Abstract
Obesity, type 2 diabetes, and non-alcoholic fatty liver disease (NAFLD) are characterized by the concepts of lipo- and glucotoxicity. NAFLD is characterized by the accumulation of different lipidic species within the hepatocytes. Bile acids (BA), derived from cholesterol, and conjugated and stored in the gallbladder, help the absorption/processing of lipids, and modulate host inflammatory responses and gut microbiota (GM) composition. The latter is the new “actor” that links the GI tract and liver in NAFLD pathogenesis. In fact, the discovery and mechanistic characterization of hepatic and intestinal farnesoid X receptor (FXR) shed new light on the gut–liver axis. We conducted a search on the main medical databases for original articles, reviews, meta-analyses of randomized clinical trials, and case series using the following keywords, their acronyms, and their associations: farnesoid X receptor, bile acids metabolism, gut microbiota, dysbiosis, and liver steatosis. Findings on the synthesis, metabolism, and conjugation processes of BAs, and their action on FXR, change the understanding of NAFLD physiopathology. In detail, BAs act as ligands to several FXRs with GM modulation. On the other hand, the BAs pool is modulated by GM, thus, regulating FXRs functioning in the frame of liver fat deposition and fibrosis development. In conclusion, BAs passed from their role of simple lipid absorption and metabolism agents to messengers between the gut and liver, modulated by GM.
Keywords: farnesoid X receptor, bile acids metabolism, gut microbiota, dysbiosis, liver steatosis
1. Introduction
Obesity, type 2 diabetes, and dyslipidemia are features of our Westernized lifestyle. Non-alcoholic fatty liver disease (NAFLD) follows the same epidemiological trends as these disorders, and has a global prevalence of 25% among the general population [1]. NAFLD is not a status, but rather a stage of conditions including liver steatosis and non-alcoholic steatohepatitis (NASH), all the way up to cirrhosis and hepatocellular carcinoma [1]. Liver fibrosis is a key factor for liver-related, and all-cause, mortality [2].
NAFLD is characterized by the accumulation of different lipidic species within the hepatocytes—a process regarded as “lipotoxicity” [3,4]. In addition, carbohydrates boost several lipogenic pathways (e.g., acetyl-CoA carboxylase, SCD-1, and fatty acid synthase) and contribute to liver steatosis [5]. Fructose is the most effective example of this detrimental interaction with lipotoxicity, and outlines the “glucotoxicity” concept. In fact, these changes are associated with reduced insulin sensitivity, strictly linked to NASH origin [6,7].
The gut is linked to the liver through the gut–liver axis [4], and gut microbiota is the link connecting these two organs. Indeed, fat accumulation in hepatocytes is accompanied by changes in the population of gut microbiota that modulate the pool of bile acids (BA) secreted by the liver and transformed/metabolized within the gut [8,9].
This review of the scientific literature aims to describe the metabolism of bile acids and their “vital” cycle, and their role in lipogenesis and lipotoxicity in the liver and NAFLD, with a special focus on the role of the farnesoid X receptor in the pathogenesis and potential treatment of liver steatosis.
2. Results
2.1. Bile Acids and Their Metabolism: The “Entero-Hepatic” Cycle
BAs are derived from cholesterol to form primary BAs (cholic acid, or CA) and chenodeoxycholic acid, or CDCA [10]. Subsequently, primary BAs are conjugated with glycine or taurine, with improved solubility, and stored in the gallbladder, ready for excretion in the gut [7,11].
The conjugation process has several functions: it minimizes the passive absorption of BAs, modulates host inflammatory responses and gut microbiota composition, and maintains the gut “eubiosis”, limiting small intestinal bacterial overgrowth (SIBO) incidence within our small bowel as a feature of “dysbiosis” [7,12].
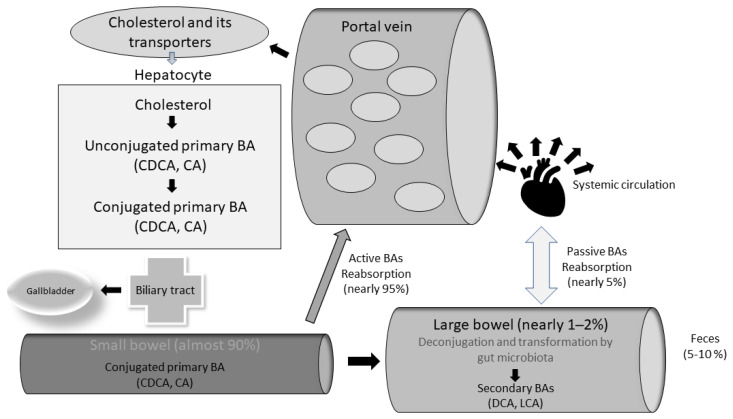
Postprandially, BAs are released from the gallbladder into the small bowel, following the secretion of GI cholecystokinin (CCK). BAs are crucial for the emulsification of dietary fat, and enhance the absorption of lipids, sterols, and vitamins [9,13]. Approximately 95% of BAs are actively reabsorbed in the terminal ileum, via the apical sodium-dependent bile acid transporter (ASBT). In the cytoplasm of the enterocyte, BAs bind to the ileal bile acid-binding protein (IBABP). Then, they are excreted into the portal circulation through the organic anion transporter polypeptide (OATPA/B), localized in the basolateral membrane of enterocytes. Further, BAs travel back into the liver through the sodium taurocholate cotransporting polypeptide (NTCP) transporter [7,8,10]. Once in the liver, free BAs are re-conjugated with taurine or glycine, before secretion into the biliary tract and intestinal lumen [10]. This enterohepatic cycle of BAs occurs almost 10 times per day, and is necessary as hepatocytes have limited capability to produce BAs [10] (Figure 1).
