Abstract
Actinomycetes secrete into their surroundings a suite of enzymes involved in the biodegradation of plant lignocellulose; these have been reported to include both hydrolytic and oxidative enzymes, including peroxidases. Reports of secreted peroxidases have been based upon observations of peroxidase-like activity associated with fractions that exhibit optical spectra reminiscent of heme peroxidases, such as the lignin peroxidases of wood-rotting fungi. Here we show that the appearance of the secreted pseudoperoxidase of the thermophilic actinomycete Thermomonospora fusca BD25 is also associated with the appearance of a heme-like spectrum. The species responsible for this spectrum is a metalloporphyrin; however, we show that this metalloporphyrin is not heme but zinc coproporphyrin. The same porphyrin was found in the growth medium of the actinomycete Streptomyces viridosporus T7A. We therefore propose that earlier reports of heme peroxidases secreted by actinomycetes were due to the incorrect assignment of optical spectra to heme groups rather than to non-iron-containing porphyrins and that lignin-degrading heme peroxidases are not secreted by actinomycetes. The porphyrin, an excretory product, is degraded during peroxidase assays. The low levels of secreted peroxidase activity are associated with a nonheme protein fraction previously shown to contain copper. We suggest that the role of the secreted copper-containing protein may be to bind and detoxify metals that can cause inhibition of heme biosynthesis and thus stimulate porphyrin excretion.
Biodegradation of lignocellulose by microorganisms plays an important role in carbon cycling, is of biotechnological interest to the paper industry, and has potential application in the field of bioremediation. Some microorganisms secrete a range of enzymes that completely degrade all the components of lignocellulose (lignin, hemicelluloses, and cellulose), while others secrete a narrower range of enzymes that only partially achieve this degradation (8, 19, 21). White rot fungi secrete both cellulolytic and ligninolytic enzymes; heme enzymes are major components of this ligninolytic activity and include the well-characterized lignin and manganese peroxidases of Phanerochaete chrysosporium (12).
Some actinomycetes, including thermophilic species and streptomycetes, secrete cellulose- and hemicellulose-degrading enzymes (2, 4, 11). Since the discovery of extracellular lignin-degrading heme peroxidases of wood-rotting fungi, considerable effort has been expended in searching for analogous enzymes in the cellulolytic actinomycetes (9, 22, 24). Indeed, some proteins secreted by the actinomycetes Streptomyces thermoviolaceus (17), Streptomyces viridosporus T7A (6, 26, 27), and Thermomonospora fusca BD25 (recently reclassified as Thermobifida fusca [35]) (3, 28) have low peroxidase activity and have been isolated and partially characterized. The peroxidase-like proteins from S. thermoviolaceus and S. viridosporus T7A were assigned as heme peroxidases on the basis of their optical spectra. However, these proteins were not shown to contain heme directly, nor were they shown to exhibit the characteristic spectral features associated with changes in the heme iron redox state, as are seen with P. chrysosporium lignin peroxidase (32–34) and other heme proteins. Electron paramagnetic resonance (EPR) spectroscopy has shown that the partially characterized metalloprotein with peroxidase activity from T. fusca BD25, although displaying heme-like spectra, exhibits no paramagnetically active heme component in any accessible redox state. Instead, it contains a nonheme iron center and copper, present in substantial molar excess over the protein (30).
Here we report that T. fusca exports into the culture medium a porphyrin, the optical spectrum of which may easily be mistaken for that of heme. The excretion of this porphyrin parallels the appearance of peroxidase activity. However, this porphyrin, although associated with the weak peroxidase activity in the culture medium, is not heme, and no true peroxidase is present. Furthermore, we confirm that the weak catalytic peroxidase activity is due to copper associated with the protein. Our results are compared with those of previous work in this area, and we conclude that no evidence has been provided to substantiate previous claims that actinomycetes secrete heme peroxidases. Thus, we propose that this class of extracellular enzymes is absent from this family.
MATERIALS AND METHODS
Both T. fusca BD25 and S. viridosporus T7A were maintained as suspensions of spores and hyphal fragments in 20% (vol/vol) glycerol at −20°C and routinely cultured on L agar plates or slants (31). T. fusca liquid cultures were grown for 7 days as described previously (31) with oat spelt xylan as the carbon source (8 g liter−1). S. viridosporus T7A was grown under identical conditions except that the growth temperature was 30°C rather than 50°C. During this period, the supernatants were monitored for peroxidase activity and for spectroscopic changes characteristic of metalloporphyrins, i.e., the appearance of a Soret absorbance band at ∼400 nm. Culture supernatants were assayed for heme directly using the pyridine hemochromogen test, i.e., 10-fold dilution of culture supernatants in 0.1 M NaOH–20% pyridine followed by the addition of a few grains of the reductant sodium dithionite (1). Solutions of the heme proteins horseradish peroxidase, myoglobin, and cytochrome c, with similar Soret absorbances, were used as positive controls. Peroxidase activity was measured spectrophotometrically at 510 nm by monitoring the formation of a Schiff base product from the reaction of 4-aminoantipyrene with oxidized 2,4-dichlorophenol. Assays were performed with defined H2O2 concentrations and with 5 mM 2,4-dichlorophenol plus 3.2 mM 4-aminoantipyrene as the colorigenic electron donor system at pH 7 and 30°C.
