Abstract
Species of Talaromyces are cosmopolitan and ubiquitous, and some are of industrial and medicinal importance. Species of Talaromyces have been successively reported in China. During our examinations of samples collected from southwestern China, two new species belonging to Talaromyces sect. Talaromyces were further discovered based on phylogenetic analyses and morphological comparisons. Talaromyces ginkgonis sp. nov., isolated from a partially colonized fruit of Ginkgo biloba, differs from closely-related fungi in the combination of conidia ellipsoidal, smooth and 3.5−4 × 2−3 μm, no growth on CYA at 37 °C and sequence divergences; T. shilinensis sp. nov. is distinguished from its related allies in the combination of smooth conidia, colonies 10−11 mm diameter on CYA at 25 °C and sequence differences. Detailed descriptions and illustrations of the new taxa are given.
Keywords: DNA barcodes, fungal biodiversity, phylogeny, taxonomy
1. Introduction
Species of Talaromyces C.R. Benj. are cosmopolitan and ubiquitous, inhabiting soil, air, indoor environments, rotten food, plant debris, healthy plant as endophytes, insects, and immunodeficient humans. The beneficial and the harmful effects of Talaromyces have been well documented [1].
Seven sections have been established and widely accepted in the genus Talaromyces: Bacillispori, Helici, Islandici, Purpurei, Subinflati, Talaromyces, and Trachyspermi [2,3]. A novel section was recently proposed as sect. Tenues [4]. A total of 171 species were compiled in the genus and listed in the latest monograph [3]. Furthermore, 26 new taxa were afterwards noted [1,4,5,6,7,8,9,10,11,12]. Twenty of them are from Asia: T. albisclerotius B.D. Sun et al., T. aspriconidius B.D. Sun et al., T. aureolinus L. Wang, T. bannicus L. Wang, T. brevis B.D. Sun et al., T. chongqingensis X.C. Wang & W.Y. Zhuang, T. guizhouensis B.D. Sun et al., T. gwangjuensis Hyang B. Lee & T.T.T. Nguyen, T. haitouensis L. Wang, T. koreana Hyang B. Lee, T. nanjingensis X.R. Sun et al., T. penicillioides L. Wang, T. rosarhiza H. Zhang & Y.L. Jiang, T. rufus B.D. Sun et al., T. sparsus L. Wang, T. teleomorpha Hyang B. Lee et al., T. tenuis B.D. Sun et al., T. wushanicus X.C. Wang & W.Y. Zhuang, T. yunnanensis Doilom & C.F. Liao, and T. zhenhaiensis L. Wang; five from Europe: T. calidominioluteus Houbraken & Pyrri, T. gaditanus (C. Ramírez & A.T. Martínez) Houbraken & Soccio, T. germanicus Houbraken & Pyrri, T. pulveris Crous, and T. samsonii (Quintan.) Houbraken & Pyrri; and one from Africa, T. africanus Houbraken et al. Talaromyces sect. Talaromyces is the largest section and now with 84 species included.
Southwestern China shows various climates, altitudes, and vegetations, and it is rich in fungal biodiversity. Two species from soil in Chongqing were just described [1]. Along with more samples isolated from the area being examined, two additional new species belonging to Talaromyces sect. Talaromyces were further discovered based on phylogenetic analyses and morphological comparisons. Detailed descriptions and illustrations of the new taxa are provided.
2. Materials and Methods
2.1. Fungal Materials
The new species were associated with fungal (Pseudocosmospora sp.) or plant (Ginkgo biloba L.) materials collected in southwestern China (Sichuan and Yunnan provinces) during 2016–2017. Dried cultures were deposited in the Herbarium Mycologicum Academiae Sinicae (HMAS, Beijing, China), and the living ex-type strains were preserved in the China General Microbiological Culture Collection Center (CGMCC, Beijing, China).
2.2. Morphological Observations
Morphological characterization was conducted following standardized methods [13]. Four standard growth media were used: Czapek yeast autolysate agar (CYA, yeast extract Oxoid, Hampshire, UK), malt extract agar (MEA, Amresco, Solon, OH, USA), yeast extract agar (YES), and potato dextrose agar (PDA). The methods for inoculation, incubation, microscopic examinations, and digital recordings followed our previous studies [1,14,15,16].
2.3. Molecular Experiments
DNA was extracted from the cultures grown on PDA for 7 days, using the Plant Genomic DNA Kit (DP305, TIANGEN Biotech, Beijing, China). Polymerase chain reaction (PCR) amplifications of the internal transcribed spacer (ITS), beta-tubulin (BenA), calmodulin (CaM), and RNA polymerase II second largest subunit (RPB2) gene regions were conducted with routine methods [1,14,15,16]. The products were purified and subjected to sequencing on an ABI 3730 DNA Sequencer (Applied Biosystems, Bedford, MA, USA). Although the ITS region is proposed as the universal DNA barcode for fungi, it is not sufficient to distinguish species of Talaromyces. The ITS sequences provided in this study might be helpful for other researchers in case of need.
2.4. Phylogenetic Analyses
The forward and the reverse sequences newly generated in this study were assembled using Seqman v. 7.1.0 (DNASTAR Inc., Madison, WI, USA). The assembled sequences were deposited in GenBank. Sequences used for phylogenetic analyses were listed in Table 1. Sequences of each of the three separate datasets (BenA, CaM, and RPB2) and those that were combined were aligned using MAFFT v. 7.221 [17], and then manually edited in BioEdit v. 7.1.10 [18] and MEGA v. 6.0.6 [19]. Maximum Likelihood (ML) analyses were determined using RAxML-HPC2 [20] on XSEDE 8.2.12 on CIPRES Science Gateway v. 3.3 [21] with the default GTRCAT model. Bayesian Inference (BI) analyses were performed with MrBayes v. 3.2.5 [22]. Appropriate nucleotide substitution models and parameters were determined by Modeltest v. 3.7 [23]. The consensus trees were viewed in FigTree v. 1.3.1 (http://tree.bio.ed.ac.uk/software/%20figtree/ accessed on 1 September 2015). The type species T. trachyspermus of Talaromyces sect. Trachyspermi served as an outgroup.
Table 1.
