Abstract
The enterococcal surface protein, Esp, is a high-molecular-weight surface protein of unknown function whose frequency is significantly increased among infection-derived Enterococcus faecalis isolates. In this work, a global structural similarity was found between Bap, a biofilm-associated protein of Staphylococcus aureus, and Esp. Analysis of the relationship between the presence of the Esp-encoding gene (esp) and the biofilm formation capacity in E. faecalis demonstrated that the presence of the esp gene is highly associated (P < 0.0001) with the capacity of E. faecalis to form a biofilm on a polystyrene surface, since 93.5% of the E. faecalis esp-positive isolates were capable of forming a biofilm. Moreover, none of the E. faecalis esp-deficient isolates were biofilm producers. Depending on the E. faecalis isolate, insertional mutagenesis of esp caused either a complete loss of the biofilm formation phenotype or no apparent phenotypic defect. Complementation studies revealed that Esp expression in an E. faecalis esp-deficient strain promoted primary attachment and biofilm formation on polystyrene and polyvinyl chloride plastic from urine collection bags. Together, these results demonstrate that (i) biofilm formation capacity is widespread among clinical E. faecalis isolates, (ii) the biofilm formation capacity is restricted to the E. faecalis strains harboring esp, and (iii) Esp promotes primary attachment and biofilm formation of E. faecalis on abiotic surfaces.
Enterococcus faecalis is a saprophytic commensal that inhabits the oral cavity and gastrointestinal flora of humans and animals, although it can behave as an opportunistic pathogen causing severe urinary tract infections, surgical wound infections, bacteremia, and bacterial endocarditis (21, 32, 47). Over the past 2 decades, E. faecalis has become responsible for up to 12% of nosocomial infections, with mortality rates for bloodstream infections ranging from 20 to 68% depending on the patient population (11). The increased incidence of E. faecalis infection has been related to the innate resistance of this microorganism to many commonly used antimicrobial agents and to its ability to become resistant to most, and in some cases to all, of the presently available antibiotics, either by mutation or by incorporation of foreign genetic material (6, 24). However, antibiotic resistance alone does not explain the prevalence of E. faecalis in enterococcal nosocomial infections, since Enterococcus faecium, a species less susceptible to commonly used antimicrobial agents, is responsible for only 20% of hospital-acquired enterococcal infections whereas E. faecalis is responsible for most of the remaining infections (33). This observation strongly supports the existence of additional virulence properties that may facilitate or enhance virulence of the E. faecalis isolates associated with infections.
Although knowledge on the pathogenic factors of E. faecalis is still limited, several virulence molecules associated preferentially with infection-derived E. faecalis strains have been described; these include cytolysin (16, 25, 26), aggregation substance (7, 28, 35), extracellular superoxide (22, 23), surface carbohydrates (17, 24), and surface proteins, such as Ace (34), EfaA (30), and Esp (43). Among these molecular species, Esp is the only one whose role in virulence has not been defined. Esp is a large surface protein of 1,873 amino acids with an N-terminal domain (amino acids 50 to 743) without significant similarity to other proteins in the database. The central core region (amino acids 744 to 1665) consists of a series of two distinct tandem repeat units encoded by nearly identical DNA sequence repeats and shows global structural similarity to C alpha and Rib proteins of group B streptococci. The C-terminal domain (amino acids 1666 to 1873) contains a membrane-spanning hydrophobic region and includes a slight variation of the LPXTGX motif found in most wall-associated surface proteins of gram-positive bacteria. It is presently hypothesized that the N-terminal region of Esp might participate in interactions with the host and that the central repeat region might serve to retract the protein from the surface, hiding the protein from the immune system.
In previous work aimed at identifying new factors involved in Staphylococcus aureus biofilm formation, we reported a high-molecular-weight cell wall-associated protein of 2,276 amino acids named Bap (9). The corresponding bap gene was found among staphylococcal species isolated from bovine mastitis but not among the human S. aureus isolates tested. Interestingly, all of the S. aureus isolates harboring Bap were strong biofilm producers, and transposon-insertional mutagenesis of bap caused the loss of the biofilm formation phenotype. Bap showed global organizational similarities to an outer membrane protein-like protein of Pseudomonas putida involved in adhesion to seeds (12), to a proline/threonine-rich protein of unknown function of Salmonella enterica serovar Typhi, and to the Esp protein of E. faecalis.
Taking into account the global structural similarity between Bap and Esp, in this work we analyzed the association between the ability of E. faecalis isolates to produce a biofilm and the presence of esp. Furthermore, we performed insertional mutagenesis of esp in different E. faecalis isolates and analyzed the effects produced in different steps of the biofilm formation process at the macroscopic and microscopic levels. A role as a putative virulence factor involved in bacterial biofilm formation was assigned to Esp.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
A total of 200 enterococcal isolates were used: 152 E. faecalis isolates, 39 E. faecium isolates, 7 E. avium isolates, 1 E. gallinarum isolate, and 1 E. durans isolate. They were isolated from patients during 1999 and 2000 at the Microbiology Department of the University Clinics of Navarra, Pamplona, Spain. Bacterial species were identified with API STREP (bioMérieux, Marcy l'Etoile, France) or GPI-Vitek (bioMérieux).
Escherichia coli XL1-Blue cells were grown in Luria-Bertani broth or on Luria-Bertani agar (Pronadisa, Madrid, Spain) with appropriate antibiotics. Enterococci were grown in brain heart infusion (BHI) broth, BHI agar, or Trypticase soy broth (TSB) supplemented with glucose (0.25 or 0.5%, wt/vol), as required. Antibiotics were used at the following concentrations: chloramphenicol, 20 μg/ml; and ampicillin, 100 μg/ml.
