Abstract
In magnetotactic bacteria, a number of specific proteins are associated with the magnetosome membrane (MM) and may have a crucial role in magnetite biomineralization. We have cloned and sequenced the genes of several of these polypeptides in the magnetotactic bacterium Magnetospirillum gryphiswaldense that could be assigned to two different genomic regions. Except for mamA, none of these genes have been previously reported to be related to magnetosome formation. Homologous genes were found in the genome sequences of M. magnetotacticum and magnetic coccus strain MC-1. The MM proteins identified display homology to tetratricopeptide repeat proteins (MamA), cation diffusion facilitators (MamB), and HtrA-like serine proteases (MamE) or bear no similarity to known proteins (MamC and MamD). A major gene cluster containing several magnetosome genes (including mamA and mamB) was found to be conserved in all three of the strains investigated. The mamAB cluster also contains additional genes that have no known homologs in any nonmagnetic organism, suggesting a specific role in magnetosome formation.
The ability of magnetotactic bacteria to migrate along magnetic field lines is based on specific intracellular structures, magnetosomes that, in most magnetotactic bacteria, are nanometer sized, membrane-bound magnetic particles consisting of the iron mineral magnetite (Fe3O4) (3, 42). The unique characteristics of bacterial magnetosomes have attracted broad interdisciplinary research interest. Their superior crystalline and magnetic properties make them potentially useful as a highly ordered biomaterial in a number of applications, e.g., in the immobilization of bioactive compounds, in magnetic drug targeting, or as a contrast agent for magnetic resonance imaging (24, 29, 45). Recently, the characteristics of bacterial magnetosomes have been used as biosignatures to identify presumptive Martian magnetofossils (15, 51).
The narrow size distributions and uniform, species-specific crystal morphologies of bacterial magnetosomes imply a high degree of biological control over the mineralization process. The biomineralization of magnetosome particles is achieved by a complex mechanism that involves the uptake and accumulation of iron and the deposition of the mineral particle with a specific size and morphology within a specific compartment provided by the magnetosome membrane (MM). In bacteria of the genus Magnetospirillum (40), the MM consists of a bilayer containing phospholipids and proteins (16, 41; D. Schüler, K. Grünberg, and B. M. Tebo, Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000, abstr H-111, p. 373, 2000). A number of proteins were identified as specifically associated with the MM in Magnetospirillum magnetotacticum and Magnetospirillum sp. strain AMB-1 (16, 25, 32). The exact role of these magnetosome-specific proteins has not been elucidated, but it has been suggested that they have specific functions in iron accumulation, nucleation of minerals, and redox and pH control (4, 16, 42). Although several genes putatively related to magnetosome formation have been identified (25, 28, 32), the genetic basis of magnetite biomineralization has remained mostly unknown. Recently, the almost complete genome sequences of two magnetotactic alpha-proteobacteria, M. magnetotacticum strain MS-1 and magnetic coccus strain MC-1, have become available (http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html), which now allows the study of magnetosome formation at the genomic level. M. magnetotacticum is a microaerophilic spirillum producing cubo-octahedral magnetite particles that are 42 nm in size (8, 40). The size of its genome is about 4.3 Mb (6). Magnetic coccus strain MC-1, which has a genome size of about 3.7 Mb (12), was reported to form pseudohexagonal prismatic magnetite crystals about 70 nm in diameter (13, 26).
The magnetotactic bacterium M. gryphiswaldense, which was isolated from a freshwater sediment (40, 46), produces up to 60 cubo-octahedral magnetosome particles that strongly resemble those found in M. magnetotacticum and other Magnetospirillum species (3, 10, 47). M. gryphiswaldense can be cultivated more readily than most other magnetotactic bacteria, which has facilitated its physiological and biochemical analysis (41, 43, 44, 48).
In this study, we have cloned and analyzed several genes encoding magnetosome proteins from M. gryphiswaldense. Except for MamA, none of these proteins have been previously reported to be related to magnetosome formation in any magnetotactic bacterium. We report here the identification and preliminary analysis of a major gene cluster that encodes a number of these magnetosome proteins and is conserved in M. gryphiswaldense, M. magnetotacticum, and magnetic coccus strain MC-1.
MATERIALS AND METHODS
Strains and growth conditions.
M. gryphiswaldense strain MSR-1 (DSM 6361) was grown microaerobically at 30°C in a growth medium containing 100 μM ferric citrate as described before (44). The batch culture was exposed to air in 100-ml, 1-liter, and 10-liter bottles containing 50 ml, 500 ml, and 5 liters of medium, respectively, and agitated at 100 rpm on a New Brunswick incubation shaker. An inoculum of 10% of the culture volume was used. Microaerobic conditions arose in the medium at higher cell densities by oxygen consumption of cells (43). Escherichia coli DH5α (GIBCO BRL) was used as the host strain for cloning experiments with pBluescriptSKII (Stratagene). For cloning of PCR products using pCR-TOPO, E. coli TOP10 (Invitrogen) was used. For E. coli strains, the culture conditions used were those described by Sambrook et al. (38).
Isolation of magnetosomes.
Approximately 10 g (wet weight) of M. gryphiswaldense cells suspended in 100 ml of 20 mM HEPES–4 mM EDTA, pH 7.4, was disrupted by three passes through a French pressure cell (20,000 lb/in2). All of the buffers used for magnetosome isolation contained 0.1 mM phenylmethylsulfonyl fluoride as a protease inhibitor. Unbroken cells and cell debris were removed from the sample by centrifugation (10 min, 680 × g). The cell extract was passed through a MACS magnetic separation column (Miltenyi Biotec). Columns were placed between two Sa-Co-magnets generating a magnetic field gradient inside the column, which caused the magnetic particles to bind to the column matrix. The absence of any black, magnetosome-like material in the cell extract after passage through the column indicated that the separation of magnetosome particles was complete. To eliminate electrostatically bound contamination, magnetic particles attached to the column were rinsed first with 50 ml of 10 mM HEPES–200 mM NaCl, pH 7.4, and subsequently with 100 ml of 10 mM HEPES, pH 7.4. After removal of the column from the magnets, magnetic particles were eluted from the column by flushing with 10 mM HEPES buffer. Finally, the magnetosome suspension was loaded on top of a sucrose cushion (55% [wt/wt] sucrose in 10 mM HEPES, pH 7.4) and subjected to ultracentrifugation (280,000 × g, 8 h, 4°C) in a swinging-bucket rotor. The magnetic particles sedimented at the bottom of the tube, whereas residual contaminating cellular material was retained by the sucrose cushion.
Isolation of nonmagnetic subcellular fractions.
After separation of magnetosomes, an aliquot of the cell extract was subjected to ultracentrifugation (330,000 × g, 1 h, 4°C). The supernatant fluid from this high-speed centrifugation contained the soluble proteins. The membrane fraction contained in the pellet was further separated by isopycnic centrifugation as described by Osborn and Munson (34).
Analytical methods.