Figure 1.
The enterohepatic bile acids cycle.
Ileal BA transport is highly efficient, but a small proportion (1–2%) of BA escapes the enterohepatic circulation, and enters the large intestine [10]. As they transit through the colon, the microbiota perform several enzymatic reactions (e.g., deconjugation, dihydroxylation, and epimerization), and form secondary BAs (namely, deoxycholic acid (DCA) from CA, and ursodeoxycholic acid (UDCA) and lithocholic acid (LCA) from CDCA) [7,14].
In detail, BAs deconjugation by gut microbiota is crucial for bile bio-transformation [15]: this small portion of BAs being microbially metabolized makes them more lipophilic, allowing the secondary BAs to be reabsorbed in the large bowel and returned to the liver, via the systemic circulation [7,11] (Figure 1).
2.2. Bacterial Bile Salt Hydrolase (BSH) Enzymes as a Sign of Bacterial Modulation of BAs Pool
Bile salt hydrolases belong to the Ntn-hydrolase superfamily of proteins [16]. In particular, microbial BSH cleaves the amide bond between the glycine and taurine portions conjugated to the steroid nucleus of bile salts [3,12]. BSH enzymes are represented across most bacterial phyla. Regarding the commensal gut microbiota, BSH activity is found in Gram-positive bacteria: Lactobacillus, Bifidobacterium [3,17,18], Clostridium spp., and Enterococcus [19,20]. However, they are also detected in some commensal Gram-negative strains, such as the Bacteroides spp [3,12,21,22].
BSH presence is also detected in pathogens. For example, the gastrointestinal Listeria monocytogenes has BSH activity, an adaptive quality guaranteeing its gut persistence [12,23,24]. Furthermore, BSH activity is also found in the environment: Xanthomonas maltophilia is present in soil [25,26], and thermophilic Brevibacillus sp in hot springs [27,28].
This adaptive quality has a horizontal transmission amongst gut bacteria, suggestive of a strong evolutionary selection for this activity [12,29].
Description of functioning, distribution, and abundance of BSH within the human gut microbiome is possible through a metagenomics approach, enabling researchers to reconstruct entire genomic libraries starting from small genomic sequences [30]. More interestingly, BSH coding sequences are found among two domains of life within the gut: bacteria and archaea. Thus, this wide distribution is suggestive of host-driven selection of BSH activity. In addition, the host seems to affect BSH activity in gut bacteria through a “host species-specific selection” of microbial BSH activities. This selective mechanism can be started by species-specific differences in host bile acid pools [31].
From a functional point of view, we can briefly describe the BSH activity as a protective shield for some bacterial species (specifically, through bile acid deconjugation) to colonize the human gut [12,32,33]. The BSH activity products (namely, glycine or taurine) can be used as a source of energy for some bacterial species [12,20,21,22,23,24,25,26,27]. For example, glycine can be metabolized to ammonia and carbon dioxide, and taurine can be metabolized to produce ammonia, carbon dioxide, and sulfate. All of them are carbon and nitrogen sources [12,19,34,35]. Furthermore, BSH regulates bacterial intracellular pH, perpetuating resistance to bile acids in low pH environments (e.g., the stomach) [12]. Finally, BSH is included as a hallmark of probiotic activity, as it allows the strain to survive gut transit [12,36,37].
2.3. Bile Acids and Their Receptors: The Emerging “FXR Case”
The concept of the gut–liver axis helps explain the complex interplay between systemic metabolism, gut hormone release, and the immune response [3]. Over the main actors in this system, the arrows making this axis efficient are operated by BAs. Indeed, the latter are also ligands for receptors that include the nuclear receptor farnesoid X receptor (FXR) and G-protein-coupled bile acid receptor 1 (or TGR5). These receptors regulate host basal metabolism and enterohepatic circulation [38].
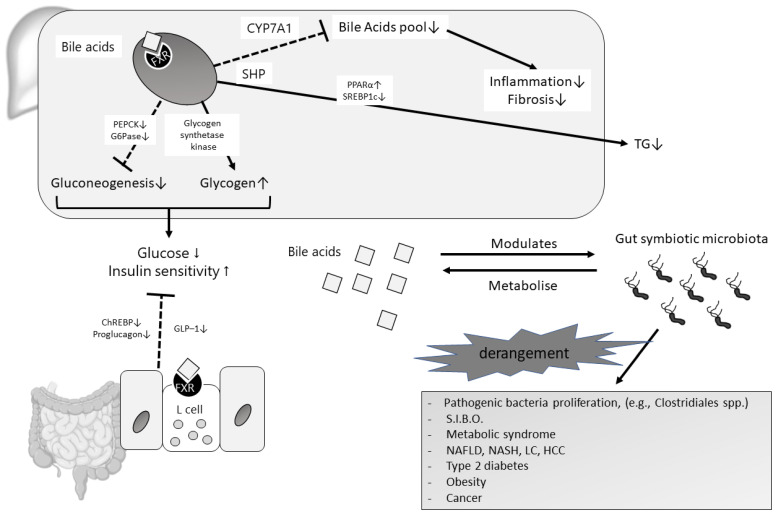
For example, the strong antimicrobial properties of BAs, and the maintenance of the eubiosis of gut microbiota, are mediated by a multifaceted mechanism of activation of FXR [3,39]. NAFLD offers a mechanistic example of dysregulation of BAs-FXR-mediated lipid and glucose metabolism [3,13]. Indeed, hepatic FXR activation mediated by BAs can induce the expression of atypical nuclear receptors small heterodimer partner (SHP), which promotes the inhibition of the sterol-regulatory element-binding protein-1c (SREBP-1c) (Figure 2), and ultimately leads to the reduced hepatic synthesis of triglycerides. Moreover, FXR can limit fat accumulation in the liver by promoting fatty acid oxidation after the activation of the hepatic NR peroxisome proliferator-activated receptor alpha (PPAR-α), and through plasma VLDL triglyceride clearance (Figure 2) [3,40,41]. FXR activation in the liver shows unidirectional effects on glucose homeostasis. It leads to the inhibition of gluconeogenesis and glycolysis through reduced expression of PEPCK phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6 phosphatase (G6Pase), with a potential protective role in insulin resistance and type II diabetes (Figure 2). However, other studies fail to confirm these actions [42,43].