DNA manipulations.
Routine DNA manipulations were performed using standard procedures (4, 41) unless otherwise stated. Plasmid DNA from E. coli was purified with a Quantum Prep Plasmid Miniprep (Bio-Rad) or Midiprep (QIAGEN) kit. Plasmids were transformed into E. faecalis by electroporation using a previously described method (48). Enterococcal transformations were enhanced by inducing chloramphenicol acetyltransferase translation with a subinhibitory concentration of chloramphenicol (0.2 μg/ml) for 2 h after electroporation. Restriction enzymes were purchased from Boehringer Mannheim and used according to the manufacturer's instructions. Oligonucleotides were obtained from Life Technologies (Table 1).
TABLE 1.
Strains, oligonucleotides, and plasmids used in this study
| E. faecalis strain, oligonucleotide, or plasmid | Relevant characteristics | Position (nucleotides)a | Strand | Source or reference |
|---|---|---|---|---|
| E. faecalis strains | ||||
| 54 | esp+, strong biofilm forming | CUNb | ||
| 54M | 54 esp::pBT2 (Cmr) | This study | ||
| 1 | esp+, strong biofilm forming | CUN | ||
| 1M | 1 esp::pBT2 (Cmr) | This study | ||
| 11279 | esp+, medium biofilm forming | CUN | ||
| 11279M | 11279 esp::pBT2 (Cmr) | This study | ||
| 11262 | esp+, weak biofilm forming | CUN | ||
| 11262M | 11262 esp::pBT2 (Cmr) | This study | ||
| 14377 | esp+, adherence++, the esp gene contains one A repeat and four C repeats | This study | ||
| 23 | esp−, non-biofilm forming (Cms) | CUN | ||
| 23C | 23 containing pTA2 (Cmr) | This study | ||
| Oligonucleotides | ||||
| esp11 | TTGCTAATGCTAGTCCACGACC | 1217–1238 | + | 43 |
| esp12 | GCGTCAACACTTGCATTGCCGAA | 2171–2149 | − | 43 |
| esp46 | TTACCAAGATGGTTCTGTAGGCAC | 2256–2279 | + | 43 |
| esp47 | CCAAGTATACTTAGCATCTTTTGG | 3169–3192 | − | 43 |
| esp2 | CAGATGGATCATCTGATGAAGT | 3254–3275 | + | 43 |
| esp5 | GTAACGTTACTGTTACATCTGC | 5338–5359 | − | 43 |
| esp3S | CGCCTTGGTATGCTAAC | 1004–1020 | + | This study |
| esp4S | GGTAAGCTTACGCCGT | 2435–2420 | − | This study |
| esp2S | GGGTCGACACTTCTATTCATCCTCT | 5707–5691 | − | This study |
| pbacSc | GCTTGCATCAAAATAAACTACATGGGTATAATAGCAATGAAATGCATTTCAAAAATATTTTGAGGAGAATTTAGTATGTTTGGAAAA | + | This study | |
| Plasmids | ||||
| pCU1 | E. coli-S. aureus shuttle vector | 3 | ||
| pBT2 | E. coli-S. aureus shuttle vector with a thermosensitive origin of replication for S. aureus | 5 | ||
| pTA1 | Vector for recombination experiments, 0.95-kb PCR fragment of esp subcloned in pBT2 | This study | ||
| pTA2 | Vector for complementation experiments; a 4.3-kb PCR fragment containing esp from E. faecalis 14377 fused with the enterococcal constitutive bacA promoter cloned in pCU1 | This study |
Nucleotide sequence positions refer to the esp sequence in the GenBank database under accession no. AF034779.
CUN, University Clinics of Navarra.
Regions -35 and -10 and the ribosome binding site from the bacA promoter according to Fujimoto et al. (15) are underlined. The ATG codon for the first amino acid of Esp is also underlined.
For Southern hybridization, chromosomal DNA was purified as previously described (31), digested with EcoRI and PstI, and analyzed by agarose gel electrophoresis. DNA fragments were transferred by alkaline capillary blotting onto nylon membranes (Hybond-N; Amersham Life Science) using standard methods (4). A 950-bp PCR fragment obtained with primers esp11 and esp12 (Table 1) was used as a DNA probe. Labeling of the probe and DNA hybridization were performed according to the protocol supplied with the PCR-DIG DNA-labeling and chemiluminescent detection kit (Boehringer Mannheim).
Epidemiology study on the presence of the esp gene and repeat number variation.
Primers esp11 and esp12 (Table 1) were used to amplify a 950-bp fragment within the N-terminal region of esp and to detect its presence in DNAs from the different enterococcal isolates. To assess the repeat number variation among the esp-positive enterococcal isolates, primers esp46 and esp47 (Table 1) were used for amplification across the A repeat region, whereas primers esp2 and esp5 (Table 1) were used for amplification across the C repeat region. According to the sequences of the esp repeat regions described by Shankar et al. (43), the number of A repeats was calculated as follows: nA = (faPCR − 182)/252, where nA is the number of A repeats and faPCR is the size of the DNA fragment after PCR amplification using primers esp46 and esp47 (Table 1). Similarly, the number of C repeats was calculated as follows: nC = (fcPCR − 384)/246, where nC is the number of C repeats and fcPCR is the size of the DNA fragment after PCR amplification using primers esp2 and esp5 (Table 1).
