Abstract
The conversion of β-glutamate to β-glutamine by archaeal and bacterial glutamine synthetase (GS) enzymes has been examined. The GS from Methanohalophilus portucalensis (which was partially purified) is capable of catalyzing the amidation of this substrate with a rate sevenfold less than the rate obtained with α-glutamate. Recombinant GS from the archaea Methanococcus jannaschii and Archaeoglobus fulgidus were considerably more selective for α-glutamate than β-glutamate as a substrate. All the archaeal enzymes were much less selective than the two bacterial GS (from Escherichia coli and Bacillus subtilis), whose specific activities towards β-glutamate were much smaller than rates with the α-isomer. These results are discussed in light of the observation that β-glutamate is accumulated as an osmolyte in many archaea while β-glutamine (produced by glutamine synthetase) is used as an osmolyte only in M. portucalensis.
The glutamate-to-glutamine conversion by glutamine synthetase (GS) provides the nitrogen donor for the first step in the biosynthesis of many amino acids, purines, pyrimidines, and amino sugars. In most bacteria and archaea GS is a multimeric enzyme consisting of 12 identical subunits, each with a mass of 50 to 55 kDa (12). Modes of regulation further classify the enzyme as GSI-α (not subject to adenylylation [4]) or GSI-β activities (as exemplified by the Escherichia coli GS [12, 23]). GS appears to have another unusual role in one particular archaeon. The halophilic methanogen Methanohalophilus portucalensis uses β-glutamine as an osmolyte when grown in media containing >2 M NaCl (10, 17, 19). Synthesis of β-glutamine in this organism has been suggested to occur via GS action on β-glutamate (16). M. portucalensis is the only organism known to date to accumulate this zwitterionic β-amino acid, although β-glutamate is a substrate for sheep brain and rat liver GS (14). The ability of GS to use β-glutamate as a substrate may be a characteristic of other archaea or of a wider range of bacteria, or it may be a property only associated with the halophilic methanogen. In order to examine this, GS from M. portucalensis was partially purified and its activity toward both α- and β-glutamate was examined. The selectivity of α- over β-glutamate is much lower for the M. portucalensis GS than for the GS from the other archaeal (cloned and purified enzymes from Methanococcus jannaschii and Archaeoglobus fulgidus) and bacterial (E. coli and Bacillus subtilis) enzyme activities examined. This enhanced relative activity toward β-glutamate is consistent with an involvement of the enzyme in the accumulation of β-glutamine in high-NaCl environments in M. portucalensis. These results are discussed in terms of the role of β-glutamate and β-glutamine as osmolytes in archaea.
MATERIALS AND METHODS
Chemicals.
Tris-HCl, MgCl2, β-mercaptoethanol, α-glutamate, β-glutamate, and NH4Cl were obtained from Sigma. Q-Sepharose Fast Flow anion exchange resin was purchased from Pharmacia.
Enzymes.
The A. fulgidus VC-16 GS structural gene was cloned and overexpressed in E. coli, and the enzyme was purified to near homogeneity by a procedure that will be described elsewhere. Similarly, the M. jannaschii glnA gene was cloned and overexpressed in E. coli, and the protein purified to homogeneity (to be reported elsewhere). GS from E. coli was purchased from Sigma, and the enzyme from B. subtilis was prepared as described previously (20).
GS assays.
Two assays for GS activity (1) were used, one measuring synthetic activity (conversion of α-glutamate to γ-glutamylhydroxamate) and the other measuring transferase activity (conversion of α-glutamine to γ-glutamylhydroxamate). The synthetic GS assay mix (0.5 ml) contained l-α-glutamate or β-glutamate (varying from 30 to 350 mM), MgCl2 (55 mM), hydroxylamine-HCl (46 mM), and imidazole (92 mM), pH 7.0. The reaction was initiated by the addition of ATP at a final concentration of 20 mM. Blanks were run in parallel, substituting water for ATP. The transferase assay mixture (0.5 ml) contained l-α-glutamine (25 mM), MnCl2 (2.5 mM), hydroxylamine-HCl (50 mM), K2HPO4 (25 mM) or Na2HAsO4, and imidazole (25 mM), pH 7.0. The GS exchange reaction was initiated with the addition of ADP to a final concentration of 4 mM (omitted in the blanks). The use of Mg2+ in the synthesis reaction and Mn2+ in the glutamyl transferase assays was based on the relative effectiveness of these two cations in assays of other GS enzymes (7) where activity in the biosynthetic assay was usually higher with Mg2+, while the glutamyl transfer activity was optimal with Mn2+. All reactions were stopped with 1 ml of an acidic FeCl3 solution (55 g of FeCl3, 20 g of trichloroacetic acid, and 21 ml of HCl per liter of solution). Absorbance of the γ-glutamylhydroxamate–iron complex was measured at 540 nm (1 μmol of complex = 0.533 units of absorbance [see reference 1]). Specific activities were calculated by normalizing activity to protein concentration determined via the Bradford assay (3) with bovine serum albumin as the standard.