The kinetic constants Km and maximum turnover number (TNmax) were determined by fitting the experimental data to the standard Michaelis-Menten equation using Kaleidagraph fitting software. Absorbance spectroscopy was carried out using a Hewlett-Packard diode-array instrument (HP8453), and porphyrin fluorescence was characterized using a Perkin-Elmer spectrofluorimeter (LS50B). Reversed-phase high-pressure liquid chromatography (HPLC) was used to isolate the porphyrin from culture supernatants; for this procedure, a Hewlett-Packard HPLC 1100 instrument equipped with a Zorbax SB-C3 300A column was used. The porphyrin was eluted with an isocratic gradient system of 35 to 40% acetonitrile in H2O and 0.1% trifluoroacetic acid (pH 2) (see Fig. 3) at a flow rate of 1 ml/min. Porphyrin concentrations during growth were quantified by HPLC fractionation of culture supernatants and integration of the major porphyrin elution peak. Reversed-phase HPLC using a Zorbax 300 SB C18 column (250 by 4.6 mm) was used to separate porphyrin standards and thus identify the T. fusca porphyrin by comparing its retention time and UV-visible spectra to those of the standards under the same conditions. T. fusca and standard porphyrins were eluted with an isocratic gradient system of 5 to 50%, 50 to 95%, and 95 to 5% acetonitrile in H2O and 0.1% trifluoroacetic acid (pH 2) (see Fig. 4) at a flow rate of 1 ml/min. Mass spectrometry was carried out by Matrix-assisted laser desorption ionization–time of flight with a Kratos Analytical Kompact Maldi II instrument.
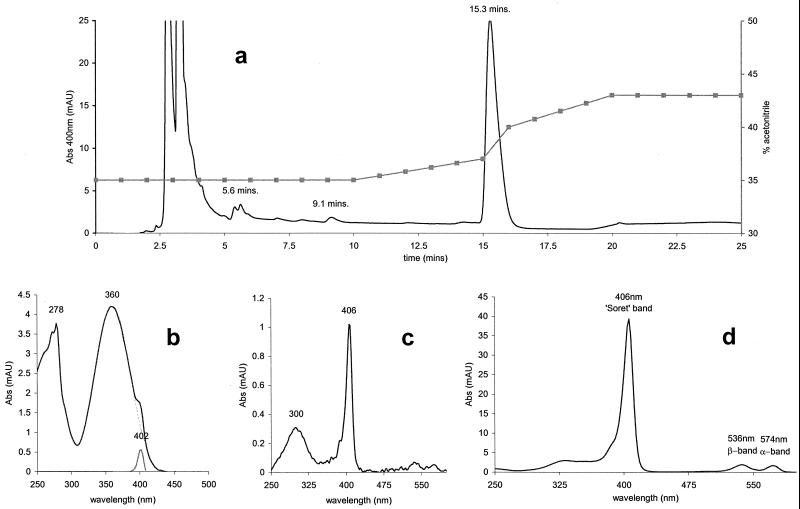
FIG. 3.
HPLC elution profile and absorbance (Abs) spectra of T. fusca porphyrin. (a) Elution profile from HPLC fractionation. Solid line, absorbance at 400 nm (milliabsorbance units [mAU]); squares, acetonitrile gradient. (b) Absorption spectrum of 400-nm elution peak at 5.6 min, shown with subtraction of simulated 360-nm absorbing species. Black line, actual spectrum; broken line, simulation of 360-nm species; grey line, spectrum minus simulation, i.e., showing the porphyrin Soret peak. (c) Absorption spectrum of 400-nm elution peak at 9.1 min. (d) Absorption spectrum of 400-nm elution peak at 15.3 min, showing α, β, and Soret absorption bands.
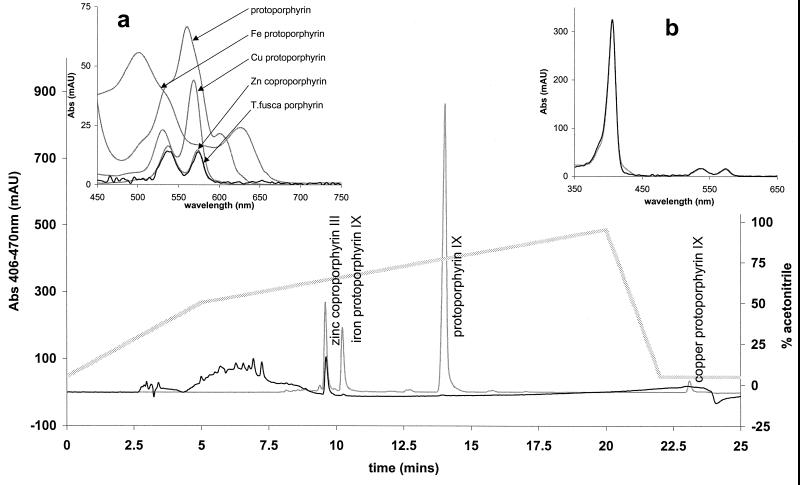
FIG. 4.
