Abstract
Recovery of mRNA from environmental samples for measurement of in situ metabolic activities is a significant challenge. A robust, simple, rapid, and effective method was developed for simultaneous recovery of both RNA and DNA from soils of diverse composition by adapting our previous grinding-based cell lysis method (Zhou et al., Appl. Environ. Microbiol. 62:316–322, 1996) for DNA extraction. One of the key differences is that the samples are ground in a denaturing solution at a temperature below 0°C to inactivate nuclease activity. Two different methods were evaluated for separating RNA from DNA. Among the methods examined for RNA purification, anion exchange resin gave the best results in terms of RNA integrity, yield, and purity. With the optimized protocol, intact RNA and high-molecular-weight DNA were simultaneously recovered from 19 soil and stream sediment samples of diverse composition. The RNA yield from these samples ranged from 1.4 to 56 μg g of soil−1 dry weight), whereas the DNA yield ranged from 23 to 435 μg g−1. In addition, studies with the same soil sample showed that the DNA yield was, on average, 40% higher than that in our previous procedure and 68% higher than that in a commercial bead milling method. For the majority of the samples, the DNA and RNA recovered were of sufficient purity for nuclease digestion, microarray hybridization, and PCR or reverse transcription-PCR amplification.
The application of culture-independent nucleic acid techniques has greatly advanced the detection and identification of microorganisms in natural environments (2, 4, 17, 39, 42, 48). However, successful application of molecular techniques relies on effective recovery of nucleic acids from environmental samples. A variety of methods have been developed and used to directly recover nucleic acids from environmental samples (1, 16, 20, 23, 24, 27, 30, 32, 38, 40, 42, 43, 45, 49), but most of the methods are not developed for recovering mRNA from environmental samples. Since RNA is not stable, recovery of intact mRNA from environmental samples is a great challenge.
The RNA/DNA ratio is an important indicator of the metabolic status of bacterial (8, 19, 21, 28, 34) and microbial (10, 11) communities. Such a ratio can allow researchers to address questions concerning whether the response of a microbial community to environmental change is due to a population increase or activity increase. To obtain a reliable RNA/DNA ratio, both RNA and DNA should be recovered from environmental samples without bias. However, unbiased recovery of both DNA and RNA is a significant challenge due to microbial heterogeneity in natural environments, variations in experimental conditions, and differences in interactions of DNA and RNA molecules with environmental matrices. Although it is difficult to eliminate all sources of variation, variation originating from microbial heterogeneity and extraction conditions can be minimized if the RNA and DNA are simultaneously extracted from the same fraction of the sample. Also, in many cases (e.g., marine sediment samples, subsurface soil and groundwater samples, and microbial samples from patients), the amount of sample is very limited, and thus recovering all of the RNA and DNA is critical for microbial analysis. Therefore, simultaneous recovery of both RNA and DNA from the same samples will have great advantages in alleviating the recovery bias and sample quantity limitations. However, no methods designed for recovery of mRNA offer the capability of simultaneous DNA recovery from environmental samples.
Because microbial cells may remain tightly bound to soil colloids, soils high in clay and/or organic matter pose particular challenges to the recovery of RNA and DNA. Thus, the effectiveness and robustness of methods for extracting RNA and DNA need to be evaluated with a variety of diverse samples. Although many methods have been developed for extracting RNA from environmental samples (1, 5, 9, 18, 27, 32, 35, 38, 43, 47), they have not been rigorously tested with a variety of soils and sediments. Thus, the general applicability of these nucleic acid recovery methods is unknown for comparative ecological studies.
An ideal procedure for recovering nucleic acids from environmental samples should meet several criteria. (i) The nucleic acid recovery efficiency should be high and not biased so that the final nucleic acids are representative of the total nucleic acids within the naturally occurring microbial community, (ii) The RNA and DNA fragments should be as large as possible so that molecular studies, such as community gene library construction and gene cloning, can be carried out. (iii) The RNA and DNA should be of sufficient purity for reliable enzyme digestion, hybridization, reverse transcription, and PCR amplification. (iv) The RNA and DNA should be extracted simultaneously from the same sample so that direct comparative studies can be performed. This will also be particularly important for analyzing samples of small size. (v) The extraction and purification protocol should be kept simple as much as possible so that the whole recovery process is rapid and inexpensive. (vi) The extraction and purification protocol should be robust and reliable, as demonstrated with many diverse environmental samples. However, none of the previous nucleic acid extraction methods have been evaluated and optimized based on all the above important criteria.
The objective of this study was to develop a robust, simple, rapid, and effective method for simultaneous recovery of intact RNA and DNA from diverse environmental samples. We adapted our previous method that was shown to offer excellent recovery of high-molecular-weight DNA from soils of diverse composition (49). We systematically developed our extraction procedure by considering all of the above criteria. Our results showed that this is a robust, reliable, and effective method for simultaneously recovering both mRNA and DNA from the same environmental sample.
MATERIALS AND METHODS
Soil collection and characterization.
Soils and stream sediments (Table 1) collected in the Great Smoky Mountains and at the Oak Ridge National Laboratory (ORNL) Environmental Research Reservation in eastern Tennessee were homogenized by manual mixing, frozen in liquid nitrogen, transported on dry ice, and stored at −40°C. Soil samples were air dried, weighed, and used for soil characterization. Soil samples used for particle size analysis were pretreated with hydrogen peroxide to remove organic material and dispersed using sodium-hexametaphosphate and sodium carbonate (14). Particle size analyses were performed using a pipette method (22). Particles of >0.053 mm in diameter were separated by wet sieving. Soil pH was determined in a 1:1 (wt/wt) soil-H2O slurry (26). Total organic carbon was determined after removing inorganic carbon in 10% HCl followed by boiling and washing in distilled water. Samples were dried, pulverized, and examined for remaining carbon and nitrogen content in a PE 2400 series II CHNS/O analyzer (Perkin-Elmer, Norwalk, Conn.).
TABLE 1.
