Abstract
The marine bryozoan, Bugula neritina, is the source of the bryostatins, a family of macrocyclic lactones with anticancer activity. Bryostatins have long been suspected to be bacterial products. B. neritina harbors the uncultivated gamma proteobacterial symbiont “Candidatus Endobugula sertula.” In this work several lines of evidence are presented that show that the symbiont is the most likely source of bryostatins. Bryostatins are complex polyketides similar to bacterial secondary metabolites synthesized by modular type I polyketide synthases (PKS-I). PKS-I gene fragments were cloned from DNA extracted from the B. neritina-“E. sertula” association, and then primers specific to one of these clones, KSa, were shown to amplify the KSa gene specifically and universally from total B. neritina DNA. In addition, a KSa RNA probe was shown to bind specifically to the symbiotic bacteria located in the pallial sinus of the larvae of B. neritina and not to B. neritina cells or to other bacteria. Finally, B. neritina colonies grown in the laboratory were treated with antibiotics to reduce the numbers of bacterial symbionts. Decreased symbiont levels resulted in the reduction of the KSa signal as well as the bryostatin content. These data provide evidence that the symbiont E. sertula has the genetic potential to make bryostatins and is necessary in full complement for the host bryozoan to produce normal levels of bryostatins. This study demonstrates that it may be possible to clone bryostatin genes from B. neritina directly and use these to produce bryostatins in heterologous host bacteria.
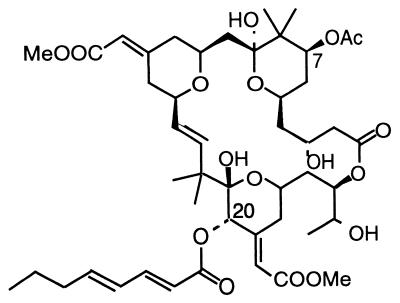
Since 1968, bryostatins in extracts from the marine bryozoan Bugula neritina have been studied for their cellular activity and therapeutic potential (24). B. neritina is the sole source of this unique family of macrolides. The bryostatins are complex polyketides based on the bryopyran ring system that are similar to bacterial secondary metabolites (25) (Fig. 1). The bryostatins, which do not originate in the diet (J. Thompson and D. Mendola, Aquaculture production of bryostatin 1: nonconfidential results of the phase 1 project, CalBioMarine Technologies, Inc.), have been proposed to be products of bacteria associated with B. neritina (2). This bryozoan harbors an uncultivated gamma proteobacterium, “Candidatus Endobugula sertula,” which is passed to the larvae prior to their release from the adult (15, 34). Bryostatins show excellent potential as therapeutic agents that act through protein kinase C (PKC) signal transduction to alter cellular activity (18, 30, 31). Phase II clinical trials of bryostatin 1 are under way for the treatment of leukemias, lymphomas, melanomas, and solid tumors (26). Bryostatin 1 also shows promise for treatment of ovarian and breast cancers and for enhancing lymphocyte survival during radiation treatment (3, 4, 5, 12, 13, 16, 20, 21, 28, 32, 33). Other bryostatins may prove to be valuable therapeutic agents as well (19). Research and development of the bryostatins are currently severely limited by inadequate availability of these compounds. If a bacterial source of bryostatins could be identified, new avenues for solving the supply problem may open, either through efforts to clone the bryostatin biosynthetic genes and then express them in a heterologous host or by culture of the bacterial source itself.
FIG. 1.
Bryostatin 1 from B. neritina.
In this article we describe evidence that the bacterial symbiont “E. sertula” is involved in the biosynthesis of bryostatins. Previously, we have shown that a difference in the types of bryostatins found in B. neritina correlated with a genetic difference (by 16S ribosomal gene sequences) in the bacterial symbiont (7). However, the chemistry differences noted in that study could be explained by a difference in the host or another unidentified bacterial symbiont. Determining the source of bryostatins in the bryozoan bacterial association presents a difficult challenge due to the inability to culture the bacterial symbiont. Earlier attempts to separate the bacterial cells from the host as a way to determine the bryostatin source proved inconclusive (6). To investigate the link between the bacterial symbiont “E. sertula” and the production of bryostatins we applied molecular techniques that do not require the culture of the bacteria. In bacteria, complex polyketides are synthesized by modular type I polyketide synthases (PKS-I). These enzymes have a set of catalytic domains, including a β-ketoacyl synthase (KS) domain, for each elongation and modification step in the synthesis of the chain. In this study, the bacterial PKS-I gene fragments from KS domains were targeted for cloning and sequencing. The objectives of the research presented here were to show (i) that the genes coding for a type I complex polyketide synthase pathway are present in the DNA obtained from the symbiotic association, (ii) that genes required in the pathway, the KS genes, are found in the symbiont “E. sertula,” and (iii) that a reduction of this symbiont results in a reduction of this detected gene and a reduction in bryostatin synthesis.
MATERIALS AND METHODS
DNA extraction.
DNA for PCR amplification of PKS-I genes using degenerate KS primers was extracted from adult B. neritina using a modification of the method of Shure et al. (29). DNA extracted from B. neritina also contains bacterial DNA, which includes that of “E. sertula” and other environmental bacteria fouling the surface of the bryozoan. Adult B. neritina (2 g) was pulverized on dry ice, and then 6 ml of extraction buffer (8 M urea, 0.35 M NaCl, 0.05 M Tris-HCl [pH 7.5], 0.02 M EDTA, 2% sarcosyl) was added, followed by a phenol-choroform extraction, DNA precipitation, and 70% ethanol wash. The DNA was dissolved in a solution of 10 mM Tris-HCl (pH 7.5) and 10 mM EDTA and then passed through a Sephadex G-200 spin column (22) to remove PCR inhibitors.