The iron content of whole cells and isolated magnetosome particles was determined by using a Perkin-Elmer 3110 atomic absorption spectrometer. Air-acetylene flame spectroscopy was used under the following conditions: wavelength, 248.6 nm; bandwidth, 0.2 nm; lamp current, 30 mA. For iron determination, the dried samples were incubated in concentrated nitric acid until digestion of the material was complete (18). The protein concentration of samples was determined by using the bicinchoninic acid protein microassay kit (Pierce) in accordance with the manufacturer's instructions.
Electron microscopy.
Purified magnetosomes were adsorbed on carbon-coated copper grids and negatively stained with 2% (wt/vol) uranyl acetate. Samples were viewed and recorded with a Philips CM10 transmission electron microscope at an accelerating voltage of 100 kV.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and determination of N-terminal and internal amino acid sequences.
Gels were prepared and run in accordance with the Laemmli procedure (20). An amount of magnetosomes equivalent to approximately 20 μg of protein was resuspended in electrophoresis sample buffer containing 2% SDS and 5% 2-mercaptoethanol. After boiling for 5 min, the samples were centrifuged for 3 min to pellet the magnetite particles. The supernatant was loaded on a 10 to 16% gradient polyacrylamide gel, which was stained with Coomassie brilliant blue after running. Digitized gels were analyzed by the ImageMaster 1D software (v.3.0; Amersham-Pharmacia). Amino-terminal protein sequence analysis was performed on an Applied Biosystems 470A amino acid sequencer by F. Lottspeich (Max-Planck-Institut für Biochemie, Martinsried, Germany) as previously described (14). Internal sequences were determined after cleavage with AspN protease (as described in reference 50).
Recombinant DNA techniques.
Total DNA of M. gryphiswaldense was isolated as described by Marmur (23). Plasmid isolation, transformation, and DNA manipulations in E. coli were essentially carried out by standard methods (38). Long oligonucleotides for hybridization used in Southern hybridization experiments were DS24 (5′-AAGCCCTCGAACATGCTGGACGAGGTGACCCTGTATACCCACTATGGCCTGTCGGTGGCC-3′) and DS33 (5′-ATGAAGTTCGAG AACTGCCGGGACTGCCGGGAAGAGGTGGTCTGGTGGGCGTTC-3′). Plasmid vectors used for cloning were pCR2.1-TOPO (Invitrogen) and pBluescriptSKII (Stratagene).
PCR amplification and DNA sequencing.
Degenerate primers for PCR amplification of a 240-bp mamC fragment were DS15F4 (5′-GCCGCBCTSGCBAAGAAYGC-3′) and DS15RV3 (5′-CGSAGYTCCTTYTCRATGAARTC-3′). For the amplification of a 960-bp mamD fragment, the primers were 1KGVDF and 4KGCR. 1KGVDF (5′-ATGTGGAGCGTCCTGGCCATG-3′) was deduced from the DNA sequence upstream of the homologous region in the genome of M. magnetotacticum. 4KGCR (5′-GCCTCAGGGTGGTGGCGGAT-3′) was deduced from the cDNA sequence close to the 3′ end of the mamC gene of M. gryphiswaldense. PCR amplification was performed with the Mastercycler Gradient (Eppendorf) by using standard protocols. Automatic sequencing of both strands of the plasmid DNA was carried out by primer walking (primers not shown).
Analysis of DNA sequence data.
Assembly of DNA sequences, identification and translation of open reading frames (ORFs), and calculation of the molecular masses of the proteins were done by the MacVector 6.5.3 software package (Oxford Molecular Ltd.). Sequence alignments were carried out by using the ClustalW algorithm (52), which is part of the same software. Protein sequences were compared to the GenBank, EMBL, and SwissProt databases by using the BLASTP program (1). Motif searches were carried out by using the Prosite program (17). Protein location was determined by the PSORT program (27). Preliminary sequence data for M. magnetotacticum MS-1 and magnetic coccus strain MC-1 was obtained from the DOE Joint Genome Institute at http: //www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html (status, 04/20/01). The amino acid sequences of the identified Mam proteins from M. gryphiswaldense were used in TBLASTN similarity searches to identify genes encoding homologous proteins in the preliminary baseline genomic assemblies of these bacteria. The identified regions of sequence homology on the respective contigs were analyzed for ORFs and translated into protein sequences.
Nucleotide sequence accession numbers.
The nucleotide sequence of the M. gryphiswaldense mamAB gene cluster has been deposited in the GenBank, EMBL, and DDBJ libraries and assigned accession number AF374354. The nucleotide sequence of the M. gryphiswaldense mamCD region has been deposited under accession number AF374355.
RESULTS
Analysis of magnetosome particles.
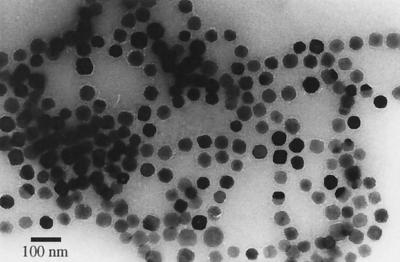
The magnetosome purification protocol resulted in 9 mg of clean magnetosomes from 1 g of magnetic cells on a dry-weight basis. Approximately 0.04 mg (dry weight) of protein was associated with 1 mg of isolated magnetosomes. The amount of MM-associated protein was equivalent to 0.07% of the total cellular protein content. Magnetosome-bound iron constituted approximately 93% of the total intracellular iron. Transmission electron microscopy indicated that isolated individual magnetite crystals were enclosed by an electron-thin layer representing the MM and were apparently free of contaminating cellular material (Fig. 1). Individual particles remained attached but were separated from each other by the membrane.
FIG. 1.
Transmission electron micrograph of purified magnetosomes from M. gryphiswaldense. Note that individual magnetosome particles are enclosed by a membrane and appear to remain attached to each other.
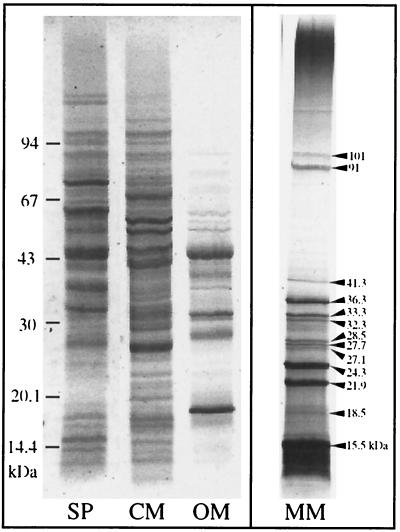
One-dimensional SDS-PAGE of solubilized proteins from purified magnetosome particles revealed 13 distinct polypeptide bands in various amounts (Fig. 2). The characteristics of the MM-specific polypeptides are shown in Table 1. According to their estimated molecular weights, they were designated MM15.5 to MM101. The most prominent polypeptide band was MM15.5. This band was prone to smearing on electrophoresis, and minor bands were frequently observed running closely below it, possibly indicating proteolytic degradation.
FIG. 2.
SDS-PAGE of the MM-associated proteins from M. gryphiswaldense compared to soluble proteins (SP) and the cytoplasmic membrane proteins (CM) and outer membrane proteins (OM). The bands were visualized by staining with Coomassie blue. Thirteen MM-specific proteins were identified in various amounts (arrowheads).