Soil and sediment characteristics
| Samplea | Collection date (mo.day.yr) | Soil colorb | Moisture (%) | Soil pH | Particle size distribution (%)
|
% C | % N | μg of DNA g of soil−1 (dry wt)c | μg of RNA g of soil−1 (dry wt)c | Soil texture | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sand | Silt | Clay | ||||||||||
| WB E-A1 | 09.03.98 | 10YR 5/2 | 6.0 | 5.57 | 51.3 | 40.2 | 8.5 | 2.4 | 0.1 | 86 ± 54 | 9.3 ± 4.0 | Loam |
| WB E-A1 | 04.28.98 | 10YR 4/3 | 22.4 | 5.76 | 78.5 | 20.3 | 1.4 | 4.6 | 0.3 | 46 ± 12 | ND | Clay loam |
| WBW-A1 | 04.28.98 | 10YR 6/3 | 57.2 | 4.26 | 50.6 | 37.3 | 12.1 | 3.9 | 0.3 | 435 ± 66 | 56.1 ± 4.5 | Sandy loam |
| ND-A3 | 08.27.98 | 10YR 5/4 | 35.4 | 3.67 | 70.9 | 17.4 | 11.7 | 2.1 | 0.1 | 157 ± 38 | 19.1 ± 17.2 | Sandy loam |
| ND-A1 | 05.12.98 | 10YR 5/4 | 42.4 | 5.05 | 66.8 | 10.3 | 22.8 | 2.6 | 0.1 | 97 ± 23 | 13.3 ± 11.5 | Sandy clay |
| MC-A1 | 05.12.98 | 10YR 5/4 | 25.4 | 4.23 | 34.3 | 62.2 | 3.5 | 5.8 | 0.2 | 43 ± 8 | 15.2 ± 2.6 | Silt loam |
| MB-A1 | 05.12.98 | 10YR 5/1 | 28.4 | 4.34 | 78.6 | 20.9 | 0.5 | 3.5 | 0.2 | 53 ± 9 | 16.7 ± 1.5 | Loamy sand |
| RC-A1 | 05.12.98 | 10YR 4/4 | 40.4 | 3.89 | 50.2 | 41.5 | 8.3 | 5.8 | 0.2 | 107 ± 38 | 20.2 ± 3.0 | Sandy loam |
| RC-A2 | 08.27.98 | 10YR 5/2 | 24.9 | 3.24 | 18.9 | 65.7 | 15.4 | 5.17 | 0.8 | 131 ± 15 | 7.8 ± 0.7 | Silt loam |
| KA | 01.09.99 | 10YR 3/3 | 25.8 | 7.69 | 28.8 | 47.8 | 23.5 | 5.1 | 0.3 | 206 ± 34 | 35.5 ± 12.7 | Loam |
| WAG-5 | 09.98 | 10YR 4/4 | 13.8 | 7.51 | 84.2 | 11.1 | 4.7 | ND | ND | 244 ± 6 | 2.6 ± 0.1 | Loamy sand |
| WBE-S1 | 05.12.98 | 10YR 5/3 | 29.9 | 5.68 | 67.9 | 24.8 | 7.2 | ND | ND | 97 ± 19 | 11.9 ± 1.0 | Sandy loam |
| WBW-S1 | 08.27.98 | 10YR 3/2 | 58.3 | 5.94 | 93.2 | 5.0 | 1.9 | ND | ND | 137 ± 57 | 22.5 ± 7.8 | Sand |
| ND-S1 | 08.27.98 | 10YR 4/4 | 29.0 | 4.94 | 98.3 | 0.8 | 0.9 | ND | ND | 23 ± 11 | 3.0 ± 2.0 | Sand |
| MC-S1 | 08.27.98 | 10YR 5/4 | 31.2 | 5.56 | 95.0 | 3.5 | 0.6 | ND | ND | 61 ± 8 | ND | Sand |
| MB-S1 | 08.27.98 | 10YR 5/4 | 22.5 | 5.28 | 96.5 | 0.9 | 2.6 | ND | ND | 49 ± 4 | 3.9 ± 0.5 | Sand |
| RCK-S1 | 08.27.98 | 10YR 5/1 | 23.4 | 4.09 | 82.7 | 6.5 | 10.8 | 13.2 | 0.4 | 41 ± 28 | 1.4 ± 1.3 | Sandy loam |
| H17-OH | 09.17.98 | 7.5YR 2/0 | 66.0 | 6.65 | ND | ND | ND | 41.2 | 4.6 | 42 ± 16 | 3.7 ± 2.5 | Loamy sand |
| CPLC-OH | 01.09.99 | 7.5YR 2/0 | 53.2 | 6.38 | 38.9 | 14.4 | 55.5 | 27.2 | 1.2 | 306 ± 254 | 12.1 ± 1.1 | Clay |
Sample designations: A, A horizon; S, stream sediment; SM, collected in the Great Smoky Mountains National Park; WBE, Walker Branch East (ORNL); WBW, Walker Branch West (ORNL); ND, Noland Divide (SM); MC, Mossy Creek (SM); MB, Minni Ball (SM); RC, Rattle Box Creek (SM); CLPC, composted leaf and clay; KA, Knoxville A horizon; ND, not determined.
Soil data were determined from the Munsell color chart as Hue (10 or 7.5), Y = yellow/R = red; lower intensity values indicate a darker soil.
Results are means ± standard deviations. The standard deviations for RNA and DNA were estimated based on three replicates.
Preparation of reagents and materials.
All stock solutions, working solutions, and water used for reagent preparation were treated with 0.1% diethylpyrocarbonate (DEPC) overnight at 37°C and autoclaved. All glassware and utensils used for nucleic acid extraction were baked at 500°C for 4 h. Nondisposable plasticware was treated with 0.1% DEPC or thoroughly cleaned with RNase Away (Life Technologies, Grand Island, N.Y.).
Optimization. (i) Optimization of buffer composition and extraction conditions.
Pure culture was used to optimize procedure components such as incubation time, solution pH, and other controllable factors that could influence the extent of RNA damage. The conditions for extracting total RNA were optimized with a pure culture Pseudomonas sp. strain KC on the basis of our previously described soil DNA extraction method (49). Cultures (2 or 4 ml) grown overnight at 30°C in 100 ml of R2A medium (12) were collected by centrifugation at 5,000 × g for 10 min, and lysed with the SDS-based lysis method without grinding or freeze-thawing cycles. The buffer was changed from pH 8.0 to pH 7.0, and the incubation time at 65°C was varied from 0.5 to 2 h in 0.5 h increments. The effect of proteinase K on the integrity of RNA was examined using concentrations of 10, 50, and 100 μg ml−1. The effects of salt concentration and detergents (N-laurylsarcosine and sodium dodecyl sulfate [SDS]) on RNA integrity were tested at pH 7.0. Total nucleic acids were electrophoresed at 2.5 or 3 V cm−1 in 1 or 1.2% agarose formaldehyde gels as described by Maniatis et al. (25). Because the size of mRNA is heterogeneous and the abundance is generally low, it is difficult to evaluate mRNA quality based on electrophoresis. However, rRNAs are abundant, and distinct bands can be visualized in agarose gels. Since 23S rRNA is bigger than 16S rRNA, the 23S band will be more intense than the 16S band if no degradation occurs. Thus electrophoretic assessment of 23S/16S rRNA stoichiometry was used as an indication of total RNA quality (25). Optimal extraction conditions determined by pure culture assays were used in all subsequent soil nucleic acid extraction analyses.
