Abstract
The gut microbiome is a unique marker for cetaceans’ health status, and the microbiome composition of their skin wounds can indicate a potential infection from their habitat. Our study provides the first comparative analysis of the microbial communities from gut regions and skin wounds of an individual Indo-Pacific finless porpoise (Neophocaena phocaenoides). Microbial richness increased from the foregut to the hindgut with variation in the composition of microbes. Fusobacteria (67.51% ± 5.10%), Firmicutes (22.00% ± 2.60%), and Proteobacteria (10.47% ± 5.49%) were the dominant phyla in the gastrointestinal tract, while Proteobacteria (76.11% ± 0.54%), Firmicutes (22.00% ± 2.60%), and Bacteroidetes (10.13% ± 0.49%) were the dominant phyla in the skin wounds. The genera Photobacterium, Actinobacillus, Vibrio, Erysipelothrix, Tenacibaculum, and Psychrobacter, considered potential pathogens for mammals, were identified in the gut and skin wounds of the stranded Indo-Pacific finless porpoise. A comparison of the gut microbiome in the Indo-Pacific finless porpoise and other cetaceans revealed a possible species-specific gut microbiome in the Indo-Pacific finless porpoise. There was a significant difference between the skin wound microbiomes in terrestrial and marine mammals, probably due to habitat-specific differences. Our results show potential species specificity in the microbiome structure and a potential threat posed by environmental pathogens to cetaceans.
Keywords: gut microbiome, Indo-Pacific finless porpoise, skin wound, pathogens
1. Introduction
The mammalian microbiome is a complex assembly of microorganisms vastly distributed in different anatomical niches of mammals. It is vital in mammalian health maintenance and disease defense [1]. Microbiome composition and structure are often organ-specific and linked to the physiological functions of the organs. The microbiome changes dynamically with host health status; minor disturbances in the microbiome may lead to its malfunction or subsequent sickness of the host. For example, Proteobacteria is one of the predominant microbiota constituents in the jejunum and is responsible for digestion, where Proteobacteria accounts for 2% of the total microbiome in the colon [2]. Dynamic changes in the richness of the microbiome, including Proteobacteria, Fusobacteria, and Bacteroidetes, were observed in morbidly obese patients [3], indicating a direct or indirect relationship between the microbiome and health status.
Additionally, other factors, including species [4,5], evolution [6], habitation [7], and dietary change [8], may also disturb the spatial and temporal composition of the microbiome. Consequently, cetaceans possess unique microbiomes in different anatomical niches, for example, the gastrointestinal tract and skin [9,10]. A previous study showed that a high similarity of function and higher-level taxonomy of the gastrointestinal tract microbiome were found between baleen whales and terrestrial herbivores; however, the protein catabolism and essential amino acid synthesis pathway in baleen whale microbiomes more closely resembled terrestrial carnivores, indicating that the diversity, structure, and function of the mammalian gut microbiome were mainly shaped by diet adaptation [10]. Moreover, differences in microbiome diversity and structure between different anatomical niches and species were found in a previous study. According to previous publications, Firmicutes, Bacteroidetes, Gammaproteobacteria, and Fusobacteria are the most abundant phyla in the midgut of the Indo-Pacific humpback dolphin—which inhabits the Pearl River Estuary of China—and short-finned pilot whales stranded in Hainan, China; however, a higher relative abundance of Fusobacteria was found in the stranded short-finned pilot whales than in the Indo-Pacific humpback dolphin [5,11]. For the gastrointestinal tract microbiome of the narrow-ridged finless porpoise, previous research found that the most abundant phylum was Gammaproteobacteria in the foregut, hindgut, and feces [12]. Both Indo-Pacific humpback dolphins and short-finned pilot whales belong to the family Delphinidae. In contrast, the narrow-ridged finless porpoise belongs to the family Phocoenidae, thus indicating that the gastrointestinal tract microbiome may be species-specific and related to evolutionary changes. In addition to the evolution-related changes and differences in species, habitations may also contribute to the differences in the microbiome of cetaceans, as different microbiome compositions were identified within captive and free-ranging common bottlenose dolphins, as well as common bottlenose dolphins living in different aquaria [7,13]. Due to complicated factors contributing to the dynamic changes in the microbiome of cetaceans, more individuals from different species should be included to better understand the microbiome in cetaceans.
Compared to the microbiome in the gastrointestinal tract, the microbiome inhabiting the skin may have a greater interaction with the mammals’ habitation/living environment [14]. Although marine mammals are constantly exposed to seawater microbiota [15,16,17], they also host their own unique microbial community, which is significantly different from the habitats on their skin [18,19,20]. The skin microbiome plays an important role in maintaining skin barrier function and the immune system while also preventing pathogen invasion. It is also important for wound healing [21,22,23,24]. A previous study demonstrated that Staphylococcus and Pseudomonadaceae are the most predominant bacteria in chronic human skin wounds [25,26,27]. Biofilms generated from Staphylococcus aureus suppress skin wound healing by inducing keratinocyte cell apoptosis or increasing cytokine expression in skin fibroblasts [28,29,30]. In contrast, Staphylococcus epidermidis was found to accelerate human skin wound healing via immune regulation [31,32], and both Staphylococcus aureus and Staphylococcus epidermidis protected the host from invasion by other pathogens through stimulation of the expression of antimicrobial peptides from human keratinocytes [33]. Previous studies have demonstrated the composition and structure of different species for the cetacean skin microbiome [19,20,28,29,34]. However, microbiome composition, structure, and function in cetacean skin wounds and their implications for cetacean health remain unclear.
Currently, more than 80 different species or subspecies of cetaceans have been found on earth; even though there is growing interest in research on the microbiome of cetaceans, little is known about the unique gastrointestinal tract microbiome or skin microbiome of cetaceans due to legal and ethical constraints. In this study, we investigated the microbiome in the gastrointestinal tract (foregut, midgut, hindgut) and skin wounds of a stranded Indo-Pacific finless porpoise (Neophocaena phocaenoides) from Eastern Guangdong waters, China. We aimed to investigate the diversity and composition of the Indo-Pacific intestinal microbial community. Additionally, we provide baseline information on the cetacean wound microbiome.