DNA samples for amplification of PKS-I genes from larvae and for gene surveys of B. neritina populations with specific KSa primers were extracted from clean larvae and small portions of adult colonies as previously described, using a QIAamp tissue kit (Qiagen, Inc., Valencia, Calif.) (7, 15). DNA extracted from larvae contains primarily B. neritina and “E. sertula” DNA, with only slight contamination of DNA from other environmental bacteria.
Amplification, cloning, and sequencing of the KS domain DNA from B. neritina.
Amplification of KS genes from B. neritina DNA was accomplished using the degenerate primers KSD1F and KSD1R (Table 1) designed from conserved regions of KS domains of bacterial PKS-I genes. Bacterial and fungal polyketide synthase and rat fatty acid synthase amino acid sequences (accession numbers M76767, P40872, Q03131, Q03132, Q03133, S41729, S43048, U24241, U31329, and U00023) were aligned and examined for conserved regions. The region targeted for amplification is downstream from the active-site cysteine. The forward primer is based on the motif HGTGT, which is conserved, but not specific, as it appears in fatty acid synthase (FAS) as well. The downstream primer is based on the motif GTNAHV, which is conserved and specific to bacterial PKS. The primers were degenerate with inosine used for positions with fourfold redundancy. PCRs contained approximately 1 ng of B. neritina DNA (either adult or larval) μl−1, a 1 μM concentration of each primer, Taq polymerase, and buffer (Boehringer Mannheim Corp., Indianapolis, Ind.). To optimize annealing of degenerate KS primers, the annealing temperature was decreased with each temperature cycle from 60 to 40°C at a rate of 2°C per cycle (11 cycles) and then maintained at 40°C (39 cycles). Cycles included denaturation (94°C; 1 min), annealing, and extension (72°C; 1 min). PCR products of approximately 300 bp were cloned using a TOPO TA Cloning kit into Invitrogen pCR 2.1-TOPO vector as described by the manufacturer (Invitrogen Corp., Carlsbad, Calif.). Recombinant clones containing inserts were sequenced and analyzed. Primers specific to an abundant clone sequence from larval DNA, KSa, were designed for specific amplification of this gene fragment (Table 1 [Ksa1F and Ksa1R]). For the KSa-specific primers, the annealing temperature was 60°C, with other conditions the same (30 cycles).
TABLE 1.
Primers for amplification of KS domains
| Primer designation |
Sequence 5′-3′ |
|---|---|
| KSD1F | ACR TGI GCR TTI GTI CC |
| KSD1R | ICA YGG IAC IGG IAC |
| KSa1F | ACG GAC AAG CGT CAT TAC |
| KSa1R | GCC AAG GCT TTA ATT CCG |
| KSa2F | GTT GTC TTT GCA GCA TCG CAT GTT ACC AC |
| KSa2R | CAC GCC CGC TAT CCC AGC ACC TAC C |
| T7KSa1F | TAA TAC GAC TCA CTA TAA CGG ACA AGC GTC ATT AC |
| T7KSa1R | TAA TAC GAC TCA CTA TAG CCA AGG CTT TAA TTC CG |
Plasmid DNA of KS clones was prepared for sequencing using the QIAprep Spin Miniprep Kit (Qiagen, Inc.). All sequencing reactions were performed using the Applied Biosystems, Inc., PRISM Ready Reaction Dye Deoxy (or BigDye) terminator cycle sequencing kit as recommended by the manufacturer. Reactions were analyzed on an ABI automated sequencer (model 373A). Cloned genes were sequenced using primers directed against the cloning vector, pCR 2.1-TOPO (Invitrogen). Sequences were assembled using Sequencher and analyzed with BLAST (1) and by alignment with PKS and FAS sequences.
Design of mRNA KSa probe and in situ hybridizations.
To determine if KSa is expressed in “E. sertula,” in situ hybridizations of a KSa-specific RNA antisense probe were performed on larval B. neritina. Briefly, 300-bp fragments from a KSa clone were amplified by PCR using the primer pairs Ksa1F-T7Ksa1R and Ksa1R-T7Ksa1F to obtain antisense and sense strands respectively (8). T7Ksa1R and T7Ksa1F are primers with a T7 promoter at the 5′ ends. PCR products were purified using a QiaQuick PCR cleanup kit (Qiagen. Inc.) and used as templates in a T7 transcription reaction using the AmpliScribe T7 high-yield transcription kit as described by the manufacturer (EpiCentre Technologies, Inc., Madison, Wis.) with the addition of biotin-16-UTP (Boehringer Mannheim, Corp.) to obtain biotinylated probes. Another transcription was carried out using fluorescein-12-UTP to generate fluorescein-labeled probes for in situ hybridizations in Escherichia coli. Fluorescein probes were used because endogenous peroxidases in E. coli interfered with the avidin-horseradish peroxidase detection system used in the larvae.
In situ hybridizations in whole larvae and E. coli bacterial cells were performed as described by Haygood and Davidson (15). The annealing temperature for the 300-bp RNA probe was 47°C, and wash temperatures were 55, 37°C, and room temperature. In situ hybridizations of the KSa mRNA probe, sense and antisense, were performed with E. coli DH5 to control for probe binding to bacterial FAS.
Settlement and antibiotic treatment of B. neritina in the laboratory.
Adult B. neritina was collected from Torrey Pines artificial reef (La Jolla, Calif., September 1996 and June 1998) and Palos Verdes (Long Beach, Calif., March 1997 and June 1998) and maintained in aquaria. Larvae were released into the aquaria, collected, rinsed in filtered (0.2-μm pore size) seawater, concentrated (approximately 8 to 10 larvae per ml), and placed in polycarbonate petri dishes (17) mounted on plastic plates (13 by 13 cm; four dishes per plate). The larvae were allowed to settle overnight (approximately 150 per dish), and then half of the petri dishes containing developing colonies were treated with seawater containing 100 μg of gentamicin sulfate ml−1 for 7 days. The other half (controls) received unamended seawater. The seawater was changed and antibiotic solutions were prepared daily. A subset of the treated colonies was additionally treated with puromycin after 3 weeks of growth in an attempt to further eliminate “E. sertula.” In preliminary studies, “E. sertula” had shown sensitivity to gentamicin, puromycin, and polymyxin B (6).
