Abstract
Gene mutations may affect the fate of many tumors including prostate cancer (PCa); therefore, the research of specific mutations associated with tumor outcomes might help the urologist to identify the best therapy for PCa patients such as surgical resection, adjuvant therapy or active surveillance. Genomic DNA (gDNA) was extracted from 48 paraffin‐embedded PCa samples and normal paired tissues. Next, gDNA was amplified and analyzed by next‐generation sequencing (NGS) using a specific gene panel for PCa. Raw data were refined to exclude false‐positive mutations; thus, variants with coverage and frequency lower than 100× and 5%, respectively were removed. Mutation significance was processed by Genomic Evolutionary Rate Profiling, ClinVar, and Varsome tools. Most of 3000 mutations (80%) were single nucleotide variants and the remaining 20% indels. After raw data elaboration, 312 variants were selected. Most mutated genes were KMT2D (26.45%), FOXA1 (16.13%), ATM (15.81%), ZFHX3 (9.35%), TP53 (8.06%), and APC (5.48%). Hot spot mutations in FOXA1, ATM, ZFHX3, SPOP, and MED12 were also found. Truncating mutations of ATM, lesions lying in hot spot regions of SPOP and FOXA1 as well as mutations of TP53 correlated with poor prognosis. Importantly, we have also found some germline mutations associated with hereditary cancer‐predisposing syndrome. gDNA sequencing of 48 cancer tissues by NGS allowed to detect new tumor variants as well as confirmed lesions in genes linked to prostate cancer. Overall, somatic and germline mutations linked to good/poor prognosis could represent new prognostic tools to improve the management of PCa patients.
Keywords: gene mutations, hereditary cancer‐predisposing syndrome, NGS, prognosis, prostate cancer, signaling pathways
1. INTRODUCTION
Prostate cancer (PCa) is the most common noncutaneous cancer of man in Europe, where the highest incidence of clinically diagnosed PCa in Northern and Western part of Europe was found (Mottet et al., 2021). In absence of early diagnosis, the mortality rate for PCa patients is very high representing about the sixth most fatal cancer in man (Dejous & Krishnan, 2020). Patients with high‐grade disease characterized by T3‐4 stage, lymph node invasion, or an extraprostatic extension have a high‐risk (most of 40%) of disease recurrence after 5–10 years from the diagnosis (Spratt et al., 2018). Currently, the main tool for PCa detection is the analysis of prostate‐specific antigen (PSA) serum levels combined with direct rectal examination (DRE). However, PSA serum detection remains one of the most controversial topics in the urologic literature, since it leads to overdiagnosis and overtreatment of positive subjects (Mottet et al., 2021). Moreover, neither overall survival (OS) nor cancer‐specific survival (CSS) benefits in patients screened by PSA were observed (Mottet et al., 2021). Prostate cancer treatments are dependent on the staging of tumor and includes active surveillance (AS), surgery, hormone therapy, radiotherapy, or a combination of these treatments (Dejous & Krishnan, 2020). Moreover, early diagnosis and disease outcome prediction are crucial points to increase patient OS (Dejous & Krishnan, 2020). Genomic alterations deeply affected cancer biology and disease course in tumors including PCa. In particular, the fusion of the genes ERG and TMPRSS2 is one of most frequent genomic alterations observed in PCa (Gasi Tandefelt et al., 2014). Moreover, somatic gene mutations linked to tumor progression such as oncogenes or tumor suppressor genes were also identified (Gandhi et al., 2018). The detection of gene mutations linked to PCa outcome might improve the knowledge of this tumor increasing prognostic tools and therapeutic options.
2. MATERIALS AND METHODS
2.1. Materials
Disposable RNAse/DNAse free plastic material was purchased by EuroClone. Ion AmpliSeq™ Custom and Community Panels, Ion AmpliSeq™ Library Kits 2.0, Ion Xpress™ Barcode Adapters 1‐96 Kits, Ion PGM™ Hi‐Q™ View OT2 Kit, Ion Sphere Quality Control Kit, Ion PGM™ Hi‑Q™ View Sequencing Kit, Ion 316™ Chip Kit v2 BC, and Qubit® dsDNA HS Assay Kit were obtained from Thermo Fisher Scientifics. Agencourt® AMPure® XP Kit was purchased from Beckman Coulter; QIAmp FFPE tissues kit was obtained from Qiagen.