2. Materials and Methods
2.1. Sample Collection
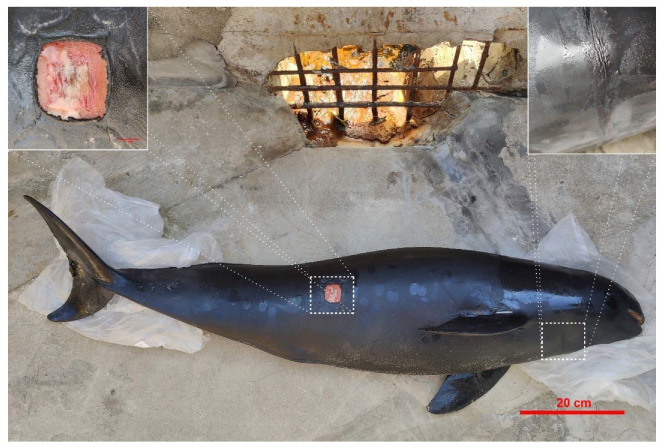
Microbiome samples were collected from an Indo-Pacific finless porpoise (Neophocaena phocaenoides) that was washed onto Nan’ao Island (Shantou, Guangdong, China) in December 2019. The carcass was examined for external lesions, and entanglement marks were found (Figure 1). The status of the carcass was evaluated using international guidelines and was shown to be in good health at the time of death. It was designated code 2 (i.e., it was a freshly dead carcass with a time of death less than 24 h as suggested by its normal appearance, the absence of swelling or discernable smell, little or no sloughing on the skin and little or no change in eye or mucus membranes). Then the carcass was weighed and measured, and age was estimated by body length [30]. For nutritional state evaluation, the blubber thickness of two anatomical locations was measured (sternal and caudoventral to the dorsal fin) [35]. Supplementary Table S1 shows age, sex, weight, and nutritional state information. Organs were examined visually, and no significant lesions were found.
Figure 1.
Image of the stranded Indo-Pacific finless porpoise.
For the intestinal tract, microbiome samples from the foregut, midgut, and hindgut contents were collected as described in a previous study [11,12]. Microbiome samples from skin wounds were collected with sterile swabs (Figure 1). All samples were collected in triplicate and stored at −80 °C until analysis. Sample collection was approved by the Regulations of the People’s Republic of China for the Implementation of Wild Aquatic Animal Protection and followed the guidelines and legal requirements in China.
2.2. Genomic DNA Extraction
Gastrointestinal tract samples were extracted with the MagPure Stool DNA KF kit B (Magen, Guangzhou, China) following the manufacturer’s instructions. For skin wound samples, collected samples were placed in TENS buffer (5 M sodium chloride, 10% SDS, Triton X-1000, Tris-HCl, EDTA), 10% SDS, and 20 mg/mL proteinase K and then incubated overnight at 55 °C. Proteins were removed by phenol/chloroform/isoamyl alcohol extractions, and DNA was precipitated with isopropanol. After washing in 75% ethanol twice, extracted DNA was resuspended in TE buffer. DNA from both gastrointestinal tract samples and skin wound samples was examined with a Qubit Fluorometer by using a Qubit dsDNA BR Assay kit (Invitrogen, Waltham, MA, USA), and the quality was further checked by gel electrophoresis.
2.3. Library Construction and Sequencing
Variable regions V3–V4 of the bacterial 16S rRNA gene were amplified with degenerate PCR primers 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Both forward and reverse primers were tagged with Illumina adapter, pad, and linker sequences. PCR enrichment was performed in a 50 μL reaction containing 30 ng template, fusion PCR primer, and PCR master mix. The PCR products were purified with Ampure XP beads and eluted in elution buffer. Libraries were qualified by an Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA, USA). The validated libraries were sequenced on an Illumina MiSeq platform (BGI, Shenzhen, China) following the standard pipelines of Illumina.
2.4. Sequencing Data Processing
After sequencing, adaptors were removed, low-quality raw reads were filtered, and then paired-end reads were merged into tags by FLASH (v1.2.11) [36]. The chimera sequences were removed after comparison with the Gold database using UCHIME (v4.2.40) [37], and tags were clustered into OTUs with a cutoff value of 97% using Usearch software (v7.0.1090) [38]. The representative OTU sequences were classified with Ribosomal Database Project (RDP) Classifier v.2.2 with a minimum confidence threshold of 0.6 [39].
2.5. Microbiome Diversity and Structure Comparison
Microbial diversity and community structure were assessed using alpha diversity estimated with observed species (Sob), Chao1, Shannon, and Simpson indices, while beta diversity was estimated using the Weight UniFrac index. The microbial diversity estimators were evaluated with MOTHUR and QIIME [40,41]. Principal coordinate analysis, based on weighted UniFrac distances, was performed to assess the bacterial taxonomic composition of the gastrointestinal tract microbiome with and without the skin wound microbiome. Analysis of similarities (ANOSIM) was conducted to estimate the potential difference in the microbiome community structure from different regions of the Indo-Pacific finless porpoise.
2.6. Statistical Analyses
Statistical analyses were carried out with R software (Vienna, Austria; http://www.R-project.org/ (accessed on 23 April 2021)). The Wilcoxon rank-sum test was applied for pairwise comparisons of the bacterial diversity between groups, and p < 0.05 was considered statistically significant. The Benjamin–Hochberg procedure was used to control the false discovery rate due to multiple testing.
3. Results
3.1. Subsection
In total, 818,397 clean reads were obtained from 12 samples, including microbiome samples from the foregut, midgut, hindgut, and skin wounds, with a mean of 68,199 sequences per sample (Supplementary Table S1). After binning the 673,809 tags, according to >97% sequence identity, 211 operational taxonomic units (OTUs) were obtained (Supplementary Figure S1B). According to the rarefaction curve, the current sequencing depth was sufficient for taxa identification (Supplementary Figure S1A).
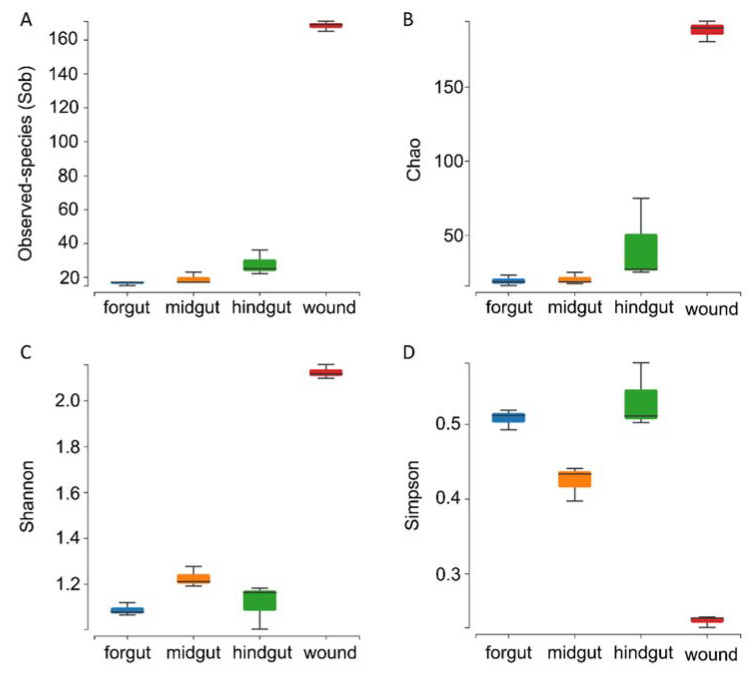
The observed species (Sobs) and Chao1 indices were applied for richness analysis for all four groups. As expected, the highest richness was found in skin wounds. No significant difference was found among the richness of the microbiomes of different gastrointestinal tract regions (p ≥ 0.1, Wilcoxon rank-sum test). However, the microbial richness index (both Sobs and Chao1) increased along the intestinal tract (foregut < midgut < hindgut) (Figure 2A,B).