(ii) Effects of lysis methods on RNA quality.
Lysis of soil microorganisms that include gram-positive bacteria and fungi is a significant challenge. The effects of four combinations of lysis methods concerning the quality and quantity of total nucleic acids were compared using 4-g samples of an A-horizon soil collected in Knoxville, Tenn. (sample KA in Table 1): (i) grinding and freeze (−70°C)-thawing (65°C) with proteinase K, (ii) grinding and freeze-thawing without proteinase K, (iii) grinding in a denaturing solution with freeze-thawing, and (iv) grinding in a denaturing solution. All soil samples were supplemented with sterilized sand and ground in liquid N2 until thawed three times. The samples for treatment iii and treatment iv were ground after adding 1 ml of denaturing solution (4 M guanidine isothiocyanate, 10 mM Tris-HCl [pH 7.0], 1 mM EDTA, 0.5% 2-mercaptoethanol). Samples for treatments i and ii were ground after adding 1 ml of extraction buffer (see below). The samples used for treatments i and iii were subjected to three freeze-thawing cycles. Each treatment had three replicates. Nucleic acid extraction was carried out as described previously (50) except for buffer pH (7.0) and incubation time (1 h at 65°C).
(iii) Separation of RNA from DNA and purification.
Total crude nucleic acids extracted from 4 g of sample KA (Table 1) were used to evaluate two different approaches for separating RNA from DNA in crude extracts. First, pH-dependent differential organic extraction was tested using Trizol LS Reagent (Life Technologies) and a procedure for the rapid extraction of RNA from mammalian tissue and gram-negative bacteria (3, 7). Second, RNA was separated from DNA using a Qiagen (Santa Clarita, Calif.) Tip 100 RNA-DNA purification system. DNA that did not bind to the column and remained in the flowthrough (flowthrough DNA) was precipitated with 0.1 volume of 5 M NaCl and 0.6 volume of isopropanol and combined with the DNA that was eluted from the column (column DNA).
(iv) Nucleic acid purification.
After separation, five commercial reagent systems were tested for further purification of the separated RNA: (i) Sephadex G-75 size exclusion resin (500 μl); (ii) Qiagen Total Nucleic Acid purification system with the Qiagen Tip 20; (iii) the SV Total RNA isolation system (Promega, Madison, Wis.); (iv) the NucleoBond system (Clontech, Palo Alto, Calif.); and (v) RNA Tack Resin (BIOTECX Laboratories, Houston, Tex.). RNA purification with different commercial systems was performed according to the manufacturer's instructions. DNA was purified twice using the Wizard DNA Clean-Up system (Promega) as described previously (49). Purified RNA was treated with RQ1 RNase-free DNase I (Promega) according to the manufacturer's instructions.
Extraction of RNA and DNA from diverse soils and sediments.
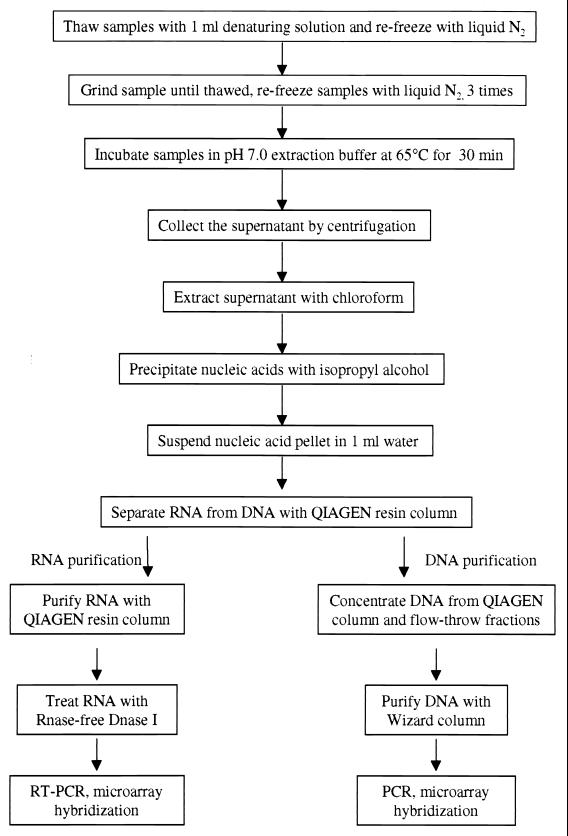
The following optimized procedure was used to simultaneously recover RNA and DNA from diverse soils and sediments (Fig. 1). Two-gram soil samples measured from frozen stocks were mixed with 2 g of baked sterile sand and transferred to an RNase-free mortar. The samples were saturated with 1 ml of denaturing solution, frozen in liquid nitrogen, and ground until thawed. The process of freezing in liquid nitrogen and grinding until thawed was repeated twice. Ground samples were transferred to 50-ml polypropylene centrifuge tubes, frozen in liquid nitrogen, and stored at −40°C until use. After addition of 9 ml of extraction buffer (100 mM sodium phosphate [pH 7.0], 100 mM Tris-HCl [pH 7.0], 100 mM EDTA [pH 8.0], 1.5 M NaCl, 1% hexadecyltrimethylammonium bromide [CTAB] and 2% SDS), the samples were incubated for 30 min at 65°C with gentle manual mixing every 10 min and centrifuged at 1,800 × g for 10 min. The supernatants were poured into prechilled tubes on ice containing 20-ml aliquots of 24:1 chloroform-isoamyl alcohol. The soil pellets were extracted two more times (optional) by adding 5 ml of extraction buffer, mixing for 10 s, incubating at 65°C for 10 min, and centrifuging as before. The combined supernatants were centrifuged at 1,800 × g for 20 min. The aqueous phase was transferred to a 50-ml Teflon Oak Ridge tube, precipitated with 0.6 volume of isopropyl alcohol for 30 min at room temperature, and centrifuged at 16,000 × g for 20 min at 20 to 25°C. The crude nucleic acid pellets were suspended in 1 ml of DEPC-treated H2O and passed through 5 ml of a Sephadex G-75 resin slurry (Pharmacia, Piscataway, N.J.) by centrifugation at 750 × g (27).