2.2. Tissue collection
Paraffin‐embedded tumor samples (23 GS6, 11 GS7, 11 GS8, and 3 GS9) from 48 patients underwent to radical prostatectomy in the years 2010–2015 were collected. The diagnosis of cancer samples was evaluated by genitourinary pathologist on hematoxiline and eosine (H&E)‐stained slides. Selected samples (both tumor and normal tissues from the same patient) were cut into 8 × 10 µm sections with the last H&E stained 4 µm sections to confirm tumor cellularity. This is a retrospective study approved by Ethics Committee (no 151095). A written consent regarding tissue analysis and outcome data for all cases enrolled was collected. This study follows the guidelines of Helsinki Declaration.
2.3. Prostate panel design
A prostate cancer‐specific Ion AmpliSeq™ Custom and Community Panel (PC panel) was designed through the AmpliSeq.com program by selecting target regions of 16 genes (APC, AR, ATM, CDK12, CHD1, COL5A1, FOXA1, MED12, KMT2D, OR5L1, PIK3CA, PTEN, RB1, SPOP, TP53, and ZFHX3) that are the more frequently mutated in prostate tumor (Frank et al., 2018; Robinson et al., 2015). The PC panel consists of two DNA primer pools (pool 1: 337 amplicons, pool 2: 331 amplicons) capable to amplified coding regions of maximum 150 bp in length to ensure optimal amplification. All gene information of PC panel was inserted in Table 1.
Table 1.
Genes related to prostate cancer.
| Gene | Name | Chromosome | Exon coverage | Protein | Function |
|---|---|---|---|---|---|
| PIK3CA | Phosphatidylinositol‐4,5‐bisphosphate 3‐kinase catalytic subunit alpha | 3 | 2,5,8,10,21 | PI3K subunit | Cell proliferation migration and survival |
| APC | Adenomatosis Polyposis Coli | 5 | 6,11 | WNT signaling pathway regulator | Tumor Suppressor, cell migration, adhesion, apoptosis |
| CHD1 | Chromodomain helicase DNA binding protein 1 | 5 | 3,12,13,14,18,29,34,35 | ATP‐dependent chromatin‐remodeling factor | Negative regulator of DNA replication |
| COL5A1 | Collagen, type V, alpha 1 | 9 | 3,7,24,33,46 | A component of type V collagen | Cellular component organization, cell adhesion |
| PTEN | Phosphatase and tensin homolog | 10 | 2,3,4,5,6,7,8 | Protein Phosphatase | Tumor suppressor, cell division regulator |
| OR5L1 | Olfactory receptor family 5 subfamily L member 1 | 11 | All coding sequence | G‐protein‐coupled receptor | Sensory transduction |
| ATM | Ataxia telangiectasia mutated | 11 | All coding sequence | Serine/threonine kinase | DNA repair, cell cycle control |
| KMT2D | Lysine methyltransferase 2D | 12 | All coding sequence | Histone methyltransferase | Tumor suppressor |
| RB1 | RB transcriptional corepressor 1 | 13 | 3,7,12,19,23 | transcription repressor | Tumor suppressor |
| FOXA1 | Forkhead box protein A1 | 14 | 2 | DNA‐binding protein | Cofactor for steroid receptor binding |
| ZFHX3 | Zinc finger homeobox 3 | 16 | 2,8,9,10 | Transcription factor | Tumor suppressor |
| TP53 | Tumor protein p53 | 17 | 2,4,5,6,7,8,9,10 | DNA repair regulator | Tumor suppressor |
| CDK12 | Cyclin dependent kinase 12 | 17 | All coding sequence | Cyclin‐dependent kinase | Transcription elongation, DNA repair, and genomic stability regulator |
| SPOP | Speckle type BTB/POZ protein | 17 | 5,6,11 | transcription regulator | Gene transcription modulator |
| AR | Androgen receptor | X | 1,4,5,8 | Hormone receptor | Androgen‐responsive gene regulator |
| MED12 | Mediator complex subunit 12 | X | 4,9,15,26,28,31 | Transcription factor binding | Mediator complex for RNA Polymerase II transcription machinery |
Note: Gene acronym, location, coverage, and function are indicated.