Figure 2.
Alpha diversity, including observed species (Sob) (A), Chao1 (B), Shannon (C), and Simpson (D) indices, of the microbiome from the GI tract and skin wounds of an Indo-Pacific finless porpoise.
For microbial species evenness, even though the highest Shannon index and lowest Simpson index were found in the skin microbiome, the midgut microbiome had the highest Shannon index and lowest Simpson index. However, there was no significant difference among the microbiomes from different anatomical niches, which indicated no significant difference in the species evenness of the four groups (Figure 2C,D).
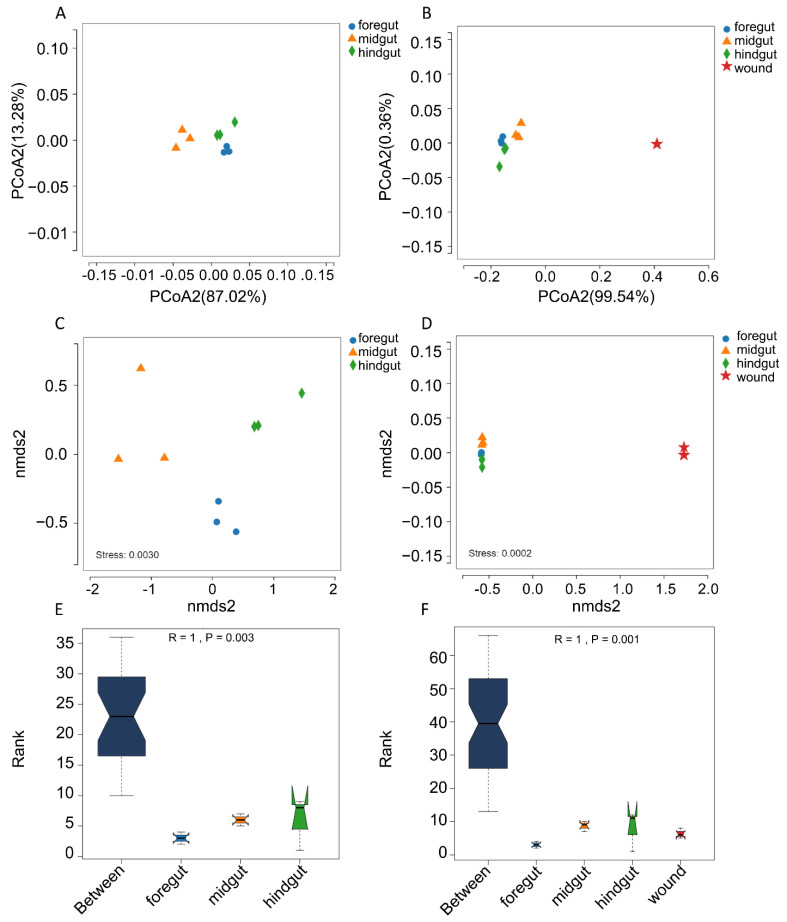
The overall microbiome composition of the microbiome from the gastrointestinal tract and skin wounds was compared using principal coordinate analysis, nonmetric multidimensional scaling analysis, and analysis of similarities (ANOSIM). For the microbiome from the gastrointestinal tract, subjects between the two groups were separated, indicating a different structure of microbiome composition (Figure 3A,C). For analysis of similarities, R = 1 and p = 0.003 were found among the gastrointestinal tract microbiomes, which indicated variation in microbiome composition in different regions of the Indo-Pacific finless porpoise (Figure 3E). All results from the principal coordinate analysis, nonmetric multidimensional scaling analysis, and ANOSIM showed significant differences in the microbial community structure between the gastrointestinal tract microbiome and the skin wound microbiome (Figure 3B,D,F). This was probably due to the different microenvironments between the gastrointestinal tract and the skin wounds.
Figure 3.
Principal coordinate analysis based on weighted UniFrac distances (A,B), nonmetric multidimensional scaling analysis (C,D) on the basis of the Bray–Curtis distance matrix, and analysis of similarities (ANOSIM) (E,F) of the microbiome from the GI tract (A,C,E) and wounds (B,D,F) of an Indo-Pacific finless porpoise.
3.2. Microbial Taxonomic Profiles
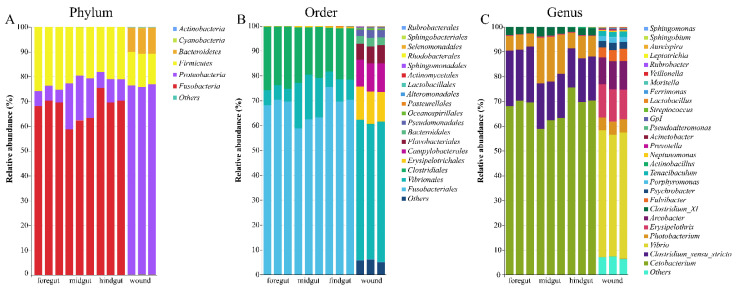
The relative abundances of the bacterial phyla, orders, and genera of the microbiome of the gastrointestinal tract and skin wounds are shown in Figure 4. Thirteen phyla, including Acidobacteria, Actinobacteria, Armatimonadetes, Bacteroidetes, Candidatus_Saccharibacteria, Chlamydiae, Cyanobacteria, Deinococcus thermmus, Firmicutes, Fusobacteria, Proteobacteria, and Verrucomicrobia, were found in all samples. For the microbiome of the gastrointestinal tract, the top three most abundant phyla were Fusobacteria (67.51% ± 5.10%), Firmicutes (22.00% ± 2.60%), and Proteobacteria (10.47% ± 5.49%). For the relative abundance in skin wounds, the dominant microbial phyla were Proteobacteria (76.11% ± 0.54%), Firmicutes (12.00% ± 2.60%), and Bacteroidetes (10.13% ± 0.49%) (Figure 3A). The microbiome in the gastrointestinal tract was predominantly composed of Fusobacteriales (58.80–75.49%), Clostridiales (17.66–25.61%), and Vibrionales (5.14–18.34%). In contrast, the microbiome in the skin wounds was predominantly composed of Vibrionales (54.24–56.34%), Erysipelotrichales (11.97–13.32%), Campylobacterales (10.84–11.52%), Flavobacteriales (6.32–7.30%), Bacteroidales (3.08–3.46%), and Pseudomonadales (2.65–3.18%) (Figure 3B). The relative abundance of the order Vibrionales in the midgut was higher than that in the foregut and hindgut, but no statistical significance was found (Wilcoxon rank-sum test, p > 0.05). Interestingly, Vibrionales increased and then decreased from the foregut through to the midgut and hindgut; the opposite occurred with Fusobacteria.