Figure 2.
The gut–liver axis representation with special focus on BAs and gut microbiota relationship in healthy and pathologic conditions.
More interestingly, the activation of FXR localized in the intestinal cells can start a crucial endocrine feedback mechanism [3,44]. In fact, the human fibroblast growth factor 19 (FGF19) is secreted into the bloodstream after ileal FXR activation, and suppresses the synthesis of BAs through the activation of the FGF receptor 4 (FGFR4) at the surface of hepatocytes [18]. Its mouse ortholog is FGF15 [3,18]. Practically, FGF 19 signaling cascade reduces liver steatosis and insulin resistance [45].
Consequently, evidence of a FXR-mediated decrease in hepatic lipid accumulation and improved insulin sensitivity is the basis for the use of its agonists in NAFLD treatment. For example, a dose of 25 mg/day of the semisynthetic BA obeticholic acid (OA) in patients with non-cirrhotic, non-alcoholic steatohepatitis was compared to placebo for 72 weeks of treatment [46]. As a primary outcome, OA improves liver histology in 46% of patients vs. 21% of patients in the placebo group; no worsening of fibrosis is observed. However, OA is observed to increase plasmatic lipids concentration, and cause pruritus. FXR agonists already show efficacy against primary cholestatic cholangitis, due to the FXR-stimulated reduction in the bile acid pool in the liver, which, in turn, reduces liver inflammation and fibrosis (Figure 2) [40]. Part of the effect of FXR agonists on NASH may be due to this physiological effect. Finally, other FXR agonists (e.g., GS9674 and LJN452) are under investigation [3,47,48] (Table 1).
Table 1.
Main FXR agonist available and under investigation.
| FXR Agonist Product Name |
Current Trials Status | Notes on Mechanism of Action | Reference/Protocol Registration |
|---|---|---|---|
| Obeticholic acid (Ocaliva®) | FDA/EMA approval for PBC in 2016 | 100 times higher affinity than CDCA | Kowdley KV et al. Hepatology. 2018 [40] |
| Efficacy to NASH (phase3) | Neuschwander-Tetri BA et al. Lancet. 2015 [39] | ||
| Bile Acid Malabsorption (phase2) | Walters JR. Aliment Pharmacol Ther. 2015 [48] | ||
| LMB763 | NAFLD (phase2) | NCT02913105 | |
| LJN452 | NAFLD (phase2) | NCT02855164 | |
| GS9674 | NAFLD (phase2) | NCT02854605 | |
| PSC (phase2) | NCT02943460 | ||
| WAY-362450 | phase1 | NCT00499629. 2007 | |
| PX20606 | phase1 | NCT01998659, NCT01998672 | |
| GW4064 | in mice | ||
| INT-767 | in mice | dual FXR and TGR5 agonist |
Table legend: PBC: primitive biliary cholangiopathies; NASH: non alcoholic steato-hepatitis; NAFLD: non alcoholic fatty liver disease; PSC: primary sclerosing cholangitis; CDCA: chenodeoxycholic acid; TGR5: G protein-coupled bile acid receptor.
The use of FGF-19 and -15 agonists is discouraged because of their correlation with hepatocyte proliferation, potentially leading to hepatocellular carcinoma (HCC) development in mice. Accordingly, human HCC cells and cirrhotic livers show increased expression of FGF19 [3,49].
These data remark the need for a better understanding of the pathways induced by FXR activation, before their clinical use.
BAs can also “ sense “ the G-protein-coupled membrane receptor, TGR5, expressed in the gallbladder, the liver, specifically Kupffer cells and endothelial cells, not in hepatocytes, the intestine, adipose tissue, spleen, and the kidneys. TGR5 regulates the composition of BAs acid pool, and modulates the immune system and metabolism [3,50,51]. Interestingly, administration of cholic acid to high-fat diet (HFD)-fed mice results in body weight reduction and enhanced energy expenditure through a TGR5–cAMP pathway characterized in brown adipose tissue and skeletal muscle [52]. BAs also activate TGR5 in enterocytes with the release of GLP-1 and peptide tyrosine PYY, with reduced food intake and normalized glucose metabolism [53].
In addition, INT-767, an FXR/TGR5 dual agonist, shows lowered lipid accumulation, inflammation, and fibrosis in various in vitro models [54,55,56], while studies on individuals with NASH are still ongoing [55] (Table 1).