TABLE 1.
Summary of protein characteristics of magnetosome-associated polypeptides separated by SDS-PAGE (Fig. 2)
| Molecular mass (kDa) | Relative amt (% of total MM protein)a | N-terminal and internal amino acid sequences |
|---|---|---|
| 101.0 | 0.8 | |
| 91.0 | 3.0 | |
| 41.8 | 2.4 | |
| 36.3 | 7.2 | MFNGDVEDGRR-S/E-NVSXGKD |
| 33.3 | 2.7 | MKFENCRDCREEVVWWAFTAD |
| 32.3 | 2.3 | |
| 28.5 | 1.6 | |
| 27.7 | 1.3 | |
| 27.1 | 1.3 | |
| 24.3 | 11.2 | KPSNMLDEVTLYTHYGLSVA |
| 21.9 | 10.0 | M-Q/A-D-L/A-F/A-L |
| 18.5 | 2.2 | |
| 15.5 | 52.4b | SFQLAPYLAKSVPGIGILGGIVGGAAALAKN |
| A-DLGVDFIEKELRHGKSAEAT-DILRDEA |
Calculated from band intensities of a densitometric scan of a Coomassie-stained gel.
Including amounts of bands representing putative degradation products.
Cloning and sequence analysis of genes encoding MM proteins in M. gryphiswaldense. (i) mamA and mamB
Based on the codon usage bias found in previously analyzed genes from Magnetospirillum species, long (50 to 65 bases), nondegenerate oligonucleotides were designed from N-terminal amino acid sequences of several major MM-specific polypeptides. These oligonucleotides were labeled and directly used as probes for hybridization. Two probes (DS24 and DS33), corresponding to the amino acid sequences of MM24.3 and MM33.3, respectively, recognized the same genomic 7.55-kb EcoRI DNA fragment in Southern blotting experiments. It was cloned into plasmid pBluescriptSKII, resulting in pDS902. Sequence analysis by primer walking of the complete 7.55-kb fragment identified eight complete and two truncated consecutive ORFs. The deduced amino acid sequences of two ORFs matched the N-terminal sequences of MM24.3 and MM33.3, respectively. Consequently, these ORFs were designated mamA and mamB (mam for MM).
The mamA gene of M. gryphiswaldense encodes the second most abundant MM protein (MM24.3). Its predicted molecular mass of 24.01 kDa is consistent with the apparent molecular mass of 24.3 kDa estimated by gel electrophoresis. Its amino acid sequence is 91% identical to that of the magnetosome-associated MAM22 protein that has been previously reported in M. magnetotacticum (32, 33). The hydropathy plot of the amino acid sequence (not shown) was indicative of a relatively hydrophilic protein that has been suggested to be electrostatically bound to the MM in M. magnetotacticum (32).
The mamB gene encodes a protein that corresponds to the N-terminal amino acid sequence of an MM-associated polypeptide band in SDS-PAGE. The 31.96-kDa molecular mass calculated from the amino acid sequence is slightly lower then that estimated by gel electrophoresis, as is frequently observed with membrane proteins. The MamB protein exhibits significant sequence similarity to members of the ubiquitous cation diffusion facilitator (CDF) family, which are involved in the transport of various heavy metals. According to secondary-structure predictions (data not shown), the MamB protein exhibits the characteristic topology of bacterial CDF family members (six transmembrane helices) and contains the family-specific signature sequence (36).
(ii) mamC and mamD
Since long, nondegenerate oligonucleotide probes derived from the MM15.5 N-terminal amino acid sequences failed to identify specific chromosomal DNA fragments in hybridization experiments, a pair of oligonucleotide primers for PCR were deduced from the N-terminal and internal amino acid sequences of this polypeptide. By using these primers, a single 240-bp fragment was amplified from genomic DNA and cloned into pCR2.1-TOPO, generating pMT1. By using the cloned 240-bp fragment as a probe, a 4.3-kb chromosomal EcoRI-fragment was identified by Southern hybridization and cloned into pBluescriptSKII, generating plasmid pKG2. Sequence analysis of the insert identified an ORF that contained the N-terminal and internal peptide sequences of the MM15.5 protein. It was designated mamC. The mamC gene encodes the most abundant polypeptide in the MM of M. gryphiswaldense (MM15.5). The calculated molecular mass of 12.24 kDa was lower than the apparent molecular mass of 15.5 kDa estimated by SDS-PAGE, as is frequently the case with hydrophobic proteins.
An incomplete ORF lacking the N-terminal portion of its corresponding protein was found on pKG2 immediately upstream of the mamC gene, suggesting a putative operon-like organization of additional genes together with mamC. To obtain the complete sequence of this ORF, a 960-bp DNA fragment was amplified by PCR using genomic DNA as the template and primers 1KGVDF and 4KGCR. The forward primer used for amplification (1KGVDF) was deduced from the DNA sequence upstream of the homologous region in the genome of M. magnetotacticum, which was previously found to be identically organized. Sequencing of the PCR product revealed that it contained the missing portion of a 942-bp-long ORF. The N terminus of its predicted protein was in close agreement with the ambiguous N-terminal amino acid sequence derived from MM21.9. We therefore concluded that another major MM polypeptide is encoded by this gene, which was designated mamD. The observed difference between the molecular mass of 29.9 kDa calculated for the predicted mamD gene product and the apparent mass of the corresponding 21.9-kDa band in SDS-PAGE might be explained by proteolytic cleavage of a substantial part of the C terminus. Hydropathy plots of the amino acid sequence (not shown) predicted a hydrophobic protein with a short hydrophilic stretch close to the C terminus. Similarity searches of databases gave no indication of the existence of known proteins homologous to MamC and MamD.
Identification and sequence analysis of genes encoding putative MM proteins in the genomes of M. magnetotacticum MS-1 and magnetic coccus strain MC-1.
Genes with significant similarity to mamA, mamB, mamC, and mamD of M. gryphiswaldense were identified in the genome sequences of both M. magnetotacticum and strain MC-1. The characteristics of the predicted mam gene products of M. magnetotacticum and strain MC-1, together with gene products of ORFs from adjacent regions, are shown in Tables 2 and 3. Generally, the homologous genes have sizes comparable to those of their respective counterparts in M. gryphiswaldense and encode proteins with characteristics very similar to theirs. Secondary-structure predictions for the equivalent genes using various algorithms gave similar results (data not shown). The alignments of Mam protein sequences are shown in Fig. 3.
TABLE 2.