Identification of T. fusca porphyrin. HPLC retention times and UV-visible absorbance (Abs) spectra of T. fusca porphyrin and porphyrin standards are shown. All spectra were recorded during HPLC separation. T. fusca traces were normalized to the scale of porphyrin standards. (Main panel) Elution profile (monitored at 406 minus 470 nm) of T. fusca porphyrin loaded as culture supernatants (black line) and porphyrin standards (grey line) and acetonitrile gradient (stippled line). mAU, milliabsorbance units. (Inset a) Visible spectrum of porphyrin standards (grey lines) and T. fusca porphyrin (black line). (Inset b) UV-visible spectra of T. fusca porphyrin (black line) and Zn coproporphyrin III standard (grey line).
All standard reagents were from Sigma Chemical Co. Samples of free porphyrins and copper, zinc, and magnesium metalloporphyrins were obtained from Porphyrin Products Inc.
RESULTS
Characterization of extracellular porphyrin.
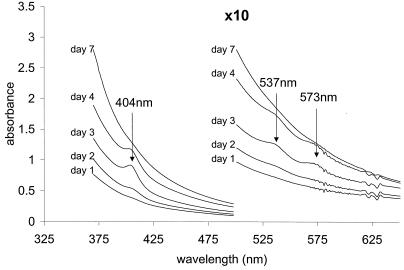
Supernatants obtained from centrifugation of T. fusca cultures possess an extracellular peroxidase activity and contain a porphyrin species. Figure 1 compares the spectra of the culture medium at intervals during growth. The Soret band at 404 nm and the two peaks in the visible region, at 537 and 573 nm, are characteristic of porphyrins, including heme (iron protoporphyrin IX). Here they are seen superimposed upon a background spectrum that we assign to melanin or a melanin-like pigment that renders the supernatant yellow. The porphyrin concentration reached a maximum on days 3 and 4, as did the peroxidase activity; thereafter, both declined (Table 1). This result suggested that a heme peroxidase enzyme could be responsible for the peroxidase activity, as reported for Streptomyces and for the white rot fungus P. chrysosporium (12). However, the species responsible for the porphyrin spectrum shown in Fig. 1 was insensitive to the powerful reductant sodium dithionite. A direct test for heme, namely, the formation of a pyridine hemochromogen when heme is treated with pyridine under alkaline conditions, gave negative results, indicating that any heme protein present must have been less than ∼1% of that expected for a heme protein displaying a Soret band of similar absorbance (data not shown). These findings eliminate the possibility that this species is a heme group.
FIG. 1.
Time course of porphyrin secretion during the growth of T. fusca. The UV-visible spectra of the culture medium on days 1, 2, 3, 4, and 7 of growth are shown (temperature, 25°C; path length, 1 cm). The Soret band at 404 nm and the two peaks in the visible region appear on day 2 and reach maxima on day 3. Thereafter, they decrease; by day 7, they are undetectable.
TABLE 1.
Time course of porphyrin secretion and peroxidase activity during mycelial growtha
| Age of culture (days) | Initial rate of product formed (nM/min) | Porphyrin concn (nM) in assay |
|---|---|---|
| 1 | 40 | 8 |
| 2 | 67 | 24 |
| 3 | 459 | 99 |
| 4 | 208 | 87 |
| 7 | 135 | 11 |
Conditions were as described in the legend to Fig. 1 and Materials and Methods.
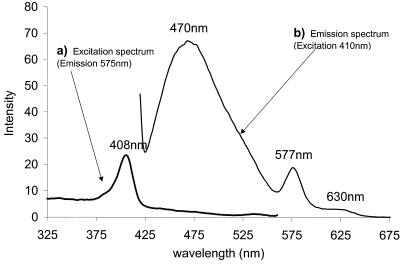
In addition, the porphyrin species was found to be fluorescent. Figure 2 shows that photoexcitation at 410 nm gives rise to three fluorescence emission bands, at 470, 577, and 630 nm. The latter two bands are characteristic of a fluorescent nonheme porphyrin. Heme groups themselves are not fluorescent. The emission band at 470 nm did not maintain a constant intensity ratio with the two porphyrin-specific bands, and we tentatively attribute this result to emission from a further fluorophore, such as melanin.
FIG. 2.
Fluorescence emission-excitation spectra. Samples of culture medium were diluted 200-fold in 100 mM potassium phosphate at pH 7.0 and 30°C. The emission and excitation band widths were set at 5 nm. (a) Excitation spectrum of T. fusca supernatant monitored at 575 nm. (b) Emission spectrum of T. fusca supernatant excited at 410 nm.
The porphyrin from T. fusca was isolated from the culture medium by HPLC. Elution was monitored optically at 400 nm, close to the Soret maximum. The total elution profile (Fig. 3A) shows one major, sharp elution band at 15.3 min (38% acetonitrile) and several minor elution bands, including those at 9.1 and 5.6 min. The absorbance spectra of the porphyrin species eluted at 5.6, 9.1, and 15.3 min are shown in Fig. 3B, C, and D, respectively. The major porphyrin eluted at 15.3 min (Fig. 3D) is spectroscopically free of protein or melanin. The porphyrin eluted at 9.1 min is complexed with a species absorbing at 300 nm, and the minor component eluted at 5.6 min contains a complex absorbing at 278 and 360 nm. The 278-nm absorbance band indicates aromatic amino acid residues of proteins, the 300-nm absorbance band indicates oxidized aromatic amino acids, and the 360-nm absorbance band indicates a melanin-like pigment.
