figure 5B).
Figure 5. Effect of different relevant factors on fos mRNA decay.
(A) HeLa TetOff cells were transfected with the reporter constructs Luc–fos, Luc–fosΔARE and Luc–β-globin along with vectors that were either empty or expressed murine HA-tagged HuR. After addition of tetracycline, mRNA decay assays were run. The Western blot on the right shows expression of murine WT HA–HuR. (B) siRNA depletion of HuR in NIH3T3 cells transfected with HuR siRNA or scrambled siRNA. Starved cells were stimulated with PDGF for 30 min, after that period cells were lysed and levels of expression of endogenous HuR and Fos proteins were visualized in Western blots. (C) A mRNA decay assay was run on extracts from cells transfected with either empty plasmid or expressing WT or a triple mutant of human HuR (T118A, S202A, S221A). The panel on the right validates expression as seen on a Western blot. (D) As in (C) but co-transfecting the reporter with vectors that express the FLAG-tagged PP2A inhibitors and HuR-binding proteins pp32 and APRIL. (E) As in (C) but in cells transfected with plasmids expressing EGFP–KSRP. Treatment with SB203580 was performed as indicated in the upper panel. In the middle panel, cells were co-transfected with the reporter and also with vectors that express EGFP–KSRP or HA–HuR. A Western blot against GFP is shown in the lower panel. (F) Endothall, a PP2A and PP1 inhibitor, was also tested at 50 μM final concentration. HeLa TetOff cells transfected with the Luc–fos reporter were incubated with endothall and/or tetracycline for different times as indicated and mRNA present in the extract was tested by RT-qPCR (real-time qPCR). In Western blots, NT corresponds to non-transfected cells. *P < 0.05; **P < 0.01; ***P < 0.001.
Protein phosphorylation is a common feature to mechanisms controlling gene expression. We tested the effect of a triple HuR mutant (T118A, S202A and S221A) lacking critical residues reported to be the target of different kinases [21–24]. Using reporter plasmids co-transfected with an HA-tagged human HuR triple mutant (HuR 3×M) or the WT control (hHuR WT) we challenged the Luc–fos reporter (Figure 5C). In contrast with WT HuR, the triple mutant was unable to stabilize the reporter.
The PP2A inhibitor proteins pp32 and APRIL are also HuR interactors [33] and we tested their effect upon mRNA stability in fos reporter assays. As shown in Figure 5(D), co-transfection with plasmids expressing either pp32 or APRIL significantly stabilized WT fos reporter mRNA. To ascertain the importance of phosphatase activity in the regulation of the decay of fos mRNA, we incubated cells transfected with WT reporters and treated them with endothall, a PP2A and PP1 pharmacological inhibitor. Results as shown in Figure 5(E) indicate that 2 h after transcription is inhibited, reporter mRNA levels drop to 50%. However, pre-incubation with endothall show no decay effect compared with control conditions strengthening the idea that phosphatase inhibition has a stabilizing effect upon fos mRNA.
