Abstract
Radiation-inducible promoters are being used in many viral vector systems to obtain spatial and temporal control of gene expression. It was previously proven that radiation-induced gene expression can also be obtained in a bacterial vector system using anaerobic apathogenic clostridia. The effect of radiation inducibility was detected using mouse tumor necrosis factor alpha (mTNF-α) as a model protein under regulation of the radiation-inducible recA promoter. In this report, experiments are described in which this recA promoter was modified in order to increase radiation responsiveness. Incorporation of an extra Cheo box in the recA promoter region resulted in an increase in mTNF-α secretion from 44% for the wild-type promoter to 412% for the promoter with an extra Cheo box after a single irradiation dose of 2 Gy. Deletion of the Cheo box in the promoter region eliminated radiation inducibility. These results prove that the Cheo box in the recA promoter is indeed the radiation-responsive element. We also tested whether we could induce the constitutive endo-β-1,4-glucanase promoter (eglA) via ionizing irradiation by introducing a Cheo box in the promoter region. While the use of the constitutive promoter did not lead to an increase in mTNF-α secretion after irradiation, the introduction of a Cheo box resulted in a 242% increase in mTNF-α secretion. Reverse transcriptase PCR of RNA samples isolated from irradiated and nonirradiated bacterial cultures demonstrated that the increase in secretion was the result of enhanced transcription of the mTNF-α gene.
In the search for new therapeutic modalities for cancer, gene therapy has attracted enormous interest over the last few years. Many strategies to apply gene therapy have been developed, and even more vectors to deliver the gene of interest have been constructed. However, one of the major pitfalls of gene therapy is still the lack of specificity of gene delivery. Developing a good gene therapy protocol involves the use of a tumor-specific vector system and gene expression limited to the tumor only. This protocol will result in a high therapeutic index: high local tumor control with low systemic side effects.
Recently, the use of bacteria as a tumor-specific protein transfer system has attracted interest. Attenuated Salmonella (18, 19), anaerobic Bifidobacterium (28), and apathogenic Clostridium (2, 5, 6) have been shown to provide selective colonization in tumors. With Clostridium, no vegetative bacteria were found in normal tissues (5). Moreover, the use of bacteria as a protein transfer system is very safe, since treatment can be stopped at any time by addition of the appropriate antibiotic (21).
Clostridium can be genetically engineered to express therapeutic proteins such as mouse tumor necrosis factor alpha (mTNF-α) locally in a tumor (23). However, to obtain spatial and temporal control of gene expression, we investigated the use of a radiation-inducible promoter in Clostridium. The use of such a promoter would ensure that the therapeutic protein would be expressed only in irradiated tumoral tissues and not in nontumoral hypoxic tissues, such as abscesses or infarcted tissues. Moreover, protein expression would occur only after radiotherapy, so that gene expression would be switched on and physicians would know from what time on the therapeutic protein would be present (3). With cytotoxic agents such as TNF-α, this protocol would mean a major advantage.
It has been found that the recA promoter, belonging to the SOS repair system of bacteria, is induced by radiotherapy at the clinically relevant dose of 2 Gy (13–15). A single dose of 2 Gy significantly increased mTNF-α secretion by recombinant clostridia, by 44%. Moreover, gene activation could be repeated with a second dose of 2 Gy, a result which makes this promoter promising for clinical use, since in patient settings, daily doses of 2 Gy are used (15). However, with this recA promoter, there is still basal activity leading to transcription and secretion of mTNF-α under nonirradiation conditions.
In the present report, we investigated whether the Cheo box in the recA promoter was responsible for the radiation induction and whether we could increase radiation responsiveness by incorporating an extra Cheo box in the promoter region.
All genes belonging to the SOS repair system are activated by the presence of DNA damage. In nonactivating conditions, a repressor called LexA (for gram-negative bacteria) or DinR (for gram-positive bacteria) binds to a specific operator sequence, called the SOS box or the Cheo box, respectively. When DNA damage occurs, the central protein, RecA, forms a complex with single-stranded DNA that stimulates autoproteolysis of the repressor and thus leads to increased transcription of the SOS genes (1, 10). These genes play a role in repairing the original DNA damage.
Our hypothesis was that the addition of a second repressor binding site in the promoter region of a radiation-inducible gene would decrease transcription under basal conditions. After radiotherapy, both binding sites would become free and repression would be absent. In this report, we describe the cloning of the different recA promoter-operator mutations and present data on mTNF-α secretion by recombinant Clostridium acetobutylicum DSM792 with and without radiation. We also investigated whether we could use the Cheo box to make a constitutive promoter radiation inducible.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
C. acetobutylicum DSM792 was grown in 2× YT medium (17) at 37°C in an anaerobic system (model 1024; Forma Scientific, Marietta, Ohio) with 90% N2 and 10% H2 and with palladium as a catalyst.