The T. fusca porphyrin was fully soluble in aqueous media at a neutral pH, whereas authentic metal-free and metallated protoporphyrins were not. Figure 4 (main panel) shows [see below] the HPLC elution profile of the porphyrin standards (Zn2+ coproporphyrin III and Fe3+, Mg2+ Cu2+, and nonmetallated protoporphyrin IX) together with the elution profile of the T. fusca porphyrin under the same conditions. Elution was monitored at 406 nm, the Soret maximum for the T. fusca prophyrin. Both the T. fusca porphyrin and the Zn2+ coproporphyrin III standard eluted at 9.6 min (64% acetonitrile). Magnesium protoporphyrin is not seen as a separate species in the elution profile because under the HPLC conditions used (pH 2), the magnesium ion coordination ligands become protonated and the magnesium is displaced; the demetallated protoporphyrin coelutes with protoporphyrin IX. The broad band eluting at between 4 and 8 min is due to melanin; porphyrin was not detected in these fractions. Figure 4, inset a, compares the spectra in the visible region of the T. fusca porphyrin to those of all the porphyrin standards. Figure 4 inset b shows the overlaid UV-visible spectra of the T. fusca porphyrin and the Zn2+ coproporphyrin III standard; these are essentially identical. All T. fusca traces have been normalized to the absorbance scale of the porphyrin standards. The acid-alkali pH dependencies of the positions and intensities of the Soret, α and β bands in the T. fusca porphyrin and the Zn2+ coproporphyrin III standard were also identical (data not shown).
Kinetics of pseudoperoxidase.
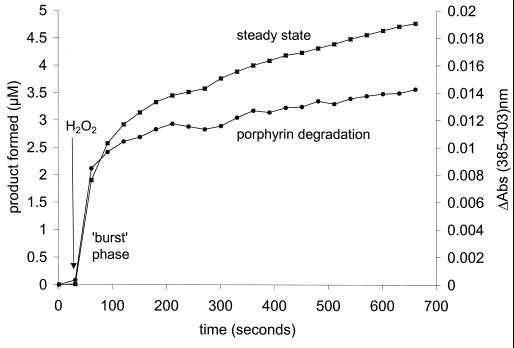
Figure 5 shows a typical time course for the peroxidase activity of the T. fusca supernatant. This time course, which monitors product formation, is biphasic. An initial burst phase is followed by a steady, slow phase. The burst phase is paralleled by a process in which the porphyrin is degraded, as demonstrated by a decrease in the absorbance at 403 nm. This result suggests that porphyrin is acting as a substrate, not as a catalyst. The amplitude of the burst phase shows that approximately 4 mol of product is formed per mol of porphyrin degraded.
FIG. 5.
Correlation between the degradation of T. fusca porphyrin and peroxidase activity during the initial burst phase. Time courses for peroxidase activity of the T. fusca supernatant (left ordinate; squares) and porphyrin degradation at an absorbance (Abs) of 385 to 403 nm (right ordinate; circles). Measurements were made with 100 mM sodium or potassium phosphate buffer at pH 7.0 and 30°C.
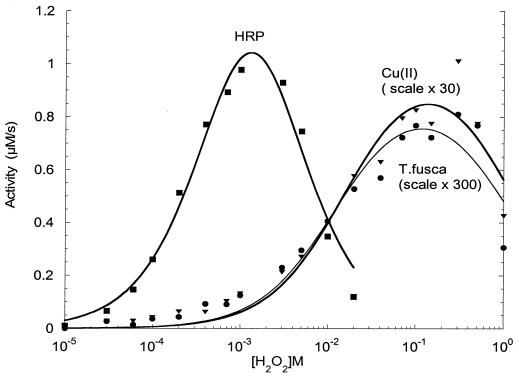
The kinetic behavior of the slow, catalytic phase was analyzed by varying the hydrogen peroxide concentration. Figure 6 compares the activity of T. fusca pseudoperoxidase with those of horseradish peroxidase and inorganic copper sulfate. The peroxide concentration is plotted logarithmically in order to display the full range utilized. From these data, the apparent Km and TNmax were determined by fitting the experimental data to the Michaelis-Menten equation (see Materials and Methods). Although the peroxidase activity of the supernatant increases with increasing peroxide concentration according to the classical hyperbolic relationship, similar behavior is seen with inorganic cupric ions as catalysts. Table 2 lists the values of the kinetic constants (Km and TNmax) obtained from the data in Fig. 6. The Km for H2O2 of 18 mM for T. fusca pseudoperoxidase is very similar to the value of 22 mM for free cupric ions and quite different from that of 0.74 mM for horseradish peroxidase, an authentic heme enzyme.
FIG. 6.
Kinetics of three peroxidase catalysts. Peroxidase activities were measured at pH 7.0 and 30°C as described in Materials and Methods. Points were fitted to a Michaelis-Menten equation with substrate inhibition. Catalyst concentrations and derived parameters are listed in Table 2. HRP, horseradish peroxidase.