Figure 4.
Relative abundance of the microbiome from the GI tract and skin wounds of an Indo-Pacific finless porpoise at the phylum level (A), order level (B), and genus level (C).
At the genus level, 101 genera were detected from twelve samples, and different taxa dominated each sample. For gastrointestinal tract samples, Cetobacterium (67.51% ± 5.10%), Clostridium_sensu_stricto (18.69% ± 2.52%), Photobacterium (10.29% ± 5.52%), and Clostridium_XI (2.91% ± 0.56%) were the dominant genera. For the microbiome in skin wounds, Vibrio (50.36% ± 1.13%), Erysipelothrix (12.79% ± 0.73%), Arcobacter (11.22% ± 0.35%), Photobacterium (5.26% ± 0.09%), Fulvibacter (4.49% ± 0.43%), Psychrobacter (2.69% ± 0.21%), Porphyromonas (2.15% ± 0.20%), and Tenacibaculum (2.13% ± 0.09%) accounted for more than 90% of the genera. The microbiome in the skin wounds contained more genera than the microbiome from the gastrointestinal tract (Figure 4C).
3.3. Identification of Key Microbes in Each Anatomical Region
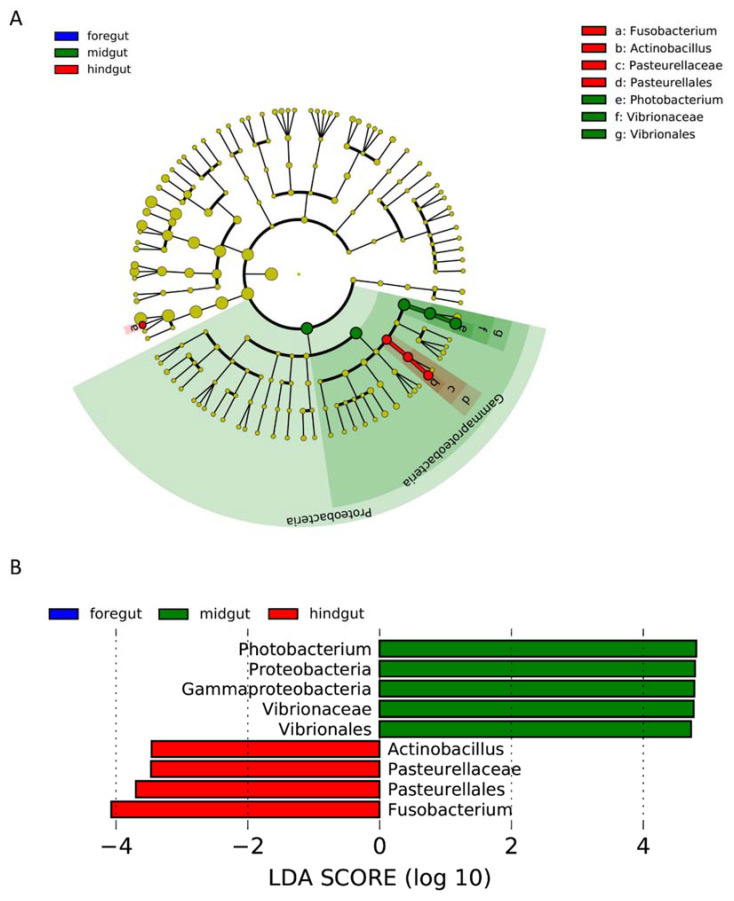
Linear discriminant analysis effect size (LEfSe) was used to compare bacterial abundances to identify the key bacteria (biomarkers) in different gastrointestinal tract regions. The key bacterial taxa driving the difference in the microbiome among different regions of the gastrointestinal tract are shown in Figure 5. For the microbiome from the gastrointestinal tract samples, key bacteria were only found in the midgut and hindgut but not the foregut (Figure 5). Within the midgut, one discriminant genus, Photobacterium, was identified. In addition, the high relative abundances of the phylum Proteobacteria, class Gammaproteobacteria, order Vibrionales, and family Vibrionaceae were significantly higher in the midgut than in the foregut and hindgut (Supplementary Figure S2). In contrast, two discriminant genera were identified in the hindgut microbiome, i.e., Actinobacillus and Pasteurellaceae (Figure 5). Meanwhile, the abundances of the order Pasteurellales and the family Pasteurellaceae were higher in the hindgut than in the foregut and midgut (Supplementary Figure S3).
Figure 5.
Linear discriminant analysis effect size (LEfSe) for key bacteria (biomarkers) identification for GI tract samples; results are shown in a cladogram (A) and histogram (B).
4. Discussion
The microbiome is not only a critical factor for host health maintenance and disease progression but also an indicator for health assessment. However, relatively few studies have investigated the composition and structure of the microbiome in the cetacean gastrointestinal tract and skin wounds. In this study, the microbiome from the foregut, midgut, hindgut, and skin wounds of an Indo-Pacific finless porpoise was studied for the first time. We found different microbiome compositions between different gastrointestinal tract regions and further identified potential pathogens from skin wounds.
4.1. Microbiome of Gastrointestinal Tract of Indo-Pacific Finless Porpoise
The microbiome’s bacterial richness, evenness, and potential function were studied and compared among the gastrointestinal tract and skin wound samples. The results show that the richness and evenness of the microbiomes from different regions of the gastrointestinal tract were similar. The alpha diversity of bacterial communities in the present study was similar to that in previous studies on the Indo-Pacific humpback dolphin [11]. However, a higher Chao index and observed richness were found in the microbiome from the gastrointestinal tract of Globicephala macrorhynchus. Similarly, the microbiome evenness results are consistent with previous studies on Indo-Pacific humpback dolphins [11], narrow-ridged finless porpoises [12], short-finned pilot whales [5], and common minke whales [42]. The bacterial richness of the gastrointestinal tract microbiome of the Indo-Pacific finless porpoise in this study is comparable to the findings in the Indo-Pacific humpback dolphin [11] and the narrow-ridged finless porpoise [12], but lower than the bacterial richness found in the common minke whale [42]. The difference in microbiome composition may be individual- or species-specific. As reported in a previous study, the difference in health status or human care may contribute to the difference in the gut microbiome of cetaceans [4,43]. Moreover, common minke whales belong to the Balaenopteridae family, while both the Indo-Pacific finless porpoise and the narrow-ridged finless porpoise belong to the Phocoenidae family [30]. All Indo-Pacific finless porpoises, Indo-Pacific humpback dolphins, and narrow-ridged finless porpoises are coastal species, while short-finned pilot whales and common minke whales are offshore species [30]. Thus, the difference in the bacterial richness of the gastrointestinal tract of different marine mammals may be due not only to individual or species differences but also to habitations. However, due to the limited sample size (a single Indo-Pacific finless porpoise) included in this study, further analysis with a bigger sample size is required to better understand the microbiome of the Indo-Pacific finless porpoise.