For primary vector construction, Escherichia coli TG1 (20) was used. This strain was grown in Luria-Bertani broth at 37°C. E. coli ER2275 was used for in vivo methylation of plasmid DNA prior to the electroporation of clostridia (8). The E. coli-Clostridium shuttle plasmid pIMP1 was used as a cloning vector (9).
mTNF-α cDNA was available on plasmid pIG2mTNF (Innogenetics, Ghent, Belgium). Plasmid pHZ117, containing the eglA gene of C. acetobutylicum P262, was a gift from H. Zappe (29). The eglA promoter and signal sequence were used to express and secrete mTNF-α. This chimeric gene construct was present on shuttle plasmid pIMP1, resulting in pIMP-eglA-mTNF-α (23). The eglA promoter in this plasmid was replaced by the C. acetobutylicum recA promoter, resulting in pIMP-recA-mTNF-α (15). Table 1 shows an overview of the plasmids used in this study. The recA promoter was isolated from chromosomal DNA as previously described (14).
TABLE 1.
Shuttle plasmids used in this studya
| Plasmid | Promoter |
|---|---|
| pIMP-eglA-mTNF-α | eglA |
| pIMP-eglACheo-mTNF-α | eglA with incorporated Cheo box |
| pIMP-recA-mTNF-α | recA |
| pIMP-recAextraCheo-mTNF-α | recA with extra Cheo box incorporated |
| pIMP-recAdeletedCheo-mTNF-α | recA with Cheo box deleted |
The eglA signal peptide was used in each case for the secretion of mTNF-α.
Media were supplemented, when applicable, with erythromycin (25 μg/ml) or ampicillin (50 μg/ml).
Mutation of the recA and eglA promoters, DNA manipulations, and transformation procedures.
Introduction and/or deletion of the Cheo box in the recA and eglA promoters was done using a Quickchange site-directed mutagenesis kit (Stratagene). Table 2 shows the sequences of the wild-type and mutated recA and eglA promoters at the 3′ region. All mutations were introduced in the pIMP1 shuttle vector containing the eglA or recA promoter followed by the eglA–mTNF-α fusion gene (Table 1).
TABLE 2.
Sequences of the parental and mutated recA and eglA promoters at the 3′ regiona
| Promoter | Type | Sequence |
|---|---|---|
| recA | Parental | 5′ TTAAGGGACTTTTATTATATTATATTGACAAATTAATAAATTACTATATAATTATATGTATAGAACAAATGTTCGAGAGAAAGGTTGGTGAACCCTTAA 3′ |
| ------------------ | ||
| Extra Cheo | 5′ TTAAGGGACTTTGAACATATGTTCTTGACAAATTAATAAATTACTATATAATTATATGTATAGAACAAATGTTCGAGAGAAAGGTTGGTGAACCCTTAA 3′ | |
| ------------------- | ||
| Deleted Cheo | 5′ TTAAGGGACTTTTTGACAAATTAATAAATTACTATATAATTATATGTATAGAACAAATGTTCGAGAGAAAGGTTGGTGAACCCTTAA 3′ | |
| eglA | Parental | 5′ GGAGGAAAAAACTATCTTTTAAAAGTTTATAGTAAATAAAAAAAAATTATTAATGTAAAAATATACTAAGTATAGAATATTTATAATAGGGGGTATTAAC 3′ |
| ------------------ | ||
| ------------------- | ||
| With Cheo | 5′ GGAGGAAAAAACTATCGAACAAAAGTTCATAGTAAATAAAAAAAAATTATTAATGTAAAAATATACTAAGTATAGAATATTTATAATAGGGGGTATTAAC 3′ |
−10 and −35 promoter elements are underlined with solid lines. Shine-Dalgarno sequences are boxed. Sequences to be mutated are underlined with a broken line. Cheo boxes are in bold.
For mutation of the eglA and recA promoters, mutagenic primers containing an extra Cheo box flanked by 10 to 15 bases of the correct sequence were designed. Similarly, mutagenic primers with the desired deletion were developed (the Cheo box is given in bold): primers used to incorporate an extra Cheo box in the recA promoter region—5′ TATATTGACAAATGAACAAATGTTCATATAATTATATG 3′ and 5′ CATATAATTATATGAACATTTGTTCATTTGTCAATATA 3′; primers used to delete the Cheo box in the recA promoter region—5′ TAATTATATGTATAdeletion 12 bpGAGAGAAAGGTTGG 3′ and 5′ CCAACCTTTCTCTCdeletion 12 bpTATACATATAATTA 3′; and primers used to introduce a Cheo box in the eglA promoter region—5′ TTTAAGGGACTTTGAACATATGTTCTTGACAAATTAAT 3′ and 5′ ATTAATTTGTCAAGAACATATGTTCAAAGTCCCTTAAA 3′.