2.4. Bile Acids, FXR, and Gut Microbiota: The Completion of the Gut-Liver Axis
The intestinal microbiota represents one of the most complex ecosystems present in nature. It accounts for over 100 trillion microorganisms [56]. Firmicutes and Bacteroidetes represent the most abundant phyla of the microbiota. Their ratio is a fundamental factor in the host’s health [57]. Among the other phyla present, we find Fusobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia [4]. Gut microbiota also encompasses viruses, protozoa, archaea, and fungi [58].
Eubiosis defines the balanced qualitative and quantitative condition of the intestinal microflora, and is essential to preserving the host’s health. On the contrary, its qualitative/quantitative perturbation, namely, dysbiosis, is associated with the development of various diseases such as NAFLD, NASH until HCC [59,60], diabetes type 2, cardiovascular disorders, and, undoubtedly, obesity [61,62] (Figure 2).
As BAs are metabolized by gut microbiota, changes in their composition influence the pathways mediated by BAs, such as FXR signaling [63]. Thus, we can imagine how BAs pool modifications induced by gut microbiota strongly influence host metabolism. In fact, the accumulation of the murine taurine-conjugated primary bile acids tauro-alfa and beta-muricholic acid antagonizes FXR signaling in the ileum in germ-free mice. On the other hand, the physiologic intestinal microbiota presence counteracts these actions, and re-establishes FXR activation with BAs synthesis [3,64] (Figure 2).
One cutting edge research, unraveling the circular interaction of gut microbiota and BAs via FXR, was led by Friedman et al. [65], through a phase 1 human study involving 17 healthy volunteers administered with OA (a BA analog and FXR agonist) at 5 mg, 10 mg, or 25 mg per day. Fecal and plasma specimens were collected at baseline (day 0) and on days 17 (end of dosing) and 37 (end of study), then analyzed by metagenomic sequencing. Thereafter, a Uniref90 high stringency genomic analysis was performed, in order to match specific genes to specific bacterial species abundant because of OA. At the same time, male C57BL/6 mice were gavaged daily with water, vehicle, or OA (10 mg/kg) for 2 weeks, and small bowel luminal contents were collected at baseline and on day 14. Mouse fecal pellets were analyzed by 16S tagged sequencing. Culture experiments allowed the researchers to determine the species-specific effects of BAs and OA on bacteria.
Interestingly, OA suppresses endogenous BA synthesis with a reversible induction of Gram-positive bacteria in the small intestine, mainly derived from diet and the oral microbiota. These strains of bacteria are normally down-regulated by the BAs pool. At a molecular level, OA treatment determines an increased activity of microbial genomic pathways involved in DNA synthesis and amino acid metabolism. Accordingly, mice treated with OA show reduced endogenous BA levels, and an increased proportion of Firmicutes in the small bowel, as compared to control mice [66]. Oppositely, BAs reduce the proliferation of these bacteria in minimum inhibitory concentration assays. In Firmicutes, there is an increase in the representation of microbial genomic pathways involved in DNA synthesis and amino acid metabolism, such as after OA treatment of healthy subjects. There are currently no data showing how BA composition and concentration affect the gut microbiota in the pathogenesis of various diseases, but it is noteworthy that the manipulation of the gut microbiota from BAs may be considered in the future to develop clinical approaches aimed at disease prevention and treatment.
Furthermore, the modulation of intestinal FXR by BAs unravels some interesting consequences for the development of obesity and liver steatosis. Indeed, experimental models of NAFLD show microbiota from obese subjects lowering the levels of BAs with higher activity of FXR. This results in the synthesis of ceramides that alter lipid metabolism at the liver level, leading to the accumulation of fatty acids [3,67]. Accordingly, FXR activation in L cells decreases proglucagon expression interfering with the glucose-responsive factor carbohydrate-responsive element-binding protein (ChREBP), and GLP-1 secretion through glycolysis inhibition [67]. Conversely, agents able to affect gut microbiota and BA composition (e.g., antibiotics, pre-, probiotics) can increase the amount of the intestinal FXR antagonist tauro-alpha-muricholic acid. Finally, the reduction in intestinal FXR activation results in the improvement of both general metabolic and specific hepatic profiles [3,30,31].
In addition, gut microbiota from obese mice has a promotional activity in favor of diet-induced obesity through FXR. Subsequently, FXR may contribute to increased adiposity through action on microbiota composition [68].
Summarizing all these pieces of evidence, we hypothesize that inhibition of FXR within the gut might protect against the development of a fatty liver. However, this hypothesis is in contrast with the assumption of FXR activation being beneficial during NAFLD [69]. Thus, intestinal FXR antagonists are considered for the treatment of metabolic diseases: the glycine-beta-muricholic acid (namely, a tauro-beta-muricholic acid derivative) is used in experimental models of NAFLD, producing a reduction in obesity, insulin resistance, and liver fibrosis/inflammation grading [70,71,72].
In conclusion, we assume that FXR regulation by BAs and gut microbiota is very complex, as is strongly influenced by the organ in which such receptors are activated.
3. Materials and Methods
We conducted a search on PubMed and Medline for original articles, reviews, meta-analyses, and case series using the following keywords, their acronyms, and their associations: farnesoid X receptor, bile acids metabolism, gut microbiota, dysbiosis, and liver steatosis. When appropriate, preliminary evidence from abstracts belonging to main national and international gastroenterological meetings (e.g., United European Gastroenterology Week, Digestive Disease Week) was also included. The papers found from the above-mentioned sources were reviewed by two of the authors (E.S. and G.S.B.), according to PRISMA guidelines [73]. The last Medline search was dated 15 May 2022.