Disruption of esp
For disruption of esp in three esp-positive biofilm-forming isolates (Table 1), a 950-bp PCR fragment within the N-terminal region of the esp gene amplified with primers esp11 and esp12 was cloned into the pGEM-T Easy vector (Promega, Madison, Wis.). The esp fragment was then cloned into the EcoRI site of the shuttle vector pBT2, and the resulting plasmid (pTA1) was transformed into E. faecalis by electroporation. After electroporation, bacterial strains were incubated for 24 to 48 h at 30°C on BHI agar with chloramphenicol. Ten milliliters of BHI broth-chloramphenicol was subsequently inoculated with a single colony which had been previously resuspended in 100 μl of BHI broth, and the culture was incubated for 24 h at 43.5°C without shaking. Tenfold serial dilutions of this culture in sterile BHI broth were plated on BHI agar-chloramphenicol and incubated for 24 h at 43.5°C. After overnight incubation, colonies were analyzed for disruption of the esp gene by colony PCR with primers esp3S and esp4S (Table 1), and the results were confirmed by Southern blot analysis.
Biofilm assay on polystyrene plates and adherence to polyvinyl chloride (PVC) plastic.
The ability of the enterococcal strains to form a biofilm on an abiotic surface was quantified essentially as described elsewhere (37). Briefly, E. faecalis strains were grown overnight in TSB with 0.25% glucose at 37°C. The culture was diluted 1:40 in TSB–0.25% glucose, and 200 μl of this cell suspension was used to inoculate sterile 96-well polystyrene microtiter plates (Iwaki, Tokyo, Japan). After 24 h at 37°C, wells were gently washed three times with 200 μl of phosphate-buffered saline (PBS), dried in an inverted position, and stained with 1% crystal violet for 15 min. The wells were rinsed again, and the crystal violet was solubilized in 200 μl of ethanol-acetone (80:20, vol/vol). The optical density at 595 nm (OD595) was determined using a microplate reader (Multiskan EX; Labsystems). Each assay was performed in triplicate and repeated three times.
Bacterial adherence to PVC plastic was studied using a phase-contrast microscope (magnification, ×1,000; Nikon Optiphot) as described elsewhere (39), with the following modifications. E. faecalis strains were grown overnight in TSB–0.25% glucose at 37°C. Subsequently, the culture was diluted 1:40 in TSB–0.25% glucose, and 200 μl of this cell suspension was inoculated into the wells of 96-well microtiter dishes containing sterile PVC disks from urine collection bags and incubated for 24 h at 37°C.
To compare biofilm formation ability and adherence to PVC of wild-type, recombinant, and complemented strains, bacteria were cultured overnight in TSB–0.25% glucose, supplemented with chloramphenicol (20 μg/ml) when appropriate, and subcultured in TSB–0.25% glucose using microtiter dishes.
Primary adherence assay.
Early adherence of E. faecalis to a polystyrene surface was determined as previously described (14), with the following modifications. E. faecalis strains were grown in TSB–0.5% glucose, supplemented with chloramphenicol (20 μg/ml) when appropriate, overnight at 37°C. Cultures were then adjusted with TSB–0.5% glucose to an OD578 of 0.1. Ten milliliters of each suspension was added to two polystyrene petri dishes. After incubation for 2 h at 37°C, petri dishes were washed three times with PBS. Cells were fixed with Bouin solution (Sigma) and Gram stained. Adherent bacterial cells were observed by oil immersion microscopy, and the mean count was determined in five microscopic fields. Each experiment was repeated three times.
Analysis of cell surface expression of Esp.
E. faecalis strains were grown overnight in TSB–0.25% glucose, supplemented with chloramphenicol (20 μg/ml) when appropriate, at 37°C. The culture was diluted 1:40 in TSB–0.25% glucose, and 200 μl of this cell suspension was used to inoculate sterile 96-well polystyrene microtiter plates (Iwaki). After 24 h at 37°C, wells were gently washed three times with 200 μl of PBS containing 0.1% Tween 20. Wells were blocked with 5% bovine serum albumin at 37°C for 1 h prior to a 2-h incubation with anti-Esp serum diluted 1:5,000 in PBS containing 0.1% Tween 20. Bound antibodies were detected with a peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (Jackson ImmunoResearch Laboratories, Inc., Bar Harbor, Maine) diluted 1:2,500.
Cell surface hydrophobicity.
The cell surface hydrophobicities of E. faecalis strains were verified as previously described (40), with the following modifications. Cells were grown overnight in TSB–0.25% glucose, supplemented with chloramphenicol (20 μg/ml) when appropriate, at 37°C. Three hundred microliters of the test hydrocarbon (n-hexadecane; Merck, Darmstadt, Germany) was added to round-bottom test tubes containing 3 ml of washed cells which had been suspended in PUM buffer (22.2 g of K2HPO4 · 3H2O, 7.26 g of KH2PO4, 1.8 g of urea, 0.2 g of MgSO4 · 7H2O, and distilled water to 1,000 ml [pH 7.1]) (39) to an OD470 of 1.0. Following a 10-min preincubation at 37°C, tubes were shaken for 30 s. The aqueous phase was carefully removed with a Pasteur pipette, and light absorbance was determined at 470 nm, using a Milton Roy 20D model spectrophotometer. The percentage of bacterial adhesion to hydrocarbon was calculated as follows: [1 − (ODF/ODI)] × 100, where ODI and ODF are the ODs of cells resuspended in PUM buffer determined at the beginning and the end of the experiment, respectively.
Complementation studies.
The esp gene was amplified with high-fidelity thermophilic DNA polymerase (Expand Long Template PCR System; Roche) from E. faecalis 14377 because the esp gene of this strain contains only one A repeat and four C repeats with primers pbacS and esp2S (Table 1). The pbacS primer includes the −35, −10, and ribosome binding sites of the constitutive promoter of the bacA gene of E. faecalis (15, 45). The chimeric pbac-esp gene was cloned into the plasmid pCU1, and the resulting plasmid, pTA2, was transformed by electroporation into E. faecalis strain 23. Stable expression of Esp was analyzed by enzyme-linked immunosorbent assay (ELISA) as described above.