To verify the insertion or deletion of the Cheo box, the DNA fragments containing the introduced mutations were subcloned in pUC19 and the DNA sequence was determined with an automated laser fluorescence ALF Express sequencer (Amersham Pharmacia BioTech). Primers used for sequencing were CY5-labeled M13 forward and reverse primers.
All general DNA manipulations in E. coli were carried out as described by Sambrook et al. (20). Restriction endonucleases and DNA-modifying enzymes were purchased from Roche Diagnostics (Brussels, Belgium), GIBCO BRL (Gaithersburg, Md.), and Eurogentec (Seraing, Belgium) and used as indicated by the suppliers.
Plasmid DNA was isolated from E. coli with a Wizard Plus SV miniprep kit (Promega Inc., Madison, Wis.).
E. coli was transformed using chemically competent cells obtained with the RbCl method. Transformation of C. acetobutylicum DSM792 was carried out by electroporation as recently described (11).
Irradiation.
Recombinant bacteria were grown until early log phase (optical density at 600 nm, ±0.3). At this time, cultures were divided into two sets, one of which was exposed to radiation while the other was mock irradiated and used as a control. Bacteria were exposed to 2 Gy with a 60Co unit at a dose rate of 0.9 Gy/min. This dose of 2 Gy was chosen because it is the dose currently used in most clinical settings. After irradiation at room temperature, bacteria were incubated anaerobically at 37°C, and samples were taken at different time intervals after exposure.
Each experiment was independently repeated three times.
Analysis of mTNF-α secretion.
The amount of mTNF-α secreted by recombinant clostridia was quantified using enzyme-linked immunosorbent assay (ELISA) kits (DiaMed EuroGen, Tessenderlo, Belgium). Supernatants taken from irradiated and nonirradiated cultures were diluted 10-fold in phosphate-buffered saline–7.5% bovine serum albumin, and 100-μl aliquots were placed in wells of a 96-well microtiter plate in duplicate. Further manipulations were done according to the manufacturer's protocol.
Concentrations of secreted mTNF-α were calculated and compared for the irradiated and nonirradiated cultures. The level of radiation-induced mTNF-α production was expressed as the fold increase in mTNF-α concentration of irradiated samples compared with that of the corresponding nonirradiated samples.
Student's t test was used for statistical analysis.
Immunoblot analysis with polyclonal rabbit anti-mTNF-α antibodies was carried out by the method of Van Mellaert et al. (25).
RT-PCR.
To prove that the induction of mTNF-α was the result of an increase in promoter activity, reverse transcriptase (RT) PCR (RT-PCR) was performed on RNA isolated from irradiated and nonirradiated bacterial cultures. One hour after radiotherapy, 4-ml aliquots of cultures were taken and RNA was extracted using an RNeasy mini kit from Qiagen (Valencia, Calif.) as previously described (12). The RNA concentration was determined spectrophotometrically. To ensure that there was no DNA contamination which could result in mTNF-α cDNA transcription from the plasmid, 1 μg of RNA was digested with Fnu4HI, which cleaves mTNF-α cDNA at positions 82 and 213, and an additional DNase treatment was carried out. After heat inactivation of the enzymes, 200 U of Moloney murine leukemia virus RT (GIBCO BRL) was added to the RNA together with a mixture containing 0.8 μl of deoxynucleoside triphosphates (5 mM each), 4 μl of 5× RT buffer, 2 μl of reverse primer (10 pmol/μl), and 2 μl of dithiothreitol (DTT) (0.1 M) in a total volume of 20 μl. After 1 h of incubation at 37°C, the resulting cDNAs were amplified using PCR. Five microliters of the RT mixture was added to a mixture containing 8.5 μl of reverse primer (10 pmol/μl), 7 μl of forward primer (10 pmol/μl), 2.5 μl of deoxynucleoside triphosphates (5 mM each), 0.5 U of JumpStart Taq DNA polymerase (Sigma Chemical Co., St. Louis, Mo.), and 4.5 μl of 10× PCR buffer in a total volume of 50 μl. After 40 PCR cycles (10 min at 95°C, 30 s at 95°C, 2 min at 40°C, 30 s at 72°C, and 5 min at 72°C), 1-μl aliquots were run on 1% agarose gels. Primers used for the amplification of mTNF-α were as follows: forward primer—5′ GTAAGATCAAGTAGTCAA 3′; and reverse primer—5′ CAGAGCAATGACTCCAAA 3′.
To verify the absence of any DNA contamination, all samples underwent the same RT-PCR procedure without the addition of Moloney murine leukemia virus RT. To ensure equal amounts of RNA in all samples, an internal fragment of C. acetobutylicum 16S rRNA was amplified using RT-PCR to function as an internal standard. Primers used for the amplification of 16S rRNA were as follows: forward primer—5′ GGAGCAAACAGGATTAGATACC 3′; and reverse primer—5′ TGCCAACTCTATGGTGTGACG 3′.