Fungal species and sequences used in phylogenetic analyses of Talaromyces sect. Talaromyces.
| Species | Strain | Locality | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|
| T. aculeatus (Raper & Fennell) Samson et al., 2011 | CBS 289.48 T | USA | textile | KF741995 | KF741929 | KF741975 | MH793099 |
| T. adpressus A.J. Chen et al., 2016 | CGMCC 3.18211 T | China: Beijing | indoor air | KU866657 | KU866844 | KU866741 | KU867001 |
| T. alveolaris Guevara-Suarez et al., 2017 | CBS 142379 T | USA | human bronchoalveolar lavage | LT558969 | LT559086 | LT795596 | LT795597 |
| T. amazonensis N. Yilmaz et al., 2016 | CBS 140373 T | Colombia | leaf litter | KX011509 | KX011490 | KX011502 | MN969186 |
| T. amestolkiae N. Yilmaz et al., 2012 | CBS 132696 T | South Africa | house dust | JX315660 | JX315623 | KF741937 | JX315698 |
| T. angelicae S.H. Yu et al., 2013 | KACC 46611 T | South Korea | dried root of Angelica gigas | KF183638 | KF183640 | KJ885259 | KX961275 |
| T. annesophieae Houbraken 2017 | CBS 142939 T | Netherlands | soil | MF574592 | MF590098 | MF590104 | MN969199 |
| T. apiculatus Samson et al., 2011 | CBS 312.59 T | Japan | soil | JN899375 | KF741916 | KF741950 | KM023287 |
| T. argentinensis Jurjević & S.W. Peterson 2019 | NRRL 28750 T | Ghana | soil | MH793045 | MH792917 | MH792981 | MH793108 |
| T. aspriconidius B.D. Sun et al., 2020 | CBS 141835 T | China: Yunnan | soil | MN864274 | MN863343 | MN863320 | MN863332 |
| T. aurantiacus (J.H. Mill. et al.) Samson et al., 2011 | CBS 314.59 T | USA | soil | JN899380 | KF741917 | KF741951 | KX961285 |
| T. aureolinus L. Wang 2021 | CGMCC 3.15865 T | China: Yunnan | soil | MK837953 | MK837937 | MK837945 | MK837961 |
| T. australis Visagie et al., 2015 | CBS 137102 T | Australia | soil under pasture | KF741991 | KF741922 | KF741971 | KX961284 |
| T. bannicus L. Wang 2021 | CGMCC 3.15862 T | China: Yunnan | soil | MK837955 | MK837939 | MK837947 | MK837963 |
| T. beijingensis A.J. Chen et al., 2016 | CGMCC 3.18200 T | China: Beijing | indoor air | KU866649 | KU866837 | KU866733 | KU866993 |
| T. brevis B.D. Sun et al., 2020 | CBS 141833 T | China: Beijing | soil | MN864269 | MN863338 | MN863315 | MN863328 |
| T. calidicanius (J.L. Chen) Samson et al., 2011 | CBS 112002 T | China: Taiwan | soil | JN899319 | HQ156944 | KF741934 | KM023311 |
| T. californicus Jurjević & S.W. Peterson 2019 | NRRL 58168 T | USA | air | MH793056 | MH792928 | MH792992 | MH793119 |
| T. cnidii S.H. Yu et al., 2013 | KACC 46617 T | South Korea | dried roots of Cnidium | KF183639 | KF183641 | KJ885266 | KM023299 |
| T. coprophilus Guevara-Suarez et al., 2020 | CBS 142756 T | Spain | herbivore dung | LT899794 | LT898319 | LT899776 | LT899812 |
| T. cucurbitiradicus L. Su & Y.C. Niu 2018 | ACCC 39155 T | China: Beijing | endophyte from root of Cucurbita moschata | KY053254 | KY053228 | KY053246 | n.a. |
| T. derxii Takada & Udagawa 1988 | CBS 412.89 T | Japan | cultivated soil | JN899327 | JX494306 | KF741959 | KM023282 |
| T. dimorphus X.Z. Jiang & L. Wang 2018 | CGMCC 3.15692 T | China: Hainan | forest soil | KY007095 | KY007111 | KY007103 | KY112593 |
| T. domesticus Jurjević & S.W. Peterson 2019 | NRRL 58121 T | USA | floor swab | MH793055 | MH792927 | MH792991 | MH793118 |
| T. duclauxii (Delacr.) Samson et al., 2011 | CBS 322.48 T | France | canvas | JN899342 | JX091384 | KF741955 | JN121491 |
| T. euchlorocarpius Yaguchi et al., 1999 | CBM PF1203 T | Japan | soil | AB176617 | KJ865733 | KJ885271 | KM023303 |
| T. flavovirens (Durieu & Mont.) Visagie et al., 2012 | CBS 102801 T | Spain | unknown | JN899392 | JX091376 | KF741933 | KX961283 |
| T. flavus (Klöcker) Stolk & Samson 1972 | CBS 310.38 T | New Zealand | unknown | JN899360 | JX494302 | KF741949 | JF417426 |
| T. francoae N. Yilmaz et al., 2016 | CBS 113134 T | Colombia | leaf litter | KX011510 | KX011489 | KX011501 | MN969188 |
| T. funiculosus (Thom) Samson et al., 2011 | CBS 272.86 T | India | Lagenaria vulgaris | JN899377 | MN969408 | KF741945 | KM023293 |
| T. fuscoviridis Visagie et al., 2015 | CBS 193.69 T | Netherlands | soil | KF741979 | KF741912 | KF741942 | MN969156 |
| T. fusiformis A.J. Chen et al., 2016 | CGMCC 3.18210 T | China: Beijing | indoor air | KU866656 | KU866843 | KU866740 | KU867000 |
| T. galapagensis Samson & Mahoney 1977 | CBS 751.74 T | Ecuador | soil under Maytenus obovata | JN899358 | JX091388 | KF741966 | KX961280 |