FIG. 1.
Schematic summary of the optimal protocol for simultaneous recovery of RNA and DNA from the same sample.
Quantification of DNA and RNA.
Nucleic acids were quantified with a PE Biosystems HTS 7000 BioAssay reader using clear flat-bottomed 96-well plates. Test plates were read from the bottom using a gain of 60 and an integration time of 60 μs. Purified RNA samples were quantified in ethidium bromide (10 μg ml−1) by comparing emission at 695 nm against that of a serial twofold dilution of yeast RNA type III (Aldrich, St. Louis, Mo.) ranging from 1 μg to 5 mg ml−1. DNA samples were quantified against a serial twofold dilution of Escherichia coli DNA type VIII (Aldrich) ranging from 1 μg to 1 mg ml−1. Background signal intensity was subtracted using the mean for three wells having only distilled water added to the ethidium bromide solution.
PCR amplification and reverse transcription.
Two micrograms of total DNase I-treated RNA was reverse transcribed using the reverse complementary primer GS1β (5′ GAT GCC GCC GAT GTA GTA 3′) for the eubacterial glnA (glutamine synthetase) gene with 200 U of Superscript II RNase H− reverse transcriptase (Life Technologies) in a total volume of 20 μl. Aliquots (2 μl) of the reverse transcription products were used for PCR amplification in PCR buffer (50 mM KCl, 10 mM Tris [pH 9.0], 0.1% Triton X-100, 2.5 mM MgCl2, 0.2 mg of bovine serum albumin ml−1) with 10 pmol each of conserved primers GS1β and GS2γ (5′ AAG ACC GCG ACC TTP ATG CC 3′), which generate a 153- or 156-bp fragment of the glnA gene. PCR amplification conditions consisted of disassociation at 94°C for 30 s, annealing at 60°C for 1 min, and extension at 72°C for 1 min for 30 cycles, followed by a final extension at 72°C for 7 min using a GeneAmp PCR System 9700 thermal cycler (PE Biosystems, Foster City, Calif.). The purity of the DNA was also examined by PCR amplification of 16S rRNA genes (49) and nitrite reductase genes (6).
Microarray hybridization.
DNA microarrays for monitoring bacteria involved in nitrogen cycling were constructed (L. Y. Wu, D. Thompson, G.-S. Li, R. Hurt, J. M. Tiedje, and J.-Z. Zhou, submitted for publication), including heme-containing nitrite reductase (nirS), copper-containing nitrite reductase (nirK), and ammonia and methane monooxygenase (amoA and pmoA) genes from pure cultures (22 nirS genes, 9 nirK genes, and 4 amoA genes), and those cloned from marine sediments (27 nirS genes, 9 nirK genes, and 18 pmoA genes); 16S rRNA genes from 10 denitrifiers and five yeast genes were used as positive and negative controls. Microarray fabrication was carried out as described elsewhere (Wu et al., submitted).
Purified RNA (2.5 μg) was labeled in a total volume of 40 μl with 1 U of RNase inhibitor (Life Technologies) μl−1; 400 U of Super Script II reverse transcriptase; 0.5 mM dATP, dGTP, and dTTP; 0.125 mM dCTP; and 1 mM Cy5-dCTP (Amersham Pharmacia Biotech, Piscataway, N.J.). The reaction mixtures were incubated for 1 h at 42°C. Labeled cDNA was purified using Sephadex G-50 molecular exclusion resin (Pharmacia), concentrated in a SpeedVac at 40°C for 1.5 h, and suspended in distilled H2O. Purified DNA (2.5 μg) was labeled with Cy5-dCTP as described elsewhere (Wu et al., submitted). Microarray hybridization, scanning, image processing, and signal quantification were performed as described elsewhere (Wu et al., submitted).
RESULTS
Soil properties.
The physical and chemical properties of the soils and sediments used in this study were diverse (Table 1). Soils and sediments were classified as loam, sandy loam, silt loam, sandy clay, or clay, with clay contents ranging from 0.5 to 55.5%. The percentage of carbon varied from 2.1 to 41.2. Soil pH ranged from 3.7 to 7.7. A-horizon soils collected at the Walker Branch, Noland Creek, Rattlebox Creek, and WAG-5 sites during August and September of 1998 had a moisture content of 20% ± 12.9%, while the A-horizon samples collected in eastern Tennessee during the spring of 1998 had a moisture content of 36% ± 19% (means ± standard deviations).
Optimization of buffer composition and extraction conditions.
Extraction conditions were optimized for RNA stability with a pure culture by varying individual components of the procedure. The intactness of extracted RNA was improved by reducing the pH of the extraction buffer from 8.0 to 7.0 and decreasing the incubation time at 65°C from 2 to 1 h (data not shown). RNA damage was observed for all treatments with various concentrations of proteinase K, indicating that the pretreatment with proteinase K did not protect RNA from degradation. None of the other modifications tested such as salt concentration and alternative detergents improved RNA quality.
Effects of cell lysis methods on RNA recovery.
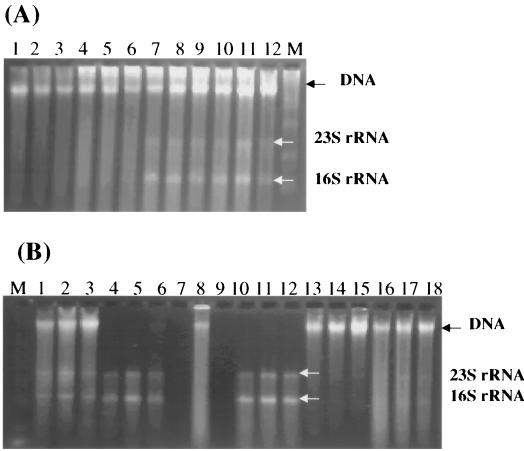
The effects of lysis methods on RNA stabilization were compared by using soil sample KA (Fig. 2A). While RNA was degraded in the treatments without a denaturing solution (lane 1 to 6), visible 16S and 23S rRNA bands were observed for the treatments with a denaturing solution (lanes 7 to 12). This suggested that the presence of denaturants during grinding is essential for RNA stabilization. Also, the quantity of recovered DNA from the treatments without denaturants (lanes 1 to 6) was considerably less than that recovered with denaturants (lanes 7 to 12). No significant differences in both RNA and DNA quality and quantity were observed between the treatments with freeze-thawing (lanes 7 to 9) and without freeze-thawing (lanes 10 to 12).