In total, we found 249 manuscripts matching our search: 69 were clinical trials/original articles, of these 47 randomized clinical trials (RCT)s; 95 reviews of the scientific literature, 1 of those a systematic review; 3 meta-analyses. We included data from 41 original articles (including in vitro and animal studies), 5 RCTs, 26 reviews of the scientific literature, excluding those not updated, and from 1 meta-analysis.
4. Conclusions
This review of the scientific literature conveys a novel concept of BAs beyond the classic concept of simple regulators of lipid absorption. They have a sophisticated mechanism of networking with cells not only in the liver, but also in the intestine. This multi-faceted net of chatting is possible through nuclear receptors such as FXR that have at least a bi-valent role in liver fat deposition and metabolism. Moreover, the gut microbiota is crucial for BAs metabolic transformation within the gut. More interesting, gut microbiota composition is modulated by BAs pool and, vice versa; it affects their forms and composition. Thus, this unconventional connection is crucial for the host’s health and survival, as BAs avoid the emergence of pathogenic microbial strains from our intestinal tract. Several factors affecting gut microbiota (e.g., diet, antibiotics) influence its composition, and bring BAs and their interactions with cells towards a dysmetabolic state (e.g., obesity, type II diabetes, NAFLD, NASH), favoring gluco- and lipotoxicity.
Indeed, BAs pool modulation is as crucial as microbial eubiosis for our survival and health. All these pieces of evidence explain the interest of researchers in nuclear receptors modulation with agonists/antagonists according to different tissue targeted in our fight against an obesity-driven Westernized world. Moreover, pre-, pro-, and post-biotics can be powerful and promising weapons to correct and heal a “fattening” BAs pool, through microbial modulation.
Acknowledgments
We thank Luigi Boccuto, Healthcare Genetics and Genomics Interdisciplinary Doctoral Program, School of Nursing, College of Behavioral, Social and Health Sciences, Clemson University, Clemson, SC, USA, for the accurate and qualified work of revision of English form of this manuscript.
Author Contributions
Conceptualization: E.S., G.S.B. and L.A. had the original idea of this manuscript; methodology: F.D.N., L.M. and H.M., performed the review of literature; validation: E.S., L.A. and G.S.B. revised and validated the literature findings; formal analysis: E.S. and M.M.; investigation: E.S. and F.D.N.; data curation: E.S., P.S. and L.A.; writing—original draft preparation: E.S., L.A. and G.S.B.; writing—review and editing: E.S., L.A. and M.M.; visualization: E.S. and H.M.; supervision: E.S., L.A. and P.S.; project administration: E.S. and L.A. All authors have read and agreed to the published version of the manuscript.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data reported in this review of literature are available online on PubMed and main national and international gastroenterological meetings websites.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kaya E., Yilmaz Y. Metabolic-associated Fatty Liver Disease (MAFLD): A Multi-systemic Disease Beyond the Liver. J. Clin. Transl. Hepatol. 2021;10:329–338. doi: 10.14218/JCTH.2021.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abeysekera K.W.M., Macpherson I., Glyn-Owen K., McPherson S., Parker R., Harris R., Yeoman A., Rowe I.A., Dillon J.F. Community pathways for the early detection and risk stratification of chronic liver disease: A narrative systematic review. Lancet Gastroenterol. Hepatol. 2022;7:770–780. doi: 10.1016/S2468-1253(22)00020-6. [DOI] [PubMed] [Google Scholar]
- 3.Marra F., Svegliati-Baroni G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J. Hepatol. 2018;68:280–295. doi: 10.1016/j.jhep.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Perry R.J., Samuel V.T., Petersen K.F., Shulman G.I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J.Y., Sohn K.H., Rhee S.H., Hwang D. Saturated fatty acids, but not unsaturated fatty acids, induce the ex-pression of cyclooxygenase-2 mediated through Toll-like receptor 4. J. Biol. Chem. 2001;276:16683–16689. doi: 10.1074/jbc.M011695200. [DOI] [PubMed] [Google Scholar]
- 6.Mota M., Banini B.A., Cazanave S.C., Sanyal A.J. Molecular mechanisms of lipotoxicity and glucotoxicity in nonalcoholic fatty liver disease. Metabolism. 2016;65:1049–1061. doi: 10.1016/j.metabol.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chabowski A., Żendzian-Piotrowska M., Konstantynowicz K., Pankiewicz W., Mikłosz A., Łukaszuk B., Górski J. Fatty acid transporters involved in the palmitate and oleate induced insulin resistance in primary rat hepatocytes. Acta Physiol. 2012;207:346–357. doi: 10.1111/apha.12022. [DOI] [PubMed] [Google Scholar]