TABLE 2.
Km and TNmax values for three catalytic systemsa
| Catalyst | Concn | Km (mM) for H2O2 | TNmax (s−1) |
|---|---|---|---|
| Horse radish peroxidase | 1 nM heme or protein | 0.74 | 2,220 |
| T. fusca protein | 30 nM protein | 18b | 0.102c |
| CuSO4 | 5 μM Cu(II) | 22b | 0.007c |
Kinetic constants Km and TNmax are from the data in Fig. 6. Conditions were as described in the legend to Fig. 6 and Materials and Methods.
Kmvalues for H2O2 for T. fusca pseudoperoxidase and free cupric ions are similar.
Turnover for the T. fusca protein is ∼14 times that of free Cu, but the protein may be associated with up to 20 mol of Cu/mol of protein (17).
Excretion of porphyrins by other microorganisms.
When the culture medium from S. viridosporus T7A was subjected to the same analytical procedures as T. fusca, spectral features identical to those shown in Fig. 1 were observed. Again, the characteristic porphyrin bands were detected and remained unchanged upon addition of the reductant sodium dithionite. No pyridine hemochromogen was formed. The spectrum of the S. viridosporus porphyrin was also identical to that of the T. fusca porphyrin after separation by HPLC (Fig. 3).
Furthermore, under certain growth conditions, this same porphyrin was also produced by Escherichia coli. The E. coli porphyrin shared the same pH dependencies of spectral band positions and intensities as the T. fusca porphyrin and the Zn2+ copropophyrin III standard. Mass spectrometry of the porphyrin from E. coli (available to us in larger amounts than the T. fusca product) revealed a molecular mass for the major fraction of 722 ± 2 Da.
DISCUSSION
Characterization of T. fusca porphyrin.
Metalloporphyrins containing Fe, Cu, Ni, or Co are nonfluorescent, whereas metal-free porphyrins and those complexed with Zn, Mg, and Cd are fluorescent (10). The fluorescence spectrum of the T. fusca porphyrin and the negative result in the pyridine hemochromogen test confirm that the porphyrin does not contain Fe and therefore is not a heme species. The UV-visible spectrum, not having four absorption bands in the visible region (Fig. 4), is not that of a metal-free porphyrin. There are three categories of metal-porphyrin bonds in metalloporphyrins, covalent type, ionic type, and intermediate type. Each type of interaction has characteristic thermodynamic stability and spectral properties. The spectral properties are related to the relative intensities of the α/β band ratios of the visible region. The covalent-type metal-porphyrin bonds, which include those of Pt2+, Ni2+, Co2+, and Cu2+, have α/β intensity ratios of ≫1; ionic types, including Mg2+, Pb2+, and Sn2+, have α/β intensity ratios of ≪1; intermediate types, which include Zn2+ and Cd2+, have α/β intensity ratios of approximately unity. Although magnesium is a commonly available metal ion, the α/β band ratio in the visible region of the spectrum excludes magnesium as the metal (25). Furthermore, magnesium porphyrins are not acid stable under the conditions used for HPLC analysis (see Results). Incorporation of metal ions into porphyrins in aqueous solutions occurs at significant rates only for Cu2+ and Zn2+ ions. The fact that Cu produces a covalent-type spectrum (Fig. 4) eliminates copper as the metal. The strong correlation of the T. fusca and zinc coproporphyrin spectral properties, i.e., the pH dependencies of peak positions and intensities, and the α/β band ratios therefore support our identification of the remaining possibility, zinc, as the metal-porphyrin bond species.
The porphyrin nucleus is hydrophobic, and the number of hydrophilic carboxyl substituents increases its solubility in aqueous media at a neutral pH. Coproporphyrin, having four carboxyl substituents, is soluble in aqueous media at a neutral pH, whereas protoporphyrin, with only two carboxyl groups, is insoluble (10, 25). The excretion of coproporphyrin III in microorganisms is well documented (7).
The mass spectrometry result obtained with the identical E. coli product is similar to that expected for zinc coproporphyrin. Its mass, 722 ± 2 Da, is similar to that of 720 Da expected for zinc coproporphyrin. In addition to the spectral properties, the chemical properties of the T. fusca porphyrin and the Zn2+ coproporphyrin III standard were also identical, as shown by the HPLC elution profile. The other water-soluble porphyrin, uroporphyrin, has eight carboxyl residues (coproporphyrin has four); it is therefore inconceivable that the chemical properties and thus the HPLC retention times would be identical for zinc coproporphyrins and zinc uroporphyrins. On the basis of spectral and chemical properties, we therefore conclude that the excreted porphyrin is zinc coproporphyrin.
The chemical and spectral characteristics of the porphyrin in the S. viridosporus culture supernatant were identical to those of the T. fusca porphyrin; we therefore conclude that this species is also likely to be zinc coproporphyrin and not heme, as previously claimed (26).
Porphyrins in other streptomycetes and actinomycetes.