Summary of features of proteins deduced from ORFs identified in chromosomal mamAB gene clusters of M. magnetotacticum, M. gryphiswaldense, and strain MC-1
| Bacterium | ORFa | Size (amino acids) | % Identity/ similarityb | Molecular mass (kDa) | Predicted locationc | Best BLASTP hit (accession no.)d | E-value | Putative function of BLAST homolog |
|---|---|---|---|---|---|---|---|---|
| M. magnetotacticum | ORF1 (mamE) | 726 | 100 | 73.2 | MM | HtrA H. infl. (A64113)g | 9e-35 | Serine protease |
| M. gryphiswaldense | NDe | |||||||
| Strain MC-1 | mamEf | 803 | 34/48 | 84.4 | MM | HtrA R. prow. (B71722)h | 2e-35 | |
| M. magnetotacticum | ORF2 | 390 | 100 | 40.3 | IM | Unknown | ||
| M. gryphiswaldense | ND | |||||||
| MC-1 | Not found | |||||||
| M. magnetotacticum | ORF3 | 347 | 100 | 37.6 | Cytoplasm | EnvB M. therm. (F69003)i | 1e-15 | Rod shape determination |
| M. gryphiswaldense | ND | |||||||
| MC-1 | ORF10f | 346 | 50/70 | 37.6 | Cytoplasm | MreB T. marit. (E72359)j | 4e-07 | Rod shape determination |
| M. magnetotacticum | ORF4 | 78 | 8.36 | IM | Unknown | |||
| M. gryphiswaldense | ND | |||||||
| MC-1 | Not found | |||||||
| M. magnetotacticum | ORF5 | 296 | 100 | 34.55 | IM | ydbO B. subt. (B69772)k | 2e-28 | Cation transport (CDF) |
| M. gryphiswaldense | ND | |||||||
| MC-1 | ORF1 | 332 | 23/43 | 36.29 | IM | MTH1893 M. therm.(F69119)i | 1e-30 | Cation transport (CDF) |
| M. magnetotacticum | ORF6 | 437 | 100 | 45.8 | IM | PH1912 P. horik. (F71205)l | 6e-33 | Cation transport |
| M. gryphiswaldense | ORF1 (fragment) | >147 | 92/94 | NAw | IM | PH1912 (F71205)lm | Cation transport | |
| MC-1 | Not found | |||||||
| M. magnetotacticum | ORF7 | 637 | 100 | 66.26 | IM | HtrA S. sonnei (BAA92745)n | 4e-14 | Serine protease |
| M. gryphiswaldense | ORF2 | 632 | 88/95 | 65.38 | IM | HtrA(BAA92745)n | 5e-12 | Serine protease |
| MC-1 | ORF2 | 671 | 34/52 | 71.8 | IM | HtrA H. pyl. (C64647)o | 1e-12 | Serine protease |
| M. magnetotacticum | ORF8 | 275 | 100 | 28.89 | IM | OrfE0 R. caps. (CAA72164)p | 0.008 | Serine protease |
| M. gryphiswaldense | ORF3 | 270 | 79/85 | 28.36 | IM | OrfE0 (CAA72164)p | 0.04 | Serine protease |
| MC-1 | ORF3 | 261 | 37/48 | 27.56 | IM | HtrA B. hens. (P54925)q | 0.002 | Serine protease |
| M. magnetotacticum | ORF9 (mamA)r | 217 | 100 | 23.97 | MM | MTH83 M. therm. (F69210)is | 3e-09 | TPR protein |
| M. gryphiswaldense | ORF4 (mamA) | 217 | 91/97 | 24.01 | MM | MTH83 (F69210)is | 3e-09 | TPR protein |
| MC-1 | ORF4 (mamA) | 219 | 37/58 | 25.08 | MM | MTH83 (F69210)is | 1e-36 | TPR protein |
| M. magnetotacticum | ORF10 | 272 | 100 | 29.95 | IM | LemA T. marit. (F72311)j | 9e-18 | Unknown |
| M. gryphiswaldense | ORF5 | 272 | 80/90 | 30.00 | IM | LemA (F72311)j | 3e-16 | Unknown |
| MC-1 | ORF6 | 308 | 32/49 | 34.85 | IM | LemA (F72311)j | 3e-17 | Unknown |
| M. magnetotacticum | ORF11 | 84 | 100 | 9.26 | IM | Unknown | ||
| M. gryphiswaldense | ORF6 | 84 | 83/93 | 9.24 | Uncertain | Unknown | ||
| MC-1 | Not found | |||||||
| M. magnetotacticum | ORF12 (mamB) | 297 | 100 | 31.87 | MM | ydfM B. subt. (C69781)k | 9e-34 | Cation transport (CDF) |
| M. gryphiswaldense | ORF7 (mamB) | 297 | 93/96 | 31.96 | MM | ydfM (C69781)k | 1e-38 | Cation transport (CDF) |
| MC-1 | ORF7 (mamB) | 285 | 44/67 | 30.03 | MM | ydfM (C69781)k | 4e-34 | Cation transport (CDF) |
| M. magnetotacticum | ORF13 | 180 | 100 | 18.74 | IM | Unknown | ||
| M. gryphiswaldense | ORF8 | 175 | 71/78 | 18.20 | IM | Unknown | ||
| MC-1 | ORF8 | 190 | 29/44 | 20.58 | IM | Unknown | ||
| M. magnetotacticum | ORF14 | 174 | 100 | 19.03 | Periplasm | Unknown | ||
| M. gryphiswaldense | ORF9 | 174 | 83/92 | 18.88 | IM | Unknown | ||
| MC-1 | ORF9 | 154 | 35/49 | 17.2 | Periplasm | Unknown | ||
| M. magnetotacticum | ORF15 | 297 | 100 | 30.9 | Cytoplasm | BmrU B. subt. (F69595)k | 1e-05 | Multidrug resistance |
| M. gryphiswaldense | ORF10 (fragment) | >144 | 74/84 | NA | Cytoplasm | BmrU (F69595)km | Multidrug resistance | |
| MC-1 | Not found | |||||||
| M. magnetotacticum | ORF16 | 331 | 100 | 34.5 | IM | ydfM B. subt. (C69781)k | 6e-26 | Cation transport (CDF) |
| M. gryphiswaldense | ND | |||||||
| MC-1 | Not foundt | |||||||
| M. magnetotacticum | Not foundu | |||||||
| M. gryphiswaldense | ND | |||||||
| MC-1 | ORF5 | 1,025 | 112.3 | Cytoplasm | MAM22 (BAA11643)v | 2e-05 | TPR protein |
ORFs are listed according to their order on M. magnetotacticum contig 3824, together with equivalent genes (closest homologs) of M. gryphiswaldense and strain MC-1. Genes that were experimentally shown to encode MM proteins are in boldface.
Identity and similarity values are with respect to the equivalent protein in M. magnetotacticum.
Location was determined by the PSORT program (27). Localization in the MM was predicted based on homology to identified MM proteins. IM, inner membrane.
Only BLASTP hits with E-values of <0.01 are shown.
ND; not determined. The N terminus of MM protein MamE of M. gryphiswaldense is homologous (16 and 18 out of 20 residues identical and similar, respectively) to the N-terminal amino acid sequence of the predicted MamE protein of M. magnetotacticum. The nucleotide sequence of the corresponding gene in M. gryphiswaldense was not determined.
A homologous gene (ORF10) is present in the genome of strain MC-1 (contig 369), but it is located outside the mamAB cluster.
From Haemophilas influenzae.
From Rickettsia prowazekii.
From Methanobacterium thermoautotrophicum.
From Thermotoga maritima.
From Bacillus subtilis.
From Pyrococcus horikoshii.