| T. ginkgonis X.C. Wang & W.Y. Zhuang sp. nov. | 10725 T | China: Sichuan | rotten fruit of Ginkgo biloba | OL638158 | OL689844 | OL689846 | OL689848 |
| T. haitouensis L. Wang 2022 | CGMCC 3.16101 T | China: Jiangsu | riverside soil | MZ045695 | MZ054634 | MZ054637 | MZ054631 |
| T. indigoticus Takada & Udagawa 1993 | CBS 100534 T | Japan | soil | JN899331 | JX494308 | KF741931 | KX961278 |
| T. intermedius (Apinis) Stolk & Samson 1972 | CBS 152.65 T | UK | swamp soil | JN899332 | JX091387 | KJ885290 | KX961282 |
| T. kabodanensis Houbraken et al., 2016 | CBS 139564 T | Iran | hypersaline soil | KP851981 | KP851986 | KP851995 | MN969190 |
| T. kendrickii Visagie et al., 2015 | CBS 136666 T | Canada | forest soil | KF741987 | KF741921 | KF741967 | MN969158 |
| T. lentulus X.Z. Jiang & L. Wang 2018 | CGMCC 3.15689 T | China: Shandong | soil | KY007088 | KY007104 | KY007096 | KY112586 |
| T. liani (Kamyschko) N. Yilmaz et al., 2014 | CBS 225.66 T | China | soil | JN899395 | JX091380 | KJ885257 | KX961277 |
| T. louisianensis Jurjević & S.W. Peterson 2019 | NRRL 35823 T | USA | air | MH793052 | MH792924 | MH792988 | MH793115 |
| T. macrosporus (Stolk & Samson) Frisvad et al., 1990 | CBS 317.63 T | South Africa | apple juice | JN899333 | JX091382 | KF741952 | KM023292 |
| T. mae X.Z. Jiang & L. Wang 2018 | CGMCC 3.15690 T | China: Shanghai | forest soil | KY007090 | KY007106 | KY007098 | KY112588 |
| T. malicola Jurjević & S.W. Peterson 2019 | NRRL 3724 T | Italy | rhizosphere of an apple tree | MH909513 | MH909406 | MH909459 | MH909567 |
| T. mangshanicus X.C. Wang & W.Y. Zhuang 2016 | CGMCC 3.18013 T | China: Hunan | soil | KX447531 | KX447530 | KX447528 | KX447527 |
| T. marneffei (Segretain et al.) Samson et al., 2011 | CBS 388.87 T | Vietnam | Rhizomys sinensis | JN899344 | JX091389 | KF741958 | KM023283 |
| T. muroii Yaguchi et al., 1994 | CBS 756.96 T | China: Taiwan | soil | MN431394 | KJ865727 | KJ885274 | KX961276 |
| T. mycothecae R.N. Barbosa et al., 2018 | CBS 142494 T | Brazil | nest of Melipona scutellaris | MF278326 | LT855561 | LT855564 | LT855567 |
| T. nanjingensis X.R. Sun et al., 2022 | CCTCC M 2012167 T | China: Jiangsu | rhizosphere soil of Pinus massoniana | MW130720 | MW147759 | MW147760 | MW147762 |
| T. neofusisporus L. Wang 2016 | CGMCC 3.15415 T | China: Tibet | leaf sample | KP765385 | KP765381 | KP765383 | MN969165 |
| T. oumae-annae Visagie et al., 2014 | CBS 138208 T | South Africa | house dust | KJ775720 | KJ775213 | KJ775425 | KX961281 |
| T. panamensis (Samson et al.) Samson et al., 2011 | CBS 128.89 T | Panama | soil | JN899362 | HQ156948 | KF741936 | KM023284 |
| T. penicillioides L. Wang 2021 | CGMCC 3.15822 T | China: Guizhou | soil | MK837956 | MK837940 | MK837948 | MK837964 |
| T. pinophilus (Hedgc.) Samson et al., 2011 | CBS 631.66 T | France | PVC | JN899382 | JX091381 | KF741964 | KM023291 |
| T. pratensis Jurjević & S.W. Peterson 2019 | NRRL 62170 T | USA | effluent of water treatment plant | MH793075 | MH792948 | MH793012 | MH793139 |
| T. primulinus (Pitt) Samson et al., 2011 | CBS 321.48 T | USA | unknown | JN899317 | JX494305 | KF741954 | KM023294 |
| T. pseudofuniculosus Guevara-Suarez et al., 2020 | CBS 143041 T | Spain | herbivore dung | LT899796 | LT898323 | LT899778 | LT899814 |
| T. purgamentorum N. Yilmaz et al., 2016 | CBS 113145 T | Colombia | leaf litter | KX011504 | KX011487 | KX011500 | MN969189 |
| T. purpureogenus (Stoll) Samson et al., 2011 | CBS 286.36 T | unknown | unknown | JN899372 | JX315639 | KF741947 | JX315709 |
| T. qii L. Wang 2016 | CGMCC 3.15414 T | China: Tibet | leaf sample | KP765384 | KP765380 | KP765382 | MN969164 |
| T. rapidus Guevara-Suarez et al., 2017 | CBS 142382 T | USA | human bronchoalveolar lavage | LT558970 | LT559087 | LT795600 | LT795601 |
| T. rosarhiza H. Zhang & Y.L. Jiang 2021 | GUCC 190040.1 T | China: Guizhou | endophyte of Rosa roxburghii | MZ221603 | MZ333143 | MZ333137 | MZ333141 |
| T. ruber (Stoll) N. Yilmaz et al., 2012 | CBS 132704 T | UK | aircraft fuel tank | JX315662 | JX315629 | KF741938 | JX315700 |
| T. rubicundus (J.H. Mill. et al.) Samson et al., 2011 | CBS 342.59 T | USA | soil | JN899384 | JX494309 | KF741956 | KM023296 |
| T. rufus B.D. Sun et al., 2020 | CBS 141834 T | China: Yunnan | soil | MN864272 | MN863341 | MN863318 | MN863331 |
| T. sayulitensis Visagie et al., 2014 | CBS 138204 T | Mexico | house dust | KJ775713 | KJ775206 | KJ775422 | MN969146 |