In order to compare the biofilm formation capacities of the wild-type strain and the complemented strain, plasmid pCU1 was introduced into the wild-type strain to render it chloramphenicol resistant, and the assays were performed in the presence of chloramphenicol (20 μg/ml).
Statistical analysis.
A nonparametric Kendall rank correlation analysis was used to assess the association between the presence of esp and biofilm formation. For analysis of primary adherence and biofilm formation, a two-tailed Student's t test was applied.
RESULTS
Relationship between biofilm formation and presence of esp.
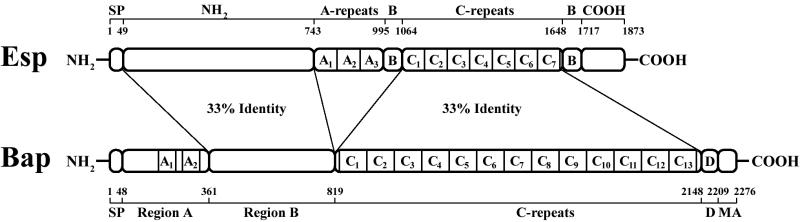
We searched eubacterial genome databases for sequence homologies to the S. aureus Bap protein using the gapped BLASTP program (1) and found that Bap and Esp share 33% sequence identity and 50% sequence similarity in an overall alignment. Individual pairwise alignments between the two proteins revealed that the N-terminal domain of Bap (region B, amino acids 361 to 819) shows 33% identity with the 694-residue N-terminal domain of Esp. Furthermore, the C repeat region of Bap, which accounts for 52% of the protein, also shows 33% identity with the C repeat region of Esp (Fig. 1).
FIG. 1.
Structural similarity between Bap and Esp. Shown is the percentage of identity between different regions of these proteins. The Esp structure is derived from strain MMH594 (43). The Bap structure is derived from the S. aureus bovine strain V329 (9). The signal peptide (SP), membrane anchor (MA), region D (D), and A and C repeats regions are shown. The N-terminal domain of Esp (residues 49 to 743) and region B of Bap (residues 361 to 819) share 33% sequence identity. The C repeat regions of Esp (residues 1064 to 1648) and Bap (residues 819 to 2148) share 33% sequence identity.
Because of this remarkable structural similarity and previous findings on the involvement of Bap in biofilm formation, we studied biofilm formation on polystyrene microtiter plates by 200 infection-derived enterococcal strains. The adherence values (OD595) obtained allowed the classification of isolates into four groups: non-biofilm forming (OD595, ≤1), weak biofilm forming (1 < OD595 ≤ 2), medium biofilm forming (2 < OD595 ≤ 3), and strong biofilm forming (OD595, >3) (Table 2). Simultaneously, the presence of the esp gene in all of the isolates was analyzed by PCR. The results indicate that biofilm formation ability is highly and significantly associated with the presence of esp (Kendall rank correlation, 0.845; P < 0.0001). The biofilm formation ability was restricted to E. faecalis isolates harboring esp, and esp was detected neither in any of the E. faecium, E. avium, or E. gallinarum isolates tested, as previously described (43), nor in any of the E. faecalis isolates unable to produce a biofilm. The majority (93.5%) of the E. faecalis esp-positive isolates were able to form biofilm, being classified as strong, medium, or weak biofilm producers (Table 2).
TABLE 2.
Distribution of enterococcal clinical isolates according to the capacity for biofilm formation on polystyrene microtiter plates and the presence or absence of esp
| esp gene status | Bacteria | No. of isolates with the following biofilm formation capacitya:
|
Total no. of isolates | |||
|---|---|---|---|---|---|---|
| − | + | ++ | +++ | |||
| Positive | E. faecalis | 6 | 26 | 18 | 43 | 93 |
| Negative | E. faecalis | 59 | 0 | 0 | 0 | 59 |
| Other enterococci | 48 | 0 | 0 | 0 | 48 | |
| Total | 113 | 26 | 18 | 43 | 200 | |
−, non-biofilm forming (OD595, ≤1); +, weak biofilm forming (1 < OD595 ≤ 2); ++, medium biofilm forming (2 < OD595 ≤ 3); +++, strong biofilm forming (OD595, > 3).
Disruption of the esp gene in E. faecalis strains with different abilities to form a biofilm.
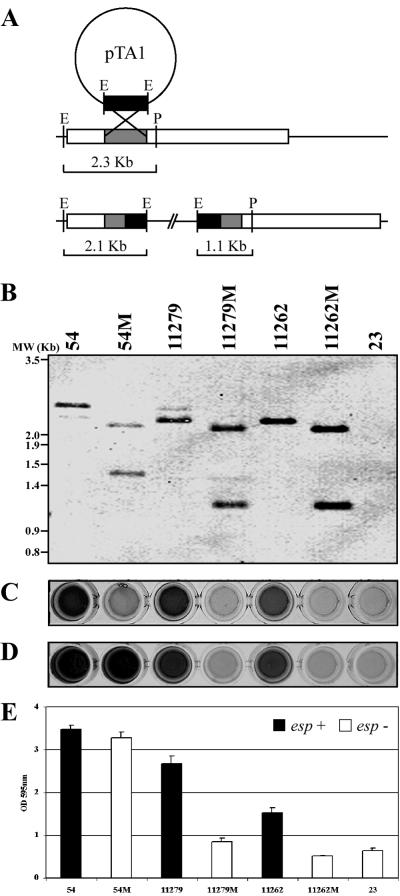
To further analyze the possible role of the Esp protein in E. faecalis biofilm formation, disruption of the esp gene was performed in strong (strain 54), medium (strain 11279), and weak (strain 11262) biofilm-forming strains. For this purpose, a 950-bp PCR fragment corresponding to an internal N-terminal region of esp was cloned into the shuttle vector pBT2, producing the pTA1 plasmid. The natural biofilm-forming strains were transformed with pTA1, and transformants were grown at a nonpermissive replicative temperature. Recombinants (Fig. 2A) were confirmed by PCR using primers esp3S and esp4S (data not shown) and Southern blotting of PstI- and EcoRI-digested chromosomal DNA (Fig. 2B).