Preparation of M. portucalensis protein extracts.
Cell pellets of M. portucalensis (grown in the presence of methanol as the substrate for methane generation in medium containing 12% NaCl as described previously [10, 19]) were resuspended in buffer (1 ml of buffer per g of cell pellet) containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 1 mM β-mercaptoethanol (buffer A) along with 1% Triton X-100. The addition of Triton X-100 led to a marked increase in the activity of crude extracts, suggesting that either the enzyme is membrane associated as has been shown for GS from Sphagnum fallax (8) or that hydrophobic cell components inhibit the enzyme upon lysis. Protease inhibitors phenylmethylsulfonyl fluoride (50 μM), Pefabloc (0.1 mM), dithiothreitol (1 mM), and pepstatin and leupeptin (each at 1 μg/ml) were added to the resolubilized pellet. The cells were then ruptured via sonication, using six 30-s intervals separated by 1 min. The protein extract was subsequently dialyzed versus 100-fold excess of buffer A containing protease inhibitors. Partial purification of the M. portucalensis GS was achieved with three chromatographic steps: passage through a Q-Sepharose Fast Flow (QFF) anion exchange column (10 ml of resin per g of cell pellet prepared in formic acid and washed with buffer A), a hydroxyapatite column (5 ml of resin per g of cell pellet, equilibrated in buffer A with the addition of 0.15 M KH2PO4), and a Blue A Dyematrex column (equilibrated in buffer A and eluted with increasing concentrations of α-glutamate). Gel filtration on Sephacryl S-300-HR was used to determine the native molecular mass of the GS. Molecular mass standards included E. coli GS (600 kDa), apoferritin (443 kDa), and ATCase holoenzyme (300 kDa) and catalytic trimer (100 kDa). Protein fractions were analyzed by the GS transferase assay and sodium dodecyl sulfate-polyacrylamide gel electrophoresis. For dilute protein samples, gels were stained in 0.1% Coomassie brilliant blue with the addition of 2% phosphoric acid and 6% ammonium sulfate, which increases the sensitivity of the Coomassie stain (13).
RESULTS
M. portucalensis action on α- and β-glutamate.
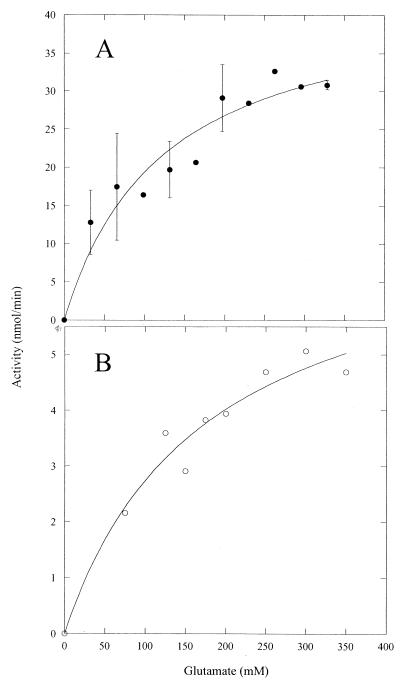
The activity of GS in crude protein extracts from M. portucalensis was measured in the transferase (α-glutamine to γ-glutamylhydroxamate) as well as synthesis (α-glutamate to γ-glutamylhydroxamate) assays (crude extracts were used prior to any purification to avoid differences in the relative amount of GS in the protein extract). In contrast to most other GS examined to date, the specific activity of M. portucalensis GS in the biosynthetic assay (0.061 μmol min−1 mg−1) was about fourfold higher than the activity in the glutamyl transferase reaction (0.015 μmol min−1 mg−1). Enzyme activity was moderately high toward both α-glutamate and β-glutamate at 37°C under conditions with 55 mM Mg2+ and 20 mM ATP. Specific activities of GS in these crude protein extracts were increased with 0.5 M potassium acetate in the buffer, so this was routinely included in assays (the intracellular concentration of K+ in