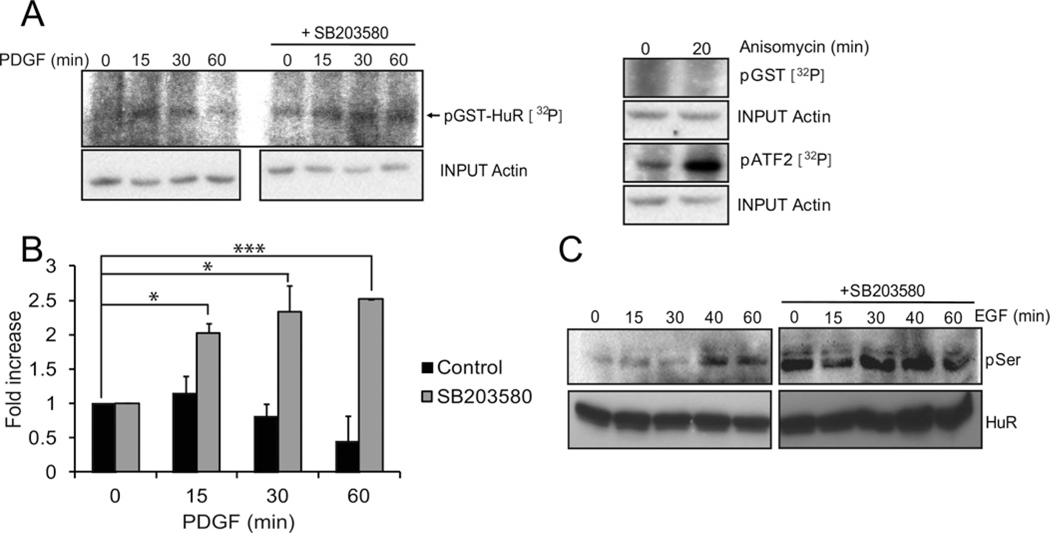
We also tested the influence of KSRP on the decay experiments using two different experimental settings. Figure 5(F) shows that in cells that are transfected with the Luc–fos reporter, overexpression of KSRP counteracts the stabilization effects elicited by SB203580 (in Figure 3E) and HuR (in Figures 5A and 5C). As seen in Figure 5(C), phosphorylation of HuR would be necessary to perform its stabilizing effect. Therefore we tested the hypothesis that p38MAPK might be affecting the phosphorylation status of HuR. Solid-phase kinase assays were performed using glutathione beads loaded with bacterially expressed GST–HuR recombinant protein. Cells treated with PDGF and SB203580 or vehicle were lysed, incubated with the beads and tested for kinase activity. Figure 6(A) shows representative gels and Figure 6(B) shows data obtained after quantification of bands from three independent experiments. Treatment with the p38 MAPK inhibitor resulted in a significant increase in HuR phosphorylation that is maximal 60 min after the addition of PDGF. Similar results were obtained by overexpression of a Strep-tagged HuR in EGF-stimulated HEK293 cells pretreated with vehicle or SB203580, to extend our analysis to other cell types. Western blots revealed with an anti-phospho-serine antibody showed an increment in the levels of phospho-serine of HuR in the absence of p38 MAPK activity (Figure 6C).
Figure 6. Determination of HuR phosphorylation state.
(A) Solid-phase assay. Proliferating NIH3T3 cells were starved for 12 h and stimulated with PDGF for the indicated times. Cell lysates were incubated overnight with bacterially expressed recombinant GST–HuR coupled with glutathione-Sepharose beads. Protein complexes bound to the beads were washed, incubated in the presence of radioactive [γ32P] ATP and this reaction mixture was run on a SDS/PAGE gel that was dried and autoradiographed. Lysates from cells treated with anisomycin or vehicle were tested for GST–ATF2 and GST phosphorylation activity, as positive and negative controls respectively. (B) Quantification of three independent solid-phase assays as in (A). (C) HEK293 cells were transfected with a vector expressing Strep-2–tagged murine HuR, stimulated with EGF and incubated with Streptactin–agarose beads. Pulled down proteins were run on SDS/PAGE gels and blotted with anti-phospho-serine antibodies. Western blot using anti-Strep-tag shows the total amount of HuR. The right panel corresponds to cells pretreated with SB203580 and the left panel shows untreated cells. *P < 0.05; **P < 0.01; ***P < 0.001.
Gel EMSAs can help reveal protein complexes bound to nucleic acids. In order to investigate the identity and dynamics of proteins binding to the fos ARE, we performed RNA EMSAs using 32P-labelled transcripts containing the fos ARE sequence. Figure 7(A) shows the main delayed complexes observed, named complex 1 (C1) and complex 2 (C2). They are composed of protein bound to the labelled probe as they disappear upon proteinase K (PK) treatment (Figure 7A). Competing with unlabelled fos ARE or β-globin oligonucleotides for complex assembly shows specificity for fos ARE sequences (Figure 7A). Labelled fos ARE probe challenged with recombinant bacterially expressed AUBPs shows that only HuR and KSRP seem to bind directly to the ARE (Figure 7B). Supershift assay performed using lysates from cells stimulated with PDGF pre-incubated with specific antibodies for the indicated proteins supports the idea of HuR binding to the fos ARE as a major component of C2 complexes (Figure 7C).