FIG. 2.
Biofilm formation phenotypes of E. faecalis esp mutants. The relationship between the expression of the Esp protein and the ability to form a biofilm in wild-type E. faecalis strains (strains 54, 11279, and 11262), their corresponding esp mutant strains (strains 54M, 11279M, and 11262M), and a natural esp-deficient strain (strain 23) is shown. (A) Schematic representation of the predicted structure after plasmid integration into esp. (B) Southern blot analysis after digestion of chromosomal DNA with PstI and EcoRI. (C) Esp production assessed by ELISA. (D) Biofilm phenotype on wells of a polystyrene microtiter plate. (E) Quantification of biofilm formation. Error bars indicate standard deviations.
ELISA analysis of the expression of the Esp protein in parental strains and isogenic mutants showed that parental strains expressed this surface protein, whereas Esp could not be detected in any of the mutants (Fig. 2C). The fact that six Esp-positive strains were not able to produce a biofilm encouraged us to determine the presence of Esp in these strains. The results showed that five out of six of these strains did not produce ELISA-detectable Esp, suggesting that these strains either did not produce Esp under our experimental conditions or produced an Esp variant which could not be recognized by the polyclonal antiserum. Accordingly, Esp could not be detected in any of the natural esp-deficient strains (data not shown).
To establish a relationship between the expression of the Esp protein and the ability to form a biofilm, parental and mutant strains were also tested for their ability to form a 24-h biofilm on polystyrene microtiter plates. Surprisingly, disruption of the esp gene in two strong biofilm-forming strains (isolates 54 and 1) did not lead to a significant decrease in this ability. In contrast, the esp mutant strains of the medium (11279M) or weak (11262M) biofilm producers hardly adhered to the surface, being unable to form a biofilm and exhibiting behavior indistinguishable from that of the natural esp-deficient strains (Fig. 2D and E).
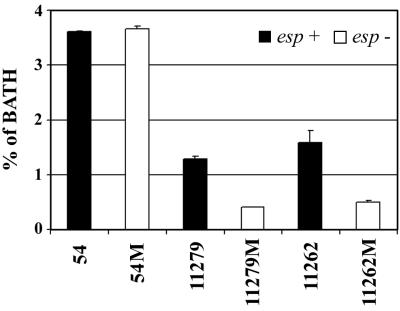
Cell surface hydrophobicities of parental and mutant strains.
Figure 3 illustrates the hydrophobic nature of the cell surfaces of parental strains and isogenic esp mutants. Parental esp-positive strains always showed similar (strain 54) or higher affinity for n-hexadecane compared with mutant strains. Remarkably, a similar decrease in hydrophobicity was observed when the hydrophobicity of a bap-positive S. aureus strain (V329) was compared with that of its bap-negative mutant (M556) (data not shown).
FIG. 3.
Differences in cell surface hydrophobicities of wild-type E. faecalis strains in relation to their corresponding esp mutants as measured by affinity to n-hexadecane. Assays were performed in triplicate. Mean values and standard deviations are shown. BATH, bacterial adhesion to hydrocarbon.
Variation in number of A and C repeats of esp in relation to biofilm formation capacity.
In order to determine whether the number of A or C repeats present in Esp was related to the intensity of the biofilm formation ability, we calculated the number of repeats in each E. faecalis strain. In agreement with previously reported results (43), the number of A repeat units ranged from 1 to 3 whereas the number of C repeat units ranged from 4 to 10, with a majority of the isolates displaying 7 C repeat units. No association was found between the number of A or C repeat units within the esp gene and the biofilm formation capacity in the E. faecalis esp-positive isolates (data not shown).
Involvement of Esp in primary attachment and biofilm formation on PVC plastic.
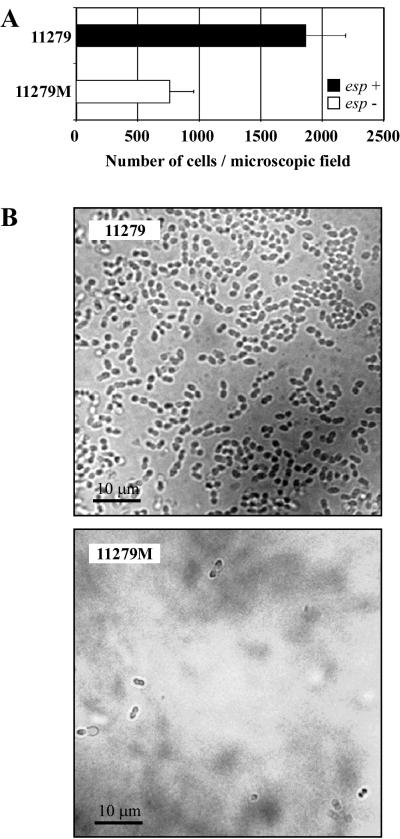
Early adherence of the wild-type E. faecalis 11279 and its isogenic esp mutant 11279M is illustrated in Fig. 4A. Strain 11279 adhered to polystyrene much more efficiently than mutant 11279M. In addition, direct observation of PVC plastic tabs from a urine collection bag using phase-contrast microscopy showed that the parental strain produced a layer of cells covering the PVC surface (Fig. 4B). In contrast, very few cells of the esp mutant were attached to PVC plastic. These results on early adherence are consistent with those described above on biofilm formation in polystyrene microtiter plates.