RESULTS
Mutation of the recA and eglA promoters.
After introduction of the wanted mutations in the vectors pIMP-eglA-mTNF-α and pIMP-recA-mTNF-α by PCR mutagenesis, mutations were verified by sequence analysis and restriction digestion. Therefore, a 605-bp fragment containing the mutated sequence in each plasmid was subcloned in pUC19 digested with HindII.
Since both eglA and recA promoters are functional in E. coli, it was possible to test the activities of the mutated promoters by determination of the expression and secretion of mTNF-α. Therefore, lysates and supernatants were analyzed by Western blot analyses using rabbit anti-mTNF-α polyclonal antibodies and alkaline phosphatase-conjugated anti-rabbit antibodies (Sigma). Using both cell lysates and supernatants, we could clearly demonstrate the presence of mTNF-α in all recombinant bacteria containing the different constructs, proving that the mutated promoters were still functional (data not shown).
After introduction of the recombinant plasmids into Clostridium by electroporation, the presence of mTNF-α in supernatants and lysates was again demonstrated by immunoblotting (data not shown).
Analysis of mTNF-α secretion.
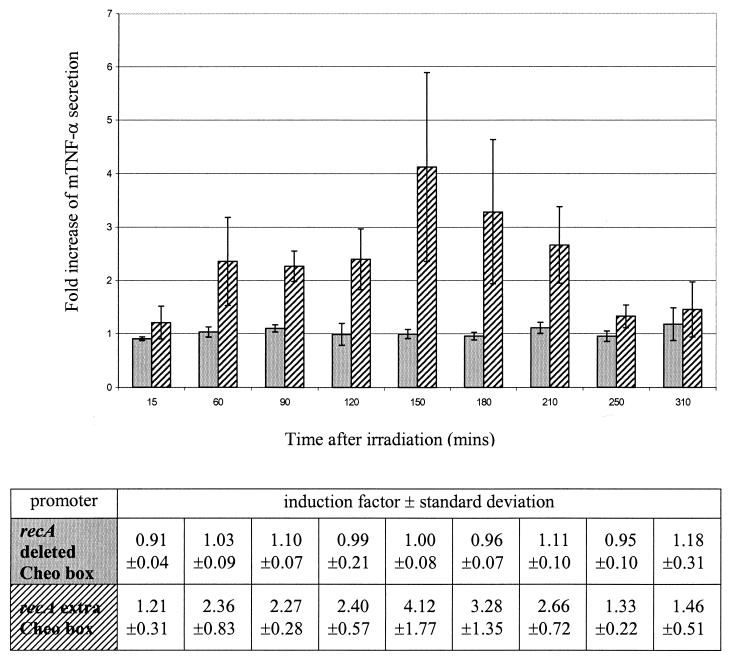
ELISA analysis was used to quantify mTNF-α secretion by recombinant clostridia. Average values can be summarized as follows. Under basal, nonirradiation conditions, bacteria containing the pIMP-recA-mTNF-α construct show basal promoter activity leading to an mTNF-α secretion level of 680 pg/ml. Bacteria containing the construct with an extra Cheo box show a basal mTNF-α secretion level of 486 pg/ml. When the Cheo box is deleted, the level of secretion increases to 756 pg/ml. This means that the addition of a Cheo box leads to a 30% decrease in basal promoter activity. Besides a decrease in basal activity, we also intended to increase the response after irradiation. As reported earlier, the wild-type recA promoter shows a 1.44-fold increase in mTNF-α secretion after a single dose of 2 Gy (15). When we deleted the Cheo box from the recA promoter region, no significant increase in mTNF-α secretion was measured after irradiation compared with the results obtained for the control samples (Fig. 1). However, when we incorporated an extra Cheo box in the recA promoter region, a 4.12-fold increase (standard deviation [SD], ±1.77) in mTNF-α secretion was seen 2.5 h after a single dose of 2 Gy (Fig. 1). At 1.5 h after irradiation, the 2.27-fold increase (SD, ±0.28) in mTNF-α secretion was significant (P < 0.02; Student's t test).
FIG. 1.
Fold increase of mTNF-α secretion in C. acetobutylicum DSM792(pIMP-recAdeletedCheo-mTNF-α) (gray bars) and C. acetobutylicum DSM792(pIMP-recAextraCheo-mTNF-α) (hatched bars) after a single dose of 2 Gy as a function of time after irradiation. The bars represent data from three independent experiments. Error bars represent standard deviations. Induction factors and standard deviations are shown in the table.