2.4. Genomic DNA extraction, sample enrichment, and NGS sequencing
Genomic DNA (gDNA) was extracted with QIAmp FFPE tissues kit (Qiagen) according to the manufacturer's instructions. gDNA quantity and quality were assessed using the Qubit ® 2.0 photometer (Thermo Fisher Scientific) and the Qubit ® dsDNA HS Assay Kit. gDNA was diluted at the final concentration of 5 ng/μl with deionized water. Libraries were prepared from 10 ng of gDNA using the PC Panel. Overall, gDNA was subjected to library preparation according with Ion Ampliseq Libreries kit 2.0 (Thermo Fisher Scientific). Target regions were initially amplified (20 PCR cycles) with a multiple PCR; after thermal cycling amplification, amplicons produced from pool 1 and pool 2 were combined and partially digested. Next, they were subjected to ligation of barcoded adapters and purified. Before sequencing, libraries were quantified using the Agilent™ 2100 Bioanalyzer™ (Agilent Genomics) and dilute to 100 pM. Barcoded libraries, combined for maximizing chip use, labor, and costs, were clonally amplified by emulsion PCR using OneTouch™ Instrument (Thermo Fisher Scientific) and enriched by the OneTouch™ ES Instrument (Thermo Fisher Scientific) using the Ion PGM™ Hi‐Q™ View OT2 Kit, following the manufacturer's instructions. Library quality control was performed using the Ion Sphere Quality Control Kit according to the manufacturer's instructions, ensuring that 10%–30% of template positive Ion Sphere particles (ISP) were targeted in the emPCR reaction. Finally, sequencing was performed on the Ion PGM™ (Thermo Fisher Scientific) with the Ion PGM™ Hi‐Q View™ Sequencing Kit (Thermo Fisher Scientific), loading barcoded samples (8 samples) into a 316 v.2 BD chip (Rothberg et al., 2011).
2.5. Data elaboration
Sequencing data analysis was conducted by using Torrent Suite software v. 5.0 (Thermo Fisher Scientific). The alignment against a reference genome (hg19) was performed by using the Torrent Mapping Alignment Program after low‐quality reads removal and adapter sequences trimming. The Torrent Variant Caller plugin was used to identify variations from the reference sequence. To identify pathogenic variations, mutations that did not affect the protein‐coding regions (intronic, 3′ and 5′ untranslated region [UTR] variations, and silent exonic mutations) were filtered out. All detected variants were manually reviewed with the Integrative Genomics Viewer (IGV V.2.1, Broad Institute). Genomic Evolutionary Rate Profiling (GERP) tool was used to predict the effect of missense mutations on the protein and calculate their conservation scores (Deshpande et al., 2018). This analysis was improved by using ClinVar and Varsome databases. For high confidence detection of somatic mutations present in heterogeneous cancer tissues, samples with coverage less than 100× and mutation frequency lower than 5% were excluded.
3. RESULTS
3.1. Detection of gene mutations by NGS analysis
Genomic sequences of 48 PCa tissues and their paired normal samples were subjected to NGS analysis for identifying disease‐causing mutations. After row data processing and the exclusion of synonymous variants, 312 mutations (5 small deletions, 1 duplication, and 306 SNVs) widespread along the exonic sequences of 16 genes related to prostate carcinoma were detected (Table S1). Three deletions were in frame, while the other two led to transcript frameshift as well as the only duplication observed in our cohort. Among the 306 SNVs, three were stop codon while the remaining 303 were missense mutations. Overall, we found 77 germline and 235 somatic mutations. Sixty‐six germline mutations were considered polymorphic variants, while the other 11 were considered possible hereditary‐causing cancer lesions. Regarding the 235 somatic mutations, 67 were classified as benign, 28 as uncertain significance, and 140 as likely pathogenic (Table S1). As shown in Figure 1, the percentage distribution of all mutations detected in genes of the PC panel was the following: KMT2D (26.45), FOXA1 (16.13), ATM (15.81), ZFHX3 (9.35), TP53 (8.06), APC (5.48), MED12 (3.23), OR5L1 (3.23), SPOP (2.58), AR (2.26), COL5A1 (1.94), CHD1 (1.94), CDK12 (1.61), RB1 (0.97), PTEN (0.65), and PIK3CA (0.32).