- 8.Ma C., Han M., Heinrich B., Fu Q., Zhang Q., Sandhu M., Agdashian D., Terabe M., Berzofsky J.A., Fako V., et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360:eaan5931. doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J., Yang D., Wang X., Asare P.T., Zhang Q., Na L., Shao L. Gut Microbiota Targeted Approach in the Management of Chronic Liver Diseases. Front. Cell. Infect. Microbiol. 2022;12:774335. doi: 10.3389/fcimb.2022.774335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A., Al-Hassi H.O., Steed H., Phipps O., Brookes M.J. Bile Acids and the Microbiome: Making Sense of This Dynamic Relationship in Their Role and Management in Crohn’s Disease. Can. J. Gastroenterol. Hepatol. 2022;2022:8416578. doi: 10.1155/2022/8416578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai J., Sun L., Gonzalez F.J. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. 2022;30:289–300. doi: 10.1016/j.chom.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inagaki T., Moschetta A., Lee Y.-K., Peng L., Zhao G., Downes M., Yu R.T., Shelton J.M., Richardson J.A., Repa J.J., et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. USA. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poland J.C., Flynn C.R. Bile Acids, Their Receptors, and the Gut Microbiota. Physiology. 2021;36:235–245. doi: 10.1152/physiol.00028.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowen A.E., Κοrman M.G., Hofmann A.F., Cass O.W., Coffin S.B. Metabolism of Lithocholate in Healthy Man. II. Enterohepatic circulation. Gastroenterology. 1975;69:67–76. doi: 10.1016/S0016-5085(19)32637-X. [DOI] [PubMed] [Google Scholar]
- 15.Urdaneta V., Casadesús J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front. Med. 2017;4:163. doi: 10.3389/fmed.2017.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long S.L., Gahan C.G., Joyce S.A. Interactions between gut bacteria and bile in health and disease. Mol. Asp. Med. 2017;56:54–65. doi: 10.1016/j.mam.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Ledesma A.E., Taranto M.P., Bustos A.Y. Characterization of substrate specificity and inhibitory mechanism of bile salt hydrolase from Lactobacillus reuteri CRL 1098 using molecular docking analysis. Biotechnol. Lett. 2021;43:1063–1073. doi: 10.1007/s10529-021-03097-y. [DOI] [PubMed] [Google Scholar]
- 18.Kumar R.S., Brannigan J.A., Prabhune A., Pundle A.V., Dodson G.G., Dodson E.J., Suresh C.G. Structural and Functional Analysis of a Conjugated Bile Salt Hydrolase from Bifidobacterium longum Reveals an Evolutionary Relationship with Penicillin V Acylase. J. Biol. Chem. 2006;281:32516–32525. doi: 10.1074/jbc.M604172200. [DOI] [PubMed] [Google Scholar]
- 19.Gopal-Srivastava R., Hylemon P.B. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J. Lipid Res. 1988;29:1079–1085. doi: 10.1016/S0022-2275(20)38464-9. [DOI] [PubMed] [Google Scholar]
- 20.Chand D., Panigrahi P., Varshney N., Ramasamy S., Suresh C. Structure and function of a highly active Bile Salt Hydrolase (BSH) from Enterococcus faecalis and post-translational processing of BSH enzymes. Biochim. et Biophys. Acta (BBA)-Proteins Proteom. 2018;1866:507–518. doi: 10.1016/j.bbapap.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Kawamoto K., Horibe I., Uchida K. Purification and characterization of a new hydrolase for conjugated bile acids, chenodeoxycholyltaurine hydrolase, from Bacteroides vulgatus. J. Biochem. 1989;106:1049–1053. doi: 10.1093/oxfordjournals.jbchem.a122962. [DOI] [PubMed] [Google Scholar]
- 22.Stellwag E., Hylemon P. Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim. et Biophys. Acta (BBA)-Enzym. 1976;452:165–176. doi: 10.1016/0005-2744(76)90068-1. [DOI] [PubMed] [Google Scholar]
- 23.Begley M., Gahan C.G., Hill C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Dussurget O., Cabanes D., Dehoux P., Lecuit M., Buchrieser C., Glaser P., Cossart P., European Listeria Genome Consortium Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 2002;45:1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- 25.Dean M., Cervellati C., Casanova E., Squerzanti M., Lanzara V., Medici A., de Laureto P.P., Bergamini C.M. Characterization of Cholylglycine Hydrolase from a Bile-Adapted Strain of Xanthomonas maltophilia and Its Application for Quantitative Hydrolysis of Conjugated Bile Salts. Appl. Environ. Microbiol. 2002;68:3126–3128. doi: 10.1128/AEM.68.6.3126-3128.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedrini P., Andreotti E., Guerrini A., Dean M., Fantin G., Giovannini P.P. Xanthomonas maltophilia CBS 897.97 as a source of new 7beta- and 7alpha-hydroxysteroid dehydrogenases and cholylglycine hydrolase: Improved biotransformations of bile acids. Steroids. 2006;71:189–198. doi: 10.1016/j.steroids.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Sridevi N., Prabhune A.A. Brevibacillus sp: A Novel Thermophilic Source for the Production of Bile Salt Hydrolase. Appl. Biochem. Biotechnol. 2008;157:254–262. doi: 10.1007/s12010-008-8326-9. [DOI] [PubMed] [Google Scholar]
- 28.Sridevi N., Srivastava S., Khan B.M., Prabhune A.A. Characterization of the smallest dimeric bile salt hydrolase from a thermophile Brevibacillus sp. Extremophiles. 2009;13:363–370. doi: 10.1007/s00792-008-0224-0. [DOI] [PubMed] [Google Scholar]
- 29.Jones B.V., Begley M., Hill C., Gahan C.G.M., Marchesi J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia B., Park D., Hahn Y., Jeon C.O. Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health. Gut Microbes. 2020;11:1300–1313. doi: 10.1080/19490976.2020.1748261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia B., Park D., Chun B., Hahn Y., Jeon C. Diet-Related Alterations of Gut Bile Salt Hydrolases Determined Using a Metagenomic Analysis of the Human Microbiome. Int. J. Mol. Sci. 2021;22:3652. doi: 10.3390/ijms22073652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Smet I., Van Hoorde L., Woestyne M.V., Christiaens H., Verstraete W. Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 1995;79:292–301. doi: 10.1111/j.1365-2672.1995.tb03140.x. [DOI] [PubMed] [Google Scholar]