M. portucalensis is 0.6 to 1.1 M, depending on external NaCl [10]). That the product produced when incubating β-glutamate with ATP and crude enzyme was indeed β-glutamine was checked by high-performance liquid chromatography analysis (10) of one of the reaction mixtures in which hydroxylamine was replaced by ammonia. The dependence of activity on glutamate concentrations is shown in Fig. 1. The ratio of specific activity for β-glutamate compared to α-glutamate ranged from 0.12 to 0.17. The apparent mean Km ± standard deviation for α-glutamate (104 ± 20 mM using all the data shown in Fig. 1A or 96 ± 36 mM as the mean of three separate kinetic experiments) was higher than the α-glutamate Km of other archaeal and bacterial GS (Table 1). The apparent Km extrapolated for β-glutamate, 175 ± 50 mM, was considerably higher (Fig. 1B). The intracellular concentration of α-glutamate for M. portucalensis is between 0.15 and 0.20 M, which is slightly above the GS Km for this substrate. In contrast to α-glutamate, intracellular β-glutamate levels are less than 0.01 M in this organism (10). The M. portucalensis GS had a Km for ATP, 6 mM, that was comparable to values for other GS enzymes (Table 1).
FIG. 1.
Dependence of M. portucalensis GS activity in crude protein extracts on α-glutamate (A) and β-glutamate (B) concentration. Assays were conducted at 37°C as described in Materials and Methods. The assays with α-glutamate as the substrate were carried out with two different preparations of protein, and the error bars show the ranges of activity for several concentrations.
TABLE 1.
Comparison of biosynthesis assay kinetic parameters for glutamine synthetase enzymes from different sources
| Enzyme source | Organism characteristic(s) | Cation |
Km (mM)
|
Vβ-glutamate/Vα-glutamate at substrate concn ofa:
|
||||
|---|---|---|---|---|---|---|---|---|
| α-Glu | β-Glu | ATP | 100 mM | 200 mM | ||||
| Archaea | ||||||||
| Methanohalophilus portucalensis | Mesophile; halophile | Mg2+ | 104 ± 20 | 175 ± 50 | 6 ± 2 | 0.14 | 0.17 | |
| Methanococcus jannaschii | Hyperthermophile; halotolerant | Mg2+ | 58 ± 8b | >200b | 0.04 | 0.016 | ||
| Archaeoglobus fulgidus | Hyperthermophile; halotolerant | Mn2+ | 3.0c | —d | 0.56c | 0.08 | 0.052 | |
| Pyrococcus KOD1 | Hyperthermophile; halotolerant | Mg2+ | 23.5e | 28.0e | ||||
| Halobacterium salinarium | Halophile | Mg2+ | 49f | 0.59f | ||||
| Bacteria | ||||||||
| Bacillus subtilis | Mesophile | Mn2+ | 0.5g | >100 | 0.4g | 0.003 | ||
| Bacillus licheniformis | Mesophile | Mn2+ | 3.6h | 0.9h | ||||
| Escherichia coli | Mesophile | Mg2+ | 2.4i | 0.68i | 4 × 10−6 | |||
| Nitrobacter agilis | Mesophile | Mg2+ | 6.3j | |||||
| Eukaryotes | ||||||||
| Sheep brain | Mg2+ | 0.18 (NH3)k | ||||||
| 0.46 (NH2OH)k | ||||||||
| Rat liver | Mg2+ | 0.18 (NH3)k | ||||||
| 0.28 (NH2OH)k | ||||||||
Ratio of GS specific activity toward β-glutamate compared to that toward α-glutamate for each substrate at 100 and 200 mM.
Determined at 60°C.
H. Schreier, unpublished results.
Observed substrate inhibition at >75 mM β-glutamate prevented any estimate of Km.
Rahman et al. (15).