Figure 1.
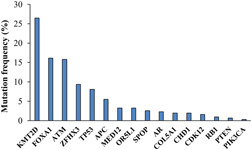
Mutation frequency of genes related to prostate cancer detected in a cohort of 48 subjects by NGS analysis. The most mutated genes are KMT2D, FOXA1, and ATM, while in RB1, PTEN, and PIK3CA few variants were detected. NGS, next‐generation sequencing
3.2. Recurrent mutations
We identified some recurrent mutations in different subjects (Figure 2). In particular, the V1822D (n = 3) and G2502S (n = 4) substitutions in APC were considered benign variants.
Figure 2.
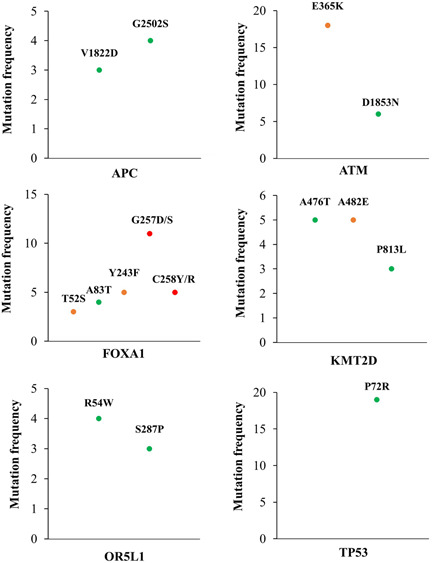
Recurrent mutations found in patients with prostate cancer. Genes with recurrent pathogenic mutations are ATM and FOXA1, while variants found in APC, OR5L1, KMT2D, and TP53 are likely benign except for the A482E substitution in KMT2D, which is considered as uncertain significance. Red dots = likely pathogenic; orange = uncertain significance; green = likely benign.
The mutation E365K (n = 18) in ATM was processed as uncertain significance and showed a high frequency in our cohort (37.5%). In this gene the benign variant D1853N (n = 6) was also identified. Recurrent mutations were also detected in FOXA1; the variants Y243F (n = 5) and T52S (n = 3) were considered as uncertain significance, while A83T (n = 4) was processed as benign. Conversely, the variants G257D/S (n = 11) and C258Y/R (n = 5) were supposedly pathological mutations.
In KMT2D, the variants A476T (n = 5) and P813L (n = 3) were benign while the mutation A482E (n = 5) was considered as uncertain significance. Finally, the benign variants R54W (n = 4) and S287P (n = 3) in OR5L1 as well as P72R in TP53 (n = 19) were also identified. Interestingly, the mutation P72R was the germline variant most frequent our cohort, which is present in approximately 40% of cases.
3.3. Hotspot mutations
We found hotspot mutations in different genes (Figure 3); in particular, the most of MED12 variants (91%) lay in the leucine‐serine‐rich domain, where three of these were close together and the others widespread along this domain. Regarding ZFHX3, hotspot mutations were detected in the protein segment between the fifth and sixth zinc finger domain and about 24% of these variants hit few codons (amino acids 789–824).
Figure 3.