BLASTP searches using the incomplete sequence of M. gryphiswaldense yielded the same hit as the complete sequence of the equivalent protein of M. magnetotacticum but with an E-value of >0.01.
From Shigella sonnei.
From Helicobacter pylori.
From Rhodobacter capsulatus.
From Bartonella henselae.
ORF9 (mamA) is identical to the mam22 gene (accession no. BAA11643) of M. magnetotacticum, which was previously described (32).
The best BLASTP hit was mam22 of M. magnetotacticum (BAA11643); therefore, the second-best hit is shown.
Has (34% identity and 51% similarity) to ORF7 (mamB) of strain MC-1.
The 222 C-terminal amino acids of ORF5 of strain MC-1 are 19% identical and 37% similar to ORF9 (mamA) of M. magnetotacticum.
From M. magnetotacticum.
NA, not applicable.
TABLE 3.
Characteristics of proteins encoded by the mamC and mamD genes of M. gryphiswaldense and their homologs in the genomes of M. magnetotacticum and strain MC-1
| Bacterium | Gene | Size (amino acids) | % Identity/ similaritya | Molecular mass (kDa) |
|---|---|---|---|---|
| M. gryphiswaldense | mamC | 125 | 100 | 12.4 |
| M. magnetotacticum | mamC | 124 | 80/90 | 12.4 |
| Strain MC-1 | mamC | 133 | 50/65 | 13.6 |
| M. gryphiswaldense | mamD | 314 | 100 | 30.2 |
| M. magnetotacticum | mamD | 314 | 81/92 | 29.9 |
| Strain MC-1 | mamD | 340 | 31/46 | 34.4 |
Identity and similarity values are with respect to the equivalent protein in M. gryphiswaldense.
FIG. 3.
Sequence alignments of identified magnetosome proteins of M. gryphiswaldense (M.g.) and their homologs from M. magnetotacticum (M.m.) and magnetic coccus strain MC-1. If applicable, the most similar homolog from a nonmagnetic organism was included. Identical amino acids are shown on a solid background, and similar amino acid are shaded. Mtherm, Methanobacterium thermoautotrophicum; Bsubt, Bacillus subtilis.
In addition to mamA to mamD, similarity searches of the genome sequence of M. magnetotacticum using the N-terminal amino acid sequence of the MM36.3 protein of M. gryphiswaldense as the query identified an ORF that encodes a predicted protein with an N terminus sharing 16 identical and 2 similar amino acids out of 20 residues with the N terminus of MM36.3 from M. gryphiswaldense. Based on the significant homology and the fact that this ORF was found to be colocated together with the mamA and mamB genes (Fig. 4), we conclude that another MM polypeptide of M. magnetotacticum is encoded by this gene, which was designated mamE. Given the high overall similarity shared by the identified mam genes of M. magnetotacticum and M. gryphiswaldense, a gene very similar to mamE is likely to occur in M. gryphiswaldense. However, the predicted molecular mass of 73.2 kDa of MamE from M. magnetotacticum contrasts with the apparent molecular mass of 36.3 kDa of the corresponding MM protein in M. gryphiswaldense, which might be the result of proteolytic cleavage of the C-terminal part of the MamE protein. A homologous gene was identified in the genome of strain MC-1. Similarity searches of databases revealed that the putative MamE proteins of M. magnetotacticum and strain MC-1 bear sequence similarity to HtrA-like serine proteases (35).
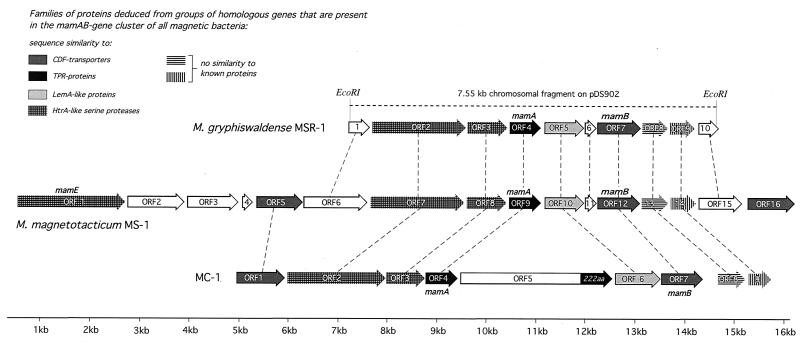
FIG. 4.
Molecular organization of the mamAB gene clusters of M. gryphiswaldense, M. magnetotacticum, and magnetic coccus strain MC-1. Arrows indicate the direction of gene transcription. Filled arrows indicate ORFs that belong to families of homologous genes shared by the mamAB clusters of the three magnetotactic bacteria investigated. The dashed lines connect equivalent genes (closest homologs).
Molecular organization of the mamAB gene cluster in M. gryphiswaldense, M. magnetotacticum MS-1, and magnetic coccus strain MC-1.
The mamB gene of M. gryphiswaldense MSR-1 was found to be located 1,120 bp downstream of mamA. As mentioned above, both genes are part of a region containing several ORFs of colinear orientation. Likewise, genes homologous to mamA and mamB were both found in the same chromosomal region in M. magnetotacticum (contig 3824) and strain MC-1 (contig 431). Since this finding was suggestive of the clustering of several genes possibly related to magnetite formation, the organization of the mamA and mamB genes, as well as the ORFs adjacent to them, was characterized in more detail. The arrangement of ORFs in the chromosomal mamAB gene clusters of M. gryphiswaldense, M. magnetotacticum, and strain MC-1 is shown in Fig. 4, and the characteristics of the corresponding predicted proteins are given in Table 2.
In M. gryphiswaldense, mamA and mamB, together with at least eight other ORFs, are arranged in a colinear fashion, implying an operon-like structure. An identical organization is present in M. magnetotacticum, which is part of a larger cluster comprising 16 consecutive ORFs with the same direction of transcription. In both organisms, the mamB gene and the two ORFs preceding it overlap by a single nucleotide, respectively.
A similar organization of mamA and mamB, together with seven consecutive ORFs extending over 11 kb, is present in magnetic coccus strain MC-1. The chromosomal mamAB clusters in the three strains are characterized by the presence of one or several members of various classes of homologous genes. Several of these classes correspond to proteins with homology to one of the following families:
(i) TPR proteins.
The mamA genes of all three strains display similarity to genes encoding TPR (tetratricopeptide repeat) proteins. The mamA gene of strain MC-1 (ORF4) is followed by ORF5, which encodes a deduced protein of 1,025 amino acids. Its C-terminal domain (222 amino acids) was also found to be similar to MAM22 of M. magnetotacticum (32, 33) (identical to MamA [this study]) and other members of the TPR family (21).
(ii) CDF transporters.
Besides mamB, two more genes (ORF5 and ORF16) in the mamAB gene cluster of M. magnetotacticum and one more in strain MC-1 (ORF1) display significant similarity to members of the CDF protein family (36). Pairwise sequence alignments revealed that ORF5 of M. magnetotacticum and ORF1 of strain MC-1 are equivalent to each other, whereas the mamB genes of the two bacteria form a group of distinct similarity (data not shown).