Manitz and Holldorf (11).
Schreier et al. (20); Wedler et al. (22).
Donohoe et al. (6).
Woolfolk et al. (23).
Kumar and Nicholas (9).
Pruisner and Stadtman (14).
Partial purification and characterization of M. portucalensis GS.
To examine in greater detail the ability of M. portucalensis GS to utilize β-glutamate, the enzyme was purified from crude extracts via a series of chromatographic steps. The enzyme did not bind to either QFF anion exchange or hydroxyapatite resins. However, elution of the protein extract through these columns resulted in the removal of more than 90% of the cellular protein (the specific activity increased from 0.061 μmol min−1 mg−1 in crude extracts to 0.11 μmol min−1 mg−1 after the QFF column and to 3.2 μmol min−1 mg−1 after chromatography on hydroxyapatite). After passage through the hydroxyapatite column, the activity of the M. portucalensis GS was no longer activated by K+. The M. portucalensis GS bound tightly to a Blue A affinity column, and while 10 mM ADP was not effective in eluting the protein, the enzyme could be eluted with 1.0 M α-glutamate. Through this step the yield was 0.015% of the original protein and a 120-fold increase in specific activity. Assuming a subunit molecular mass of 50 kDa, the M. portucalensis GS obtained from the Blue A column would represent 5 to 10% of the total protein at this stage in the purification. Kinetic analyses showed that the relative rate of this enzyme preparation toward β-glutamate compared to α-glutamate remained 0.14. The specific activity of this material at 37°C with 164 mM α-glutamate, 20 mM ATP, 46 mM hydroxylamine, and 55 mM Mg2+ in imidzaole, pH 7.0, was 7.3 μmol min−1 mg−1. This indicates a specific activity of the pure protein of 70 to 140 μmol min−1 mg−1. Sephacryl S-300-HR gel filtration chromatography indicated that the native molecular mass of M. portucalensis GS was approximately 550 kDa, which is consistent with the dodecameric structure found for other archaeal and bacterial GS enzymes. Subjecting the partially purified preparations to repeated chromatographic steps (most notably a second passage through the Blue A column) improved the extent of purification but decreased the yield of material. Eventually small amounts (<20 μg) of relatively pure protein from 5 g of cell pellet could be obtained. Sodium dodecyl sulfate-polyacrylamide gels showed a single protein band with a molecular mass of 50 kDa. The specific activity of this material was determined to be 150 ± 50 μmol min−1 mg−1, an unusually high value compared to GS enzymes from other sources, but consistent with the values estimated for the material eluted through a single Blue A Dyematrex column.
Glycine and alanine, feedback inhibitors of the GS biosynthesis assay from several sources (5, 12), were tested for their ability to inhibit the M. portucalensis GS. Both amino acids were effective in reducing the M. portucalensis enzyme activity when present at a concentration of 10 mM in the synthetic assay. Alanine inhibited 26% and glycine inhibited 37% of the M. portucalensis GS activity (conditions included 164 mM α-glutamate and 37°C). For comparison, 5 mM alanine or glycine inhibited the archaeal GS enzymes from Methanobacterium ivanovi (2) and A. fulgidus (H. Schreier, unpublished results) to a similar extent. The M. portucalensis GS was also completely inhibited by the transition state analog methionine sulfoximine (1 mM), consistent with the amidation reaction proceeding via a glutamylphosphate intermediate as it does for the well-characterized GS from E. coli and B. subtilis (12). The combination of subunit size, native molecular mass, and feedback inhibition suggests that the M. portucalensis GS is a GSI-type enzyme.
β-Glutamate as a substrate for other archaeal GS.
The unusual characteristics of the M. portucalensis GS, reduced selectivity of α- over β-glutamate and higher activity in the synthesis versus transferase assay, may be common to GS from other archaea. Alternatively, the GS from this halophilic methanogen could be unique since, to date, only M. portucalensis strains have been shown to synthesize and accumulate β-glutamine in response to osmotic stress. With this in mind, two other archaeal GS were examined for their ability to utilize the two glutamate isomers. Both M. jannaschii and A. fulgidus are thermophiles; neither one accumulates β-glutamine as an osmolyte, although M. jannaschii accumulates β-glutamate as its major compatible solute (18).
