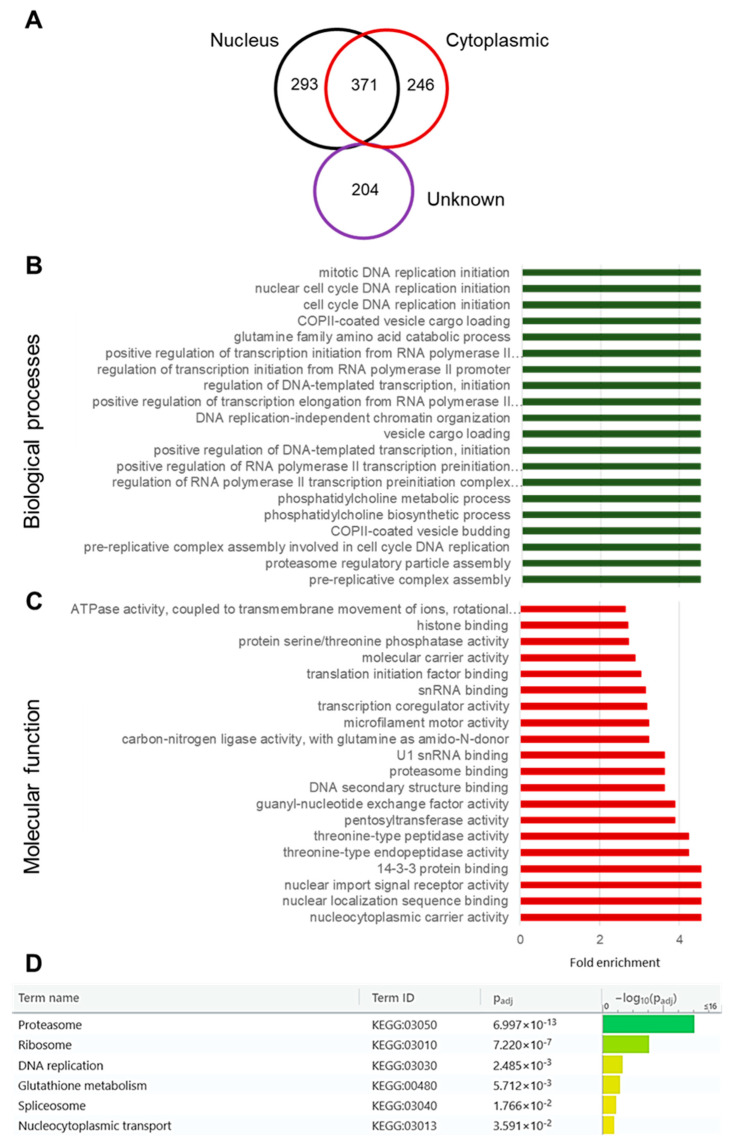
Figure 5.
Functional prediction analysis of putative interaction partners of PfSAMS. A total of 1114 identified potential interactors of PfSAMS were matched to the PlasmoDB database entries for their cellular localization, biological processes, and molecular function based on GO term enrichment analyses of both computed and curated data at a p-value of 0.01. (A) Venn diagram showing the predicted nuclear and cytoplasmic localization of the identified interactors, including proteins of unknown localization. The GO categories “Nucleus” (GO:0005634) and “Cytoplasm” (GO:0005737) were considered. (B,C) Interactors of the 20 highest scoring biological processes (B) and 20 highest scoring molecular functions (C) as predicted by GO enrichment analysis, sorted by fold enrichment. (D) KEGG analysis of PfSAMS interactors.