According to the PCoA, NMDS, and ANOSIM analysis results, the microbial community compositions of the microbiomes from the foregut, midgut, and hindgut were clustered and significantly different from each other (Figure 3). Previous studies also found that the intestinal microbiome composition of cetaceans, such as Indo-Pacific humpbacks, short-finned pilot whales, and common minke whales, is significantly different among different regions of the gastrointestinal tract [5,11,42]. However, the microbiome structure of the narrow-ridged finless porpoise is similar along the gastrointestinal tract. It cannot be separated by NMDS, even though the narrow-ridged finless porpoise belongs to the same family, Phocoenidae, as the Indo-Pacific finless porpoise [12].
Fusobacteria (67.51% ± 5.10%), Firmicutes (22.00% ± 2.60%), and Proteobacteria (10.47% ± 5.49%) were the predominant phyla in the microbiome of the foregut, midgut, and hindgut, which is highly similar to the composition of the microbiome of short-finned pilot whales [5]. Unlike the composition of the microbiome of the Indo-Pacific finless porpoise in our study, a previous study showed that the predominant phyla in the Indo-Pacific humpback dolphin included not only Fusobacteria (14.44%), Firmicutes (47.05%), and Proteobacteria (14.82%) but also the phylum Bacteroidetes (23.63%) [11]. It has been reported that the phyla Bacteroidetes and Firmicutes were predominant in the microbiome of baleen whales, which is similar to the composition of the microbiome of terrestrial herbivores [10]. According to a previous report, the phylum Bacteroidetes plays an important role in polysaccharide degradation and is regarded as an important marker of health status in humans [44,45]. Disease progression may reduce the abundance of the phylum Bacteroidetes and increase the abundance of the phylum Firmicutes [45]. Thus, the reduced abundance of the phylum Bacteroidetes may also be due to the progression of disease in the Indo-Pacific finless porpoise stranded in Shantou.
The genus Photobacterium is vastly distributed in environmental media and marine animals [46,47]. According to previous studies, Photobacterium may cause gastroenteritis in humans [48]. Photobacterium infection normally occurs in immune-compromised individuals and is induced by consuming seafood, such as fish [46], which is the major prey of cetaceans. The discriminant genus Photobacterium was identified from the midgut and found in all gastrointestinal tract regions of the Indo-Pacific finless porpoise, indicating sickness (gastroenteritis) of the stranded Indo-Pacific finless porpoise. The genus Photobacterium was also found in other cetaceans, such as striped dolphins [49] and short-finned pilot whales [5], suggesting that the genus Photobacterium may not be a species-specific microbe in cetaceans. A previous study identified the genus Photobacterium from L. vannamei obtained from the local aquaculture farm of Shantou [50]. Photobacterium was also identified within the blowhole, mouth, tongue, and stomach of striped dolphins [49]. These results suggest that cetaceans may become infected by prey and highlight the importance of monitoring the local environmental microbiome for cetacean health management and conservation [51].
The genera Haemophilus, Pasteurella, and Actinobacillus belong to the family Pasteurellaceae, and both Haemophilus and Pasteurella were found in marine mammals, whereas the genus Actinobacillus was seldom found in marine mammals [52,53]. However, the genus Actinobacillus is a species-specific microbe of the Indo-Pacific finless porpoise which is still not clear. Meanwhile, the genus Actinobacillus will induce multiple organ infections or septicemia in terrestrial mammals [54,55], but the possible pathological mechanism in cetaceans is still not clear considering the special immune system of cetaceans [56].
4.2. Microbiome on the Skin Wounds of the Indo-Pacific Finless Porpoise
Similar to the human skin microbiome, the phyla Proteobacteria, Firmicutes, Fusobacteria, Bacteroidetes, and Actinobacteria are commonly found on the skin of cetaceans [9,20,29,32,57]. The composition of the skin microbiome is different between terrestrial and marine mammals, such as humans and cetaceans. The phylum Actinobacteria accounts for approximately 52% of the human skin microbiome but only accounts for approximately 8% of the skin microbiome of captive bottlenose dolphins and killer whales [20,58]. For the phyla Proteobacteria and Bacteroidetes, the percentage in the human skin microbiome is much lower than that in free-ranging humpback whales (16.5% vs. 42.2–61.1%, 6.3% vs. 30–82.2%) [20,28,29,57]. In contrast, the proportion of the phylum Firmicutes is much higher in humans than in cetaceans (24.4% vs. 7.3%) [20,59]. The difference in the skin microbiome between terrestrial and marine mammals may be due to the difference in the microbiome of the two habitats [6,10,18].
In this study, we identified a higher percentage of the phylum Firmicutes and a lower abundance of the phylum Bacteroidetes in skin wounds of the Indo-Pacific finless porpoise. In addition, a lower abundance of the phylum Actinobacteria was found in skin wounds of Indo-Pacific finless porpoises than in captive bottlenose dolphins and killer whales [20].
The genera Vibrio (50.36% ± 1.13%) and Erysipelothrix (12.79% ± 0.73%), considered potential cetacean pathogens, were identified from skin wounds of the stranded Indo-Pacific finless porpoise. Vibrio owensii accounted for 90% of the genus Vibrio in the skin wounds; however, its pathogenicity to mammals is still unclear. Vibrio owensii induces acute hepatopancreatic necrosis disease in invertebrates, such as shrimp and lobsters, but the possible infection cycle and pathogenicity in mammals are still unknown [60,61]. The high abundance of the genus Vibrio may be due to the indirect contact of the Indo-Pacific finless porpoise with invertebrates in the local habitat. Erysipelothrix rhusiopathiae, which is infectious to both livestock and humans [59,62], was the dominant species in the genus Erysipelothrix on the skin wounds of the Indo-Pacific finless porpoise. A previous study found that severe symptoms, including pulmonary edema, organ failure, and serosanguineous effusion, may occur in cetaceans due to the acute inflammatory reactions induced by Erysipelothrix rhusiopathiae [63]. The genera Photobacterium, Tenacibaculum, and Psychrobacter may also induce severe wound infection in animals [64,65,66]; however, the possible pathology in marine mammals is still unclear.