FIG. 2.
Optimization of nucleic acid recovery protocol. All of the treatments were carried out in triplicate. (A) Effects of cell lysis treatments on RNA integrity. Lanes: 1 to 3, grinding and freeze-thawing with proteinase K; 4 to 6, grinding and freeze-thawing without proteinase K; 7 to 9, grinding and freeze-thawing with denaturing solution; 10 to 12, grinding and denaturing solution without freeze-thawing; M, RNA ladder. (B) Comparison of methods for RNA and DNA separation. Lanes: M, RNA ladder; 1 to 3, total crude nucleic acids from KA soil; 4 to 6, RNA fraction from Trizol; 7 to 9, DNA fraction from Trizol; 10 to 12, RNA fraction from Qiagen system; 13 to 15, flowthrough DNA fraction from Qiagen system; 16 to 18, column DNA from Qiagen system.
Separation of RNA from DNA and purification.
Total crude extracts from the KA soil sample (Fig. 2A, lanes 10 to 12) were selected for evaluating separation procedures because the dark color of the crude nucleic acid suspension suggested a high organic contaminant content. While Trizol reagent yielded reasonable RNA recovery (Fig. 2B, lanes 4 to 6), the band intensity of RNA obtained with the Qiagen system was greater (Fig. 2B, lanes 10 to 12). Also, most of the visible discoloring organic contaminants cofractionated with the RNA by the Trizol method, but much less cofractionated with the Qiagen RNA-DNA isolation system.
Substantially less DNA was recovered with Trizol reagent than with the Qiagen system (Fig. 2B, lanes 7 to 9 and 13 to 18). DNA recovered using Trizol was difficult to resuspend, so that no or poorly soluble DNA was obtained (lanes 7 to 9). Although more DNA was recovered with the Qiagen system than with Trizol reagent, much of the high-molecular-weight DNA eluted with the flowthrough that is discarded according to the manufacturer's protocol. This is likely due to the interference of organic contaminants with DNA binding to anion exchange resins. The DNA in the flowthrough was recovered by precipitation and combined with the DNA eluted from the column. Based on both RNA and DNA quality and quantity, the Qiagen system was selected for later studies.
It was not clear whether RNA contaminated with organic matter could be effectively separated from DNA using the Qiagen system. The DNA fraction that bound to Qiagen resin (Fig. 2B, lanes 13 to 18) contained low-molecular-weight nucleic acids that could be sheared DNA or organic contaminants complexed with RNA. We examined both the flowthrough and column DNA for sensitivity to nucleases. RNase-free DNase I treatment completely eliminated the nucleic acid present in both the flowthrough and resin-binding fractions, while treatment with DNase-free RNase A resulted in no change in the appearance of the nucleic acids from either fraction indicating that the low molecular weight nucleic acids were primarily DNA.
RNA purification.
Five different methods were compared for further RNA purification. Both Qiagen and SV total RNA purification systems removed the majority of the brown color from the soil RNA fractions. The RNA Tack, NucleoBond, and Sephadex G-75 resins were less effective. The soil RNA purified using the Qiagen system and the SV Total RNA isolation system supported reverse transcription and PCR amplification (data not shown). However, the quantity of RNA recovered using the SV Total RNA isolation system was substantially less than that recovered using the Qiagen system. Therefore, the Qiagen system was selected for RNA purification.
Extraction of RNA and DNA from diverse soils and sediments.
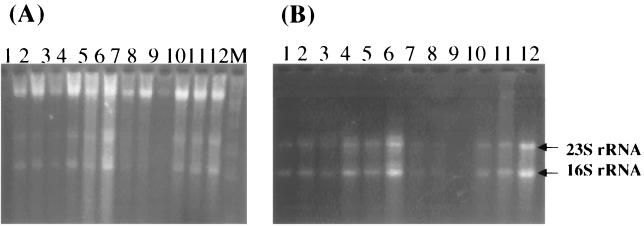
The optimized method was evaluated with 19 soils and sediments of diverse composition (Table 1). 23S and 16S bands were observed for the majority of the soil samples, indicating that the soil RNA was effectively protected from enzymatic degradation (Fig. 3 [not all data are shown]). However, the 23S bands were not more intense than the 16S band, as would be expected for optimal rRNA stoichiometry, suggesting that minor degradation of RNA had occurred for these samples. Similar to our previous study (49), most of the soil DNA fragments were larger than 23 kb.
FIG. 3.
Nucleic acids recovered from soils and stream sediments. (A) Total crude nucleic acids. 1% of the total nucleic acid yield from 2 g of A-horizon soils and 2.5% of the total yield from 4 g of stream sediments were loaded. Lanes: 1, ND-A1; 2, MB-A1; 3, MC-A1; 4, RC-A1; 5, WBE-A1; 6, WBW-A; 7, ND-S1; 8, MB-S1; 9, MC-S1; 10, RC-S1; 11, WBE-S1; 12, WBW-S1. (B) Purified RNAs. 2% of total RNA samples from 2 g of A-horizon soil samples and 5% of total purified RNA from 4 g of stream sediment samples were loaded. The lane numbers of the samples are the same as shown in panel A.
Because fluorometric and absorbance measurements of nucleic acid samples contaminated with humic acid-like materials are unreliable (8, 23, 27, 32, 35), the concentrations listed in Table 1 were measured after purification of the samples. The average DNA yields ranged from 23 to 435 μg of DNA g (dry weight) of soil−1. The average RNA yield for samples varied from 1.4 to 56.1 μg of RNA g of soil−1. The variation in DNA and RNA recovery among these samples was as high as onefold for some soils. This could be due to the heterogeneity of microbial populations and/or to losses during fractionation and purification. Less variation was observed for stream sediments. Among the A-horizon soil samples, significant relationships were observed between RNA yield and moisture content (R = 0.71; P = 0.02), and between RNA yield and DNA yield (R = 0.71; P = 0.02).