After the antibiotic treatment, the dishes were suspended vertically in tanks (25 to 40 liters) with aerated seawater maintained at 18 to 20°C and changed every 48 h with addition of non axenic phytoplankton (50% Rhodomonas sp. and equal portions of Chlorella ellipsoidea, Nannochloris, Isochrysis, and Monochrysis spp.) The colonies were maintained in fresh, nonsterile seawater which had been passed through a series of sand filters. Preliminary experiments had shown that it was not necessary to maintain the antibiotic-treated B. neritina in sterile seawater to prevent “E. sertula” from repopulating the colonies.
The cultures were examined weekly by eye and less frequently with a dissection microscope to monitor health and growth. Several dishes and at least 25 to 35 colonies from each treatment were examined. Periodically, healthy B. neritina colonies were removed from the dishes at the base using forceps for use in symbiont and bryostatin assays. Healthy colonies that contained live zooids were selected and pooled together for the assays.
Analysis of “E. sertula” levels, microbial community differences, and KSa signal.
After growth in nonsterile seawater for 2 to 3 months, the B. neritina colonies had been colonized by bacteria from the seawater (growing on their surfaces and ingested). Denaturing gradient gel electrophoresis (DGGE) was used to compare the levels of both the “E. sertula” and the total bacterial communities between the control and treated colonies (11, 23). A portion of the small-subunit (SSU) rRNA gene was amplified using a conserved bacterial primer 1055f, and a universal primer with a GC clamp, 1392r-GC clamp (11). For amplification from B. neritina, 100 ng of DNA was used in 50-μl reaction mixtures with Taq polymerase and buffer (Boehringer Mannheim) as described by Haygood et al. (14), with 30 cycles of 94°C (1 min), 50°C (1 min), and 72°C (1 min). PCRs for each sample were run in triplicate and pooled prior to separation by DGGE. To make an “E. sertula” standard, symbiont SSU rRNA genes were amplified from B. neritina DNA using symbiont-specific primers as described by Haygood and Davidson (15). The “E. sertula” standard was then amplified with the primers described above from symbiont SSU rRNA gene amplification products and used to locate the “E. sertula” bands on gels.
Polyacrylamide gels (6.5%; 1:37 bis/acrylamide ratio) were prepared and run in TAE buffer (20 mM Tris base, 10 mM sodium acetate, and 0.5 mM EDTA, pH adjusted to 7.4) at 60°C with the CBS Scientific Inc. DGGE 2000 system at a running voltage of 200 V. An optimal gradient of 20 to 55% denaturant in the gel was used to separate the rRNA gene fragments. The DNA bands were visualized with 1× SYBR Gold (Molecular Probes, Eugene, Oreg.) on a UV transilluminator and were cut and extracted from the gel (11) for reamplification of DNA to be sequenced. Sequences from the suspected “E. sertula” band were compared with the “E. sertula” SSU rRNA sequence to confirm identity. The overall community was compared by examining the banding pattern from the control and treated samples.
Specific KSa primers (KSa1F and KSa1R) described above were used to amplify genes from equal amounts of DNA from the control and treated B. neritina to assess the level of this gene signal in treated colonies lacking most of their symbionts.
Bryostatin activity assays: tritiated phorbol dibutyrate displacement from protein kinase C (PKC) on rat brain liposomes.
To measure bryostatin activity in small samples a method was adapted from the protocol described by Schaufelberger et al. (27) for detecting bryostatin activity in extracts (9, 10, 27). Bryostatins bind to PKC at the same site as phorbol ester, but with a much higher affinity under the assay conditions in which they irreversibly displace the phorbol from the binding site on PKC (9). This assay is not specific to bryostatins but indicates the presence of PKC binding and correlates well with bryostatin in B. neritina as determined by high-performance liquid chromatography (HPLC) fractionation of extracts from B. neritina (Mary Koleck [SAIC, Frederick, Md.] personal communication). The bryozoan Scrupocellaria sp. does not contain bryostatins and occasionally grows together with B. neritina. Scrupocellaria was used as a negative control for phorbol displacement activity in bryozoan tissue extracts.
To prepare B. neritina extracts, fresh or frozen (−80°C) samples were lyophilized and then homogenized in 95% ethanol for a final concentration of 12 μg ml−1 (dry weight to volume). Debris was pelleted by centrifugation, and the supernatants were used in assays. Each sample dose tested was run in triplicate to generate displacement curves.
A liposome suspension enriched in PKC was prepared by homogenization of one Sprague-Dawley rat brain in 30 ml of 10 mM Tris-HCL/buffer, pH 7.4, using a tissue tearor set at maximum (position 5, 1 min). The suspension was centrifuged at 16,000 × g for 10 min, and the supernatant was discarded. The process was repeated until the pellet was a buff color. The pellet was homogenized in 30 ml of 10 mM Tris buffer for 20 s on full speed with the tissue tearor, and then aliquots (1 to 2 ml) were stored at −80°C. Just before use, this suspension was thawed, homogenized, and centrifuged in a tabletop centrifuge at 14,000 rpm (20,800 × g) for 10 min. and then resuspended in the binding buffer (50 mM Tris-HCl [pH 7.4], bovine serum albumin [2 mg ml−1]).