Nonheme porphyrins have been found in culture supernatants of the soil streptomycetes Streptomyces misakiensis and Streptomyces viridochromogenes (23). The UV-visible spectra of these culture supernatants showed weak 400-nm porphyrin and 280-nm protein absorption bands, which appeared, as in the T. fusca spectra, superimposed on a broad melanin-like absorption band that swept down from the UV spectrum to the visible spectrum. Although the porphyrin type was not determined, EPR spectroscopy identified the presence of copper, chelated by ligands possibly donated by the porphyrin. This result does not exclude the possibility that zinc porphyrin was also present in the culture supernatants, as Zn2+ is EPR silent. EPR spectroscopy did not detect copper porphyrin in the T. fusca culture medium. The central metal ion in excreted porphyrins may depend upon the ions available in the growth medium. In any case, the data for S. misakiensis and S. viridochromogenes show that the presence of a distinct Soret band cannot be assumed to indicate the presence of heme.
Protein-free fluorescent coproporphyrin III has also been found in the culture medium of the marine actinomycete Arthrobacter aurescens RS-2; its excretion was enhanced by aluminium and nickel (29). The porphyrin was excreted together with a protein, possibly protective in nature and of approximately the same size as the pseudoperoxidase of T. fusca (28).
Porphyrin excretion in other organisms.
The porphyrin that appears in T. fusca and S. viridosporus cultures is not unique to actinomycetes; its formation in E. coli indicates that the porphyrin may be a pathological excretory product rather than a functional secretory product.
Aberrant porphyrin excretion has been seen in photosynthetic bacteria (5, 14), although in these organisms the porphyrin was complexed with proteins, suggesting a role in porphyrin biosynthesis; again, the major porphyrin excreted was coproporphyrin III. Excretion or accumulation of coproporphyrin III seems to be typical of blocked porphyrin synthesis in microorganisms. The conversion of coproporphyrinogen III to protoporphyrin IX is a rate-limiting step in bacterial heme biosynthesis (29).
Zinc coproporphyrin excretion is also not peculiar to microorganisms; it is also recognized clinically as a feature of heme metabolism in neonates, whose earliest stools (meconium) have been shown to contain porphyrins, the major component of which is zinc coproporphyrin (13, 16). It is recognized hematologically as one of the most common by-products of heme biosynthesis (20), a consequence of its chemical stability and aqueous solubility.
Secreted and intracellular proteins and enzymes.
Streptomycetes are characterized by containing genes for a remarkable set of families of catalases and peroxidases, only some of which may be expressed at any time. The Streptomyces coelicolor genome project (18; http://www.sanger.ac.uk/Projects/S_coelicolor/) has already revealed the presence of at least one catalase-peroxidase gene and at least three paralogues of classical catalase genes (P. Nicholls et al., unpublished data). In addition, these organisms contain a unique family of nonmetal fatty acid-dependent peroxidases apparently derived evolutionarily from hydrolytic ancestors and whose X-ray crystal structures have been determined (15). The streptomycete catalases and peroxidases can exist in more than one form related by proteolytic degradation. The original holoenzymes contain not only the heme group characteristic of catalases and peroxidases but also a C-terminal fragment that contains manganese and that can engage in manganese-dependent peroxidation (36). However, there is no evidence that this enzyme, either in its holoenzyme form or in proteolytically degraded forms, can be secreted by streptomycetes. Moreover, the EPR study of the T. fusca protein showed no manganese, an element with very characteristic EPR signals (30).
The emerging genetic analysis of T. fusca (http://www.jgi.doe.gov/JGI_microbial/html/thermobifida/thermob_homepage.html) also shows this microorganism to contain several intracellular catalase and peroxidase enzymes; these appear to include the classical Micrococcus-like catalase, manganese catalase, glutathione peroxidase, and a putative nonheme peroxidase. Preliminary results have shown the occurrence of classical catalase activity in broken T. fusca cells (Nicholls et al., unpublished). This activity, however, does not appear in the growth medium, unless and until severe damage to the cells and breakdown of their walls and membranes have occurred.
The weak peroxidase activity found in the T. fusca culture medium is biphasic; the initial, burst phase correlates with porphyrin degradation, while the slower, steady phase is comparable to the peroxidase activity of free cupric ions. T. fusca copper-containing peroxidase has up to 20 copper atoms per protein molecule (30), and the total [Cu2+] is therefore 20 times the molar concentration of the protein (Table 2). This information implies that approximately 120 nM bound Cu was present in the assay system of Fig. 6. Both the apparent Km and the turnover per Cu atom are therefore similar for the “enzyme” and for free CuSO4.
The heme peroxidase reported for S. viridosporus (26) was not shown directly to contain heme. Our results show that this, too, is a nonheme porphyrin associated with melanin and protein, like that in T. fusca. Similarly, the reported heme peroxidase of S. thermoviolaceus, which has a specific peroxidase activity comparable to that of T. fusca, may also owe its peroxidase activity to transition metal ions associated, either specifically or adventitiously, with protein. An authentic type III heme peroxidase (members of the catalase-peroxidase family are classified as type I) has yet to be demonstrated in a prokaryote.