(iii) HtrA.
The mamE gene (ORF1) is located at the 5′ end of the mamAB gene cluster in M. magnetotacticum and is most similar to the mamE gene of strain MC-1. However, in strain MC-1, this gene is located outside the mamAB cluster. Additional genes with similarity to htrA genes were identified in the mamAB regions of M. gryphiswaldense (ORF2), M. magnetotacticum (ORF7), and strain MC-1 (ORF2). In all three organisms, it is immediately followed by an ORF that also bears weak similarity to htrA-like genes.
(iv) lemA
In all three magnetotactic strains, an ORF with sequence similarity to lemA-like genes (M. gryphiswaldense, ORF5; M. magnetotacticum, ORF10; strain MC-1, ORF6) is situated between the mamA and mamB genes. lemA-like genes have been identified in the genomes of a number of bacteria and are of unknown function. The LemA protein was first identified as an epitope in the bacterial pathogen Listeria monocytogenes (22).
Two more classes of genes have counterparts in the mamAB cluster of each of the magnetotactic strains (M. gryphiswaldense, ORF8 and ORF9; M. magnetotacticum, ORF13 and ORF14; strain MC-1, ORF8 and ORF9), but their predicted products display no significant sequence similarity to any known proteins from databases. In addition, there is a set of genes that are part of the mamAB cluster in M. gryphiswaldense (ORF1, ORF6, and ORF10) and M. magnetotacticum (ORF1, ORF2, ORF3, ORF4, ORF6, ORF11, and ORF15) but are absent from the homologous chromosomal region in strain MC-1. Respective homologs to ORF1 and ORF3 of M. magnetotacticum were identified in a different region of the strain MC-1 chromosome (contig 369), while no genes with similarity to ORF2, ORF4, ORF6, ORF11, and ORF15 of M. magnetotacticum and ORF1, ORF6, and ORF10 of M. gryphiswaldense could be detected in strain MC-1.
Organization of the mamC and mamD genes in M. gryphiswaldense, M. magnetotacticum, and strain MC-1.
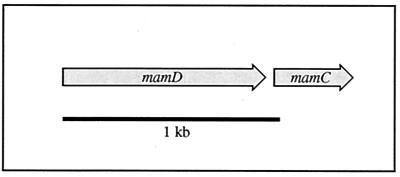
The genes encoding MM proteins MamC and MamD in M. gryphiswaldense and their respective homologs in M. magnetotacticum and strain MC-1 are not closely linked to the mamAB gene cluster. In M. gryphiswaldense and M. magnetotacticum, mamD is immediately followed by mamC (Fig. 5). In the genome of strain MC-1, the identified homologous genes are not linked (mamC, contig 369; mamD, contig 431).
FIG. 5.
Molecular organization of the mamC and mamD genes of M. gryphiswaldense. An equivalent arrangement of genes is present in M. magnetotacticum.
DISCUSSION
The purification protocol reported in this study allowed the efficient isolation of magnetosome particles from M. gryphiswaldense. The isolated magnetosomes of M. gryphiswaldense exhibited characteristics (i.e., size, morphology, presence of the membrane, etc.) similar to those of the magnetosomes from M. magnetotacticum and Magnetospirillum sp. strain AMB-1, as previously described (16, 29). The tendency of isolated magnetosome particles to maintain their chainlike alignment might suggest that individual particles are attached to each other by specific interactions. A total of 13 polypeptide bands could be identified in Coomassie-stained SDS-polyacrylamide gels of the solubilized MM of M. gryphiswaldense, although the possibility cannot be excluded that proteins loosely attached to the MM were lost during preparation or that additional proteins are present below the level of detection by Coomassie staining.
In this study, the genes for four major MM proteins from M. gryphiswaldense were cloned and analyzed. In addition, a gene encoding a putative MM protein in M. magnetotacticum was identified based on sequence data from a homologous MM protein in M. gryphiswaldense. Four of the newly identified genes (mamB, mamC, mamD, and mamE) have not been previously reported to encode MM-specific proteins in other magnetotactic bacteria. None of these genes or neighboring genes from the mamAB cluster in the three magnetotactic bacteria investigated display substantial similarity to the magA and mpsA genes of Magnetospirillum sp. strain AMB-1, which were previously reported to encode MM-associated proteins (25, 28). Genes sharing homology with magA and mpsA of strain AMB-1 were identified in different chromosomal regions of both M. magnetotacticum and strain MC-1 in a preliminary analysis (unpublished data), indicating that these genes are not linked to chromosomal regions comprising the mamAB or mamCD genes. Likewise, the bacterioferritin-encoding gene (bfr) of M. magnetotacticum, which has been speculated to be involved in magnetite biomineralization (5), is also located in a distant genomic region. These findings suggest that the genetic determination of magnetosome formation is complex and involves several different genomic sites in addition to the mamAB and mamCD chromosomal regions identified in this study.
Comparative analysis of the mam gene sequences from M. gryphiswaldense with the almost completed genomic assemblies of M. magnetotacticum and strain MC-1 allowed us to identify homologous genes in the latter organisms. Generally, the Mam proteins of M. gryphiswaldense and M. magnetotacticum have nearly identical sequences (91 to 97% similarity) while the amino acid sequence similarity between the Magnetospirillum species and strain MC-1 is 46 to 67%. Although the biochemical composition of the MM remains to be analyzed in the latter bacterium, the extensive sequence similarity shared by the Mam proteins of all three of these magnetotactic bacteria implies that they are likely to be functionally equivalent. For Mam proteins with homology to known protein families from databases, namely, MamA, MamB, and MamE, the similarity between the equivalent proteins from the magnetotactic bacteria was generally found to be significantly higher than to database homologs from other organisms.
The arrangement of the mamAB genes, as well as the genetic organization of the flanking regions, was found to be conserved in all three magnetotactic strains. In bacteria, functionally related genes are often located close to each other. Therefore, the operon-like arrangement of genes in the conserved mamAB region suggests that the neighboring genes might be related to the formation of magnetosomes. Interestingly, most of the genes identified in the mamAB cluster encode putative membrane proteins, several of them with sizes consistent with the molecular masses of protein bands observed in MM preparations from various Magnetospirillum species (this study; 16, 31, 32). Hence, several of the products of genes from the mamAB cluster might correspond to these unidentified proteins but also could have other functions related to magnetite biomineralization, such as the uptake and transport of iron into the cell and intracellular differentiation during MM formation. In addition to genes that are specific for either the Magnetospirillum species or strain MC-1, the mamAB cluster is characterized by a set of genes found in all three magnetotactic bacteria. These genes can be assigned to six different homology classes. In addition to two unknown classes, four classes of genes correspond to proteins with homology to one of the following families: (i) TPR proteins, (ii) CDF transporters, (iii) HtrA-like serine proteases, and (iv) LemA-like proteins.