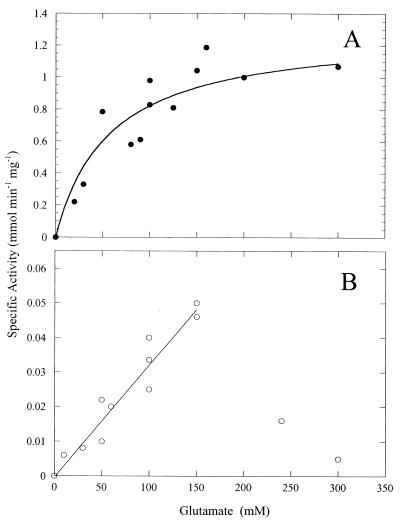
The M. jannaschii GS displays characteristics similar to most GSI class enzymes previously studied: it is a dodecamer of 51-kDa subunits, is feedback inhibited by glycine and alanine, and is inhibited by low concentrations of methionine sulfoximine (P. Robinson and M. F. Roberts, unpublished results). Kinetic parameters for this recombinant enzyme were measured at 37 and 50°C. With α-glutamine as the substrate for the hydroxylamine transferase reaction, the Km was 23 ± 1 mM and the Vmax was estimated to be 3.5 ± 0.1 μmol min−1 mg−1 at 37°C. At 50°C Vmax increased to 26.7 ± 4.5 μmol min−1 mg−1, while the Km increased by about a factor of two (∼40 mM). Like most GS enzymes, the biosynthetic activity of this GS using α-glutamate and Mg2+ as the optimal cation was considerably lower than that in the transferase reaction (with Mn2+ as the optimal metal ion). At 60°C, M. jannaschii GS exhibited a Km for α-glutamate of 58 ± 8 mM and a Vmax of 1.3 ± 0.2 μmol min−1 mg−1 (Fig. 2A). Low biosynthetic activity was also observed for recombinant Pyrococcus sp. GS (15). The dependence of M. jannaschii GS activity on β-glutamate concentration was linear up to 150 mM, implying a high Km. At higher concentrations, the activity of this GS toward β-glutamate decreased so that a Km could not be determined (Fig. 2B). This substrate inhibition was significant since at 300 mM β-glutamate very little GS activity was detectable (while activity with α-glutamate was easily measured). If the specific activity of the M. jannaschii GS toward β-glutamate is compared to that toward α-glutamate at different glutamate concentrations, one sees that at a maximum it is 0.04. Therefore, for this archaeal GS, β-glutamate is a much poorer substrate than α-glutamate.
FIG. 2.
Dependence of M. jannaschii GS activity at 60°C on the concentration of α-glutamate (A) and β-glutamate (B). Note the steep decrease in activity for β-glutamate concentrations above 200 mM.
Another way to assess the binding affinities of the two glutamate isomers to GS is to examine the inhibitory effect of each on the α-glutamine transferase reaction. At 50 mM α-glutamine (50°C), there is a 50% decrease in GS activity in the presence of 100 mM α-glutamate. This is consistent with a Ki of 47 mM (assuming competitive inhibition by α-glutamate in the transferase assay), not too far off from the Km of 58±8 mM in the forward direction assay. β-Glutamate at 100 mM had no effect on the GS transferase at 50°C with 50 or 25 mM α-glutamine. With 10 mM α-glutamine as substrate, there was detectable inhibition by 100 mM β-glutamate consistent with a Ki of 300 mM (assuming competitive inhibition under these conditions). Higher concentrations of β-glutamate were more inhibitory than expected, again consistent with the observed substrate inhibition seen with high concentrations of β-glutamate as the substrate in the forward direction assay.
A. fulgidus grows optimally at 83°C with 2% NaCl. The GS from this organism also catalyzed the conversion of β-glutamate to β-glutamine. The specific activity with β-glutamate (100 mM) as the substrate was 0.080 of the specific activity for α-glutamate (100 mM). For 200 mM β-glutamate, GS specific activity was 0.052 that toward α-glutamate. No GS activity toward β-glutamate was detected at β-glutamate concentrations above 300 mM; enzyme activity toward α-glutamate was still measurable at comparable α-glutamate concentrations (although somewhat inhibited).
β-Glutamate as a substrate for bacterial GS.