| T. shilinensis X.C. Wang & W.Y. Zhuang sp. nov. | XCW_SN259 T | China: Yunnan | associated with Pseudocosmospora sp. | OL638159 | OL689845 | OL689847 | OL689849 |
| T. siamensis (Manoch & C. Ramírez) Samson et al., 2011 | CBS 475.88 T | Thailand | forest soil | JN899385 | JX091379 | KF741960 | KM023279 |
| T. soli Jurjević & S.W. Peterson 2019 | NRRL 62165 T | USA | soil | MH793074 | MH792947 | MH793011 | MH793138 |
| T. sparsus L. Wang 2021 | CGMCC 3.16003 T | China: Beijing | soil | MT077182 | MT083924 | MT083925 | MT083926 |
| T. stellenboschiensis Visagie & K. Jacobs 2015 | CBS 135665 T | South Africa | soil | JX091471 | JX091605 | JX140683 | MN969157 |
| T. stipitatus (Thom) C.R. Benj. 1955 | CBS 375.48 T | USA | rotting wood | JN899348 | KM111288 | KF741957 | KM023280 |
| T. stollii N. Yilmaz et al., 2012 | CBS 408.93 T | Netherlands | AIDS patient | JX315674 | JX315633 | JX315646 | JX315712 |
| T. striatoconidium (R.F. Castañeda & W. Gams) Houbraken et al., 2020 | CBS 550.89 T | Cuba | leaf litter of Pachyanthus poirettii | MN431418 | MN969441 | MN969360 | MT156347 |
| T. thailandensis Manoch et al., 2013 | CBS 133147 T | Thailand | forest soil | JX898041 | JX494294 | KF741940 | KM023307 |
| T. tumuli Jurjević & S.W. Peterson 2019 | NRRL 62151 T | USA | soil from prairie | MH793071 | MH792944 | MH793008 | MH793135 |
| T. veerkampii Visagie et al., 2015 | CBS 500.78 T | Columbia | soil | KF741984 | KF741918 | KF741961 | KX961279 |
| T. verruculosus (Peyronel) Samson et al., 2011 | NRRL 1050 T | USA | soil | KF741994 | KF741928 | KF741944 | KM023306 |
| T. versatilis Bridge & Buddie 2013 | IMI 134755 T | UK | unknown | MN431395 | MN969412 | MN969319 | MN969161 |
| T. viridis (Stolk & G.F. Orr) Arx 1987 | CBS 114.72 T | Australia | soil | AF285782 | JX494310 | KF741935 | JN121430 |
| T. viridulus Samson et al., 2011 | CBS 252.87 T | Australia | soil | JN899314 | JX091385 | KF741943 | JF417422 |
| T. wushanicus X.C. Wang & W.Y. Zhuang 2021 | CGMCC 3.20481 T | China: Chongqing | soil | MZ356356 | MZ361347 | MZ361354 | MZ361361 |
| T. xishaensis X.C. Wang et al., 2016 | CGMCC 3.17995 T | China: Hainan | soil | KU644580 | KU644581 | KU644582 | MZ361364 |
| T. yunnanensis Doilom & C.F. Liao 2020 | KUMCC 18-0208 T | China: Yunnan | rhizosphere soil | MT152339 | MT161683 | MT178251 | n.a. |
| T. zhenhaiensis L. Wang 2022 | CGMCC 3.16102 T | China: Zhejiang | mudflat soil | MZ045697 | MZ054636 | MZ054639 | MZ054633 |
| T. trachyspermus (Shear) Stolk & Samson 1973 | CBS 373.48 T | USA | unknown | JN899354 | KF114803 | KJ885281 | JF417432 |
GenBank accession numbers in bold indicating the newly generated sequences. Full names of the culture collection centers: ACCC (Agricultural Culture Collection of China); CBS (Centraalbureau voor Schimmelcultures, now Westerdijk Fungal Biodiversity Institute); CCTCC (China Center for Type Culture Collection); CGMCC (China General Microbiological Culture Collection); GUCC (Culture Collection at Department of Plant Pathology, Agriculture College, Guizhou University); IMI (CABI Bioscience); KACC (Korean Agricultural Culture Collection); KUMCC (Kunming Institute of Botany Culture Collection); NRRL (USDA-ARS Culture Collection).
3. Results
3.1. Phylogenetic Analysis
To infer the phylogeny of Talaromyces sect. Talaromyces and to determine the positions of the new species, three separate datasets (BenA, CaM and RPB2) and those that were combined were compiled and analyzed. Detailed characteristics of the datasets are listed in Table 2.
Table 2.
Detailed characteristics of datasets of Talaromyces sect. Talaromyces.
| Gene Fragment | No. of Seq. | Length of Alignment (bp) | No. of Variable Sites | No. of Parsimony-Informative Sites | Model for BI |
|---|---|---|---|---|---|
| BenA | 87 | 643 | 246 | 200 | K81uf + I + G |
| CaM | 87 | 581 | 305 | 260 | SYM + I + G |
| RPB2 | 85 | 978 | 359 | 319 | TVM + I + G |
| BenA + CaM + RPB2 | 87 | 2202 | 910 | 779 | GTR + I + G |
Full names of the used models: GTR + I + G (General Time Reversible model with unequal base frequencies with Invariable sites and Gamma distribution); K81uf + I + G (Kimura 3-parameter model with unequal base frequencies with Invariable sites and Gamma distribution); SYM + I + G (Symmetrical model with Invariable sites and Gamma distribution); TVM + I + G (Transversion model with Invariable sites and Gamma distribution).