The crude extracts containing bryostatins together with tritiated phorbol dibutyrate (3H-PDBu) were added to the suspension of rat brain liposomes rich in PKC. Reactions (1 ml total) were prepared in binding buffer with addition of extracts (between 2 and 15 μl), 10 μl of 3H-PDBu (10 nM; 10 μCi), and 100 μl of the rat brain liposome suspension. Reactions with 10 μl of 20 μM unlabeled PDBu or no added sample were run as the positive and negative control solutions respectively. The negative control was run in duplicate for each assay and used as the total signal bound. Reactions were incubated for 1 h at 30°C and then passed through glass fiber filters (Whatman GF/B) on a vacuum manifold to capture liposomes, rinsed with 2 ml of 10 mM Tris-HCl buffer (pH 7.4), and placed in scintillation vials with 4 ml of scintillation cocktail. Vials were shaken for 30 min prior to reading on a scintillation counter. The reduction in tritium signal relative to the total tritium signal in the negative control was calculated and expressed as a percentage. The amount of crude extract required to displace 50% of the bound tritiated phorbol was the 50% inhibitory concentration (IC50), which was the value compared between samples. Bryostatin 1 standard curves were generated to give the IC50 for bryostatin 1 and allowed an estimation of bryostatin levels (expressed as bryostatin 1 equivalents) in the samples.
Nucleotide sequence accession number.
The nucleotide sequence of KSa from B. neritina (Fig. 2) has been assigned GenBank accession no. AF283572.
FIG. 2.
Alignment of amino acid sequences of KSa from B. neritina with representative KS domains of PKS-I and FAS. Residues conserved among PKS-I are indicated above the B. neritina sequence.
RESULTS
Amplification, cloning, and sequencing of KS domain DNA from B. neritina.
Degenerate primers (KSD1F and KSD1R) were used in a step-down PCR protocol to amplify a 300-bp fragment from B. neritina DNA. Since bacterial PKS-I enzymes are modular, PCR products represent a pool of KS fragments from different modules and different bacteria present in the sample. Two clone libraries were prepared, one from adult B. neritina DNA and one from larval DNA. Twenty-seven clones were sequenced, and nine unique sequences were identified, designated KSa to KSi. Four unique sequences, KSa (13 clones), KSb (1 clone), KSe (3 clones), and KSh (1 clone), were obtained from the larval DNA library, and six sequences were obtained from the adult library, KSb, KSc, KSd (two clones each), KSf, KSg, and KSi (one clone each). Only the KSb sequence was found in both libraries. Cloned DNA sequences were analyzed by using the BLAST (Basic Local Alignment Search Tool) server of the National Center for Biotechnology Information, accessed over the Internet (1). All of these sequences have the expected conserved signature regions for KS and show highest similarity in BLAST searches to bacterial PKS-I (Fig. 2). The most abundant sequence, KSa, was present in 13 out of 19 clones from the larval library (GenBank accession number AF283572) and was targeted for further study. An alignment of 85 amino acids of the clones, unalignable regions excluded, revealed identities between 32 and 58% relative to KSa.
Survey of B. neritina for KSa content and in situ hybridization.
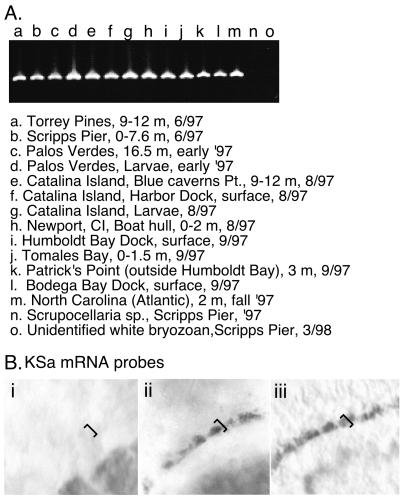
The KSa gene fragment was amplified by PCR from 10 samples of DNA from adult B. neritina colonies found in a wide range of locations in California from Humbolt Bay to San Diego, and included Catalina Island. Various habitat types and depths were sampled (boat docks and piers, 1 to 2 m; submerged rocks, 16 to 25 m). In addition, B. neritina from North Carolina was included. All of these samples had been shown previously to contain bryostatins (7). All samples produced ample product when KSa was amplified with the KSa-specific primers (Fig. 3A, lanes A to M). Two samples of other bryozoans collected with B. neritina did not yield products (Fig. 3A, lanes N and O). In similar experiments with the other eight KS sequences cloned, KSb to KSi, bands were only observed in a few samples, suggesting that these sequences originated in environmental bacteria.
FIG. 3.
KSa detection in B. neritina and in bacteria of the larvae. (A) Amplification of a KSa gene fragment from B. neritina DNA using specific primers. Lanes A to L show amplification of the KSa gene fragment in a variety of B. neritina samples, including both adults and larvae, collected throughout the year from different depths and habitats in California, and lane M shows one sample from North Carolina. Lanes N, Scrupocellaria sp., and O, unidentified white bryozoan, show lack of KSa amplification from two other species of bryozoa collected with B. neritina from the Scripps Pier. (B) In situ hybridization of KSa RNA probe in whole B. neritina larvae. The pallial sinus containing bacterial cells is indicated by a bracket. (i) sense KSa probe (negative control); (ii) antisense KSa probe; (iii) 16S rRNA probe showing location of “E. sertula” bacterial cells in the pallial sinus.
An antisense KSa RNA probe bound to the bacterial cells in the pallial sinus of the larvae, indicating that the “E. sertula” symbiont expresses the KSa message (Fig. 3B, panel ii). This probe did not bind to E. coli (not shown). The sense probe did not bind to bacterial cells in the larvae (Fig. 3B, panel i).
Growth of B. neritina colonies after antibiotic treatments.
After antibiotic treatments, the growth of the treated and that of the control B. neritina colonies were similar, showing no apparent detrimental effects from either the gentamicin treatments (up to 150 μg ml−1) or the reduction of symbiont population. The zooids, lophophores, and growth form of the colonies appeared normal relative to the control colonies. In all cases the range of colony sizes increased as growth progressed. The range of sizes and average size were similar between the control and the gentamicin-treated colonies (Fig. 4A) within a settlement. Between settlements (1996, 1997, and 1998) there was some difference in growth rates and final colony sizes reached; however, in every case the matched control and treated batches within a settlement had similar growth rates and final colony sizes. The differences between the settlements were likely due to variability of the larvae and environmental conditions, since both the control and treated B. neritina behaved in a similar manner.