β-Glutamate was also examined as a substrate for GS from two bacteria, B. subtilis and E. coli (Table 1). The B. subtilis GS exhibited a 300-fold lower specific activity when β-glutamate was present as the substrate at 200 mM than it did with α-glutamate as the substrate (1.2 nmol min−1 mg−1 versus 370 nmol min−1 mg−1). GS specific activity increased linearly with increasing β-glutamate, implying a very high Km for that β-amino acid. The E. coli GS had very low activity toward β-glutamate; the relative rate with 200 mM substrate was 10−6 that obtained using α-glutamate.
DISCUSSION
β-Glutamine is synthesized and accumulated as an osmolyte in M. portucalensis, an organism that is among the most halophilic of the methanogenic archaea. Previous work using 13C label incorporation (16) suggested that biosynthesis of β-glutamine from α-glutamate could arise either (i) via an activity that generated β-glutamate (aminomutase or other activity) and then conversion of that β-amino acid to β-glutamine via GS or (ii) by an aminomutase activity on α-glutamine to convert it to β-glutamine (in this case β-glutamate would be a poor substrate for the M. portucalensis GS). While bacterial glutamate mutase enzymes are known, these convert glutamate to methylaspartate (21) and would not be responsible for conversion of α- to β-glutamate; there is no precedent for a glutamine mutase activity. For the second pathway, the small amounts of β-glutamate used by several methanogens as an osmolyte with minimal turnover under normal growth conditions (16–19) would be generated by β-glutamine hydrolysis. The work described here clearly shows that GS activity is capable of converting β-glutamate to β-glutamine in M. portucalensis. In attempting to determine how β-glutamate is synthesized in archaea, we (D. D. Martin and M. F. Roberts, unpublished results) have incubated protein extracts from various methanogens with either α- or β-glutamate or α-glutamine and various cofactors needed (or postulated) for other aminomutase activities (pyridoxal phosphate, glutathione, ferrous ammonium sulfate, sodium dithionite, S-adenosylmethionine, and acetyl coenzyme A) under anaerobic as well as aerobic conditions and never detected isomerization of α-glutamate to β-glutamate and vice versa (as judged by nuclear magnetic resonance and high-performance liquid chromatography). Also, in assays with partially purified M. portucalensis GS, β-glutamine is only generated from β-glutamate when ATP, NH3 (NH2OH in the case of the colorimetric assay), and Mg2+ are all present. Omission of any one of these blocks production of β-glutamine. Thus, it is likely the M. portucalensis GS is the source of synthesizing β-glutamine from β-glutamate in these cells.
GS from M. portucalensis is of the GSI variety similar to the other archaeal GSs characterized thus far. However, there are a number of significant differences between this enzyme and most other GSI-α enzymes. One notable difference is the effectiveness of the biosynthetic assay (α-glutamate conversion to γ-glutamylhydroxamate) compared to the glutamyl transfer assay (α-glutamine conversion to γ-glutamylhydroxamate). Most GS enzymes purified to date display at least fourfold-higher activity in transferase assays than in biosynthesis assays of γ-glutamylhydroxamate formation (7), although the exact ratio depends on the metal ion (Mn2+ or Mg2+) used in the biosynthetic assay and the assay pH. The reverse was observed with the M. portucalensis GS. Conversion of α-glutamate was four times faster than conversion of α-glutamine to γ-glutamylhydroxamate (using Mg2+ or Mn2+). Of the many bacterial GSs examined, only Bacillus licheniformis GS showed an eightfold-greater biosynthesis than transfer activity (6). The M. portucalensis GS high Km for glutamate isomers in the biosynthesis assay is also striking. Since M. portucalensis is a halophile, this may reflect adaptation to the very high intracellular α-glutamate. The Km for α-glutamine, 23 mM, is comparable to the α-glutamine Km (22.7 mM) observed with GS purified from M. ivanovi (2). Another difference is the relatively high rate of synthesis of β-glutamine, an osmolyte in M. portucalensis, from β-glutamate. Given the specific activities observed in crude extracts and the observation that once made, β-glutamine turns over slowly (17), one can estimate that to synthesize 1 μmol of β-glutamine per mg of cell protein (an average value for this organism) it would take about 4 h, well under the 24-h doubling time (19) of the organism under standard growth conditions.