It also improbable that the peroxidase activity of the T. fusca protein is the function of a true ligninolytic enzyme. Unlike the relatively classical peroxidases of the lignin-degrading fungi, its turnover is very low and its relative affinity for peroxide is also rather poor. In a marine Vibrio sp. (14), copper induces the excretion of extracellular proteins whose function may be to bind to and detoxify external copper ions. The role of the extracellular Cu protein in T. fusca (30) is thus uncertain. It may be a peroxidase of relatively low activity, it may be a detoxifying Cu-binding protein, or it may have a function in the complex process of lignin degradation that has yet to be determined.
The appearance of porphyrins into which iron has not been inserted is a consequence of metal ion inhibition of heme biosynthesis. The protein that we have identified as containing copper may have a role in metal detoxification. Thus, during rapid growth in the presence of metals, such as copper, porphyrins are excreted as waste products from incomplete heme synthesis; a protective copper-binding protein is also secreted. The peroxidase activity may merely be the consequence of binding of the redox-active metal, copper, to this protein. Our view, therefore, is that there is no evidence to support the contention that actinomycetes secrete heme peroxidases and that the spectra recorded from culture media are due to zinc (or another metal, e.g., Cu)-containing porphyrins and not to heme.
ACKNOWLEDGMENTS
We thank Neil Barnard for carrying out the mass spectrometry and Chris Cooper for discussions concerning peroxidase activity and structure.
This work was supported by BBSRC grants to M.T.W. and to A.S.B. and by a BBSRC scholarship to M.G.M.
REFERENCES
- 1.Antonini E, Brunori M. Hemoglobin and myoglobin in their reactions with ligands. Amsterdam, The Netherlands: North-Holland Publishing Co.; 1971. [Google Scholar]
- 2.Ball A S, McCarthy A J. Saccharification of straw by actinomycete enzymes. J Gen Microbiol. 1988;134:2139–2147. [Google Scholar]
- 3.Ball A S, Trigo C. Characterization of a novel non-heme-containing extracellular peroxidase from Thermomonospora fusca. Biochem Soc Trans. 1995;23:272–276. doi: 10.1042/bst0230272. [DOI] [PubMed] [Google Scholar]
- 4.Ball A S, Trigo C. The role of actinomycetes in plant litter decomposition. Recent Res Dev Soil Biol Biochem. 1997;1:9–20. [Google Scholar]
- 5.Biel A J. Characterization of a coproporphyrin-protein complex from Rhodobacter capsulatus. FEMS Microbiol Lett. 1991;81:43–48. doi: 10.1016/0378-1097(91)90468-p. [DOI] [PubMed] [Google Scholar]
- 6.Burke N S, Crawford D L. Use of azo dye ligand chromatography for the partial purification of a novel extracellular peroxidase from Streptomyces viridosporus T7A. Appl Microbiol Biotechnol. 1998;49:523–530. doi: 10.1007/s002530051208. [DOI] [PubMed] [Google Scholar]
- 7.Burnham B F, Bachmann R C. Enzymatic syntheses of porphyrins. In: Dolphin D, editor. The porphyrins. VI. New York, N.Y: Academic Press, Inc.; 1979. pp. 233–256. [Google Scholar]
- 8.Deobald L A, Crawford D L. Lignocellulose biodegradation. In: Hurst C J, editor. Manual of environmental microbiology. Washington, D.C.: American Society for Microbiology; 1997. pp. 730–737. [Google Scholar]
- 9.Falcon M A, Rodriguez A, Carnicero A, Regalado V, Perestelo F, Milstein O, Delafuente G. Isolation of micro-organisms with lignin transformation potential from soil of Tenerife Island. Soil Biol Biochem. 1995;27:121–126. [Google Scholar]
- 10.Falk J E. Chemistry and biochemistry of porphyrins and metalloporphyrins. Compr Biochem. 1963;9:3–33. [Google Scholar]
- 11.Godden B, Legon T, Helvenstein P, Penninckx M. Regulation of the production of hemicellulolytic and cellulolytic enzymes by a streptomyces sp growing on lignocellulose. J Gen Microbiol. 1989;135:285–292. doi: 10.1099/00221287-135-2-285. [DOI] [PubMed] [Google Scholar]
- 12.Gold M H, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gourley R G, Kreamer B, Arend R. Excremental studies in human neonates. Identification of zinc coproporphyrin as a marker for meconium. Gastroenterology. 1990;99:1705–1709. doi: 10.1016/0016-5085(90)90476-h. [DOI] [PubMed] [Google Scholar]
- 14.Harwoodsears V, Gordon A S. Copper-induced production of copper-binding supernatant proteins by the marine bacterium Vibrio alginolyticus. Appl Environ Microbiol. 1990;56:1327–1332. doi: 10.1128/aem.56.5.1327-1332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann B, Tolzer S, Pelletier I, Altenbuchner J, vanPee K H, Hecht H J. Structural investigation of the cofactor-free chloroperoxidases. J Mol Biol. 1998;279:889–900. doi: 10.1006/jmbi.1998.1802. [DOI] [PubMed] [Google Scholar]