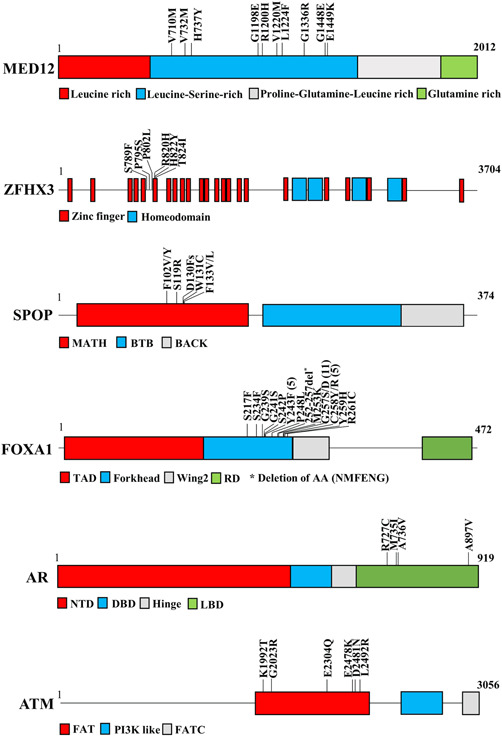
Hotspot regio.ns detected in MED12, ZFHX3, SPOP, FOXA1, AR, and ATM. Genes are represented as bars or boxes not to scale; protein domains and mutations are indicated.
Hotspot mutations in SPOP were also discovered; approximatively 87% of these variants lay very close together in the MATH domain. Interestingly, all the seven lesions found in the MATH domain were considered pathogenic, while the only one detected outside (E334D) was a polymorphism. Most of FOXA1 mutations (62%) were clustered in a short protein segment (AA 217–261) of the Forkhead domain. In particular, all lesions were classified as pathogenic except the S217F and Y243F substitutions that were considered as uncertain significance.
We found that about 66% of mutations in AR were located in the ligand‐binding domain (LBD) and were characterized as pathogenic lesions. Three of these were close together, while the fourth was located at the end of LBD. Finally, we discovered several lesions (12%) localized in the FRAP‐ATM‐TRRAP (FAT) domain of ATM, where three of these variants lay very close together while the others were spread along this motif.
3.4. Linkage between gene mutation and disease outcome
Mutations found in our cohort were matched with patient follow‐up data. As shown in Figure 4, the percentage of mutated genes between the group with good and poor prognosis was different. The mutation frequency of MED12, AR, CHD1, OR5L1, and KTM2D was lower in patients with poor prognosis. In particular, lesions found in KMT2D were much more common in the group of patient with good prognosis. Conversely, mutations detected in FOXA1, SPOP, ATM, and TP53 were mainly found in patients with poor prognosis, while the mutation percentage of APC, COL5A1, ZFHX3, and CDK12 was substantially unchanged. In more detail, different FOXA1 variants laying in the forkhead domain were linked to biochemical recurrence as well as those found in SPOP. Moreover, the truncating lesions R805X and L2692X as well as the substitution R3008H in ATM were associated with poor prognosis. Similarly, lesions in TP53 such as Y163H, T172Ifs, and R267P were associated with both higher Gleason score and tumor progression (Table 2).
Figure 4.
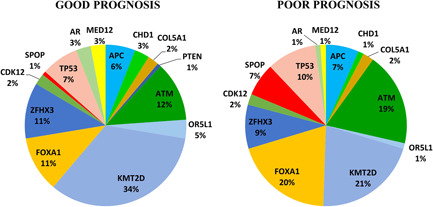
Pie chart showing the percentage of mutated genes in PCa patients with good (n = 20) or poor (n = 19) prognosis. Gene mutation percentage in PCa patients with poor or good prognosis is indicated. In more detail, patients with poor prognosis show increased mutation frequency in FOXA1, SPOP, TP53, and ATM. Conversely, the mutation frequency in KMT2D, OR5L1, CHD1, AR, and MED12 is lower. The percentage of mutated genes for each group (good or poor) was calculated in comparison with all mutations (100%) detected in patients of that group. Mutations in RB1 and PIK3CA were excluded because they are found only in patients without follow‐up data. PCa, prostate cancer
Table 2.