FIG. 4.
Comparisons at different steps of the biofilm formation process of a wild-type strain (11279) and its esp-deficient mutant (11279M). (A) Primary attachment assay. Significant differences (P < 0.0001) between 11279 and 11279M were detected. Adherent bacterial cell mean values and standard deviations are shown. (B) Attachment to PVC plastic. The capacity of strains 11279 and 11279M to form a 24-h biofilm on PVC plastic was observed by phase-contrast microscopy (magnification, ×1,000).
Esp allows primary attachment to an abiotic surface.
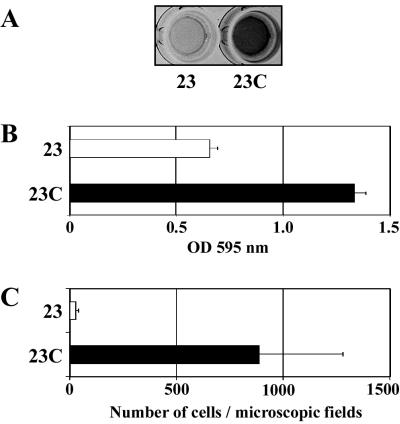
Complementation tests were performed to determine whether overexpression of Esp was able to confer biofilm formation ability to an E. faecalis esp-negative strain. Since the nucleotide sequence upstream of the ATG codon of the esp gene (accession number AF034779) is very short, we were uncertain whether the promoter of esp was included in this sequence. To solve this problem, esp was overexpressed under the control of a constitutive promoter in E. faecalis, as described in Materials and Methods. The expression of the Esp protein in the complemented strain 23C measured by ELISA is shown in Fig. 5A. The complemented strain showed an enhanced capacity to form a biofilm on a polystyrene surface (Fig. 5B). However, the complemented strain was always classified into the group of weak biofilm-forming strains (1 < OD595 ≤ 2). The number of cells bound to polystyrene was quantified in order to determine whether the enhanced capacity for biofilm formation in the complemented strain was due to a modification of primary adherence to polystyrene. It was observed that the number of attached cells in the complemented strain was at least 20-fold higher than that in the wild-type strain (P < 0.0001), demonstrating that Esp highly enhances primary binding to polystyrene (Fig. 5C). In addition, we assessed the ability of 23C to attach to PVC plastic from urinary bags using phase-contrast microscopy. The complemented strain 23C showed multiple groups of cells adhered to the PVC plastic. In contrast, very few individual cells were attached to the plastic in the case of the wild-type strain (data not shown).
FIG. 5.
Complementation studies. (A) Esp production assessed by ELISA in E. faecalis 23, a wild-type esp-deficient strain, and in this strain complemented with pTA2 (23C), (B) Biofilm formation capacities of 23 and 23C on polystyrene plates. Assays were performed in triplicate; mean values and standard deviations are shown. (C) Capacity of these strains for primary attachment in polystyrene plates. The number of attached cells of the complemented strain 23C was significantly higher (P < 0.0001) than that of the wild-type strain. Assays were performed in triplicate; adherent bacterial cell mean values and standard deviations are shown.
DISCUSSION
Like other gram-positive microorganisms, enterococci are able to produce biofilms on abiotic surfaces (27), increasing their high innate resistance to antibiotics (13), yet the factors controlling enterococcal biofilm formation and maintenance remain unknown (36). The initial step in the colonization of catheters and biofilm formation is bacterial adherence to the biomaterial (20). To date, mutagenesis studies on the initial attachment of gram-positive bacteria to abiotic surfaces have been performed mainly with Staphylococcus epidermidis (19, 42, 46) and have shown that a variety of bacterial surface proteins are involved in the process. Among these, the best characterized protein is AtlE, a cell surface-localized autolysin which has also vitronectin-binding activity and therefore is involved in binding to both biotic and abiotic surfaces (19). Since not all S. epidermidis strains harboring the atlE gene produce a biofilm in vitro, the presence of atlE appears to be necessary but insufficient to induce primary attachment to abiotic surfaces.
The esp gene has been identified in a random sequencing project on the genome of a clinical strain of E. faecalis (43). It has been preferentially detected in infection-derived E. faecalis strains but not in other less pathogenic enterococcal species, strongly suggesting a role of Esp in the pathogenesis of E. faecalis (43). Further supporting this hypothesis are the results obtained in this study on the highly significant association (P < 0.0001) between the presence of the esp gene and the ability to produce a biofilm in vitro. Eighty-seven out of 93 E. faecalis strains harboring esp were able to produce a biofilm in vitro, whereas none of the 59 esp-deficient strains tested were able to produce a biofilm. Furthermore, Esp expression determined by ELISA revealed that five out of the six esp-positive, biofilm-negative E. faecalis isolates analyzed did not produce Esp, strongly suggesting that the absence of biofilm formation in these strains is associated with the lack of Esp expression. These results strongly suggest that esp is involved in the biofilm formation process of E. faecalis.
Insertional mutagenesis of esp has shown no significant effect on biofilm formation in the strongest biofilm-producing E. faecalis strain analyzed (strain 54). This could be attributed to additional surface adhesins that might mediate the initial attachment to the abiotic surface in the absence of esp. On the other hand, the finding that none of the esp-defective E. faecalis strains was able to produce a biofilm suggests the existence of a genetic association between the presence of esp and the presence of these adhesins. A striking common feature between esp and bap genes, both of which are associated with abiotic surface attachment, is that they are flanked by a sequence similar to that of the transposase IS905 in the case of esp (24) and IS431 in the case of bap (our unpublished results). Based on this observation and in agreement with other authors (10, 44) it is tempting to speculate that esp could be part of a pathogenicity island (PAI). It is known that adhesins, which mediate the capacity of bacteria to attach to specific eukaryotic receptor molecules, are major virulence factors encoded by PAIs (18). The possibility that additional adhesins might flank esp and bap and constitute a PAI is under study.