Taken together, we investigated the microbiome of a single Indo-Pacific finless porpoise from three different intestinal regions and skin wounds. Our results reveal the different compositions of the microbiome in different intestinal regions of the stranded Indo-Pacific finless porpoise. Meanwhile, we found a remarkable difference in the microbiome of infected skin wounds of the Indo-Pacific finless porpoise compared with that in terrestrial mammals, which may be due to the vast pathogens originating on land and the unique microbes from the sea within the habitations of cetaceans. However, due to the limitations of using only one individual porpoise in this study, whether the unique composition of the microbiome of Indo-Pacific finless porpoises is species-specific still remains unclear. Importantly, we found the possible pathogenic genera Photobacterium, Actinobacillus, Vibrio, Erysipelothrix, Tenacibaculum, and Psychrobacter from different anatomical niches of the Indo-Pacific finless porpoise. However, the pathogenesis of these pathogens in cetaceans is still not clear. Additionally, several studies have shown that the gut microbiome in mammals can change over time following death, which could have influenced the emergence and distribution of potential pathogens [67,68]. Thus, further studies of the microbiome, possible pathogens, and potential pathogenesis of pathogens in marine mammals should be conducted to better understand the mutual interactions of microbes and marine mammals and changes in microbiome structure following death.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10071295/s1, Figure S1: Composition and abundance of microbiome from GI tract and skin wound. (A) The rarefaction curve during the sequencing. (B) Venn diagram of OTUs; Figure S2: Relative abundance of class Gammaproteobacteria, order Vibrionales, family Vibrionaceae, and genera Photobacterium of GI tract samples; Figure S3: Relative abundance of genera Photobacterium (A) and Actinobacillus (B) of GI tract samples. Table S1: Health status assessment by post-mortem examinations.
Author Contributions
Conceptualization, C.L., H.X., B.L. and W.L.; methodology, C.L., H.X. and B.L.; software, C.L., Y.Z. and B.L.; validation, Y.Z. and J.L.; formal analysis, H.X., Z.T. and B.L.; investigation, C.L. and H.X.; resources, E.S., B.L. and W.L.; data curation, P.L. and X.C.; writing—original draft preparation, C.L., H.X. and Y.S.; writing—review and editing, E.S., B.L. and W.L.; visualization, J.L. and L.Y.; supervision, B.L. and W.L.; project administration, P.L. and W.L.; funding acquisition, B.L. and W.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The animal study protocol was approved by the Animal Ethics Committee of Shantou University Medical College and declared exempt of our project (project: Possible causes of death of marine mammals, from 2022.01-2027.12, date of approval: 12 May 2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data were available from corresponding authors under reasonable request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
The authors gratefully acknowledge the financial support from the National Science Foundation for Young Scientists of China (Grant No. 42006105), the Key Program of Marine Economy Development (Six Marine Industries) Special Foundation of Department of Natural Resources of Guangdong Province (GDNRC[2022]48) and Ministry of Agriculture (Chinese White Dolphin Conservation Action), CNOOC Foundation, and the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (grant number GML2019ZD0606), and the Science and Technology Plan Projects of Guangdong Province (Grant No. 2021B1212050025, STKJ2021125).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cho I., Blaser M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundin O.H., Mendoza-Ladd A., Zeng M., Diaz-Arevalo D., Morales E., Fagan B.M., Ordonez J., Velez P., Antony N., McCallum R.W. The human jejunum has an endogenous microbiota that differs from those in the oral cavity and colon. BMC Microbiol. 2017;17:160. doi: 10.1186/s12866-017-1059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutierrez-Repiso C., Moreno-Indias I., Martin-Nunez G.M., Ho-Plagaro A., Rodriguez-Canete A., Gonzalo M., Garcia-Fuentes E., Tinahones F.J. Mucosa-associated microbiota in the jejunum of patients with morbid obesity: Alterations in states of insulin resistance and metformin treatment. Surg. Obes. Related Dis. Off. J. Am. Soc. Bariat. Surg. 2020;16:1575–1585. doi: 10.1016/j.soard.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Bai S., Zhang P., Zhang C., Du J., Du X., Zhu C., Liu J., Xie P., Li S. Comparative Study of the Gut Microbiota Among Four Different Marine Mammals in an Aquarium. Front. Microbiol. 2021;12:769012. doi: 10.3389/fmicb.2021.769012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai S., Zhang P., Lin M., Lin W., Yang Z., Li S. Microbial diversity and structure in the gastrointestinal tracts of two stranded short-finned pilot whales (Globicephala macrorhynchus) and a pygmy sperm whale (Kogia breviceps) Integr. Zool. 2021;16:324–335. doi: 10.1111/1749-4877.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry L.P., Bruijning M., Forsberg S.K.G., Ayroles J.F. The microbiome extends host evolutionary potential. Nat. Commun. 2021;12:5141. doi: 10.1038/s41467-021-25315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki A., Segawa T., Sawa S., Nishitani C., Ueda K., Itou T., Asahina K., Suzuki M. Comparison of the gut microbiota of captive common bottlenose dolphins Tursiops truncatus in three aquaria. J. Appl. Microbiol. 2019;126:31–39. doi: 10.1111/jam.14109. [DOI] [PubMed] [Google Scholar]
- 8.Salazar N., Valdes-Varela L., Gonzalez S., Gueimonde M., de Los Reyes-Gavilan C.G. Nutrition and the gut microbiome in the elderly. Gut Microb. 2017;8:82–97. doi: 10.1080/19490976.2016.1256525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo C.D., Weller D.W., Nelson K.E., Chivers S.J., Torralba M., Grimes D.J. Bacterial Species Identified on the Skin of Bottlenose Dolphins Off Southern California via Next Generation Sequencing Techniques. Microb. Ecol. 2018;75:303–309. doi: 10.1007/s00248-017-1071-2. [DOI] [PubMed] [Google Scholar]
- 10.Sanders J.G., Beichman A.C., Roman J., Scott J.J., Emerson D., McCarthy J.J., Girguis P.R. Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nat. Commun. 2015;6:8285. doi: 10.1038/ncomms9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan X., Li J., Cheng Z., Ao M., Tian R., McLaughlin R.W., Zheng J., Wang D. The intestinal microbiome of an Indo-Pacific humpback dolphin (Sousa chinensis) stranded near the Pearl River Estuary, China. Integr. Zool. 2021;16:287–299. doi: 10.1111/1749-4877.12477. [DOI] [PubMed] [Google Scholar]