Figure 7. Protein complexes binding to fos AREs.
(A) A radioactive RNA probe containing the fos ARE sequence was incubated with NIH3T3 cell protein lysates, run on a non-denaturing gel and specific complex formation was studied by autoradiography. Increasing amounts of unlabelled probes containing the fos ARE or β-globin sequences were added as specific and non-specific controls respectively. An additional control was performed using PK. The two major complexes studied are showed with arrows as C1 (complex 1) and C2 (complex 2). (B) A similar experiment was performed using bacterially expressed recombinant GST (HuR, TTP and BRF1) or 6× histidine fusion proteins (KSRP) incubated with the ARE probe. (C) Presence of putative protein candidates in the complexes assembled at ARE sequences was tested by incubating cell lysates with the probe in the presence of antibodies against representative AUBP family members. A supershift is indicated by an arrow. (D) The same experiment was performed using lysates of NIH3T3 cells that were stimulated with PDGF over a time course. The intensity of two major complexes [C2 containing (HuR) or not, C1] was quantified and depicted in the graph. (E) Intensity of the HuR containing complexes was tested in lysates from cells stimulated with PDGF after pre-incubation with the p38 MAPK inhibitor SB203580 or vehicle. *P < 0.05; **P < 0.01; ***P < 0.001.
Over a time course performing EMSAs using lysates of cells treated with PDGF, we observed that C2–HuR complex levels increase after addition of the stimulus, whereas intensity of bands corresponding to C1 complexes concomitantly decrease, both effects being maximal at 30 min, then C2 levels start to decrease and C1 levels augment, restoring original values after 2 h (Figure 7D). A similar experiment performed with the addition of SB203580 and quantifying bands corresponding to the C2–HuR complex shows a statistically significant increase in the levels of C2–HuR complex formation in the presence of the p38 MAPK inhibitor (Figure 7E).
DISCUSSION
As a representative early responsive gene, fos is rapidly and transiently induced upon the onset of environmental conditions that imply changes in cell fate commitment. In resting cells, fos expression is maintained at very low levels. Upon stimulation of cells with growth factors acting on either tyrosine kinase or G-protein-coupled receptors the fos promoter is rapidly induced [34]. Total mRNA levels peak shortly 30 min after stimulation and sharply decrease thereafter. The translated Fos protein is part of the AP-1 transcription factor complexes that bind enhancers at different gene promoters and contribute to a second wave of gene expression, which is responsible of cell fate commitment changes. Subsequently, post-translational modifications target the Fos protein for degradation. This rapid and transient pattern of Fos expression is observed in a variety of cells and under different stimuli. This fact provides compelling evidence that both up- and down-regulation of the fos gene are important in cellular homoeostasis.
Abundant information has been gathered on the regulation of gene promoter activity [35,36]. However, mechanisms regulating the decay of mature mRNAs are still poorly understood. Our work focused on the pathways affecting fos mRNA turnover. Particularly AREs present in the fos 3′-UTR have already been proved to affect the half-life of the fos mRNA. Fusing the fos 3′-UTR into stable reporter mRNAs induced decay, but mRNA stability was regained when its ARE sequence was deleted [9]. Even though these regulatory elements are present in 8% of transcripts including a variety of early-responsive genes [8], the variable features of their sequences and the mechanisms by which they contribute to the stability of mRNAs bearing them is still mostly unknown. Identifying the biochemical pathways that are responsible to transduce the signal from membrane receptors to effector proteins that regulate mRNA decay events then becomes particularly important.