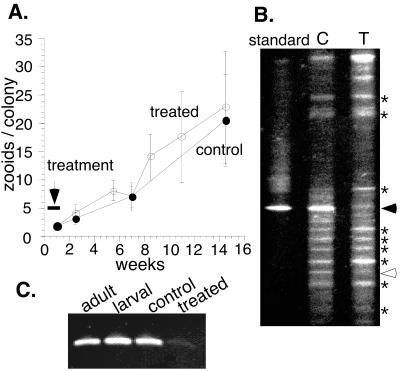
FIG. 4.
Growth, bacterial content and KSa gene detection in control and treated B. neritina. (A) Growth of colonies in number of zooids per colony. Thickbar; period of antibiotic treatment error bars, standard deviation. (B) Denaturing gradient gel showing bacterial SSU rRNA fragment pattern from treated and control colonies. Standard, purified “E. sertula”; solid arrow, “E. sertula”; ∗, common fragments; open arrow, band appearing in control diminished in treated B. neritina DNA. (C) PKS KSa gene fragments amplified from DNA of B. neritina wild adults, larvae, and laboratory-cultured control and antibiotic-treated colonies.
“E. sertula” DNA levels and microbial community analysis of treated and control colonies.
After an initial treatment with 100 μg of gentamicin sulfate ml−1 for 7 days, colonies were allowed to grow without further antibiotic treatments, except for a subset of the 1998 settlement which underwent a second antibiotic treatment. Symbiont levels were analyzed at 1 month and 2 to 3 months. Several replicate analyses of treated and control samples were conducted with consistent results. A representative DGGE gel is shown in Fig. 4B.
DGGE of 16S rRNA amplification products showed that the banding patterns were similar between the control and treated colony DNA amplifications, except for the “E. sertula” band (Fig. 4B). The “E. sertula” signal was by far the strongest in the control colony DNA, suggesting that this was the most-abundant bacterial species associated with B. neritina. The “E. sertula” signal was greatly reduced in the treated-colony DNA. In addition, a few bands appeared more strongly in the treated-colony DNA lane than they appear in the control lane. All of the amplification products that appeared in the controls also appeared in the treated-colony lane, with the exception of the “E. sertula” band and one additional faint band that was also reduced. This second amplification product was not consistently detected in wild specimens (6). This pattern indicated that the only major bacterial species reduced by the gentamicin treatments was the symbiont “E. sertula.” The decrease in the “E. sertula” population was confirmed by comparing the PCR products amplified with “E. sertula” -specific primers from a dilution series of quantified treated-colony DNA with dilutions of quantified control colony DNA. These comparisons showed a 50- to 100-fold reduction of “E. sertula” DNA in treated colonies relative to controls (250 ng of treated-colony DNA yielded product comparable to between 2.5 and 5 ng of control-colony DNA [6]).
Detection of polyketide synthase gene KSa.
PCR gene amplification using KSa-specific primers from wild adult, larvae, and cultured control B. neritina colony DNA all yielded strong signals for the PKS KSa gene fragment. Amplification from the treated-colony DNA resulted in only a small amount of product (Fig. 4C), indicating a substantially reduced level of KSa present in treated colonies.
Bryostatin activity assays.
Extracts of B. neritina colonies from three gentamicin-treated and control settlements were tested for bryostatin activity using the tritiated phorbol displacement assay. In addition, colonies of Scrupocellaria sp. that had fortuitously settled in the dishes along with the B. neritina larvae were assayed for activity. The Scrupocellaria sp. extracts showed no phorbol displacement activity (Fig. 5A).
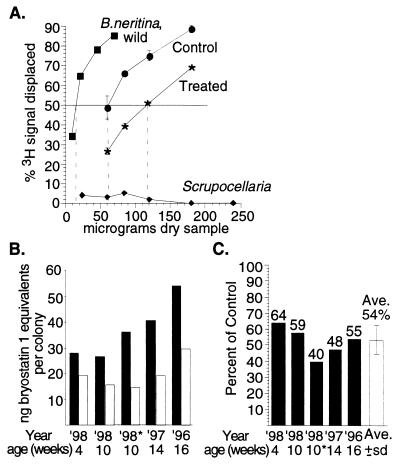
FIG. 5.
Bryostatin activity. (A) Representative 3H-PDBu displacement curve used to determine relative bryostatin activity levels in B. neritina. Standard deviations (error bars) are shown for each point; however, for most points the variation is only as wide as or smaller than the point symbol. Dashed lines dropped to the x axis indicate IC50s (dose giving 50% displacement). Scrupocellaria sp. extracts were tested as a negative control. (B) Bryostatin 1 equivalents in laboratory-grown B. neritina control (solid bars) and treated (open bars) colonies from settlements from 1996 to 1998 (the bryostatin 1 PDBu displacement curve gave an IC50 of 13 ng). (C) The percentage of bryostatin activity remaining in treated colonies relative to controls for all treatments. The bryostatin activity levels in controls represent the 100% values. For all values, variation was less than 1.5 percentage points. Open bar, the average bryostatin level remaining in treated colonies for all settlements and ages, with the standard deviation (error bar) shown.