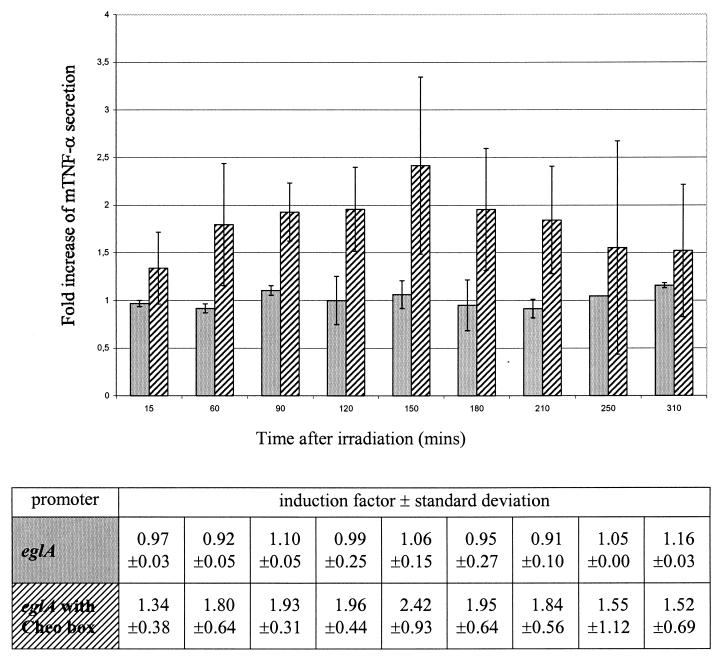
When we irradiated the recombinant bacteria containing the pIMP-eglA-mTNF-α construct, no increase in mTNF-α secretion was seen, confirming the constitutive properties of the eglA promoter (Fig. 2). However, when a Cheo box was incorporated in the eglA promoter region, an increase of 2.42-fold (SD, ±0.9) was seen 2.5 h after 2-Gy irradiation (Fig. 2). Again, at 1.5 h after irradiation, the 1.93-fold increase (SD, ±0.31) in mTNF-α secretion was significant (P < 0.05; Student's t test).
FIG. 2.
Fold increase of mTNF-α secretion in C. acetobutylicum DSM792(pIMP-eglA-mTNF-α) (gray bars) and C. acetobutylicum DSM792(pIMP-eglACheo-mTNF-α) (hatched bars) after a single dose of 2 Gy as a function of time after irradiation. The bars represent data from three independent experiments. Error bars represent standard deviations. Induction factors and standard deviations are shown in the table.
RT-PCR.
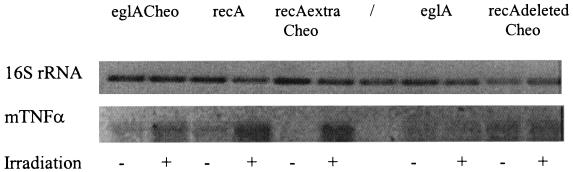
RT-PCR was carried out to prove that the increase in mTNF-α secretion was the result of an increase in promoter activity. One microliter of the PCR mixture was placed on a gel (Fig. 3). The upper panel in Fig. 3 represents the 650-bp internal fragment of 16S rRNA which was amplified to ensure that equal amounts of RNA were used in each PCR. The lower panel in Fig. 3 represents the 470-bp internal fragment of mTNF-α which was amplified. Equal amounts of RNA were used in all reactions (Fig. 3). When mTNF-α was amplified, for the constructs containing the eglA promoter with a Cheo box introduced (Fig. 3, first and second lanes), the wild-type recA promoter (third and fourth lanes), and the recA promoter with an extra Cheo box (fifth and sixth lanes), the nonirradiated samples showed a weaker band than the irradiated samples, indicating that more mRNA was present in the irradiated samples. For the constitutive eglA promoter (Fig. 3, eighth and ninth lanes) and the recA promoter with a deletion of the Cheo box (tenth and eleventh lanes), no difference was seen between the irradiated and the nonirradiated samples. For the control samples, both the recA promoter with an extra Cheo box and the eglA promoter containing a Cheo box showed a weaker band than the corresponding wild-type promoters. This weaker signal can be attributed to lower transcription levels because of higher repression levels under noninducing conditions. The reverse was seen for the recA promoter with a deletion of the Cheo box: a stronger signal in the nonirradiated samples for the mutated promoter than for the wild-type promoter. This stronger signal is the result of the absence of repression.
FIG. 3.
RT-PCR of irradiated and nonirradiated RNAs extracted from C. acetobutylicum DSM792. The upper panel represents the amplification of a 650-bp internal fragment of 16S rRNA which functions as an internal standard to ensure that equal amounts of RNA were used in each RT reaction. The lower panel represents the amplification of a 470-bp internal fragment of mTNF-α. RNA extracted from C. acetobutylicum DSM792 was transformed with pIMP-eglACheo-mTNF-α (first and second lanes), pIMP-recA-mTNF-α (third and fourth lanes), pIMP-recAextraCheo-mTNF-α (fifth and sixth lanes), pIMP-eglA-mTNF-α (eighth and ninth lanes), and pIMP-recAdeletedCheo-mTNF-α (tenth and eleventh lanes). The seventh lane shows a positive control for 16S rRNA (PCR performed on chromosomal DNA from C. acetobutylicum).