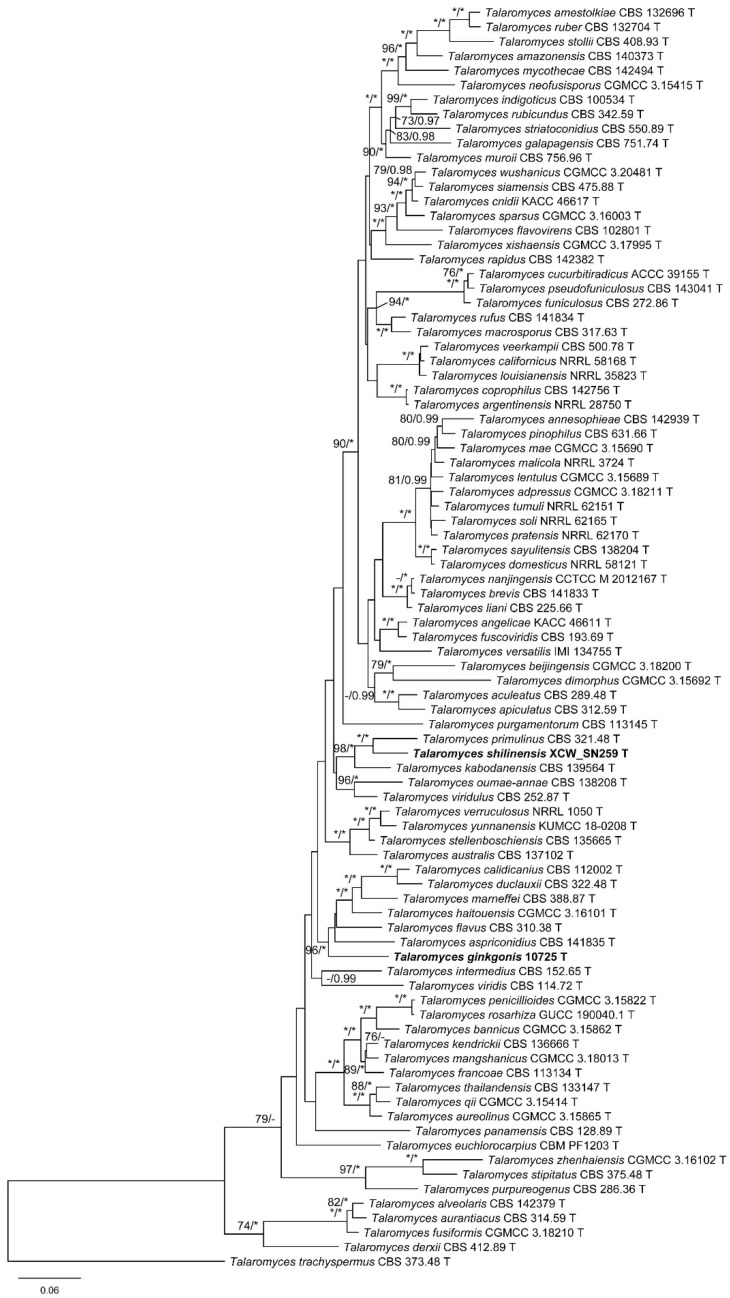
In the BenA phylogeny (Figure S1), the strain 10725 was clustered with T. aspriconidius, T. calidicanius, T. duclauxii, T. flavus, T. haitouensis, and T. marneffei; and XCW_SN259 was grouped with T. kabodanensis and T. primulinus. In the CaM tree (Figure S2), 10725 showed as a distinct lineage, while XCW_SN259 was a sister taxon of T. primulinus. In the RPB2 phylogeny (Figure S3), the position of 10725 was similar to that shown in the BenA phylogeny with relatively weak supports, while the sister relationship between T. primulinus and XCW_SN259 was confirmed as that in the CaM phylogeny. In the phylogenetic tree of the combined three-gene dataset (Figure 1), the position of 10725 was identical with the BenA and RPB2 trees and that of XCW_SN259 was consistent in of all the trees (Figure 1 and Figures S1–S3).
Figure 1.
ML phylogeny of Talaromyces sect. Talaromyces inferred from the combined (BenA + CaM + RPB2) dataset. Bootstrap values ≥70% (left) or posterior probability values ≥0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability.
3.2. Taxonomy
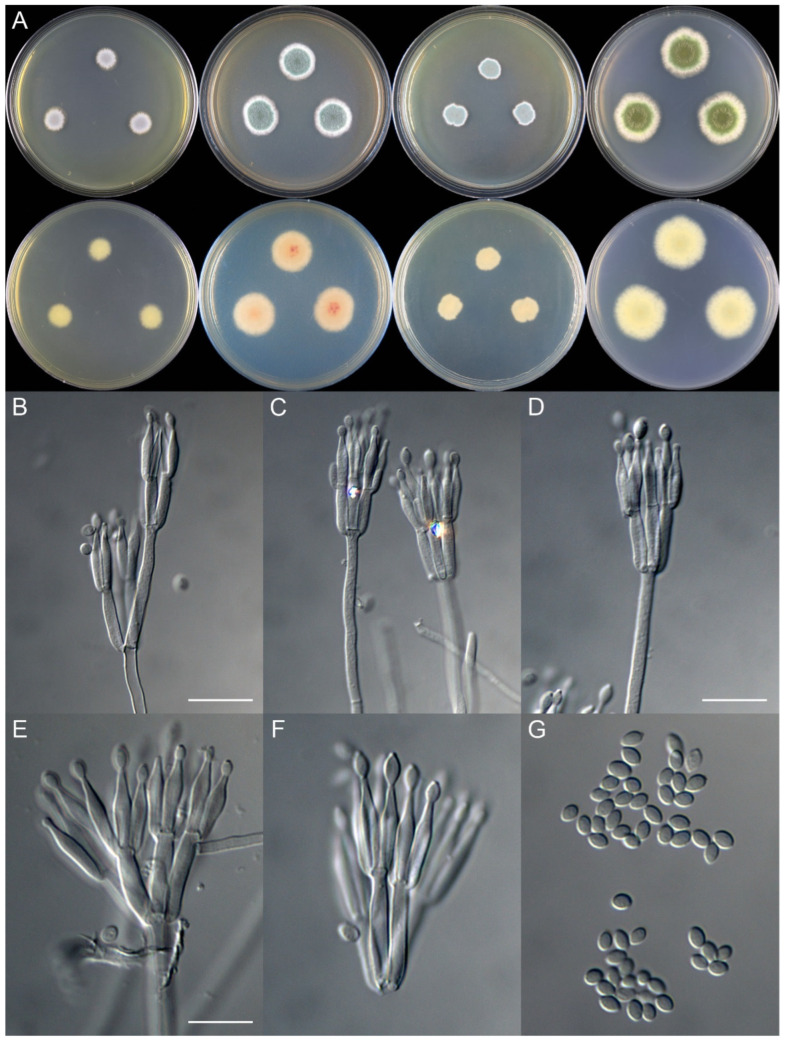
Talaromyces ginkgonis X.C. Wang & W.Y. Zhuang, sp. nov. Figure 2
Figure 2.
Colonial and microscopic morphology of Talaromyces ginkgonis (10725). (A) Colony phenotypes (25 °C, 7 days; top row left to right, obverse CYA, MEA, YES, and PDA; bottom row left to right, reverse CYA, MEA, YES, and PDA); (B–F) Conidiophores; (G) Conidia. Bars: B = 15 µm, applies to C; D = 12.5 µm; E = 10 µm, applies to F and G.