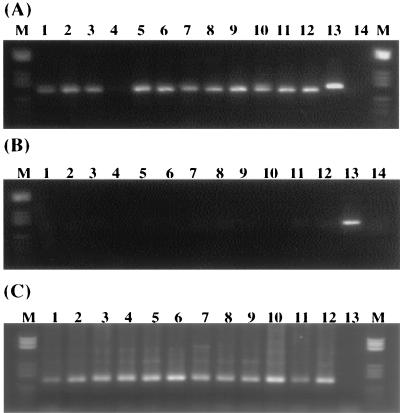
The mRNA extracted and purified from most of the samples supported reverse transcription and PCR amplification. A 153- and/or 156-bp PCR product of the bacterial glutamine synthetase gene glnA (Fig. 4A) was routinely reverse transcribed and amplified from all of the soil and sediment RNA samples except for stream sediment MC-S1 and the ND-A3 and RC-A2 A-horizon soils. Measurable glnA amplification did not occur for purified RNA that was not reverse transcribed (Fig. 4B). Densitometry showed that the extent of product formation in the negative control reactions (Fig. 4B) could contribute to no more than 0.8% of the signal intensities recovered by reverse transcription-PCR shown in Fig. 4A. Primers for the copper nitrite reductase gene (nirK) also produced the expected PCR product after reverse transcription. All DNA samples purified by this method have supported PCR amplification of the glnA gene (Fig. 4C), the nirK gene, the nirS gene, the amoA gene, the sulfite reductase gene, and the 16S gene (data not shown).
FIG. 4.
PCR amplification of the purified nucleic acids with the primers specific to the ginA gene. (A) Reverse transcription and PCR amplification of the purified RNAs. (B) PCR amplification directly from RNA without reverse transcription. (C) PCR amplification of the purified DNA. The lane numbers, 1 to 12, correspond to the samples in Fig. 3A. Lane 13, 174-bp positive control; lane 14, negative control; lane M, marker V.
The quality of the DNA and RNA extracted from the soil and stream sediment samples shown in Fig. 3 was further examined using microarray hybridization. The purified DNA and cDNA prepared from isolated RNA hybridized well with functional gene microarrays for all of the samples of different physical and chemical characteristics. The hybridization patterns between the DNA (gene content) and RNA (gene activity) were similar but were different with respect to specific spot intensity (data not shown). No hybridization was observed with the negative yeast gene controls on the microarrays. These data provide strong evidence indicating that the recovered mRNA can support reverse transcription and that recovered RNA and DNA were both of suitable purity for microarray hybridization analysis.
Comparison of different methods for DNA recovery efficiency.
The DNA yields from soils and sediments reported here are substantially higher than those reported in other studies (9, 23, 30, 40, 49). This could be due to differences in cell lysis efficiency, protection of nucleic acids from nuclease degradation, and/or the biological activity of the samples used in the analysis. To determine whether the current procedure is more effective for recovering nucleic acids, DNA was extracted from the KA soil sample by the current method, our previous DNA extraction method (49), and a bead milling procedure (MoBio, Solana Beach, Calif.). The DNA yield by our current extraction procedure was 205.9 ± 34.4 μg g of soil−1, and by our previous DNA extraction procedure the yield was 154.6 ± 59.2 μg g of soil−1. Also, the current procedure yielded an average of 68% more DNA than a commercial bead milling method (122.6 ± 44.4 μg g of soil−1). DNA recovered by the bead milling procedure ranged in size from 4 to 9 kb, whereas the bulk of the DNA recovered by our method is larger than 23 kb. These results suggest that this method is more effective than our previous method and the commercially available bead milling method with regard to total nucleic acids yield.
DISCUSSION
A key step for culture-independent nucleic acid approaches is the direct extraction of nucleic acids from environmental matrices. The parameters critical to effective recovery of nucleic acids include the efficiency of cell lysis, efficiency of nucleic acid recovery after lysis, and purification from contaminating humic acid-like organic matter (41, 46, 49). Although many methods have been published and successfully used, the effective recovery of nucleic acids from environmental samples, particularly from soils, is still a challenge.
Advantages of the method.
Compared to other methods, the main advantages of this method are as follows.
(i) RNA stabilization.
Obtaining a high quality RNA sample (i.e., mRNA) is critical for using RNA as an indicator of activity. The method we have presented here allows us to obtain high-quality RNA. The obtained RNA is pure enough for enzymatic manipulation.
(ii) Simultaneous extraction.
Most of the current methods recover DNA and RNA separately either from replicate soil samples (5, 32) or from separate fractions of crude extracts (9, 47) using RNase or DNase treatments. Although these methods were described as simultaneous, they are not really simultaneous, because to obtain RNA (or DNA), the DNA (or RNA) in the fraction of the crude extract is destroyed by enzymes and, thus, lost. Such methods could not be used when the samples are limited in quantity. Although CsCl-based ultracentrifugation procedures (38) allow recovery of both mRNA and DNA from the same fraction of crude extract, the methods are time-consuming, and the RNA fractions were not demonstrated to routinely support enzymatic manipulation.
(iii) High-molecular-weight DNA.
High-molecular-weight DNA is one of the important criteria for environmental studies because sheared community DNA is not suitable for cloning and can cause PCR amplification artifacts (17, 39). In contrast to the commercial bead milling-based methods, the method presented here is consistently able to recover high-molecular-weight DNA (>23 kb).
(iv) DNA yield.
The DNA yields from soils and sediments reported in Table 1 are substantially higher than those reported in other studies (9, 23, 30, 40, 49). Comparative studies with the same soil samples indicated that the method presented in this study is more effective than our previous method (50) and a commercially available bead milling method.
(v) Robustness and reliability.
This extraction and purification protocol has been evaluated with more than 20 diverse soil and sediment samples presented in this study and with more than 50 samples that were not presented.
(vi) Comprehensiveness.
The presented methods were developed systematically by considering all of the criteria mentioned in the introduction. The method presented here does meet these requirements, although not perfectly. To our knowledge, no other method has been evaluated and developed based on all the above-mentioned important criteria and with so many different samples.