The absence of any band in the samples to which no RT was added confirmed that there was no DNA contamination in any of the samples (data not shown).
DISCUSSION
When DNA damage occurs, bacteria have at their disposal more than 20 genes to repair the original damage. All these genes belong to the so-called SOS repair system (10). These genes have in their promoter region a specific operator sequence, called the Cheo box for gram-positive bacteria and the SOS box or LexA binding site for gram-negative bacteria, to which a repressor binds (1, 7). This repressor is called LexA for gram-negative bacteria and DinR for gram-positive bacteria. When DNA damage is present, RecA will form a complex with single-stranded DNA, and this complex will stimulate autoproteolysis of LexA or DinR, resulting in increased transcription of the SOS repair genes. Both LexA and DinR bind to their operator sequence as dimers (4, 27). The consensus sequence for the Cheo box in gram-positive bacteria is 5′ GAAC-N4-GTTC 3′ (1). This consensus sequence is positioned within promoter regions such that the regulatory molecule LexA bound at these sites can interfere with the initiation of transcription by RNA polymerase. Several genes can be found which have two or more putative Cheo boxes, and for those in which repressor binding is proven, the distance between the two boxes is 15 to 16 bp (27).
We investigated whether the Cheo box in the recA promoter of C. acetobutylicum DSM792 was responsible for induction after ionizing irradiation. We deleted the Cheo box and found that there was no increase in mTNF-α secretion after irradiation, in contrast to the results obtained with the wild-type recA promoter when radiation induction was present. When we incorporated a second Cheo box 50 bp upstream of the first, we could increase the radiation responsiveness of the promoter from a 44% increase in the secretion of mTNF-α for the wild-type promoter to 412% for the mutated promoter, in comparison with the results obtained without irradiation. We chose to insert a second Cheo box 50 bp upstream of the first to ensure that there was no spherical hindrance between the two dimers. These results thus demonstrate that the Cheo box in the promoter region of recA is indeed the radiation-responsive element and can be used to increase the response after ionizing irradiation. The addition of a Cheo box also led to a 30% decrease in basal promoter activity.
Moreover, we proved that the introduction of a Cheo box in the constitutive eglA promoter caused the mutated promoter to respond to ionizing radiation, in contrast to the results obtained with the wild-type eglA promoter. From the literature it is known that the Cheo box is normally located between bp −42 and bp −106 from the start codon (27). Therefore, we introduced a Cheo box 71 bp upstream of the ribosome binding site.
RT-PCR demonstrated that the increase in secretion was the result of increased promoter activity, since higher concentrations of mRNA were present in the irradiated samples. Increased secretion of therapeutic proteins such as mTNF-α in Clostridium after irradiation is thus the result of increased activity at the transcriptional level. RT-PCR also demonstrated that under nonirradiation conditions (resembling basal conditions), the addition of a Cheo box resulted in a lower level of transcription and the deletion of a Cheo box resulted in a higher level of transcription after irradiation. These results prove that the Cheo box functions as a repressor binding site which becomes free after DNA damage caused by, for example, ionizing irradiation, leading to removal of repression and increased transcription.
Several publications have shown that the Cheo box or LexA binding site is responsible for activation of the SOS repair genes after DNA damage (1). Van der Lelie et al. have demonstrated that the addition of a second LexA binding site in the E. coli recN promoter increases inducibility after treatment with genotoxic agents (24).
To our knowledge, this is the first report which proves that the Cheo box is responsible for increased transcription of the recA gene after ionizing irradiation in Clostridium and can be used to further increase inducibility. Moreover, that fact that we could use radiation to induce the strong eglA promoter by introducing a Cheo box in the promoter region implies that the secretion of high doses of therapeutic proteins such as TNF-α can be controlled by ionizing irradiation.
Since the Cheo box is functional in the eglA promoter, independent of its natural sequence context, it seems possible to use radiation to induce other clostridial promoters which might be even stronger. We tested only the presence of two Cheo boxes in one promoter, but the addition of more boxes might even increase inducibility and decrease basal activity further.