Fungal Names: FN570954
Etymology: The specific epithet refers to the substrate of the fungus
in Talaromyces sect. Talaromyces
Typification: CHINA. Sichuan Province, Chengdu City, Dujiangyan City, Mount Qingcheng, 30°54′8″ N 103°33′40″ E, on a partially colonized fruit of Ginkgo biloba L., 22 August 2016, Xin-Cun Wang 10725, cultured by Xin-Cun Wang (holotype HMAS 247853, ex-type strain CGMCC 3.20698)
DNA barcodes: ITS OL638158, BenA OL689844, CaM OL689846, RPB2 OL689848
Colony diam., 7 days, 25 °C (unless stated otherwise): CYA 9–13 mm; CYA 37 °C no growth; MEA 19–21 mm; YES 12–13 mm; PDA 15–27 mm
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, plain; margins moderately wide, fimbriate; mycelia colorless; texture velutinous; sporulation moderately dense; conidia en masse greyish green; soluble pigments absent; exudates absent; reverse greenish white.
On MEA 25 °C, 7 days: Colonies nearly circular, plain; margins wide, fimbriate; mycelia white; texture velutinous; sporulation dense; conidia en masse vivid green; soluble pigments absent; exudates absent; reverse buff but pink at centers and white at margins.
On YES 25 °C, 7 days: Colonies irregular, plain; margins narrow, fimbriate; mycelia white; texture velutinous; sporulation dense; conidia en masse bluish green; soluble pigments absent; exudates absent; reverse buff at centers, green at periphery, and white at margins.
On PDA 25 °C, 7 days: Colonies nearly circular to irregular, plain; margins wide, irregular; mycelia white; texture velutinous; sporulation dense; conidia en masse yellowish green to vivid green; soluble pigments absent; exudates absent; reverse usually pink at centers, green to buff at periphery, and white at margins.
Micromorphology: Conidiophores biverticillate, rarely terverticillate; stipes smooth-walled, 150–360 × 2.0–3.0 μm; metulae 3–5, 11.0–22.5 × 2.0–3.5 μm; phialides acerose, tapering into very thin neck, 3–5 per metula, 12.0–15.0 × 2.0–3.0 μm; conidia ellipsoidal to fusiform, smooth, 3.5–4.0 × 2.0–3.0 μm
Note: This species is phylogenetically related to T. aspriconidius, T. calidicanius, T. duclauxii, T. flavus, T. haitouensis, and T. marneffei, with strong support in the combined three-gene tree (Figure 1). Morphologically, it differs from T. aspriconidius and T. calidicanius in the smooth conidia; from T. marneffei in the ellipsoidal conidia; and from T. duclauxii, T. flavus, and T. haitouensis in the slower growth rate on MEA and YES at 25 °C (Table 3).
Table 3.
Morphological comparisons of new Talaromyces species and their closely related species.
| Species | CYA 25 °C (mm) | CYA 37 °C (mm) | MEA (mm) | YES (mm) | Conidiophore | Conidia Shape | Conidia Wall | Conidia Size (µm) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| T. aspriconidius | 22–23 | 22–23 | 36–37 | 28–29 | biverticillate | globose | strikingly roughened | 3–4 | [4] |
| T. calidicanius | 27–30 | no growth | 47–48 | 40–41 | biverticillate | ellipsoidal to fusiform | finely rough to rough with spiral striations | 2.5–4.5 × 2–3 | [2] |
| T. duclauxii | 25–27 | 3–4 | 48–50 | 43–44 | biverticillate | ellipsoidal | smooth to finely rough | 3–4 × 1.5–3.5 | [2] |
| T. flavus | 9–10 | 19–20 | 31–32 | 24–26 | monoverticillate | ellipsoidal | smooth | 2–3 × 1.5–2.5 | [2] |
| T. haitouensis | 22–25 | 18–20 | 48–51 | 25–28 | biverticillate | pyriform to ellipsoidal | smooth | 2.5–3 × 2–2.5 | [10] |
| T. marneffei | 13–25 | 5–10 | 15–27 | 17–25 | mono- to biverticillate | subglobose | smooth | 2.5–4 × 2–3 | [2] |
| T. ginkgonis | 9–13 | no growth | 19–21 | 12–13 | biverticillate | ellipsoidal to fusiform | smooth | 3.5–4 × 2–3 | This study |
| T. kabodanensis | 15–25 | no growth | 37–44 | 28–35 | biverticillate | ovoidal to fusiform | finely rough to rough with spiral striations | 2.5–3.5 × 1.5–2.5 | [24] |
| T. primulinus | 5–6 | no growth | 20–25 | 8–10 | biverticillate | ellipsoidal to fusiform | smooth to finely rough | 2–4 × 1.5–3 | [2] |
| T. shilinensis | 10–11 | no growth | 36–38 | 18–19 | biverticillate | ellipsoidal to broad fusiform | smooth | 2.5–3.5 × 2–2.5 | This study |
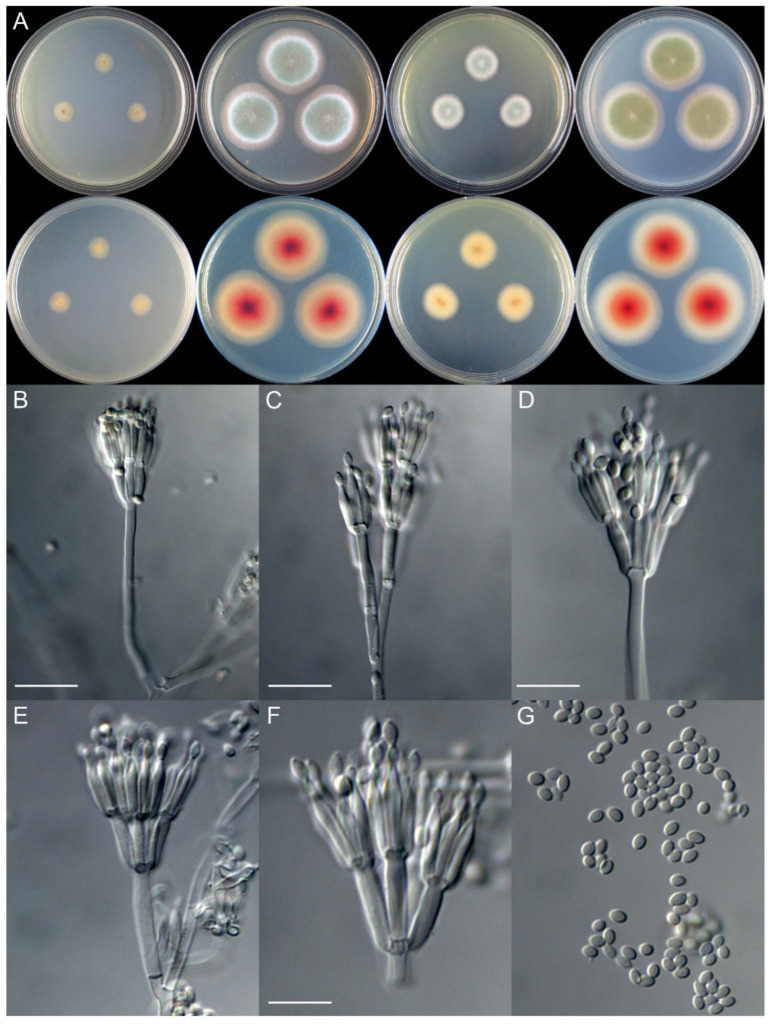
Talaromyces shilinensis X.C. Wang & W.Y. Zhuang, sp. nov. Figure 3
Figure 3.