A representative graph of the percentage of tritiated phorbol displacement plotted as a function of the amount of sample added is shown in Fig. 5A. The IC50s calculated from the binding curves were expressed as IC50−1 (activity per microgram of sample). The IC50s indicated that reduction of the “E. sertula” population was consistently correlated with reduced bryostatin levels (22 to 60% less bryostatin activity by weight). The least difference in bryostatin activity between control and treated B. neritina was observed when the colonies had been growing quickly and the level of bryostatins was low in the control colonies (1998 settlement). It is possible that the levels of bryostatin production could not keep up with the rate of bryozoan colony growth. The greatest differences in bryostatin activity were observed when growth had slowed, as in the 1996 end point measurements and the subset of 1998 settlement treated a second time with antibiotics. A likely explanation is that the bryostatins were accumulating in the slowly growing colonies but accumulated at a faster rate in the controls. Since the measurements are by weight, if growth slows and the production of bryostatins remains the same, then bryostatins will accumulate faster in colonies with a slow growth rate. The highest bryostatin levels by weight and the greatest difference between the control and treated-colony bryostatin activities occurred with the 1998 double antibiotic treatment, in which the growth was stunted but the colonies appeared healthy and were feeding. In this treatment the growth of both the control and treated colonies slowed due to the removal from the main tank for 1 week. The sizes of both control and treated colonies were similar.
The IC50s were transformed into bryostatin 1 equivalents by comparing the IC50s of the colony extracts to that of bryostatin 1. Since the PDBu assay measures binding to PKC by all bryostatins present in the sample, this number is referred to as the bryostatin 1 equivalent and does not represent the actual amount of bryostatin 1. The amounts by weight were converted into amount per zooid from determined dried zooid weights. The total amount of bryostatin per colony was estimated based on colony size (zooid number). We found that the average of the three treatments showed that 54% of the bryostatin activity remained in treated colonies when comparing bryostatin 1 equivalents per colony.
DISCUSSION
The biosynthetic capability for production of bryostatins must reside in the bryozoan B. neritina, its specific bacterial symbiont “E. sertula,” another associated organism, or a combination of these organisms. In this article, we describe evidence that “E. sertula” has the genetic ability to produce complex PKS-I and that the symbiont is required for bryostatin synthesis, suggesting that “E. sertula” is the biosynthetic source of bryostatins.
Gene fragments from the KS domain, which is present in all modules of bacterial PKS-I genes, were amplified, cloned, and sequenced from both larval and adult B. neritina DNA. The most abundant sequence, KSa, was found in 13 out of 19 larval clones (Fig. 2). The abundance of KSa sequence among the clones obtained from DNA of the larvae suggest “E. sertula” as the source of this sequence. Larvae should have primarily “E. sertula” bacterial DNA since the larvae have a monoculture of the symbiont in the pallial sinus (15) and do not have a gut. PCR using primers (Table 1) specific to the KSa clone amplified this gene fragment from all adult and larval B. neritina samples tested but did not result in amplification from other bryozoans growing in proximity to B. neritina (Fig. 3A). This survey included samples from the Atlantic; Northern and Southern California; boat docks; submerged rocks; and different seasons, temperatures, and depths. The presence of the Ksa gene fragment in all B. neritina samples showed that the sequence is specific to and always present in B. neritina. If KSa originated from a contaminant found in the environment we would expect the presence of this gene fragment to have varied in different samples, as was the case with the other eight KS sequences cloned, and to have appeared in other bryozoans living in the same milieu as B. neritina.
An RNA antisense probe synthesized from a KSa clone was used for in situ hybridization to label KSa message within the larvae. This probe bound to bacterial cells within the pallial sinus, indicating that the mRNA for KSa was expressed in the “E. sertula” cells of the pallial sinus of B. neritina (Fig. 3B). The negative control sense probe did not bind. The KSa probe did not bind to closely related bacteria, indicating that the probe was not binding to an FAS gene but was specific to “E. sertula” (not shown). Although we do not know whether these PKS genes are involved in the bryostatin synthesis pathway, the presence of these genes in “E. sertula” shows that they have the potential for synthesis of complex polyketides such as bryostatin.
To test the hypothesis that “E. sertula” is involved in bryostatin production in the host bryozoan, and to confirm by a third method that KSa originated from “E. sertula,” we attempted to eliminate the symbiont from B. neritina reared in the laboratory and assayed for bryostatin and KSa gene levels. To date it has not been possible to eliminate the symbiont from the host entirely. However, the levels of “E. sertula” were substantially reduced (an estimated 5% remaining) in gentamicin-treated B. neritina as shown by DGGE comparative analysis of total SSU rRNA genes amplified from control and treated colony DNA (Fig. 4B). This result was confirmed by specific amplification of “E. sertula” SSU rRNA genes from a dilution series of control and treated-colony DNA (6). Although these methods are not strictly quantitative, the results from several amplifications were consistent and confirmed that the “E. sertula” was substantially reduced by antibiotic treatment.
The bacteria that colonize B. neritina after settlement are not likely specific symbionts but environmental bacteria that colonize surfaces and pass through the gut tract of the bryozoan. DGGE analysis of SSU rRNA genes confirmed that the total microbial community, excluding “E. sertula,” was similar in treated and control colonies (Fig. 4B). “E. sertula” yielded the dominant signal in control colonies and was greatly reduced in the treated-colony bacterial community SSU rRNA profiles. SSU rRNA genes amplified from the control colonies were also present in the treated colonies, indicating that similar microbial species were present in both. Although one additional minor product was detected in the controls that was reduced in the treated colonies, this product has not consistently appeared in DGGE analyses of wild populations (6). This sporadic appearance indicated that this bacterium was not a candidate for the bryostatin producer. To date DGGE analyses of wild B. neritina have shown that only “E. sertula” was consistently found associated with B. neritina (6), although it is possible that there is another symbiont present that is not detectable by PCR of the SSU rRNA genes. PCR amplification of the KSa gene using specific primers yielded a strong signal from control B. neritina colonies but produced a weak signal from the treated colonies with reduced “E. sertula” (Fig. 4C). In light of the DGGE evidence, the reduction of KSa in the treated B. neritina is due to the reduction of “E. sertula” numbers.