Systemic administration of therapeutic proteins such as TNF-α is limited due to hepatotoxicity and life-threatening hypotension as major side effects (16). Many groups have been exploring the means to obtain the selective expression of genes locally in a tumor (reviewed in reference 26). Limiting the expression of toxic agents to a tumor cell is extremely important if damage to the surrounding normal tissues is to be avoided. Anaerobic bacteria selectively colonize the hypoxic-necrotic areas of a solid tumor which are absent in healthy normal tissues, and genetically engineered bacteria secrete therapeutic proteins locally in the tumor (22, 23). The use of a radiation-inducible promoter will ensure that no protein is secreted in other necrotic tissues outside the tumor and that secretion will increase after irradiation of the tumor (3). In this manner, the combination of radiotherapy, one of the standard treatment modalities in cancer, and Clostridium as a tumor-specific protein transfer system can increase concentrations of therapeutic agents locally in the tumor due to both spatial and temporal control of protein expression. This combination can result in higher tumor control rates with lower systemic side effects. One advantage of our system is the potential to increase the effectiveness of the recA promoter by the addition of Cheo boxes in order to increase responsiveness to ionizing irradiation. When, in the future, more potent clostridial promoters are discovered, the insertion of a Cheo box could make them radiation responsive, increasing the potential use of Clostridium as a vector for cancer therapy.
ACKNOWLEDGMENTS
We acknowledge financial support from Het Fonds voor Wetenschappelijk Onderzoek-Vlaanderen, Verkennende Internationale Samenwerking, and Het K.U.Leuven Onderzoeksfonds. S. Nuyts is a research fellow of I.W.T. (Vlaams Instituut voor de Bevordering van het Wetenschappelijk-Technologisch Onderzoek in de Industrie).
We thank Raf Berghmans (DiaMed EuroGen) for providing the ELISA kits.
REFERENCES
- 1.Cheo D L, Bayles K W, Yasbin R E. Cloning and characterization of DNA damage-inducible promoter regions from Bacillus subtilis. J Bacteriol. 1991;173:1696–1703. doi: 10.1128/jb.173.5.1696-1703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox M E, Lemmon M J, Mauchline M L, Davis T O, Giaccia A J, Minton N P, Brown J M. Anaerobic bacteria as delivery system for cancer gene therapy: in vitro activation of 5-fluorocytosine by genetically engineered clostridia. Gene Ther. 1996;3:173–178. [PubMed] [Google Scholar]
- 3.Hallahan D E, Mauceri H J, Sueng L P, Dunphy E J, Wayne J D, Hanna N D, Toledano A, Hellman S, Kufe D W, Weichselbaum R R. Spatial and temporal control of gene therapy using ionizing irradiation. Nat Med. 1995;1:786–791. doi: 10.1038/nm0895-786. [DOI] [PubMed] [Google Scholar]
- 4.Kim B, Little J W. Dimerization of a specific DNA-binding protein on the DNA. Science. 1992;255:203–205. doi: 10.1126/science.1553548. [DOI] [PubMed] [Google Scholar]
- 5.Lambin P, Theys J, Landuyt W, Rijken P, van der Kogel A, van der Schueren E, Hodgkiss R, Fowler J, Nuyts S, Anné J. Colonisation of Clostridium in the body is restricted to hypoxic and necrotic areas of tumours. Anaerobe. 1998;4:183–188. doi: 10.1006/anae.1998.0161. [DOI] [PubMed] [Google Scholar]
- 6.Lemmon M J, van Zijl P, Fox M E, Mauchline M L, Giaccia A J, Minton N P, Brown J M. Anaerobic bacteria as a gene delivery system that is controlled by the tumor microenvironment. Gene Ther. 1997;4:791–796. doi: 10.1038/sj.gt.3300468. [DOI] [PubMed] [Google Scholar]
- 7.Little J W, Mount D W, Yanisch-Perron C R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci USA. 1981;78:4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mermelstein L D, Papoutsakis E T. In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1993;59:1077–1081. doi: 10.1128/aem.59.4.1077-1081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mermelstein L D, Welker N E, Bennett G E, Papoutsakis E T. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC824. Bio/Technology. 1992;10:190–195. doi: 10.1038/nbt0292-190. [DOI] [PubMed] [Google Scholar]
- 10.Miller R, Kokjohn T. General microbiology of recA: environmental and evolutionary significance. Annu Rev Microbiol. 1990;44:365–394. doi: 10.1146/annurev.mi.44.100190.002053. [DOI] [PubMed] [Google Scholar]
- 11.Nakotte S, Schaffer M, Böhringer M, Dürre P. Electroporation of, plasmid isolation from and plasmid conservation in Clostridium acetobutylicum DSM792. Appl Microbiol Biotechnol. 1998;50:564–567. doi: 10.1007/s002530051335. [DOI] [PubMed] [Google Scholar]