Colonial and microscopic morphology of Talaromyces shilinensis (XCW_SN259). (A) Colony phenotypes (25 °C, 7 days; top row left to right, obverse CYA, MEA, YES, and PDA; bottom row left to right, reverse CYA, MEA, YES, and PDA); (B–F) Conidiophores; (G) Conidia. Bars: B = 15 µm; C = 12.5 µm; D = 10 µm, applies to E and G; F = 7.5 µm.
Fungal Names: FN570955
Etymology: The specific epithet refers to the type locality
in Talaromyces sect. Talaromyces
Typification: CHINA. Yunnan Province, Kunming City, Shilin Yi Autonomous County, Gui Mountain National Forest Park, 24°38′15″ N 103°35′49″ E, isolated from a rotten twig associated with ascomata of Pseudocosmospora sp., 26 September 2017, Yi Zhang, Yu-Bo Zhang and Huan-Di Zheng 11,825, cultured by Yu-Bo Zhang, XCW_SN259 (holotype HMAS 247854, ex-type strain CGMCC 3.20699)
DNA barcodes: ITS OL638159, BenA OL689845, CaM OL689847, RPB2 OL689849
Colony diam., 7 days, 25 °C (unless stated otherwise): CYA 10–11 mm; CYA 37 °C no growth; MEA 36–38 mm; YES 18–19 mm; PDA 35–37 mm
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, plain; margins wide, entire; mycelia colorless; texture velutinous; sporulation sparse; conidia en masse light yellowish green; soluble pigments absent; exudates absent; reverse almost colorless but light brown at centers
On MEA 25 °C, 7 days: Colonies nearly circular, plain, slightly protuberant at centers; margins very wide, entire; mycelia colorless and white; texture velutinous, funiculose at central areas; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse buff but pink to reddish brown at centers.
On YES 25 °C, 7 days: Colonies nearly circular, plain, slightly protuberant at centers; margins moderately wide, entire; mycelia colorless; texture velutinous; sporulation dense; conidia en masse greyish green; soluble pigments absent; exudates absent; reverse buff but light brown at centers.
On PDA 25 °C, 7 days: Colonies nearly circular, plain, slightly protuberant at centers; margins very wide, entire; mycelia colorless; texture velutinous; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse white, pink to reddish brown at centers.
Micromorphology: Conidiophores biverticillate, rarely quaterverticillate; stipes smooth-walled, 50–110 × 2.0–3.0 μm; metulae 4–6, 8.5–12.5 × 2.5–3.0 μm; phialides acerose, tapering into very thin neck, 4–5 per metula, 9.0–13.0 × 1.8–2.5 μm; conidia ellipsoidal to broad-fusiform, smooth, 2.5–3.5 × 2.0–2.5 μm
Note: This species is a sister of T. primulinus with strong support in the phylogenies inferred from all datasets (Figure 1 and Figures S1–S3), and it also related to T. kabodanensis in the BenA and combined trees (Figure 1 and Figure S1). It has 27 pairwise nucleotide differences from T. primulinus and 23 bp from T. kabodanensis in the BenA dataset; 29 nucleotide differences from T. primulinus in CaM; and 45 nucleotide differences from T. primulinus in RPB2. Morphologically, it differs from T. kabodanensis in the smooth conidia and from T. primulinus in the faster growth rate on CYA, MEA, and YES at 25 °C (Table 3).
4. Discussion
Forty-three species of the Talaromyces have been reported as new to science based on materials collected from China. They are distributed all over the country, especially in southwestern regions, for example, T. chongqingensis and T. wushanicus are from Chongqing, T. albisclerotius, T. guizhouensis, T. penicillioides, T. resinae, T. rosarhiza, and T. tenuis are from Guizhou, T. ginkgonis is from Sichuan, T. neofusisporus and T. qii are from Tibet, and T. aspriconidius, T. aureolinus, T. bannicus, T. rufus, T. shiliensis, and T. yunnanensis are from Yunnan [1,4,6,9,10,11]. This proves that southwestern China is one of the global biodiversity hotspots. In northern China, 13 species were recorded from Beijing; in eastern parts of the country, 9 were from Jiangsu, Shandong, Shanghai, Taiwan, and Zhejiang; and a few species were occasionally found in the south, central, and northeast. This might be due to the frequency of investigations, climates, richness of plants, as well as human activities. We certainly expect to discover more species of the group in unexplored regions and even in surveyed areas in different seasons.
Along with the joining of the two new species, Talaromyces sect. Talaromyces currently possesses up to 86 species around the world. Forty species were originally described as being from Asia, of which 29 are from China, four are from Japan, two are from South Korea and Thailand, respectively, and only one was reported in India, Iran, and Vietnam; 18 taxa are from North America, including 14 from the USA and a single one from Canada, Cuba, Mexico, and Panama; 12 species are distributed in Europe (France, Italy, Netherlands, Spain, UK); six are reported in South America (Brazil, Colombia, and Ecuador); five are from Africa (Ghana and South Africa); and four are from Oceania (Australia and New Zealand). Concerning the known distribution of the genus, one may easily imagine that the biodiversity of Talaromyces may have been underrated, although it is well recognized in areas of East Asia and North America, intensive excursions covering a broad range of areas in the world should be suggested to have a better understanding of the biodiversity of this group.