The high degree of conservation of the nucleotide sequences of A and C repeats present in both esp (43) and bap (9) strongly suggests that these repeats could have an important role in the function of this protein. Although our data reveal that the number of repeats is not related to the amount of biofilm produced, we found, in agreement with a previous report (43), that none of the esp-positive strains exhibited a complete loss of either A or C repeats, strongly suggesting that both regions are important for the functionality of the protein. These A and C repeats are not the only structural feature shared by Esp and Bap proteins. Analysis of their amino acid sequence reveals the presence of dimerization domains and calcium binding motifs in both of them. We are presently evaluating the contribution of these domains to the function of Esp and Bap.
Esp exhibits characteristics of surface protein receptors designated microbial surface components recognizing adhesive matrix molecules (38) that adhere to components of the host to initiate colonization. Many of these proteins have a modular design and contain a number of tandem repeat domains that probably arise from a series of recombination and/or duplication events. Although we have no evidence for the presence of domains for binding to host factors in Esp, we cannot exclude for Esp under in vivo conditions a direct ligand-binding activity to the extracellular matrix or an indirect role modulating ligand-binding activity of other molecules. If this is the case, Esp-mediated adherence of E. faecalis to plastic biomaterials would not be the primary role of this protein in the host-bacterium interaction. It has been proposed that the presence of Esp could increase cell surface hydrophobicity and facilitate hydrophobic interactions (43). In contrast with previous results (27) where it was observed that adherence of E. faecalis to urinary catheters was not related to bacterial hydrophobicity, our results demonstrate that the presence of Esp in the cell surface increases hydrophobicity, adherence to abiotic surfaces, and biofilm formation. This apparent discrepancy could be at least partially explained if the E. faecalis clinical strains analyzed were esp deficient, which would likely result in a low hydrophobicity.
Routinely, antibiotic susceptibility is determined in clinical laboratories using the broth microdilution susceptibility test. However, it is well established that antimicrobials directed to planktonic cells may not be efficient against biofilm cells (8, 29). In E. faecalis, improvement of the selection of effective antimicrobial agents against recalcitrant infections is urgently needed. Taking into account the strong correlation between the presence of esp and the ability to produce a biofilm, it may be possible to screen for putative biofilm-forming E. faecalis strains by testing for the presence of the esp gene by PCR or for its product by ELISA. The genetic (presence of esp) rather than phenotypic (adherence or biofilm formation) nature of this screening is advantageous in that it allows preliminary identification of strains which are highly adherent and are thus good candidates for antibiotic susceptibility testing in biofilms (2).
It has been estimated that over 65% of nosocomial infections in the developed world are derived from biofilm-related infections. This represents a warning signal, since in the near future the use of medical implants is likely to increase. In this context, bacterial molecules involved in attachment mediated by host proteins and others, such as Esp, involved in adherence to abiotic surfaces and biofilm formation could become promising therapeutic targets in control programs for eradicating persistent enterococcal infections associated with the presence of biofilms.
ACKNOWLEDGMENTS
Alejandro Toledo-Arana and Jaione Valle contributed equally to this work.
We express our gratitude to V. Shankar for providing the antibodies against Esp, to F. Götz for plasmid pCU1, and to R. Brückner for plasmid pBT2.
This work was supported by grant BIO99-0285 from the Comisión Interministerial de Ciencia y Tecnología and grants from the Cardenal Herrera-CEU University and from the Departamento de Educación y Cultura del Gobierno de Navarra. Alejandro Toledo-Arana is a predoctoral Mutis program fellow from Agencia Española de Cooperación Internacional (AECI), Spain. Jaione Valle is a predoctoral fellow from the Ministerio de Ciencia y Tecnologia (FPI), Spain. C. Cucarella is a predoctoral fellow from the Cardenal Herrera-CEU University.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amorena B, Gracia E, Monzón M, Leiva J, Oteiza C, Pérez M, Alabart J, Hernandez-Yago J. Antibiotic susceptibility of Staphylococcus aureus in biofilms developed in vitro. J Antimicrob Chemother. 1999;44:43–55. doi: 10.1093/jac/44.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Augustin J, Rosenstein R, Wieland B, Schneider U, Schnell N, Engelke G, Entian K D, Gotz F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992;204:1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 5.Bruckner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 6.Cetinkaya Y, Falk P, Mayhall C G. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow J W, Thal L A, Perri M B, Vazquez J A, Donabedian S M, Clewell D B, Zervos M J. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37:2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 9.Cucarella C, Solano C, Valle J, Amorena B, Lasa I I, Penades J R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton T J, Gasson M J. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edmond M B, Ober J F, Dawson J D, Weinbaum D L, Wenzel R P. Vancomycin-resistant enterococcal bacteremia: natural history and attributable mortality. Clin Infect Dis. 1996;23:1234–1239. doi: 10.1093/clinids/23.6.1234. [DOI] [PubMed] [Google Scholar]
- 12.Espinosa-Urgel M, Salido A, Ramos J L. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol. 2000;182:2363–2369. doi: 10.1128/jb.182.9.2363-2369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foley I, Gilbert P. In-vitro studies of the activity of glycopeptide combinations against Enterococcus faecalis biofilms. J Antimicrob Chemother. 1997;40:667–672. doi: 10.1093/jac/40.5.667. [DOI] [PubMed] [Google Scholar]