Studying fos expression in response to the activation of different MAPK pathways we found that only ERK1/2 signalling induced fos promoter activity. In a different approach, using pharmacological inhibitors we observed that, to our surprise, inhibiting the activation of the p38 MAPK increased the amount of fos mRNA, although p38 MAPK did not affect fos promoter activity. We therefore focused on the study of p38 MAPK-dependent events regulating mRNA levels in the absence of potential promoter effects. Experiments to test the stability or instability of mRNA in different cellular conditions tend to be designed using general transcription inhibitors (e.g. actinomycin D or α-amanitin). These effective reagents are also strong stress inducers and deleterious to the life of cells. Therefore, in order to avoid secondary effects when studying regulation of fos mRNA decay mediated by its 3′-UTR, we designed a vector tool that allows us to selectively control mRNA expression of a reporter. Our construct expresses a luciferase gene reporter in which we cloned the entire 3′-UTR of fos downstream from the luciferase coding region. This vector allows for controlled transcription with a tetracycline-responsive (TetOff) promoter. Using this reporter, we avoid unnecessary secondary stress effects as induction of any stress-activated protein kinases (SAPKs), including p38MAPK or inhibition of de novo synthesis of participating factors. Transcripts produced by this reporter construct showed clearly that they are under an ARE-dependent mRNA decay control.
In accordance with the data obtained studying endogenous fos expression, we found that inhibition of the p38 MAPK signalling pathway by using either dominant-negative forms of the MEK3 dual-specificity kinase (a well-known p38 MAPKK) or the p38 inhibitor SB203580, extended the half-life of the Luc–fos reporter mRNA. This effect may seem contradictory to previous reports in which p38 MAPK activation correlates with the stabilization of mRNAs [24,32,45]. However, these reports are based on different genes and cellular contexts. For instance, p38 MAPK activation in response to γ-radiation and consequent HuR phosphorylation by this kinase was shown to be responsible for the stabilization of p21cip mRNA [24]. On the other hand, p38 MAPK stabilizes cytokine and tumour necrosis factor (TNF)-α mRNAs during an inflammatory response [19,37,38]. Particularly p38–MK2 signalling prevents TTP binding to target mRNAs and degradation extending the half-life of TNF-α mRNA [39]. However the effect of p38 MAPK on mRNA stability does not need to be unique, as p38-dependent and -independent effects have already been reported [40]. It is accepted that the particular environment in which a given mRNA is transcribed and the identity and post-translational status of the RNA-binding proteins present and involved can actually determine the fate of mRNAs. Our studies are focused on a normal proliferating context, in which no stresses are applied.
HuR, KSRP and TTP are representative ARE-binding proteins or AUBPs. By means of a LC–MS sequencing approach and using a biotinylated probe corresponding to the fos ARE sequence (40-bp-long) that includes the three consensus sequences AUUUA we searched for associated proteins. Several experiments rendered similar results: AUBPs HuR and KSRP were isolated as major components of fos ARE-binding complexes. This information was validated by Western blots using specific antibodies and by RNA EMSAs performed with a radioactive probe containing fos ARE sequences and bacterially expressed recombinant proteins. Whereas HuR was also detected in endogenous cell lysates by supershift using a specific antibody, we found no evidence of TTP binding to the fos ARE by any of these approaches.
HuR stabilizing effects on mRNA stability have already been observed for several genes [20,41,42] and in particular for fos [8]. Our experiments show that co-transfection of HuR-expressing plasmids prevents decay of a reporter mRNA containing fos regulatory ARE regions. Accordingly, when we knocked down HuR expression by siRNAs we found a remarkable reduction in fos protein expression. An antagonistic role for HuR and KSRP in controlling ARE containing mRNA decay has already been reported for the stability of the iNOS (inducible nitric oxide synthase, NOS2) mRNA [19]. Accordingly, we report in the present study that overexpression of KSRP prevents fos reporter mRNA stabilization by HuR. In addition, EMSAs performed using a fos ARE probe and increasing amounts of either recombinant protein (KSRP and HuR) showed that they form mutually exclusive complexes, similar to that reported for the iNOS ARE [25] (results not shown). The negative effect of KSRP on fos mRNA stability is also supported by a partial but significant reversion of the stabilizing effect of SB203580.