Acknowledgments
The authors would like to thank Huan-Di Zheng, Yu-Bo Zhang, and Yi Zhang of the Institute of Microbiology, Chinese Academy of Sciences for jointly collecting the samples for this study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8070647/s1, Figure S1: ML phylogeny of Talaromyces sect. Talaromyces inferred from BenA dataset. Bootstrap values ≥70% (left) or posterior probability values ≥0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability; Figure S2: ML phylogeny of Talaromyces sect. Talaromyces inferred from CaM dataset. Bootstrap values ≥70% (left) or posterior probability values ≥0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability; Figure S3: ML phylogeny of Talaromyces sect. Talaromyces inferred from RPB2 dataset. Bootstrap values ≥70% (left) or posterior probability values ≥0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability.
Author Contributions
Conceptualization, W.-Y.Z. and X.-C.W.; methodology, X.-C.W.; software, X.-C.W.; validation, X.-C.W. and W.-Y.Z.; formal analysis, X.-C.W.; investigation, X.-C.W.; resources, X.-C.W. and W.-Y.Z.; data curation, X.-C.W.; writing—original draft preparation, X.-C.W.; writing— review and editing, W.-Y.Z. and X.-C.W.; visualization, X.-C.W.; supervision, W.-Y.Z.; project administration, W.-Y.Z.; funding acquisition, W.-Y.Z. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences newly generated in this study have been submitted to the GenBank database.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This project was supported by the National Natural Science Foundation of China (31750001) and the Key Research Program of Frontier Science, Chinese Academy of Sciences (QYZDY-SSW-SMC029).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang Z.K., Wang X.C., Zhuang W.Y., Cheng X.H., Zhao P. New species of Talaromyces (Fungi) isolated from soil in southwestern China. Biology. 2021;10:745. doi: 10.3390/biology10080745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yilmaz N., Visagie C.M., Houbraken J., Frisvad J.C., Samson R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houbraken J., Kocsube S., Visagie C.M., Yilmaz N., Wang X.C., Meijer M., Kraak B., Hubka V., Bensch K., Samson R.A., et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun B.D., Chen A.J., Houbraken J., Frisvad J.C., Wu W.P., Wei H.L., Zhou Y.G., Jiang X.Z., Samson R.A. New section and species in Talaromyces. MycoKeys. 2020;68:75–113. doi: 10.3897/mycokeys.68.52092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crous P.W., Cowan D.A., Maggs-Kolling G., Yilmaz N., Larsson E., Angelini C., Brandrud T.E., Dearnaley J.D.W., Dima B., Dovana F., et al. Fungal Planet description sheets: 1112–1181. Persoonia. 2020;45:251–409. doi: 10.3767/persoonia.2020.45.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei S., Xu X., Wang L. Four new species of Talaromyces section Talaromyces discovered in China. Mycologia. 2021;113:492–508. doi: 10.1080/00275514.2020.1853457. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen T.T.T., Frisvad J.C., Kirk P.M., Lim H.J., Lee H.B. Discovery and extrolite production of three new species of Talaromyces belonging to sections Helici and Purpurei from freshwater in Korea. J. Fungi. 2021;7:722. doi: 10.3390/jof7090722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pyrri I., Visagie C.M., Soccio P., Houbraken J. Re-evaluation of the taxonomy of Talaromyces minioluteus. J. Fungi. 2021;7:993. doi: 10.3390/jof7110993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H., Wei T.P., Mao Y.T., Ma M.X., Ma K., Shen Y., Zheng M.J., Jia W.Y., Luo M.Y., Zeng Y., et al. Ascodesmis rosicola sp. nov. and Talaromyces rosarhiza sp. nov., two endophytes from Rosa roxburghii in China. Biodivers. Data J. 2021;9:e70088. doi: 10.3897/BDJ.9.e70088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han P.J., Sun J.Q., Wang L. Two new sexual Talaromyces species discovered in estuary soil in China. J. Fungi. 2022;8:36. doi: 10.3390/jof8010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doilom M., Guo J.W., Phookamsak R., Mortimer P.E., Karunarathna S.C., Dong W., Liao C.F., Yan K., Pem D., Suwannarach N., et al. Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: Four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front. Microbiol. 2020;11:585215. doi: 10.3389/fmicb.2020.585215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun X.R., Xu M.Y., Kong W.L., Wu F., Zhang Y., Xie X.L., Li D.W., Wu X.Q. Fine identification and classification of a novel beneficial Talaromyces fungal species from Masson pine rhizosphere soil. J. Fungi. 2022;8:155. doi: 10.3390/jof8020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visagie C.M., Houbraken J., Frisvad J.C., Hong S.B., Klaassen C.H., Perrone G., Seifert K.A., Varga J., Yaguchi T., Samson R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X.C., Chen K., Xia Y.W., Wang L., Li T.H., Zhuang W.Y. A new species of Talaromyces (Trichocomaceae) from the Xisha Islands, Hainan, China. Phytotaxa. 2016;267:187–200. doi: 10.11646/phytotaxa.267.3.2. [DOI] [Google Scholar]
- 15.Wang X.C., Chen K., Qin W.T., Zhuang W.Y. Talaromyces heiheensis and T. mangshanicus, two new species from China. Mycol. Prog. 2017;16:73–81. doi: 10.1007/s11557-016-1251-3. [DOI] [Google Scholar]
- 16.Wang X.C., Chen K., Zeng Z.Q., Zhuang W.Y. Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Sci. Rep. 2017;7:8233. doi: 10.1038/s41598-017-08697-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 19.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 21.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 22.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posada D., Crandall K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 24.Crous P.W., Wingfield M.J., Burgess T.I., Hardy G.E., Crane C., Barrett S., Cano-Lira J.F., Le Roux J.J., Thangavel R., Guarro J., et al. Fungal Planet description sheets: 469–557. Persoonia. 2016;37:218–403. doi: 10.3767/003158516X694499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences newly generated in this study have been submitted to the GenBank database.