- 14.Fournier B, Hooper D C. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J Bacteriol. 2000;182:3955–3964. doi: 10.1128/jb.182.14.3955-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto S, Ike Y. pAM401-based shuttle vectors that enable overexpression of promoterless genes and one-step purification of tag fusion proteins directly from Enterococcus faecalis. Appl Environ Microbiol. 2001;67:1262–1267. doi: 10.1128/AEM.67.3.1262-1267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore M S, Segarra R A, Booth M C, Bogie C P, Hall L R, Clewell D B. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol. 1994;176:7335–7344. doi: 10.1128/jb.176.23.7335-7344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzman C A, Pruzzo C, LiPira G, Calegari L. Role of adherence in pathogenesis of Enterococcus faecalis urinary tract infection and endocarditis. Infect Immun. 1989;57:1834–1838. doi: 10.1128/iai.57.6.1834-1838.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacker J, Kaper J B. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 19.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 20.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 21.Hunt C P. The emergence of enterococci as a cause of nosocomial infection. Br J Biomed Sci. 1998;55:149–156. [PubMed] [Google Scholar]
- 22.Huycke M M, Gilmore M S. In vivo survival of Enterococcus faecalis is enhanced by extracellular superoxide production. Adv Exp Med Biol. 1997;418:781–784. doi: 10.1007/978-1-4899-1825-3_184. [DOI] [PubMed] [Google Scholar]
- 23.Huycke M M, Joyce W A, Wack M F. Augmented production of extracellular superoxide production by blood isolates of Enterococcus faecalis J. Infect Dis. 1996;173:743–746. doi: 10.1093/infdis/173.3.743. [DOI] [PubMed] [Google Scholar]
- 24.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ike Y, Clewell D B, Segarra R A, Gilmore M S. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J Bacteriol. 1990;172:155–163. doi: 10.1128/jb.172.1.155-163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ike Y, Hashimoto H, Clewell D B. High incidence of hemolysin production by Enterococcus (Streptococcus) faecalis strains associated with human parenteral infections. J Clin Microbiol. 1987;25:1524–1528. doi: 10.1128/jcm.25.8.1524-1528.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joyanes P, Pascual A, Martinez-Martinez L, Hevia A, Perea E J. In vitro adherence of Enterococcus faecalis and Enterococcus faecium to urinary catheters. Eur J Clin Microbiol Infect Dis. 2000;19:124–127. doi: 10.1007/s100960050443. [DOI] [PubMed] [Google Scholar]
- 28.Kreft B, Marre R, Schramm U, Wirth R. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells Infect. Immun. 1992;60:25–30. doi: 10.1128/iai.60.1.25-30.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis K. Riddle of biofilm resistance. Antimicrob Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe A M, Lambert P A, Smith A W. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect Immun. 1995;63:703–706. doi: 10.1128/iai.63.2.703-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mengaud J, Dramsi S, Gouin E, Vazquez-Boland J A, Milon G, Cossart P. Pleiotropic control of Listeria monocytogenes virulence factors by a gene which is autoregulated. Mol Microbiol. 1991;5:2273–2283. doi: 10.1111/j.1365-2958.1991.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 32.Moellering R C., Jr Emergence of Enterococcus as a significant pathogen. Clin Infect Dis. 1992;14:1173–1176. doi: 10.1093/clinids/14.6.1173. [DOI] [PubMed] [Google Scholar]
- 33.Mundy L M, Sahm D F, Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev. 2000;13:513–522. doi: 10.1128/cmr.13.4.513-522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nallapareddy S R, Qin X, Weinstock G M, Hook M, Murray B E. Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun. 2000;68:5218–5224. doi: 10.1128/iai.68.9.5218-5224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olmsted S B, Dunny G M, Erlandsen S L, Wells C L. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J Infect Dis. 1994;170:1549–1556. doi: 10.1093/infdis/170.6.1549. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole G, Kaplan H B, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 37.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 38.Patti J M, Hook M. Microbial adhesins recognizing extracellular matrix macromolecules. Curr Opin Cell Biol. 1994;6:752–758. doi: 10.1016/0955-0674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 39.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg M, Gutnick D, Rosenberg E. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett. 1980;9:29–33. [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Schumacher-Perdreau F, Heilmann C, Peters G, Gotz F, Pulverer G. Comparative analysis of a biofilm-forming Staphylococcus epidermidis strain and its adhesion-positive, accumulation-negative mutant M7. FEMS Microbiol Lett. 1994;117:71–78. doi: 10.1111/j.1574-6968.1994.tb06744.x. [DOI] [PubMed] [Google Scholar]
- 43.Shankar V, Baghdayan A S, Huycke M M, Lindahl G, Gilmore M S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shankar V, Gilmore M S. Characterization of the Enterococcus faecalis alpha C protein homolog. Evidence for the expression of alternate forms in commensal and infection derived isolates. Adv Exp Med Biol. 1997;418:1045–1048. [PubMed] [Google Scholar]
- 45.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J Bacteriol. 1997;179:7843–7855. doi: 10.1128/jb.179.24.7843-7855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veenstra G J, Cremers F F, van Dijk H, Fleer A. Ultrastructural organization and regulation of a biomaterial adhesin of Staphylococcus epidermidis. J Bacteriol. 1996;178:537–541. doi: 10.1128/jb.178.2.537-541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodford N. Glycopeptide-resistant enterococci: a decade of experience. J Med Microbiol. 1998;47:849–862. doi: 10.1099/00222615-47-10-849. [DOI] [PubMed] [Google Scholar]
- 48.Xu Y, Singh K V, Qin X, Murray B E, Weinstock G M. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect Immun. 2000;68:815–823. doi: 10.1128/iai.68.2.815-823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]