The notion that phosphorylation of HuR plays a crucial role in connecting extracellular signal inputs to a specific response is a complex subject of increasing interest [43]. We report in the present study that, unlike the WT version, a triple HuR mutant with relevant phosphorylation sites mutated to alanines (T118A, S202A, S221A) is unable to prevent decay of the reporter Luc–fos construct, highlighting the importance of functional phosphorylation sites in HuR. However, and in contrast with reports obtained working under different conditions [24], we have found no evidence of direct phosphorylation of HuR by p38 MAPK. In addition, in our system, inhibition of p38 MAPK signalling significantly increases HuR phosphorylation both in solid-phase and in Strep-tag purification assays. Although counterintuitive, association of HuR with the PP2A inhibitors pp32 and APRIL has been described [33]. This interaction modulates HuR’s ability to bind its target mRNAs in living cells and suggests a role in connecting HuR to pathways controlling the stability of ARE-containing mRNAs. Co-transfection of vectors expressing pp32 and APRIL significantly stabilized the Luc–fos reporter transcript. Accordingly incubation of cells with a pharmacological phosphatase inhibitor had a similar stabilizing effect. These results highlight the involvement of a phosphatase activity in the promotion of fos mRNA decay.
HuR binding to specific sequences is necessary to stabilize target mRNAs. As shown by our own data and the literature, HuR phosphorylation might have a positive contribution to this effect [22,24]. Using a radioactive probe designed with fos ARE sequences, we performed RNAEMSAs that revealed the assembly of a complex (C2) that contains HuR as an important component according to mobility supershift assays. Time course experiments after the addition of PDGF showed a peak of intensity in the formation of C2 complexes that is remarkably coincident in time with a peak in fos mRNA expression. Interestingly, C2 complexes remained assembled longer upon incubation of cells with a p38 MAPK pharmacological inhibitor. Taken together, these results and the phosphorylation assays suggest a role for p38 MAPK in HuR dephosphorylation and consequent disassembling of HuR–fos mRNA complexes.
Summarizing, our results are consistent with the following sequence of events as depicted in Figure 8. Upon growth factor stimulation of resting cells, a variety of MAPK pathways are activated showing two distinctive temporal patterns as shown in Supplementary Table S1. Whereas ‘early-activated MAPKs’ (such as ERK2 and ERK5) attain maximal activity at 5 min after stimulation, ‘late-activated MAPKs’ (JNKs and p38s) display activity at later times and sometimes for longer intervals. Early activation of ERK2 is consistent with its role as an activator of the fos promoter. Once translated, Fos protein is phosphorylated by different MAPKs [1,4] and as a member of AP-1 transcription factors conveys signals further to other gene promoters. By the time enough Fos protein has been produced and activated, fos mRNA might no longer be needed and p38 MAPKs promote its decay. Phosphorylated HuR prevents fos mRNA decay and might become the subject of p38 MAPK-dependent dephosphorylation. This event might involve the release of phosphatase inhibitors and facilitate changes in the repertoire of proteins bound to the fos ARE. Lower amounts of HuR bound to the fos mRNA might therefore allow for the assembly of a protein complex of different composition at the ARE sequences, most probably including KSRP and targeting the fos mRNA molecules to degradation. This AUBP exchange at regulatory sites in target mRNAs has already been described for other target mRNAs [25,44]. The idea of a phosphorylation-regulated AUBP exchange [45] presents an interesting model for switching between situations that promote conditions of stability or instability for specific mRNAs and contribute to gene expression regulation.
Figure 8. Concerted role for early and late activated MAPKs in the rapid and transient expression of the fos early responsive gene.
Our current view of the model shows that, whereas early-activated MAPKs (ERK2) trigger fos promoter induction, late-activated MAPKs (p38) regulate mRNA decay. After mitogenic receptor stimulation, once a time interval for peak Fos protein translation and phosphorylation has been completed, low levels of fos mRNA are restored, through phosphorylation-dependent events. The assembly of a protein complex of different composition at the ARE sequence, probably including KSRP, targets the fos mRNA to degradation.