- 33.Farooqui N., Elhence A., Shalimar A Current Understanding of Bile Acids in Chronic Liver Disease. J. Clin. Exp. Hepatol. 2021;12:155–173. doi: 10.1016/j.jceh.2021.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kastl A., Zong W., Gershuni V.M., Friedman E.S., Tanes C., Boateng A., Mitchell W.J., O’Connor K., Bittinger K., Terry N.A., et al. Dietary fiber-based regulation of bile salt hydrolase activity in the gut microbiota and its relevance to human disease. Gut Microbes. 2022;14:2083417. doi: 10.1080/19490976.2022.2083417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W., Wu Z., Dai Z., Yang Y., Wang J., Wu G. Glycine metabolism in animals and humans: Implications for nutrition and health. Amino Acids. 2013;45:463–477. doi: 10.1007/s00726-013-1493-1. [DOI] [PubMed] [Google Scholar]
- 36.Begley M., Sleator R.D., Gahan C.G., Hill C. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 2005;73:894–904. doi: 10.1128/IAI.73.2.894-904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinto M.G.V., Franz C.M., Schillinger U., Holzapfel W.H. Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Int. J. Food Microbiol. 2006;109:205–214. doi: 10.1016/j.ijfoodmicro.2006.01.029. [DOI] [PubMed] [Google Scholar]
- 38.Bertolini A., Fiorotto R., Strazzabosco M. Bile acids and their receptors: Modulators and therapeutic targets in liver inflammation. Semin. Immunopathol. 2022;44:547–564. doi: 10.1007/s00281-022-00935-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parséus A., Sommer N., Sommer F., Caesar R., Molinaro A., Ståhlman M., Greiner T.U., Perkins R., Bäckhed F. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017;66:429–437. doi: 10.1136/gutjnl-2015-310283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe M., Houten S.M., Wang L., Moschetta A., Mangelsdorf D.J., Heyman R.A., Moore D.D., Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Investig. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pineda Torra I., Claudel T., Duval C., Kosykh V., Fruchart J.C., Staels B. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol. Endocrinol. 2003;17:259–272. doi: 10.1210/me.2002-0120. [DOI] [PubMed] [Google Scholar]
- 42.Savkur R.S., Bramlett K.S., Michael L.F., Burris T.P. Regulation of pyruvate dehydrogenase kinase expression by the farnesoid X receptor. Biochem. Biophys. Res. Commun. 2005;329:391–396. doi: 10.1016/j.bbrc.2005.01.141. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Lee F.Y., Barrera G., Lee H., Vales C., Gonzalez F.J., Willson T.M., Edwards P.A. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc. Natl. Acad. Sci. USA. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C.L., McDonald J.G., Luo G., Jones S.A., Goodwin B., Richardson J.A., et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez-Sola G., Uriarte I., Latasa M.U., Fernandez-Barrena M.G., Urtasun R., Elizalde M., Barcena-Varela M., Jiménez M., Chang H.C., Barbero R., et al. Fibroblast growth factor 15/19 (FGF15/19) protects from diet-induced hepatic steatosis: Development of an FGF19-based chimeric molecule to promote fatty liver regeneration. Gut. 2017;66:1818–1828. doi: 10.1136/gutjnl-2016-312975. [DOI] [PubMed] [Google Scholar]
- 46.Neuschwander-Tetri B.A., Loomba R., Sanyal A.J., Lavine J.E., Van Natta M.L., Abdelmalek M.F., Chalasani N., Dasarathy S., Diehl A.M., Hameed B., et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen T., Shi A., Wei Y., Luo X., Xi L. Farnesoid X Receptor as a Promising Therapeutic Target for Nonalcoholic Fatty Liver Disease (NAFLD) and the Current Development of Its Agonists. Discov. Med. 2021;32:113–121. [PubMed] [Google Scholar]
- 48.Kowdley K.V., Luketic V., Chapman R., Hirschfield G.M., Poupon R., Schramm C., Vincent C., Rust C., Parés A., Mason A., et al. A randomized trial of obeticholic acid monotherapy in patients with primary biliary cholangitis. Hepatology. 2017;67:1890–1902. doi: 10.1002/hep.29569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uriarte I., Latasa M.U., Carotti S., Fernandez-Barrena M.G., Garcia-Irigoyen O., Elizalde M., Urtasun R., Vespasiani-Gentilucci U., Morini S., De Mingo A., et al. Ileal FGF15 contributes to fibrosis-associated hepatocellular carcinoma development. Int. J. Cancer. 2014;136:2469–2475. doi: 10.1002/ijc.29287. [DOI] [PubMed] [Google Scholar]
- 50.Takeda S., Kadowaki S., Haga T., Takaesu H., Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett. 2002;520:97–101. doi: 10.1016/S0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- 51.Jiao T.-Y., Ma Y.-D., Guo X.-Z., Ye Y.-F., Xie C. Bile acid and receptors: Biology and drug discovery for nonalcoholic fatty liver disease. Acta Pharmacol. Sin. 2022;43:1103–1119. doi: 10.1038/s41401-022-00880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe M., Houten S., Mataki C., Christoffolete M., Kim B.W., Sato H., Messaddeq N., Harney J.W., Ezaki O., Kodama T., et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 53.Thomas C., Gioiello A., Noriega L., Strehle A., Oury J., Rizzo G., Macchiarulo A., Yamamoto H., Mataki C., Pruzanski M., et al. TGR5-Mediated Bile Acid Sensing Controls Glucose Homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anfuso B., Tiribelli C., Adorini L., Rosso N. Obeticholic acid and INT-767 modulate collagen deposition in a NASH in vitro model. Sci. Rep. 2020;10:1699. doi: 10.1038/s41598-020-58562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Comeglio P., Cellai I., Mello T., Filippi S., Maneschi E., Corcetto F., Corno C., Sarchielli E., Morelli A., Rapizzi E., et al. INT-767 prevents NASH and promotes visceral fat brown adipogenesis and mitochondrial function. J. Endocrinol. 2018;238:107–127. doi: 10.1530/JOE-17-0557. [DOI] [PubMed] [Google Scholar]