Our previous studies (49) showed that the combination of mechanical, physical, and chemical methods efficiently recovers high-molecular-weight DNA from soils of diverse composition. Although some comparative tests such as direct bacterial counts have not been performed in this study, the current procedure was demonstrated to be at least as effective as our previous extraction method with regard to cell lysis, and such comparative tests had been performed for the previous extraction method (49). Because nuclease activities are minimized at the beginning of the extraction process, the present protocol should be more effective than our previous method with regard to nucleic acids stabilization.
The key difference between the present procedure and our previous method is that sample grinding is performed at a temperature below 0°C in a denaturing solution to immediately inactivate both cellular (36) and extracellular nuclease activities. Moreover, both our current and previous grinding-SDS-based extraction procedures are more effective than the bead milling method. One reason could be that grinding is more efficient than bead milling for homogenizing soils, releasing bacteria bound to soil colloids, and mechanically disrupting the cell wall. Because grinding seems to be the most-efficient lysis treatment available today (13), the combination of grinding-based mechanical lysis and SDS-based chemical lysis methods should be effective for most nucleic acid-based environmental studies, especially for the studies in which microbial community diversity or structure is of particular interest. The DNA from all of the soils and sediments examined can be amplified and hybridized. Although RNA purified from most of these samples could be reverse transcribed and amplified, some variation in RNA purity was observed. For example, the RNA isolated from samples ND-A3, RC-A1, and RC-A2 was difficult to purify, and the purified RNA samples could not be routinely reverse transcribed and amplified. We found that a second purification using a Qiagen column improved the purity but resulted in some RNA loss.
Similar to the results reported by Picard et al. (33), small amounts of nucleic acids were still recovered after the second and even the third wash. Thus, repeated washings of the soil pellets appeared to be necessary to obtain maximum recovery. However, this may depend on soil types. For some soils, repeated washings could be omitted. Also, if the samples are heavily contaminated with humic substances, as indicated by the dark color of the samples, effective RNA separation from DNA might be difficult using Qiagen anion exchange resin. Additional steps before RNA and DNA separation may be necessary. After suspending the RNA-DNA pellets in 1 ml of water, we found that passing the samples through Sephadex G-75 and then suspending the samples in extraction buffer (1:9, vol/vol), followed by chloroform extraction and alcohol precipitation, facilitated the separation of RNA from DNA and improved the purity of the final product. This is likely because humic substances may interfere with RNA and DNA binding to the resin.
For separating RNA from DNA, anion exchange resins worked better than pH-based differential organic extraction. Because organic contaminants may inhibit DNA binding to Qiagen resin, a substantial proportion of the DNA remains in the flowthrough even if the resin binding capacity is not exceeded. However, the DNA that remained in the flowthrough could be recovered by precipitation. Damage to the RNA fraction was minimal with the Qiagen RNA-DNA isolation system as judged by a comparison of 16S and 23S band intensities in agarose gels before and after separation (Fig. 2). If quantitative recovery of the DNA fraction is not particularly important, Trizol reagent is useful, because it is quicker and less expensive than commercially available anion exchange reagent systems.
The method for simultaneous recovery of RNA and DNA described here offers the potential for using mRNA/DNA ratios (11, 28, 33) for comparing the differences of microbial activities among different samples. For measuring gene expression levels in environmental samples, the recovery efficiency of RNA and DNA from a sample must be known. However, evaluating DNA and mRNA recovery from indigenous soils is extremely difficult and challenging with currently available technology, because nucleic acid recovery depends on soil texture, composition, contaminants, and the interactions between soils and microorganisms, which may lead to differential nucleic acids adsorption (11, 15, 31, 37). There are two main approaches to evaluate nucleic acid recovery from soils. One is a 32P-based radioactive approach. One can seed a known amount of radiolabeled DNA and mRNA prior to extraction and then evaluate how much labeled nucleic acid is recovered to estimate the recovery efficiency of nucleic acids from the samples. However, this approach is not amenable to this extraction procedure because of the difficulty with effective shielding and contamination of equipment such as mortars, pestles, and spatulas that are used during the grinding process. Also, due to safety issues, the recovered DNA and mRNA with 32P-labeled nucleic acids have very limited use and could not be used routinely. An alternative approach is to use fluorescence detection methods by seeding known amounts of fluorescence-labeled nucleic acids and then calculating the recovery efficiency based on the recovered fluorescence-labeled nucleic acids. Because of the interference of humic acid-like materials and stability of the dyes, the results are generally not reliable and reproducible. A reliable robust procedure for routinely estimating recovery efficiency needs be developed and validated.
ACKNOWLEDGMENTS
This research was supported by the Department of Energy Natural and Accelerated Bioremediation Research and Biotechnology Investigations-Ocean Margins Programs; ORNL is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
REFERENCES
- 1.Alm E W, Stahl D A. Critical factors influencing the recovery and integrity of rRNA extracted from environmental samples: use of an optimized protocol to measure depth-related biomass distribution in freshwater sediments. J Microbiol Methods. 2000;40:153–162. doi: 10.1016/s0167-7012(00)00120-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman J, Triplett E W. Rapid and direct method for extraction of RNA from soil. Soil Biol Biochem. 1997;29:1621–1624. [Google Scholar]
- 6.Braker G, Zhou J-Z, Wu L-Y, Devol A H, Tiedje J M. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in marine sediment communities. Appl Environ Microbiol. 2000;66:2096–2104. doi: 10.1128/aem.66.5.2096-2104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium isothiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Dell' Anno A, Fabiano M, Duineveld G C A, Kok A, Danovaro R. Nucleic acid (DNA, RNA) quantification and RNA/DNA ratio determination in marine sediments: comparison of spectrophotometric, fluorometric, and high-performance liquid chromotography methods and estimation of detrietal DNA. Appl Environ Microbiol. 1998;64:3238–3245. doi: 10.1128/aem.64.9.3238-3245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duarte G F, Rosado A S, Seldin L, Keijzer-Wolters A C, van Elsas J D. Extraction of ribosomal RNA and genomic DNA from soil for studying the diversity of the indigenous bacterial community. J Microbiol Methods. 1998;32:21–29. [Google Scholar]