TPR motifs, which have been identified across the biological kingdom in a large number of proteins with diverse functions, are known to mediate protein-protein interactions (for a review, see reference 21). Proteins with multiple copies of TPR motifs function as scaffolding proteins and coordinate the assembly of proteins into multisubunit complexes (11, 49). TPR proteins are represented by the mamA genes in all three strains and ORF5 of strain MC-1. MamA of M. gryphiswaldense shares extensive similarity with the previously identified MAM22 protein of M. magnetotacticum (32). Since the nomenclature of this protein does not reflect its actual molecular mass of 24 kDa and its gene was found to be part of a putative operon containing additional mam genes, we propose to reassign the mam22 gene to mamA as in M. gryphiswaldense. By analogy to TPR function in many eukaryotic proteins, Okuda et al. suggested that MAM22 localized in the MM may act as a receptor interacting with proteins from the cytoplasm (32, 33). Alternatively, the function of the MamA proteins in the MM may involve the formation of multiprotein complexes within the MM or between the individual magnetosome particles.
CDF proteins occur ubiquitously in eukaryotes, bacteria, and archaea and are involved in the transport of various heavy metals. CDF proteins are represented by the MamB protein and additional CDF homologs present in the mamAB region of M. magnetotacticum and strain MC-1. Several members of this family are known to confer resistance to Cu, Cd, and Zn (30, 36). Although members of the CDF protein family have not yet been demonstrated to be involved in iron transport, its specific location in the MM suggests that MamB might participate in the transport of iron into the MM vesicle.
Members of the HtrA protein family are widely distributed in nature. In E. coli and other bacteria, they are heat shock-induced serine proteases that are active in the periplasm, where their main function is the degradation of misfolded proteins. Different HtrA proteins have distinct regulatory and housekeeping functions in the cell (9, 35). Besides mamE, several additional, highly divergent genes with sequence similarity to htrA-like genes were identified in the mamAB regions of all three magnetotactic bacteria. The reported N terminus of the 66.2-kDa MM protein from Magnetospirillum sp. strain AMB-1 (25) has no homology to predicted products of the mamAB gene cluster identified in this study but does bear similarity to HtrA-like proteins (unpublished data). Although these findings suggest that HtrA-like proteins are constituents of the MM in several magnetotactic bacteria, their role is not apparent. In addition to the presence of a catalytic domain characteristic of trypsin-like serine proteases, profile searches of the Prosite database with each of the two homologous MamE sequences identified two PDZ domains characteristic of HtrA proteins in the MamE sequences of M. magnetotacticum and strain MC-1, respectively (data not shown) (37, 39). It is generally believed that the role of PDZ domains is to position ion channels, receptors, or other signaling molecules in the correct spatial arrangement (7). Hence, it might be speculated that HtrA-like proteins fulfill similar functions in the MM.
Since magnetosome formation in magnetotactic bacteria is under strict biological control, it has been assumed that a number of different gene functions are involved in this complex process (19). Our data suggest that several of these functions might be contributed by genes with homology to ubiquitous families. In addition to those, there is a set of genes represented by mamC, mamD, and ORF8 and ORF9 of the mamAB cluster of M. gryphiswaldense, whose predicted products lack recognizable homology to any prokaryotic or eukaryotic proteins from databases but are present in all magnetotactic bacteria. Hence, it can be speculated that genes of unknown function are involved in magnetosome formation. Functional studies are required to elucidate the specific role of these candidate genes in bacterial magnetite biomineralization.
ACKNOWLEDGMENTS
This study was supported by grants from the DFG and the BMBF.
We thank F. Lottspeich for determination of N-terminal amino acid sequences, M. Bauer for advice on sequence analysis, and E. Bäuerlein and M. Hildebrand for helpful discussions. Preliminary sequence data for M. magnetotacticum MS-1 and magnetic coccus strain MC-1 was obtained from the DOE Joint Genome Institute at http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R, Rossello-Mora R, Schüler D. Phylogeny and in situ identification of magnetotactic bacteria. In: Baeuerlein E, editor. Biomineralization. Weinheim, Germany: Wiley-VCH; 2000. pp. 47–60. [Google Scholar]
- 3.Balkwill D, Maratea D, Blakemore R P. Ultrastructure of a magnetotactic spirillum. J Bacteriol. 1980;141:1399–1408. doi: 10.1128/jb.141.3.1399-1408.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazylinski D. Structure and function of the bacterial magnetosome. ASM News. 1995;61:337–343. [Google Scholar]
- 5.Bertani L E, Huangl J S, Weir B A, Kirschvink J L. Evidence for two types of subunits in the bacterioferritin of Magnetospirillum magnetotacticum. Gene. 1997;201:31–36. doi: 10.1016/s0378-1119(97)00424-1. [DOI] [PubMed] [Google Scholar]
- 6.Bertani L E, Weko J, Phillips K V, Gray R F, Kirschvink J L. Physical and genetic characterization of the genome of Magnetospirillum magnetotacticum, strain MS-1. Gene. 2001;264:257–263. doi: 10.1016/s0378-1119(01)00331-6. [DOI] [PubMed] [Google Scholar]
- 7.Bezprozvanny I, Maximov A. PDZ domains: more than just a glue. Proc Natl Acad Sci USA. 2001;98:787–789. doi: 10.1073/pnas.98.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blakemore R P, Maratea D, Wolfe R S. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J Bacteriol. 1979;140:720–729. doi: 10.1128/jb.140.2.720-729.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boucher J C, Martinez-Salazar J, Schurr M J, Mudd M H, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosain cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess J G, Kawaguchi R, Sakaguchi T, Thornhill R H, Matsunaga T. Evolutionary relationships among Magnetospirillumstrains inferred from phylogenetic analysis of 16S-rRNA sequences. J Bacteriol. 1993;175:6689–6694. doi: 10.1128/jb.175.20.6689-6694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A K, Cohen P W, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean A J, Bazylinski D A. Genome analysis of several marine, magnetotactic bacterial strains by pulsed-field gel electrophoresis. Curr Microbiol. 1999;39:219–225. doi: 10.1007/s002849900448. [DOI] [PubMed] [Google Scholar]
- 13.DeLong E F, Frankel R B, Bazylinski D A. Multiple evolutionary origins of magnetotaxis in bacteria. Science. 1993;259:803–806. doi: 10.1126/science.259.5096.803. [DOI] [PubMed] [Google Scholar]
- 14.Eckerskorn C, Mewes W, Goretzki H, Lottspeich F. A new siliconized-glass fiber as support for protein-chemical analysis of electroblotted proteins. Eur J Biochem. 1988;176:509–519. doi: 10.1111/j.1432-1033.1988.tb14308.x. [DOI] [PubMed] [Google Scholar]