- 12.Wan X.L., McLaughlin R.W., Zheng J.S., Hao Y.J., Fan F., Tian R.M., Wang D. Microbial communities in different regions of the gastrointestinal tract in East Asian finless porpoises (Neophocaena asiaeorientalis sunameri) Sci. Rep. 2018;8:14142. doi: 10.1038/s41598-018-32512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki A., Akuzawa K., Kogi K., Ueda K., Suzuki M. Captive environment influences the composition and diversity of fecal microbiota in Indo-Pacific bottlenose dolphins, Tursiops aduncus. Marine Mammal Sci. 2021;37:207–219. doi: 10.1111/mms.12736. [DOI] [Google Scholar]
- 14.Callewaert C., Ravard Helffer K., Lebaron P. Skin Microbiome and its Interplay with the Environment. Am. J. Clin. Dermatol. 2020;21:4–11. doi: 10.1007/s40257-020-00551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuang M., Sanganyado E., Li P., Liu W. Distribution of microbial communities in metal-contaminated nearshore sediment from Eastern Guangdong, China. Environ. Pollut. 2019;250:482–492. doi: 10.1016/j.envpol.2019.04.041. [DOI] [PubMed] [Google Scholar]
- 16.Hong H., Qiu J., Liang Y. Environmental factors influencing the distribution of total and fecal coliform bacteria in six water storage reservoirs in the Pearl River Delta Region, China. J. Environ. Sci. 2010;22:663–668. doi: 10.1016/S1001-0742(09)60160-1. [DOI] [PubMed] [Google Scholar]
- 17.Chen L., Tsui M.M.P., Lam J.C.W., Hu C., Wang Q., Zhou B., Lam P.K.S. Variation in microbial community structure in surface seawater from Pearl River Delta: Discerning the influencing factors. Sci. Total Environ. 2019;660:136–144. doi: 10.1016/j.scitotenv.2018.12.480. [DOI] [PubMed] [Google Scholar]
- 18.Bik E.M., Costello E.K., Switzer A.D., Callahan B.J., Holmes S.P., Wells R.S., Carlin K.P., Jensen E.D., Venn-Watson S., Relman D.A. Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat. Commun. 2016;7:10516. doi: 10.1038/ncomms10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apprill A., Miller C.A., Van Cise A.M., U’Ren J.M., Leslie M.S., Weber L., Baird R.W., Robbins J., Landry S., Bogomolni A., et al. Marine mammal skin microbiotas are influenced by host phylogeny. R. Soc. Open. Sci. 2020;7:192046. doi: 10.1098/rsos.192046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiarello M., Villeger S., Bouvier C., Auguet J.C., Bouvier T. Captive bottlenose dolphins and killer whales harbor a species-specific skin microbiota that varies among individuals. Sci. Rep. 2017;7:15269. doi: 10.1038/s41598-017-15220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khazaei T., Williams R.L., Bogatyrev S.R., Doyle J.C., Henry C.S., Ismagilov R.F. Metabolic multistability and hysteresis in a model aerobe-anaerobe microbiome community. Sci. Adv. 2020;6:eaba0353. doi: 10.1126/sciadv.aba0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomic-Canic M., Burgess J.L., O’Neill K.E., Strbo N., Pastar I. Skin Microbiota and its Interplay with Wound Healing. Am. J. Clin. Dermatol. 2020;21:36–43. doi: 10.1007/s40257-020-00536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson T.R., Gomez B.I., McIntyre M.K., Dubick M.A., Christy R.J., Nicholson S.E., Burmeister D.M. The Cutaneous Microbiome and Wounds: New Molecular Targets to Promote Wound Healing. Int. J. Mol. Sci. 2018;19:2699. doi: 10.3390/ijms19092699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y., Dunaway S., Champer J., Kim J., Alikhan A. Changing our microbiome: Probiotics in dermatology. Br. J. Dermatol. 2020;182:39–46. doi: 10.1111/bjd.18659. [DOI] [PubMed] [Google Scholar]
- 25.Gardner S.E., Hillis S.L., Heilmann K., Segre J.A., Grice E.A. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes. 2013;62:923–930. doi: 10.2337/db12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price L.B., Liu C.M., Frankel Y.M., Melendez J.H., Aziz M., Buchhagen J., Contente-Cuomo T., Engelthaler D.M., Keim P.S., Ravel J., et al. Macroscale spatial variation in chronic wound microbiota: A cross-sectional study. Wound Rep. Regen. 2011;19:80–88. doi: 10.1111/j.1524-475X.2010.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolcott R.D., Hanson J.D., Rees E.J., Koenig L.D., Phillips C.D., Wolcott R.A., Cox S.B., White J.S. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Rep. Regen. 2016;24:163–174. doi: 10.1111/wrr.12370. [DOI] [PubMed] [Google Scholar]
- 28.Bierlich K.C., Miller C., DeForce E., Friedlaender A.S., Johnston D.W., Apprill A. Temporal and Regional Variability in the Skin Microbiome of Humpback Whales along the Western Antarctic Peninsula. Appl. Environ. Microbiol. 2018;84:e02574-02517. doi: 10.1128/AEM.02574-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apprill A., Robbins J., Eren A.M., Pack A.A., Reveillaud J., Mattila D., Moore M., Niemeyer M., Moore K.M., Mincer T.J. Humpback whale populations share a core skin bacterial community: Towards a health index for marine mammals? PLoS ONE. 2014;9:e90785. doi: 10.1371/journal.pone.0090785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jefferson T.A., Webber M.A., Pitman R.L. Marine Mammals of the World a Comprehensive Guide to Their Identification. Elsevier; Amsterdam, The Netherlands: 2015. [Google Scholar]
- 31.Naik S., Bouladoux N., Linehan J.L., Han S.J., Harrison O.J., Wilhelm C., Conlan S., Himmelfarb S., Byrd A.L., Deming C., et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linehan J.L., Harrison O.J., Han S.J., Byrd A.L., Vujkovic-Cvijin I., Villarino A.V., Sen S.K., Shaik J., Smelkinson M., Tamoutounour S., et al. Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell. 2018;172:784–796.e718. doi: 10.1016/j.cell.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christaki E., Giamarellos-Bourboulis E.J. The complex pathogenesis of bacteremia: From antimicrobial clearance mechanisms to the genetic background of the host. Virulence. 2014;5:57–65. doi: 10.4161/viru.26514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Cise A.M., Wade P.R., Goertz C.E.C., Burek-Huntington K., Parsons K.M., Clauss T., Hobbs R.C., Apprill A. Skin microbiome of beluga whales: Spatial, temporal, and health-related dynamics. Anim. Microb. 2020;2:39. doi: 10.1186/s42523-020-00057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siebert U., Wünschmann A., Weiss R., Frank H., Benke H., Frese K. Post-mortem Findings in Harbour Porpoises (Phocoena phocoena) from the German North and Baltic Seas. J. Comp. Pathol. 2001;124:102–114. doi: 10.1053/jcpa.2000.0436. [DOI] [PubMed] [Google Scholar]