PDBu displacement assays indicated an average reduction of 46% in bryostatin activity from treated colonies relative to controls (Fig. 5C). This reduction, however, was not proportional to the loss of bacterial symbiont (46% loss in activity compared to 95% estimated loss of “E. sertula”). This finding may seem to contradict the hypothesis that the bacterial symbionts produce bryostatins, but there are reasons not to expect a strict linear relationship between the two. The synthesis of bryostatins may be under the control of the symbiont or the host and may be up-regulated when bacterial numbers are reduced. An alternative explanation is that there are other compounds in the bryozoan or from another undetected symbiont that bind PKC. The data currently available do not support either of these alternative explanations. Extracts of other bryozoans have not shown activity in the PKC binding assay. HPLC peaks from B. neritina extracts that test positive in this assay have been identified as bryostatins. B. neritina compounds have been investigated for their PKC binding ability and bioactivity for the past 30 years, and no other compounds besides bryostatins have been found. Considering these factors, it is unlikely that there are other compounds that react in the PKC binding assay to the level we observed (54% of the activity). The possibility of an undetected symbiont has been investigated by bacterial population surveys of B. neritina from different locations, and data to date do not support a second unidentified bacterial symbiont. In addition, the larvae contain one transmitted symbiotic species, “E. sertula,” and contain bryostatins.
Since “E. sertula” was the only significantly reduced bacterial species after the antibiotic treatments and since the growth and morphology of treated colonies showed no detrimental effects, differences in KSa gene signal and bryostatin activity can be attributed to the reduction of “E. sertula.”
In summary, an abundant KS sequence, KSa, was (i) shown by PCR to be present in all populations of B. neritina available, (ii) shown by mRNA hybridization to be expressed in the “E. sertula” cells in the pallial sinus of the larvae, and (iii) shown to be reduced in concert with reduction of “E. sertula” levels. These data indicate that the KSa sequence originates in the bacterial symbiont “E. sertula.” In addition, the observation that reduction of “E. sertula” levels reduced bryostatin production suggests that “E. sertula” is the likely biosynthetic source of the bryopyran ring and possibly the entire bryostatin molecule. This finding has important implications for understanding the function of the symbiont in this association between a bryozoan and a bacterium. Although the benefit of bryostatins to the bryozoan remains untested, bryostatins likely serve as a form of chemical defense that is provided by the bacterial symbionts. These findings also illustrate how molecular methods may be used to solve some of the problems associated with advancing marine natural products from discovery to use as therapies. If symbiotic bacteria or their genes can be harnessed for production of bryostatins or novel analogues thereof, lack of supply may no longer be a barrier for the development and use of this important family of compounds.
ACKNOWLEDGMENTS
CalBioMarine Technologies, Inc. (Carlsbad, Calif.) contributed larvae and expertise regarding the culturing of B. neritina. Carolyn Sheehan provided technical assistance. Thanks also go to Ron McConnaughey, who assisted with collections of B. neritina adults and helped set up aquaria for growth experiments.
This work was supported by a California Sea Grant College grant (R/MP-61) and the National Sea Grant Technology Program (R/MP-84A). S. W. Allen was supported on a Small Business Innovation Research grant (R44 CA58158-02A3) from the National Cancer Institute, awarded to CalBioMarine Technologies, Inc. G. E. Lim was supported by a Howard Hughes Medical Institute Fellowship.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthoni U, Nielsen P H, Perieira M, Christophersen C. Comp. Biochem Physiol. 1990;96B:431–437. [Google Scholar]
- 3.Baldwin N G, Rice C D, Tuttle T M, Bear H D, Hirsch J I, Merchant R E. Ex vivo expansion of tumor-draining lymph node cells using compounds which activate intracellular signal transduction. I. Characterization and in vivo anti-tumor activity of glioma-sensitized lymphocytes. J Neurooncol. 1997;32:19–28. doi: 10.1023/a:1005719700570. [DOI] [PubMed] [Google Scholar]
- 4.Basu A. The involvement of novel protein kinase C isozymes in influencing sensitivity of breast cancer MCF-7 cells to tumor necrosis factor-alpha. Mol Pharmacol. 1998;53:105–111. doi: 10.1124/mol.53.1.105. [DOI] [PubMed] [Google Scholar]
- 5.Correale P, Caraglia M, Fabbrocini A, Guarrasi R, Pepe S, Patella V, Marone G, Pinto A, Bianco A R, Tagliaferri P. Bryostatin 1 enhances lymphokine activated killer sensitivity and modulates the beta 1 integrin profile of cultured human tumor cells. Anticancer Drugs. 1995;6:285–90. doi: 10.1097/00001813-199504000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Davidson S K. The biology of the bryostatins in the marine bryozoan Bugula neritina. Dissertation. La Jolla, Calif: University of California, San Diego; 1999. [Google Scholar]
- 7.Davidson S K, Haygood M G. Identification of sibling species of the bryozoan Bugula neritina that produce different anticancer bryostatins and harbor distinct strains of the bacterial symbiont “Candidatus Endobugula sertula.”. Biol Bull. 1999;196:273–280. doi: 10.2307/1542952. [DOI] [PubMed] [Google Scholar]
- 8.DeLong E F, Taylor L T, Marsh T L, Preston C M. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeVries D J, Herald C L, Pettit G R, Blumberg P M. Demonstration of sub-nanomolar affinity of bryostatin 1 for the phorbol ester receptor in rat brain. Biochem Pharmacol. 1988;37:4068–4073. doi: 10.1016/0006-2952(88)90097-4. [DOI] [PubMed] [Google Scholar]