- 12.Nuyts S, Van Mellaert L, Lambin P, Anné J. Efficient isolation of total RNA from Clostridium without DNA contamination. J Microbiol Methods. 2001;44:235–238. doi: 10.1016/s0167-7012(01)00219-6. [DOI] [PubMed] [Google Scholar]
- 13.Nuyts S, Theys J, Landuyt W, Van Mellaert L, Lambin P, Anné J. Increasing specificity of anti-tumor therapy: cytotoxic protein delivery by non-pathogenic clostridia under regulation of radio-induced promoters. Anticancer Res. 2001;21:857–862. [PubMed] [Google Scholar]
- 14.Nuyts S, Van Mellaert L, Theys J, Landuyt W, Lambin P, Anné J. The use of radio-induced bacterial promoters in anaerobic conditions: a means to control gene expression in Clostridium-mediated therapy for cancer. Radiat Res. 2001;155:716–726. doi: 10.1667/0033-7587(2001)155[0716:tuorib]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Nuyts S, Van Mellaert L, Theys J, Landuyt W, Bosmans E, Anné J, Lambin P. Radio-responsive recA promoter significantly increases TNFα production in recombinant clostridia after 2 Gy irradiation. Gene Ther. 2001;8:1197–1201. doi: 10.1038/sj.gt.3301499. [DOI] [PubMed] [Google Scholar]
- 16.Old L J. Tumour necrosis factor. Science. 1985;230:630–636. doi: 10.1126/science.2413547. [DOI] [PubMed] [Google Scholar]
- 17.Oultram J M, Loughlin, Swinfield T J, Brehm J K, Thompson D E, Minton N P. Introduction of plasmids into whole cells of Clostridium acetobutylicum by electroporation. FEMS Microbiol Lett. 1988;56:83–88. [Google Scholar]
- 18.Pawelek J M, Brooks K, Bermudes D. Tumor-targeted Salmonella as novel anticancer vector. Cancer Res. 1997;54:4537–4544. [PubMed] [Google Scholar]
- 19.Platt J, Sodi S, Kelley M, Rockwell S, Bermudes D, Low K B, Pawelek J. Antitumour effects of genetically engineered Salmonella in combination with radiation. Eur J Cancer. 2000;36:2397–2402. doi: 10.1016/s0959-8049(00)00336-1. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Theys J, Landuyt W, Nuyts S, Van Mellart L, Bosmans E, Rijnders A, Van den Bogaert W, van Oosterom A, Anné J, Lambin P. Improvement of Clostridium tumour targeting vectors evaluated in rat rhabdomyosarcomas. FEMS Immunol Med Microbiol. 2001;30:37–41. doi: 10.1111/j.1574-695X.2001.tb01547.x. [DOI] [PubMed] [Google Scholar]
- 22.Theys, J., W. Landuyt, S. Nuyts, L. Van Mellaert, P. Lambin, and J. Anné. Specific targeting of cytosine deaminase to solid tumors by engineered Clostridium acetobutylicum. Cancer Gene Ther., in press. [DOI] [PubMed]
- 23.Theys J, Nuyts S, Landuyt W, Van Mellaert L, Dillen C, Böhringer M, Dürre P, Lambin P, Anné J. Stable Escherichia coli-Clostridium acetobutylicum shuttle vector for secretion of murine tumor necrosis factor alpha. Appl Environ Microbiol. 1999;65:4295–4300. doi: 10.1128/aem.65.10.4295-4300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Lelie D, Regniers L, Borremans B, Provoost A, Verschaeve L. The VITOTOX test, an SOS bioluminescence Salmonella typhimurium test to measure genotoxicity kinetics. Mutat Res. 1997;389:279–290. doi: 10.1016/s1383-5718(96)00158-1. [DOI] [PubMed] [Google Scholar]
- 25.Van Mellaert L, Dillen C, Proost P, Sablon E, Deleys R, Van Broekhoven A, Heremans H, Van Damme J, Eyssen H, Anné J. Efficient secretion of biologically active mouse tumor necrosis factor α by Streptomyces lividans. Gene. 1994;150:153–158. doi: 10.1016/0378-1119(94)90876-1. [DOI] [PubMed] [Google Scholar]
- 26.Vile R G, Russell S J, Lemoine N R. Cancer gene therapy: hard lessons and new courses. Gene Ther. 2000;7:2–8. doi: 10.1038/sj.gt.3301084. [DOI] [PubMed] [Google Scholar]
- 27.Winterling K W, Chafin D, Hayes J F, Sun J, Levine A S, Yasbin R E, Woodgate R. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J Bacteriol. 1998;180:2201–2211. doi: 10.1128/jb.180.8.2201-2211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yazawa K, Fujimori M, Amano J, Kano Y, Tanuguchi S. Bifidobacterium longum as a delivery system for cancer gene therapy: selective localization and growth in hypoxic tumors. Cancer Gene Ther. 2000;7:269–274. doi: 10.1038/sj.cgt.7700122. [DOI] [PubMed] [Google Scholar]
- 29.Zappe H, Jones W A, Jones D T, Woods D R. Structure of an endo-β-1,4-glucanase gene from Clostridium actebutylicum P262 showing homology with endoglucanase genes from Bacillus spp. Appl Environ Microbiol. 1988;54:1289–1292. doi: 10.1128/aem.54.5.1289-1292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]