- 16.Horiuchi K, Adachi K, Fujise Y, Naruse H, Sumimoto K, Kanayama N, Terao T. Isolation and characterization of zinc coproporphyrin I: a major fluorescent component in meconium. Clin Chem. 1991;37:1173–1177. [PubMed] [Google Scholar]
- 17.Iqbal M, Mercer D K, Miller P G G, McCarthy A J. Thermostable extracellular peroxidases from Streptomyces thermoviolaceus. Microbiology. 1994;140:1457–1465. [Google Scholar]
- 18.Kieser T K, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical streptomyces genetics. Colney, Norwich, England: John Innes Foundation. John Innes Centre, Norwich Research Park; 2000. [Google Scholar]
- 19.Kirk T K, Farrell R L. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 20.Labbe R F, Tettmer R L. Zinc protoporphyrin: a product of iron-deficient erythropoiesis. Semin Hematol. 1989;26:40–46. [PubMed] [Google Scholar]
- 21.Leonowicz A, Matuszewska A, Luterek J, Ziegenhagen D, WojtasWasilewska M, Cho N S, Hofrichter M, Rogalski J. Biodegradation of lignin by white rot fungi. Fungal Genet Biol. 1999;27:175–185. doi: 10.1006/fgbi.1999.1150. [DOI] [PubMed] [Google Scholar]
- 22.Magnuson T S, Roberts M A, Crawford D L, Hertel G. Immunological relatedness of extracellular ligninases from the actinomycetes Streptomyces viridosporus T7A and Streptomyces badius 252. Appl Biochem Biotechnol. 1991;28:433–443. doi: 10.1007/BF02922623. [DOI] [PubMed] [Google Scholar]
- 23.Mangrich A S, Lermen A W, Santos E J, Gomes R C, Coelho R R R, Linhares L F, Senesi N. Electron paramagnetic resonance and ultraviolet-visible spectroscopic evidence for copper porphyrin in actinomycete melanins. Biol Fertil Soils. 1998;26:341–345. [Google Scholar]
- 24.Mercer D K, Iqbal M, Miller P G G, McCarthy A J. Screening actinomycetes for extracellular peroxidase activity. Appl Environ Microbiol. 1996;62:2186–2190. doi: 10.1128/aem.62.6.2186-2190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips J N. Physico-chemical properties of porphyrins. Compr Biochem. 1963;9:35–72. [Google Scholar]
- 26.Ramachandra M, Crawford D, Hertel G. Characterization of an extracellular lignin peroxidase of the lignocellulolytic actinomycete Streptomyces viridosporus. Appl Environ Microbiol. 1988;54:3057–3063. doi: 10.1128/aem.54.12.3057-3063.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramachandra M, Crawford D L, Pometto A L. Extracellular enzyme activities during lignocellulose degradation by Streptomyces spp.—a comparative study of wild-type and genetically manipulated strains. Appl Environ Microbiol. 1987;53:2754–2760. doi: 10.1128/aem.53.12.2754-2760.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rob A, Ball A S, Tuncer M, Wilson M T. Thermostable novel non-haem extracellular glycosylated peroxidase from Thermomonospora fusca BD25. Biotechnol Appl Biochem. 1996;24:161–170. [Google Scholar]
- 29.Scharf R, Zimmels Y, Kimchie S. Metal-induced extracellular protein excretion in Arthrobacter aurescens. FEMS Microbiol Lett. 1993;109:139–144. [Google Scholar]
- 30.Svistunenko D A, Rob A, Ball A, Torres J, Symons M C R, Wilson M T, Cooper C E. The electron paramagnetic resonance characterisation of a copper-containing extracellular peroxidase from Thermomonospora fusca BD25. Biochim Biophys Acta Protein Struct Mol Enzymol. 1999;1434:74–85. doi: 10.1016/s0167-4838(99)00163-6. [DOI] [PubMed] [Google Scholar]
- 31.Tuncer M, Ball A S, Rob A, Wilson M T. Optimization of extracellular lignocellulolytic enzyme production by a thermophilic actinomycete Thermomonospora fusca BD25. Enzyme Microb Technol. 1999;25:38–47. [Google Scholar]
- 32.Wariishi H, Gold M H. Lignin peroxidase compound III. Mechanism of formation and decomposition. J Biol Chem. 1990;265:2070–2077. [PubMed] [Google Scholar]
- 33.Wariishi H, Huang J, Dunford H B, Gold M H. Reactions of lignin peroxidase compound I and compound II with veratryl alcohol—transient-state kinetic characterization. I Biol Chem. 1991;266:20694–20699. [PubMed] [Google Scholar]
- 34.Wariishi H, Marquez L, Dunford H B, Gold M H. Lignin peroxidase compound II and compound III—spectral and kinetic characterization of reactions with peroxides. J Biol Chem. 1990;265:11137–11142. [PubMed] [Google Scholar]
- 35.Zhang Z S, Wang Y, Ruan J S. Reclassification of Thermomonospora and Microtetraspora. Int J Syst Bacteriol. 1998;48:411–422. doi: 10.1099/00207713-48-2-411. [DOI] [PubMed] [Google Scholar]
- 36.Zou P J, Borovok I, Lucana D O D, Muller D, Schrempf H. The mycelium-associated Streptomyces reticuli catalase-peroxidase, its gene and regulation by FurS. Microbiology. 1999;145:549–559. doi: 10.1099/13500872-145-3-549. [DOI] [PubMed] [Google Scholar]