- 36.Magoc T., Salzberg S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian J., Du J., Lu Z., Han J., Wang Z., Li D., Guan X., Wang Z. Distribution of microbiota across different intestinal tract segments of a stranded dwarf minke whale, Balaenoptera acutorostrata. Microbiologyopen. 2020;9:e1108. doi: 10.1002/mbo3.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You L., Ying C., Liu K., Zhang X., Lin D., Yin D., Zhang J., Xu P. Changes in the fecal microbiome of the Yangtze finless porpoise during a short-term therapeutic treatment. Open. Life Sci. 2020;15:296–310. doi: 10.1515/biol-2020-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cockburn D.W., Koropatkin N.M. Polysaccharide Degradation by the Intestinal Microbiota and Its Influence on Human Health and Disease. J. Mol. Biol. 2016;428:3230–3252. doi: 10.1016/j.jmb.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 45.Fang X., Wang X., Yang S., Meng F., Wang X., Wei H., Chen T. Evaluation of the Microbial Diversity in Amyotrophic Lateral Sclerosis Using High-Throughput Sequencing. Front. Microbiol. 2016;7:1479. doi: 10.3389/fmicb.2016.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Labella A.M., Arahal D.R., Castro D., Lemos M.L., Borrego J.J. Revisiting the genus Photobacterium: Taxonomy, ecology and pathogenesis. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2017;20:1–10. doi: 10.2436/20.1501.01.280. [DOI] [PubMed] [Google Scholar]
- 47.Labella A.M., Castro M.D., Manchado M., Lucena T., Arahal D.R., Borrego J.J. Photobacterium malacitanum sp. nov., and Photobacterium andalusiense sp. nov., two new bacteria isolated from diseased farmed fish in Southern Spain. Syst. Appl. Microbiol. 2018;41:444–451. doi: 10.1016/j.syapm.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Altekruse S.F., Bishop R.D., Baldy L.M., Thompson S.G., Wilson S.A., Ray B.J., Griffin P.M. Vibrio gastroenteritis in the US Gulf of Mexico region: The role of raw oysters. Epidemiol. Infect. 2000;124:489–495. doi: 10.1017/S0950268899003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Godoy-Vitorino F., Rodriguez-Hilario A., Alves A.L., Gonçalves F., Cabrera-Colon B., Mesquita C.S., Soares-Castro P., Ferreira M., Marçalo A., Vingada J., et al. The microbiome of a striped dolphin (Stenella coeruleoalba) stranded in Portugal. Res. Microbiol. 2017;168:85–93. doi: 10.1016/j.resmic.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X., Sun Z., Zhang X., Zhang M., Li S. Hemolymph Microbiomes of Three Aquatic Invertebrates as Revealed by a New Cell Extraction Method. Appl. Environ. Microbiol. 2018;84:e02824-17. doi: 10.1128/AEM.02824-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee R.J., Rangdale R.E. 22-Tracing pathogens in molluscan shellfish production chains. In: Brul S., Fratamico P.M., McMeekin T.A., editors. Tracing Pathogens in the Food Chain. Woodhead Publishing; Sawston, UK: 2011. pp. 527–547. [DOI] [Google Scholar]
- 52.Acevedo-Whitehouse K., Rocha-Gosselin A., Gendron D. A novel non-invasive tool for disease surveillance of free-ranging whales and its relevance to conservation programs. Anim. Conserv. 2010;13:217–225. doi: 10.1111/j.1469-1795.2009.00326.x. [DOI] [Google Scholar]
- 53.Hansen M.J., Bertelsen M.F., Christensen H., Bisgaard M., Bojesen A.M. Occurrence of Pasteurellaceae bacteria in the oral cavity of selected marine mammal species. J. Zoo Wild. Med. 2012;43:828–835. doi: 10.1638/2011-0264R1.1. [DOI] [PubMed] [Google Scholar]
- 54.Tobias S., Lee J.H., Tomford J.W. Rare Actinobacillus infection of the cavernous sinus causing painful ophthalmoplegia: Case report. Neurosurgery. 2002;51:807–809. doi: 10.1097/00006123-200209000-00037. discussion 809–810. [DOI] [PubMed] [Google Scholar]
- 55.Liven E., Larsen H.J., Lium B. Infection with actinobacillus suis in pigs. Acta Veter. Scand. 1978;19:313–315. doi: 10.1186/BF03547636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beineke A., Siebert U., Baumgartner W. The immune system of marine mammals. Part 1: Immune response, cytokines and immunotoxicity. Tieraerztliche Prax. Ausg. Kleintiere Heimtiere. 2006;34:118–126. [Google Scholar]
- 57.Apprill A., Mooney T.A., Lyman E., Stimpert A.K., Rappe M.S. Humpback whales harbour a combination of specific and variable skin bacteria. Environ. Microbiol. Rep. 2011;3:223–232. doi: 10.1111/j.1758-2229.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 58.Grice E.A., Kong H.H., Conlan S., Deming C.B., Davis J., Young A.C., Program N.C.S., Bouffard G.G., Blakesley R.W., Murray P.R., et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wattrang E., Eriksson H., Jinnerot T., Persson M., Bagge E., Söderlund R., Naghizadeh M., Dalgaard T.S. Immune responses upon experimental Erysipelothrix rhusiopathiae infection of naïve and vaccinated chickens. Veter. Res. 2020;51:114. doi: 10.1186/s13567-020-00830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu L., Xiao J., Zhang M., Zhu W., Xia X., Dai X., Pan Y., Yan S., Wang Y. A Vibrio owensii strain as the causative agent of AHPND in cultured shrimp, Litopenaeus vannamei. J. Invert. Pathol. 2018;153:156–164. doi: 10.1016/j.jip.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Goulden E.F., Hall M.R., Bourne D.G., Pereg L.L., Høj L. Pathogenicity and infection cycle of Vibrio owensii in larviculture of the ornate spiny lobster (Panulirus ornatus) Appl. Environ. Microbiol. 2012;78:2841–2849. doi: 10.1128/AEM.07274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reboli A.C., Farrar W.E. Erysipelothrix rhusiopathiae: An occupational pathogen. Clin. Microbiol. Rev. 1989;2:354–359. doi: 10.1128/CMR.2.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ceccolini M.E., Wessels M., Macgregor S.K., Deaville R., Perkins M., Jepson P.D., John S.K., Guthrie A. Systemic Erysipelothrix rhusiopathiae in seven free-ranging delphinids stranded in England and Wales. Dis. Aquat. Organ. 2021;145:173–184. doi: 10.3354/dao03609. [DOI] [PubMed] [Google Scholar]
- 64.Pogoreutz C., Gore M.A., Perna G., Millar C., Nestler R., Ormond R.F., Clarke C.R., Voolstra C.R. Similar bacterial communities on healthy and injured skin of black tip reef sharks. Anim. Microb. 2019;1:9. doi: 10.1186/s42523-019-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faílde L.D., Losada A.P., Bermúdez R., Santos Y., Quiroga M.I. Evaluation of immune response in turbot (Psetta maxima L.) tenacibaculosis: Haematological and immunohistochemical studies. Microb. Pathog. 2014;76:1–9. doi: 10.1016/j.micpath.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 66.Hundenborn J., Thurig S., Kommerell M., Haag H., Nolte O. Severe Wound Infection with Photobacterium damselae ssp. damselae and Vibrio harveyi, following a Laceration Injury in Marine Environment: A Case Report and Review of the Literature. Case Rep. Med. 2013;2013:610632. doi: 10.1155/2013/610632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeBruyn J.M., Hauther K.A. Postmortem succession of gut microbial communities in deceased human subjects. PeerJ. 2017;5:e3437. doi: 10.7717/peerj.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hauther K.A., Cobaugh K.L., Jantz L.M., Sparer T.E., DeBruyn J.M. Estimating Time Since Death from Postmortem Human Gut Microbial Communities. J. Forensic Sci. 2015;60:1234–1240. doi: 10.1111/1556-4029.12828. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were available from corresponding authors under reasonable request.