In general, the role of p38 MAPK as a mediator of mRNA stability seems to be highly dependent upon the gene model under study and the environmental context of the cellular system. Most reports pointing to p38 MAPK as a stabilizing mRNA pathway describe genes involved in inflammatory responses under stress conditions of myogenic differentiation, whereas our data show a destabilizing role for p38 MAPK on fos mRNA, in the context of proliferative stimulation. Ultimately, as for promoter activity, extracellular originated signalling regulates gene expression at the mRNA decay level through a variety of molecular components. The final outcome is of fascinating complexity. Our results show how concerted early (ERK) or late (p38 and JNK) MAPK activation can contribute to both rapid and transient activation of the early responsive gene fos. Our working model provides a blueprint for the events taking place, identifying additional components of the complex assembled at the fos ARE (including proteins and miRNAs). Its precise regulation by p38 MAPK, including the activation of a putative HuR phosphatase, constitute current challenges that warrant further investigation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the members of the IFIBYNE - CONICET and Departamento de Química Biológica FCEN - UBA for sharing equipment and for their support and thoughtful advice. We thank Dr Didier Picard and his staff (UNIGE-Geneva, Switzerland) for their useful suggestions. We thank Dr Joan Steitz (Yale-HHMI, U.S.A.) for providing APRIL, pp32 and SET constructs, Dr Gary Brewer (Rutgers-RWJMS, U.S.A.) for the AUF1 vectors, Dr Angel Nebreda (IRB-Barcelona, Spain) for the human cDNA of WT and mutated versions of HuR and Dr Hartmut Kleinert (Uni-Mainz, Germany) for the KSRP constructs. We also thank Daniela Capiatti (INGEBI-UBA, Argentina) for the PP2A inhibitor endothall.
FUNDING
This work was supported by the University of Buenos Aires [grant number 20020100100949]; the Consejo Nacional de Ciencia y Tecnica [grant number PIP 11220110100573CO]; and the Agencia Nacional de Promocion Cientifica y Tecnologica [grant number PICT 2011 # 1637]; the Union for International Cancer [grant numbers ICR-08-120 and ICR-11-008]; and the Miami CFAR/ Sylvester Cancer Center-Argentine Network for Translational Research in AIDS Malignancies which is supported by NCI/OHAM Supplements to Miami CFAR [grant P30AI073961].
Abbreviations:
- AP-1
activator protein 1
- APRIL
acidic leucine-rich nuclear phosphoprotein 32 family member B, ANP32B
- ARE
AU-rich element
- AUBP
AU-rich binding protein
- Bi–RNA
5-end biotin-labelled RNA oligonucleotide
- DMEM
Dulbecco’s modified Eagle’s medium
- EGF
epidermal growth factor
- EMSA
electrophoretic mobility-shift assay
- ERK
extracellular-signal-regulated kinase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HA
haemagglutinin
- HEK
human embryonic kidney
- HuR
Hu-antigen 1, Elav-like protein 1, ELAVL1
- IEG
immediate-early responsive gene
- iNOS
inducible nitric oxide synthase, NOS2
- JNK
c-Jun N-terminal kinase
- KSRP
far upstream element-binding protein 2, KHSRP
- Luc
luciferase
- MAPK
mitogen-activated protein kinase
- MAPKK
mitogen-activated protein kinase kinase
- MK2
mitogen-activated protein kinase-activated protein kinase 2
- PDGF
platelet-derived growth factor
- PEI
polyethyleneimine
- PK
proteinase K
- PP
protein phosphatase
- PPI
protein–protein interaction
- qPCR
quantitative PCR
- TNF
tumour necrosis factor
- TTP
tristetraprolin, ZFP36
- UTR
untranslated region
- WT
wild-type
REFERENCES
- 1.Tanos T, Marinissen MJ, Coluccio-Leskow F, Hochbaum D, Martinetto H, Gutkind JS and Coso OA (2005) Phosphorylation of c-Fos by members of the p38 MAPK family. Role in the AP-1 response to UV light. J. Biol. Chem 280, 18842–18852 [DOI] [PubMed] [Google Scholar]
- 2.Cochran BH, Zullo J, Verma IM and Stiles CD (1984) Expression of the c-fos gene and of a fos-related gene is stimulated by platelet-derived growth factor. Science 226, 1080–1082 [DOI] [PubMed] [Google Scholar]
- 3.Coso OA, Chiarello M, Kalinec G, Kyriakis JM, Woodgett J and Gutkind JS (1995) Transforming G protein-coupled receptors potently activate JNK (SAPK). Evidence for a divergence from the tyrosine kinase signaling pathway. J. Biol. Chem 270, 5620–5624 [DOI] [PubMed] [Google Scholar]