Is the ability of the M. portucalensis GS to synthesize β-glutamate unique to this organism or a general characteristic of other archaea? The two archaeal GS enzymes that were examined (Table 1) exhibited poorer activity toward β-glutamate. The maximal rate with β-glutamate as a substrate was 0.04 to 0.08 that for the comparable concentration of α-glutamate. However, for both bacterial GS enzymes, β-glutamate was a much poorer substrate (relative activity of <0.003 that for α-glutamate), with a very high Km. The only other GS enzymes known to convert β-glutamate to β-glutamine with good efficiency are the GS from sheep brain and rat liver (14), although the relative rates for α- and β-glutamate depend on the nitrogen donor. Vmax for sheep brain amidation of β-glutamate is 46% that observed for α-glutamate with hydroxylamine and 18% when ammonia is used; partially purified rat liver GS displays a similar sensitivity to the nitrogen donor, but with lower rates for β-glutamate compared to α-glutamate (14). What makes β-glutamate a reasonable substrate for the M. portucalensis and mammalian GS but not for other GS enzymes is not clear at this stage.
The lower activities of M. jannaschii and A. fulgidus GS toward β-glutamate are consistent with the observation that neither of these hyperthermophilic organisms accumulates β-glutamine as an osmolyte. In M. jannaschii, α-glutamate concentrations are in the range of 0.04 to 0.12 M, while the intracellular concentration of β-glutamate is 0.4 to 0.6 M, depending on the external NaCl concentration (H. Meekins and M. F. Roberts, unpublished results). Since M. jannaschii GS activity decreased for β-glutamate at concentrations of ≥0.3 M, there would be little conversion of β-glutamate to β-glutamine under these intracellular conditions. α-Glutamate, with its much lower Km, would still be converted to α-glutamine for nitrogen assimilation and for use in protein synthesis. The observation that β-glutamate is a poor substrate for these archaeal GSs may be a general trend in halotolerant organisms in which α-glutamate (or β-glutamate) concentrations are high and anions are used for osmotic balance. If the β-glutamate were a good substrate, then it would be converted to β-glutamine, a zwitterion, and alter the cell potential (unless intracellular K+ also decreased).
In summary, the GS from M. portucalensis was shown to have unique kinetic characteristics consistent with its role in generating high concentrations of the osmolyte β-glutamine. In two other archaea, one of which uses β-glutamate as an osmolyte, β-glutamate was a poorer substrate. Further investigations of what is responsible for these kinetic differences will require structural analyses of these diverse GS.
ACKNOWLEDGMENTS
This work has been supported, in part, by grant DE-FG02-91ER20025 (to M.F.R.) from the Department of Energy Biosciences Division and by a grant from the National Science Foundation (H.J.S.) and the Wallenburg Foundation VIRTUE program (H.J.S.).
REFERENCES
- 1.Bender R A, Janseen K, Resnick A, Blumenberg-Foor M, Magasanik B. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol. 1977;129:1001–1009. doi: 10.1128/jb.129.2.1001-1009.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatnagar L, Zeikus J, Aubert J. Purification and characterization of glutamine synthetase from the archaebacterium Methanobacterium ivanovi. J Bacteriol. 1985;165:638–643. doi: 10.1128/jb.165.2.638-643.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the determination of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Brown J R, Masuchi Y, Robb F T, Doolittle A W F. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J Mol Evol. 1994;38:566–576. doi: 10.1007/BF00175876. [DOI] [PubMed] [Google Scholar]
- 5.Deuel T F, Pruisiner S. Regulation of glutamine synthetase from Bacillus subtilis by divalent cations, feedback inhibitors, and L-glutamine. J Biol Chem. 1974;249:257–264. [PubMed] [Google Scholar]