- 10.DeVries D J, Rao K S, Willis R H. Application of a radioreceptor assay to the screening and characterisation of compounds from marine organisms with activity at the phorbol ester binding site of protein kinase C. Toxicon. 1997;35:347–354. doi: 10.1016/s0041-0101(96)00179-1. [DOI] [PubMed] [Google Scholar]
- 11.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming M D, Bear H D, Lipshy K, Kostuchenko P J, Portocarero D, McFadden A W, Barrett S K. Adoptive transfer of bryostatin-activated tumor-sensitized lymphocytes prevents or destroys tumor metastases without expansion in vitro. J Immunother Emphasis Tumor Immunol. 1995;18:147–55. doi: 10.1097/00002371-199510000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Grant S, Traylor R, Pettit G R, Lin P S. The macrocyclic lactone protein kinase C activator, bryostatin 1, either alone, or in conjunction with recombinant murine granulocyte-macrophage colony-stimulating factor, protects Balb/c and C3H/HeN mice from the lethal in vivo effects of ionizing radiation. Blood. 1994;83:663–667. [PubMed] [Google Scholar]
- 14.Haygood M, Distel D, Herring P. Polymerase chain reaction and 16S rRNA gene sequences from the luminous bacterial symbionts of two deep-sea anglerfishes. J Mar Biol Assoc U K. 1992;71:149–159. [Google Scholar]
- 15.Haygood M G, Davidson S K. Small subunit ribosomal RNA genes and in situ hybridization of the bacterial symbionts in the larvae of the bryozoan Bugula neritina and proposal of “Candidatus Endobugula sertula.”. Appl Env Microbiol. 1997;63:4612–4616. doi: 10.1128/aem.63.11.4612-4616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson M D, Torri J A, Lippman M E, Dickson R B. Regulation of motility and protease expression in PKC-mediated induction of MCF-7 breast cancer cell invasiveness. Exp Cell Res. 1999;247:105–113. doi: 10.1006/excr.1998.4336. [DOI] [PubMed] [Google Scholar]
- 17.Keough M J. Dispersal of the bryozoan Bugula neritina and effects of adults on newly metamorphosed juveniles. Mar Ecol Prog Ser. 1989;57:163–171. [Google Scholar]
- 18.Kraft A S, Smith J B, Berkow R L. Bryostatin, an activator of the calcium phospholipid-dependent protein kinase, blocks phorbol ester-induced differentiation of human promyelocytic leukemia cells HL-60. Proc Natl Acad Sci USA. 1986;83:1334–1338. doi: 10.1073/pnas.83.5.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraft A S, Woodley S, Pettit G R, Gao F, Coll J C, Wagner F. Comparison of the antitumor activity of bryostatins 1, 5, and 8. Cancer Chemother Pharmacol. 1996;37:271–278. doi: 10.1007/BF00688328. [DOI] [PubMed] [Google Scholar]
- 20.Lind D S, Tuttle T M, Bethke K P, Frank J L, McCrady C W, Bear H D. Expansion and tumor specific cytokine secretion of bryostatin-activated T-cells from cryopreserved axillary lymph nodes of breast cancer patients. Surg Oncol. 1993;2:273–282. doi: 10.1016/s0960-7404(06)80002-2. [DOI] [PubMed] [Google Scholar]
- 21.Lipshy K A, Kostuchenko P J, Hamad G G, Bland C E, Barrett S K, Bear H D. Sensitizing T-lymphocytes for adoptive immunotherapy by vaccination with wild-type or cytokine gene-transduced melanoma. Ann Surg Oncol. 1997;4:334–341. doi: 10.1007/BF02303584. [DOI] [PubMed] [Google Scholar]
- 22.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 23.Muyzer G, Waal E C D, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettit G R, Herald C L, Doubek D L, Herald D L. Isolation and structure of bryostatin 1. J Am Chem Soc. 1982;104:6846–6848. [Google Scholar]
- 25.Pettit G R. The bryostatins. Prog Chem Org Nat Prod. 1991;57:153–195. doi: 10.1007/978-3-7091-9119-4_3. [DOI] [PubMed] [Google Scholar]
- 26.Pluda J M, Cheson B D, Phillips P H. Clinical trials referral resource. Clinical trials using bryostatin-1. Oncology (Huntington) 1996;10:740–742. [PubMed] [Google Scholar]
- 27.Schaufelberger D E, Alvarado A B, Andrews P, Beutler J A. Detection and quantitation of bryostatin 1 and 2 in Bugula neritina by combined high-performance liquid chromatography and 3H-phorbol dibutyrate displacement. J Liq Chromatography. 1990;13:583–598. [Google Scholar]
- 28.Scheid C, Prendiville J, Jayson G, Crowther D, Fox B, Pettit G R, Stern P L. Immunomodulation in patients receiving intravenous bryostatin 1 in a phase I clinical study: comparison with effects of bryostatin 1 on lymphocyte function in vitro. Cancer Immunol Immunother. 1994;39:223–230. doi: 10.1007/BF01525985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shure M, Wessler S, Fedoroff N. Molecular identification and isolation of the waxy locus in maize. Cell. 1983;35:225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- 30.Smith J B, Smith L, Pettit G R. Bryostatins: potent, new mitogens that mimic phorbol ester tumor promoters. Biochem Biophys Res Commun. 1985;132:939–945. doi: 10.1016/0006-291x(85)91898-4. [DOI] [PubMed] [Google Scholar]
- 31.Steube K G, Drexler H G. Differentiation and growth modulation of myeloid leukemia cells by the protein kinase C activating agent bryostatin-1. Leuk Lymphoma. 1993;9:141–148. doi: 10.3109/10428199309148517. [DOI] [PubMed] [Google Scholar]
- 32.Sung S J, Lin P S, Schmidt-Ullrich R, Hall C E, Walters J A, McCrady C, Grant S. Effects of the protein kinase C stimulant bryostatin 1 on the proliferation and colony formation of irradiated human T-lymphocytes. Int J Radiat Biol. 1994;66:775–783. [PubMed] [Google Scholar]
- 33.Taylor L S, Cox G W, Melillo G, Bosco M C, Espinoza-Delgado I. Bryostatin-1 and IFN-gamma synergize for the expression of the inducible nitric oxide synthase gene and for nitric oxide production in murine macrophages. Cancer Res. 1997;57:2468–73. [PubMed] [Google Scholar]
- 34.Woollacott R M. Association of bacteria with bryozoans larvae. Mar Biol. 1981;65:155–158. [Google Scholar]