- 10.Fabiano M, Donavaro R, Crisafi E, La Feria R, Povero P, Acosta Pomar L. Particulate matter composition and bacterial distribution in Terra Nova Bay (Antarctica) during summer 1989-1990. Polar Biol. 1995;15:393–400. [Google Scholar]
- 11.Fleming J T, Sanseverino J, Sayler G S. Quantitative relationship between naphthalene catabolic gene frequency and expression in predicting PAH degradation in soils at town gas manufacturing sites. Environ Sci Technol. 1993;27:1068–1074. [Google Scholar]
- 12.Fries M R, Zhou J-Z, Chee-Sanford J C, Tiedje J M. Isolation, characterization and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frostegard A, Courtois S, Ramisse V, Clerc S, Bernillon D, Le Gall F, Jeannin P, Nesme X, Simonet P. Quantification bias related to the extraction of DNA directly from soils. Appl Environ Microbiol. 1999;65:5409–5420. doi: 10.1128/aem.65.12.5409-5420.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gee G W, Bauder J W. Particle size analysis. Agron Monogr. 1986;9:383–411. [Google Scholar]
- 15.Greaves M P, Wilson M J. The adsorption of nucleic acids by montmorillonite. Soil Biol Biochem. 1969;1:317–323. [Google Scholar]
- 16.Holben W H, Jansson J K, Chelm B K, Tiedje J M. DNA probe method for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol. 1988;54:703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffrey W H, Nazaret S, Von Haven R. Improved method for recovery of mRNA from aquatic samples and its application to detection of mer expression. Appl Environ Microbiol. 1994;60:1814–1821. doi: 10.1128/aem.60.6.1814-1821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffrey W H, Von Haven R, Hoch M P, Coffin R B. Bacterioplankton RNA, DNA, protein content and relationships to rates of thymidine and leucine incorporation. Aquat Microb Ecol. 1996;10:87–95. [Google Scholar]
- 20.Johnston W H, Stapelton R, Sayler G S. Direct extraction of microbial DNA from soils and sediments. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 1.3.2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 1–9. [Google Scholar]
- 21.Kerkhof L, Ward B B. Comparison of nucleic acid hybridization and flourometry for measurement of the relationship between RNA/DNA ratio and growth rate in a marine bacterium. Appl Environ Microbiol. 1993;59:1303–1309. doi: 10.1128/aem.59.5.1303-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilmer V J, Alexander L T. Methods of making mechanical analyses of soils. Soil Sci. 1949;68:15–24. [Google Scholar]
- 23.Lovell C R, Piceno Y. Purification of DNA from estuarine sediments. J Microbiol Methods. 1994;20:161–174. [Google Scholar]
- 24.Malik M, Kain J, Pettigrew C, Ogram A. Purification and molecular analysis of microbial DNA from compost. J Microbiol Methods. 1994;20:183–196. [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 26.McLean E O. Soil and pH in lime requirement. Agron Monogr. 1982;9:199–224. [Google Scholar]
- 27.Moran M A, Torsvik V L, Torsvik T, Hodson R E. Direct extraction and purification of rRNA for ecological studies. Appl Environ Microbiol. 1993;59:915–918. doi: 10.1128/aem.59.3.915-918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muttray A F, Mohn W W. Quantitation of the population size and metabolic activity of a resin acid degrading bacterium in activated sludge using slot-blot hybridization to measure the rRNA:rDNA ratio. Microb Ecol. 1999;38:348–357. doi: 10.1007/s002489901005. [DOI] [PubMed] [Google Scholar]
- 29.Ogram A V, Mathot M L, Harsh J B, Boyle J, Pettigrew C A., Jr Effects of DNA polymer length on its adsorption to soils. Appl Environ Microbiol. 1994;60:393–396. doi: 10.1128/aem.60.2.393-396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogram A, Sayler G S, Barkay T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1987;7:57–66. [Google Scholar]
- 31.Ogram A, Sayler G S, Gustin D, Lewis R J. DNA adsorption to soils and sediments. Environ Sci Technol. 1988;22:982–984. doi: 10.1021/es00173a020. [DOI] [PubMed] [Google Scholar]
- 32.Ogram A, Sun W, Brockman F J, Fredrickson J K. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl Environ Microbiol. 1995;61:763–768. doi: 10.1128/aem.61.2.763-768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picard C, Ponsonnet E, Padget E, Nesme X, Simonet P. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1992;58:2717–2722. doi: 10.1128/aem.58.9.2717-2722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pichard S L, Paul J H. Gene expression per gene dose, a specific measure of gene expression in aquatic microorganisms. Appl Environ Microbiol. 1993;59:451–457. doi: 10.1128/aem.59.2.451-457.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purdy K J, Embley T M, Takaii S, Nedwell D B. Rapid extraction of DNA and rRNA from sediments by a novel hydroxyapatite spin-column method. Appl Environ Microbiol. 1996;62:3905–3907. doi: 10.1128/aem.62.10.3905-3907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauhut R, Klug G. mRNA degradation in bacteria. FEMS Microbiol Rev. 1999;23:353–370. doi: 10.1111/j.1574-6976.1999.tb00404.x. [DOI] [PubMed] [Google Scholar]
- 37.Romanowski G, Lorenz M G, Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol. 1991;57:1057–1061. doi: 10.1128/aem.57.4.1057-1061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selenska S, Klingmüller W. Direct recovery and molecular analysis of DNA and RNA from soil. Microb Releases. 1992;1:41–46. [PubMed] [Google Scholar]
- 39.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by SSU rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 40.Steffan R J, Goksoyr J, Bej A K, Atlas R M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988;54:2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiedje J M, Zhou J-Z, Nusslein K, Moyer C L, Fulthorpe R R. Extent and patterns of soil microbial diversity. In: Martins M T, et al., editors. Progress in microbial ecology. Sao Paulo, Brazil: Brazilian Society for Microbiology; 1997. pp. 35–41. [Google Scholar]
- 43.Tsai Y-L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai Y-L, Park M J, Olson B H. Rapid method for direct extraction of mRNA from Seeded Soils. Appl Environ Microbiol. 1991;57:765–768. doi: 10.1128/aem.57.3.765-768.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vesico P A, Nierzwicki-Bauer S A. Extraction and purification of PCR amplifiable DNA from lacustrine subsurface sediments. J Microbiol Methods. 1995;21:225–233. [Google Scholar]
- 46.Wintzingerode F V, Gohel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 47.Yu Z, Mohn W W. Killing two birds with one stone: simultaneous extraction of DNA and RNA from activated sludge biomass. Can J Microbiol. 1999;45:269–272. [Google Scholar]
- 48.Zhou J-Z, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]
- 49.Zhou J-Z, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]