- 6.Donohoe T J, Bernlohr R W. Properties of the Bacillus licheniformis A5 glutamine synthetase purified from cells grown in the presence of ammonia or nitrate. J Bacteriol. 1981;147:589–601. doi: 10.1128/jb.147.2.589-601.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbard J S, Stadtman E R. Regulation of glutamine synthetase. II. Patterns of feedback inhibition in microorganisms. J Bacteriol. 1967;93:1045–1055. doi: 10.1128/jb.93.3.1045-1055.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahl S, von Berswordt-Wallrabe P, Heeschen V, Schmidt H, Rudolph H. A membrane bound enzyme in Sphagnum fallax. J Plant Physiol. 1998;153:270–275. [Google Scholar]
- 9.Kumar S, Nicholas D J D. Purification, properties, and regulation of glutamine synthetase from Nitrobacter agilis. J Gen Microbiol. 1984;130:959–966. [Google Scholar]
- 10.Lai M-C, Sowers K R, Robertson D E, Roberts M F, Gunsalus R P. Distribution of compatible solutes in the halophilic methanogenic archaebacteria. J Bacteriol. 1991;173:5352–5358. doi: 10.1128/jb.173.17.5352-5358.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manitz B, Holldorf A W. Purification and properties of glutamine synthetase from the archaebacterium Halobacterium salinarium. Arch Microbiol. 1993;159:90–97. [Google Scholar]
- 12.Meister A. Catalytic mechanism of glutamine synthetase: overview of glutamine metabolism. In: Mora J, Palacios R, editors. Glutamine metabolism, enzymology and regulation. New York, N.Y: Academic Press; 1980. pp. 1–40. [Google Scholar]
- 13.Neuhoff V, Stamm R, Eibl H. Clear background and highly sensitive protein staining with Coomassie blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis. 1985;6:427–428. [Google Scholar]
- 14.Pruisner S, Stadtman E R. The enzymes of glutamine metabolism. New York, N.Y: Academic Press, Inc.; 1973. [Google Scholar]
- 15.Rahman R N Z A, Jongsareejit B, Fujiwara S, Imanaka T. Characterization of recombinant glutamine synthetase from the hyperthermophilic archaeon Pyrococcus sp. strain KOD1. Appl Environ Microbiol. 1997;63:2472–2476. doi: 10.1128/aem.63.6.2472-2476.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts M F, Lai M-C, Gunsalus R P. Biosynthetic pathways of the osmolytes Nɛ-acetyl-β-lysine, β-glutamine, and betaine in Methanohalophilus strain FDF1 suggested by nuclear magnetic resonance analyses. J Bacteriol. 1992;174:6688–6693. doi: 10.1128/jb.174.20.6688-6693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson D E, Lai M-C, Gunsalus R P, Roberts M F. Composition, variation, and dynamics of major osmotic solutes in Methanohalophilus strain FDF1. Appl Environ Microbiol. 1992;58:2438–2443. doi: 10.1128/aem.58.8.2438-2443.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson D, Roberts M F, Belay N, Stetter K O, Boone D. Occurrence of β-glutamate, a novel osmolyte, in methanogenic bacteria. Appl Environ Microbiol. 1990;56:1504–1508. doi: 10.1128/aem.56.5.1504-1508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson P M, Roberts M F. Effects of osmolyte precursors on the distribution of compatible solutes in Methanohalophilus portucalensis. Appl Environ Microbiol. 1997;63:4032–4038. doi: 10.1128/aem.63.10.4032-4038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreier H J, Rostkowski C A, Kellner E M. Altered regulation of the glnRA operon in a Bacillus subtilis mutant that produces methionine sulfoximine-tolerant glutamine synthetase. J Bacteriol. 1993;175:892–897. doi: 10.1128/jb.175.3.892-897.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tollinger M, Konrat R, Hilbert B H, Marsh E N, Krautler B. How a protein prepares for B12 binding: structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium tetanomorphum. Structure. 1998;6:1021–1033. doi: 10.1016/s0969-2126(98)00103-8. [DOI] [PubMed] [Google Scholar]
- 22.Wedler F C, Hoffmann F M, Kenney R, Carfi J. Maintenance of specificity, information, and thermostability in thermophilic Bacillus sp. glutamine synthetases. Experentia Suppl. 1976;26:187–197. doi: 10.1007/978-3-0348-7675-9_15. [DOI] [PubMed] [Google Scholar]
- 23.Woolfolk C A, Shapiro B, Stadtman E R. Regulation of glutamine synthetase. I. Purification and properties of glutamine synthetase from Escherichia coli. Arch Biochem Biophys. 1966;116:177–192. doi: 10.1016/0003-9861(66)90026-9. [DOI] [PubMed] [Google Scholar]