- 56.Roth J.D., Feigh M., Veidal S.S., Fensholdt L.K., Rigbolt K.T., Hansen H.H., Chen L.C., Petitjean M., Friley W., Vrang N., et al. INT-767 improves histopathological features in a diet-induced ob/ob mouse model of biopsy-confirmed non-alcoholic steatohepatitis. World J. Gastroenterol. 2018;24:195–210. doi: 10.3748/wjg.v24.i2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walters J.R.F., Johnston I.M., Nolan J.D., Vassie C., Pruzanski M.E., Shapiro D.A. The response of patients with bile acid diarrhoea to the farnesoid X receptor agonist obeticholic acid. Aliment. Pharmacol. Ther. 2014;41:54–64. doi: 10.1111/apt.12999. [DOI] [PubMed] [Google Scholar]
- 58.Abenavoli L., Scarpellini E., Colica C., Boccuto L., Salehi B., Sharifi-Rad J., Aiello V., Romano B., De Lorenzo A., Izzo A.A., et al. Gut Microbiota and Obesity: A Role for Probiotics. Nutrients. 2019;11:2690. doi: 10.3390/nu11112690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheykhsaran E., Abbasi A., Leylabadlo H.E., Sadeghi J., Mehri S., Mazraeh F.N., Feizi H., Baghi H.B. Gut microbiota and obesity: An overview of microbiota to microbial-based therapies. Postgrad. Med. J. 2022 doi: 10.1136/postgradmedj-2021-141311. [DOI] [PubMed] [Google Scholar]
- 60.Scarpellini E., Ianiro G., Attili F., Bassanelli C., De Santis A., Gasbarrini A. The human gut microbiota and virome: Potential therapeutic implications. Dig. Liver Dis. 2015;47:1007–1012. doi: 10.1016/j.dld.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vera-Barajas A., Abenavoli L., Scarpellini E., Méndez-Sánchez N., Valencia-Rodríguez A., Ponciano-Rodríguez G., Wang D.Q.-H. The mechanism of dysbiosis in alcoholic liver disease leading to liver cancer. Hepatoma Res. 2020;6:5. doi: 10.20517/2394-5079.2019.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aggarwal V., Sunder S., Verma S.R. Disease-associated dysbiosis and potential therapeutic role of Akkermansia muciniphila, a mucus degrading bacteria of gut microbiome. Folia Microbiol. 2022;May 20:1–14. doi: 10.1007/s12223-022-00973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maggio R., Viscomi C., Andreozzi P., D’Ettorre G., Viscogliosi G., Barbaro B., Gori M., Vullo V., Balsano C. Normocaloric low cholesterol diet modulates Th17/Treg balance in patients with chronic hepatitis C virus infection. PLoS ONE. 2014;9:e112346. doi: 10.1371/journal.pone.0112346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dao M.C., Everard A., Aron-Wisnewsky J., Sokolovska N., Prifti E., Verger E.O., Kayser B.D., Levenez F., Chilloux J., Hoyles L., et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut. 2016;65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 65.Friedman E., Li Y., Shen T.-C.D., Jiang J., Chau L., Adorini L., Babakhani F., Edwards J., Shapiro D., Zhao C., et al. FXR-Dependent Modulation of the Human Small Intestinal Microbiome by the Bile Acid Derivative Obeticholic Acid. Gastroenterology. 2018;155:1741–1752.e5. doi: 10.1053/j.gastro.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wahlström A., Sayin S.I., Marschall H.-U., Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 67.Sayin S.I., Wahlström A., Felin J., Jäntti S., Marschall H.U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Li F., Jiang C., Krausz K.W., Li Y., Albert I., Hao H., Fabre K.M., Mitchell J.B., Patterson A., Gonzalez F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trabelsi M.-S., Daoudi M., Prawitt J., Ducastel S., Touche V., Sayin S.I., Perino A., Brighton C.A., Sebti Y., Kluza J., et al. Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine L cells. Nat. Commun. 2015;6:7629. doi: 10.1038/ncomms8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y., Hou H., Wang X., Dai X., Zhang W., Tang Q., Dong Y., Yan C., Wang B., Li Z., et al. Diammonium Glycyrrhizinate Ameliorates Obesity Through Modulation of Gut Microbiota-Conjugated BAs-FXR Signaling. Front. Pharmacol. 2021;12:796590. doi: 10.3389/fphar.2021.796590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian S.-Y., Chen S.-M., Pan C.-X., Li Y. FXR: Structures, biology, and drug development for NASH and fibrosis diseases. Acta Pharmacol. Sin. 2022;43:1120–1132. doi: 10.1038/s41401-021-00849-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gonzalez F.J., Jiang C., Xie C., Patterson A.D. Intestinal Farnesoid X Receptor Signaling Modulates Metabolic Disease. Dig. Dis. 2017;35:178–184. doi: 10.1159/000450908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P.A., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions: Explanation and Elaboration. PLoS Med. 2009;6:e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data reported in this review of literature are available online on PubMed and main national and international gastroenterological meetings websites.