- 4.Monje P, Marinissen MJ and Gutkind JS (2003) Phosphorylation of the carboxyl-terminal transactivation domain of c-Fos by extracellular signal-regulated kinase mediates the transcriptional activation of AP-1 and cellular transformation induced by platelet-derived growth factor. Mol. Cell. Biol 23, 7030–7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang SH, Sharrocks AD and Whitmarsh AJ (2003) Transcriptional regulation by the MAP kinase signaling cascades. Gene 320, 3–21 [DOI] [PubMed] [Google Scholar]
- 6.O’Donnell A, Odrowaz Z and Sharrocks AD (2012) Immediate-early gene activation by the MAPK pathways: what do and don’t we know? Biochem. Soc. Trans 40, 58–66 [DOI] [PubMed] [Google Scholar]
- 7.Gingerich TJ, Feige JJ and LaMarre J (2004) AU-rich elements and the control of gene expression through regulated mRNA stability. Anim. Health Res. Rev 5, 49–63 [DOI] [PubMed] [Google Scholar]
- 8.Barreau C, Paillard L and Osborne B (2006) AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shyu AB, Greenberg ME and Belasco JG (1989) The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 3, 60–72 [DOI] [PubMed] [Google Scholar]
- 10.Chen CA, Xu N and Shyu AB (1995) mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol 15, 5777–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma WJ, Cheng S, Campbell C, Wright A and Furneaux H (1996) Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem 271, 8144–8151 [DOI] [PubMed] [Google Scholar]
- 12.Kajita Y, Nakayama J, Aizawa M and Ishikawa F (1995) The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. Common modular structure and binding properties of the 2xRBD-Gly family. J. Biol. Chem 270, 22167–22175 [DOI] [PubMed] [Google Scholar]
- 13.Chou CF, Mulky A, Maitra S, Lin WJ, Gherzi R, Kappes J and Chen CY (2006) Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell. Biol 26, 3695–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carballo E, Lai WS and Blackshear PJ (1998) Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281, 1001–1005 [DOI] [PubMed] [Google Scholar]
- 15.Stoecklin G, Colombi M, Reineri I, Leuenberger SA, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T and Moroni C (2002) Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21, 4709–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Prujin GJM, Stoecklin G, Moroni C, Mann M and Karin M (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107, 451–464 [DOI] [PubMed] [Google Scholar]
- 17.Zhu W, Brauchle MA, Di Padova F, Gram H, New L, Ono K, Downey JS and Han J (2001) Gene suppression by tristetraprolin and release by the p38 pathway. Am. J. Physiol. Lung Cell. Mol. Physiol 281, L499–L508 [DOI] [PubMed] [Google Scholar]
- 18.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WFC, Blackwell TK and Anderson P (2004) MK2-induced tristetraprolin:14–3–3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahtani KR, Brook M, Dean JLE, Sully G, Saklatvala J and Clark AR (2001) Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol 21, 6461–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CA, Xu N and Shyu AB (2002) Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol. Cell. Biol 22, 7268–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdelmohsen K, Pullman R, Lal A, Kim HH, Galban S, Yang X, Beltrow JD, Walker M, Shubert J, Gillespie DA et al. (2007) Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25, 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HH, Abdelmohsen K, Lal A, Pullman R, Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM and Gorospe M (2008) Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 22, 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doller A, Winkler C, Azrilian I, Schulz S, Hartman S, Pfeilsschifter J and Eberhardt W (2011) High-constitutive HuR phosphorylation at Ser 318 by PKC{delta} propagates tumor relevant functions in colon carcinoma cells. Carcinogenesis 32, 676–685 [DOI] [PubMed] [Google Scholar]