- 15.Friedmann E I, Wierzchos J, Ascaso C, Winklhofer M. Special feature: chains of magnetite crystals in the meteorite ALH84001: evidence of biological origin. Proc Natl Acad Sci USA. 2001;98:2176–2181. doi: 10.1073/pnas.051514698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorby Y A, Beveridge T J, Blakemore R P. Characterization of the bacterial magnetosome membrane. J Bacteriol. 1988;170:834–841. doi: 10.1128/jb.170.2.834-841.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbard J A, Lewandowska K B, Hughes M N, Poole R K. Effects of iron-limitation of E. colion growth, the respiratory chains and gallium uptake. Arch Microbiol. 1986;146:80–86. doi: 10.1007/BF00690163. [DOI] [PubMed] [Google Scholar]
- 19.Kirschvink J L, Hagadorn J W. A grand unified theory of biomineralization. In: Baeuerlein E, editor. Biomineralization. Weinheim, Germany: Wiley-VCH; 2000. pp. 139–149. [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lamb J R, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 22.Lenz L L, Dere B, Bevan M J. Identification of an H2–M3-restricted Listeriaepitope: implications for antigen presentation by M3. Immunity. 1996;5:63–72. doi: 10.1016/s1074-7613(00)80310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 24.Matsunaga T, Takeyama H. Biomagnetic nanoparticle formation and application. Supramol Sci. 1998;5:391–394. [Google Scholar]
- 25.Matsunaga T, Tsujimura N, Okamura H, Takeyama H. Cloning and characterization of a gene, mpsA, encoding a protein associated with intracellular magnetic particles from Magnetospirillumsp. strain AMB-1. Biochem Biophys Res Commun. 2000;268:932–937. doi: 10.1006/bbrc.2000.2236. [DOI] [PubMed] [Google Scholar]
- 26.Meldrum F C, Mann S, Heywood B R, Frankel R B, Bazylinski D A. Electron microscopy study of magnetosomes in a cultured coccoid magnetotactic bacterium. Proc. R. Soc. Lond. Ser. B: Biol. Sci. 1993;251:231–236. [Google Scholar]
- 27.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura C, Burgess J G, Sode K, Matsunaga T. An iron-regulated gene. magA, encoding an iron transport protein of Magnetospirillumsp. strain AMB-1. J Biol Chem. 1995;270:28392–28396. doi: 10.1074/jbc.270.47.28392. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura N, Hashimoto K, Matsunaga T. Immunoassay method for the determination of immunoglobulin G using bacterial magnetic particles. Anal Chem. 1991;63:268–272. doi: 10.1021/ac00003a015. [DOI] [PubMed] [Google Scholar]
- 30.Nies D H, Silver S. Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol. 1995;14:186–199. doi: 10.1007/BF01569902. [DOI] [PubMed] [Google Scholar]
- 31.Okamura Y, Takeyama H, Matsunaga T. Two-dimensional analysis of proteins specific to the bacterial magnetic particle membrane from Magnetospirillumsp. AMB-1. Appl Biochem Biotechnol. 2000;84–86:441–446. doi: 10.1385/abab:84-86:1-9:441. [DOI] [PubMed] [Google Scholar]
- 32.Okuda Y, Denda K, Fukumori Y. Cloning and sequencing of a gene encoding a new member of the tetratricopeptide protein family from magnetosomes of Magnetospirillum magnetotacticum. Gene. 1996;171:99–102. doi: 10.1016/0378-1119(95)00008-9. [DOI] [PubMed] [Google Scholar]
- 33.Okuda Y, Y, Fukumori H. Expression and characterization of a magnetosome-associated protein, TPR-containing MAM22, in Escherichia coli. FEBS Lett. 2001;491:169–173. doi: 10.1016/s0014-5793(01)02178-0. [DOI] [PubMed] [Google Scholar]
- 34.Osborn M J, Munson R. Separation of the inner (cytoplasmic) and outer membranes of gram-negative bacteria. Methods Enzymol. 1974;XXXI:642–652. doi: 10.1016/0076-6879(74)31070-1. [DOI] [PubMed] [Google Scholar]
- 35.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 36.Paulsen I T, Saier M H. A novel family of ubiquitous heavy metal ion transport proteins. J Membr Biol. 1997;156:99–103. doi: 10.1007/s002329900192. [DOI] [PubMed] [Google Scholar]
- 37.Ponting C P. Evidence for PDZ domains in bacteria, yeast, and plants. Protein Sci. 1997;6:464–468. doi: 10.1002/pro.5560060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sassoon N, Arie J P, Betton J M. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol Microbiol. 1999;33:583–589. doi: 10.1046/j.1365-2958.1999.01505.x. [DOI] [PubMed] [Google Scholar]
- 40.Schleifer K H, Schüler D, Spring S, Weizenegger M, Amann R, Ludwig W, Köhler M. The genus Magnetospirillum gen. nov., description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticumcomb. nov. Syst Appl Microbiol. 1991;14:379–385. [Google Scholar]
- 41.Schüler D. Characterization of the magnetosome membrane in Magnetospirillum gryphiswaldense. In: Baeuerlein E, editor. Biomineralization. Weinheim, Germany: Wiley-VCH; 2000. pp. 109–118. [Google Scholar]
- 42.Schüler D. Formation of magnetosomes in magnetotactic bacteria. J Mol Microbiol Biotechnol. 1999;1:79–86. [PubMed] [Google Scholar]
- 43.Schüler D, Baeuerlein E. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J Bacteriol. 1998;180:159–162. doi: 10.1128/jb.180.1.159-162.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schüler D, Baeuerlein E. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch Microbiol. 1996;166:301–307. doi: 10.1007/s002030050387. [DOI] [PubMed] [Google Scholar]
- 45.Schüler D, Frankel R B. Bacterial magnetosomes: microbiology, biomineralization and biotechnological applications. Appl Microbiol Biotechnol. 1999;52:464–473. doi: 10.1007/s002530051547. [DOI] [PubMed] [Google Scholar]
- 46.Schüler D, Köhler M. The isolation of a new magnetic spirillum. Zentralbl Mikrobiol. 1992;147:150–151. [Google Scholar]
- 47.Schüler D, Spring S, Bazylinski D A. Improved technique for the isolation of magnetotactic spirilla from a freshwater sediment and their phylogenetic characterization. Syst Appl Microbiol. 1999;22:466–471. doi: 10.1016/S0723-2020(99)80056-3. [DOI] [PubMed] [Google Scholar]
- 48.Schüler D, Uhl R, Baeuerlein E. A simple light-scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol Lett. 1995;132:139–145. [Google Scholar]
- 49.Tang Y P, Dallas M M, Malamy M H. Characterization of the Batl (Bacteroides aerotolerance) operon in Bacteroides fragilis: isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivomodel systems. Mol Microbiol. 1999;32:139–149. doi: 10.1046/j.1365-2958.1999.01337.x. [DOI] [PubMed] [Google Scholar]
- 50.Tempst P, Link A J, Riviere L R, Fleming M, Elicone C. Internal sequence analysis of proteins separated on polyacrylamide gels at the submicrogram level: improved methods, applications and gene cloning strategies. Electrophoresis. 1990;11:537–553. doi: 10.1002/elps.1150110704. [DOI] [PubMed] [Google Scholar]
- 51.Thomas-Keprta K L, Clemett S J, Bazylinski D A, Kirschvink J L, McKay D S, Wentworth S J, Vali H, Gibson E K, McKay M F, Romanek C S. Special feature: truncated hexa-octahedral magnetite crystals in ALH84001: presumptive biosignatures. Proc Natl Acad Sci USA. 2001;98:2164–2169. doi: 10.1073/pnas.051500898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]