- 24.Lafarga V, Cuadrado A, Lopez de Silanes I, Bengochea R, Fernandez-Capetillo O and Nebreda AR (2009) p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)S checkpoint. Mol. Cell. Biol 29, 4341–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J and Kleinert H (2005) Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 33, 4813–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallouzi I, Brennan CM and Steitz JA (2001) Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA 7, 1348–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naipauer J, Gatelli A, Degese M, Slomiansky V, Werthmeier E, LaMarre J, Castilla L, Abba M, Kordon E and Coso OA (2013) The use of alternative polyadenylation sites renders integrin β1 (Itgb1) mRNA isoforms with differential stability during mammary gland development. Biochem. J 454, 345–357 [DOI] [PubMed] [Google Scholar]
- 28.You Y, Chen CA and Shyu AB (1992) Two cellular proteins bind specifically to a purine-rich sequence necessary for the destabilization function of a c-fos protein-coding region determinant of mRNA instability. Mol. Cell. Biol 12, 2931–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Echeverria PC, Forafonov F, Pandey DP, Mulebach G and Picard D (2011) Detection of changes in gene regulatory patterns, elicited by perturbations of the Hsp90 molecular chaperone complex, by visualizing multiple experiments with an animation. BioData Min. 4, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cline M, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B et al. (2007) Integration of biological networks and gene expression data using cytoscape. Nat. Protoc 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Echeverria PC, Bernthaler A, Dupuis P, Mayer B and Picard D (2011) An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine. PLoS One 6, e26044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Briata P, Forcales SP, Ponassi M, Corte G, Chen CY, Karin M, Puri PL and Gherzi R (2005) p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 20, 891–903 [DOI] [PubMed] [Google Scholar]
- 33.Brennan CM, Gallouzi I and Steitz JA (2000) Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol 151, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fromm C, Coso OA, Montaner S, Xu N and Gutkind JS (1997) The small GTP-binding protein Rho links G protein-coupled receptors and Galpha12 to the serum response element and to cellular transformation. Proc. Natl. Acad. Sci. U.S.A 94, 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spitz F and Furlong E (2012) Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet 13, 613–626 [DOI] [PubMed] [Google Scholar]
- 36.Johnson D and Dent S (2013) Chromatin: receiver and quarterback for cellular signals. Cell 152, 685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otkjaer K, Holtman H, Kragstrup TW, Paludan SR, Johansen C, Gaesthel M, Kragballe K and Iversen L (2010) The p38 MAPK regulates IL-24 expression by stabilization of the 3′ UTR of IL-24 mRNA. PLoS One 5, e8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winzen R, Kracht M, Ritter B, Wilhem A, Chen CA, Shyu AB, Muller M, Gaesthel M, Resch K and Holtmann H (1999) The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18, 4969–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A and Gaesthel M (2006) Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenineuridine-rich element. Mol. Cell. Biol 26, 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frevel MAE, Bakheet T, Silva AM, Hissong JG, Khabar KSA and Williams BRG (2003) p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol 23, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan XC and Steitz JA (1998) Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17, 3448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez de Silanes I, Lal A and Gorospe M (2005) HuR: post-transcriptional paths to malignancy. RNA Biol. 2, 11–13 [DOI] [PubMed] [Google Scholar]
- 43.Eberhardt W, Doller A and Pfeilsschifter J (2012) Regulation of the mRNA-binding protein HuR by posttranslational modification: spotlight on phosphorylation. Curr. Protein Pept. Sci 13, 380–390 [DOI] [PubMed] [Google Scholar]
- 44.Doller A, Schlepckow K, Schwalbe H, Pfeilsschifter J and Eberhardt W (2010) Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol. Cell. Biol 30, 1397–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A and Gaesthel M (2012) The p38MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet. 8, e1002977 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.