Clinical parameters of 48 prostate cancer patients.
| Acronym | Age | Diagnosis | PSA value | Surgical resection | Outcome | Therapy | Mutated genes | Notes |
|---|---|---|---|---|---|---|---|---|
| B4536 | 62 | Prostate adenocarcinoma Gs 6 (3 + 3) | 0.41 | 2012 | Biochemical recurrence | Rescue radiation therapy | ATM, FOXA1 | None |
| B4972 | 66 | Prostate adenocarcinoma Gs 6 (3 + 3) | 0.02 | 2012 | No metastasis | None | KMT2D | Very low mutation frequency |
| B6393 | Deceased | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2012 | Deceased for oligodendroglioma no PCa metastasis | None | ATM, APC | Very low mutation frequency |
| B6998 | 70 | Prostate adenocarcinoma Gs 6 (3 + 3) and colon adenocarcinoma | Undetectable | 2012 | Abdominal lymph node metastasis | Chemotherapy | ZFHX3 | Very low mutation frequency |
| B3059 | 71 | Prostate adenocarcinoma Gs 6 (3 + 3) | NA | 2012 | Biochemical recurrence | Rescue radiation therapy | ATM, ZFHX3 | Very low mutation frequency |
| B1845 | 71 | Prostate adenocarcinoma Gs 6 (3 + 3) | NA | 2013 | No follow up | Unavailable data | none | No pathological mutations were detected |
| B992 | 80 | Prostate adenocarcinoma Gs 6 (3 + 3) and urothelial carcinoma | NA | 2013 | No follow up | Unavailable data | ATM, KMT2D, TP53 | ATM germline |
| B2726 | 75 | Prostate adenocarcinoma Gs 6 (3 + 3) and colon adenocarcinoma | 0.01 | 2013 | Biochemical recurrence | Rescue radiation therapy | KMT2D, SPOP | SPOP (hot spot) |
| B3082 | 77 | Prostate adenocarcinoma Gs 6 (3 + 3) | NA | 2013 | Biochemical recurrence | Rescue radiation therapy | FOXA1 | Very low mutation frequency |
| B2508 | 72 | Prostate adenocarcinoma Gs 6 (3 + 3) | NA | 2013 | No follow up | Unavailable data | none | No pathological mutations were detected |
| B501 | 73 | Prostate adenocarcinoma Gs 6 (3 + 3) | NA | 2011 | No follow up | Unavailable data | CHD1, APC, OR5L1, ATM, KMT2D, RB1, FOXA1, ZHFX3, SPOP, AR, MED12 | Multiple gene mutations |
| B8935 | 72 | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2011 | No metastasis | None | MED12, FOXA1 | Multiple mutations of FOXA1 gene |
| B1387 | 81 | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2011 | No metastasis | None | CHD1, COL5A1, KMT2D, ZFHX3, MED12, FOXA1 | Multiple gene mutations |
| B1753 | 68 | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2011 | No metastasis | None | ATM, KMT2D, ZFHX3, CDK12 | Very low mutation frequency |
| B1806 | 77 | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2011 | Biochemical recurrence | Rescue radiation therapy | ATM, KMT2D, FOXA1 | None |
| B8502 | 63 | Prostate adenocarcinoma Gs 6 (3 + 3) | NA | 2011 | No metastasis | None | APC, ATM, KTM2D, CDK12, FOXA1 | Multiple mutations of KTM2D |
| B6265 | 78 | Prostate adenocarcinoma Gs 6 (3 + 3) | NA | 2011 | No follow up | Unavailable data | KTM2D, FOXA1, ATM | None |
| B7149 | 85 | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2011 | No metastasis | None | none | No pathological mutations were detected |
| B7595 | 67 | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2011 | No metastasis | None | KMT2D | Very low mutation frequency |
| B7487 | 78 | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2011 | No metastasis | None | FOXA1 | None |
| B7756 | 75 | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2011 | No metastasis | None | none | No pathological mutations were detected |
| B7360 | 81 | Prostate adenocarcinoma Gs 6 (3 + 3) | Undetectable | 2011 | No metastasis | None | ATM, TP53, MED12 | None |
| B8752 | 78 | Prostate adenocarcinoma Gs 7 (3 + 4) | Undetectable | 2011 | No metastasis | Rescue radiation therapy | ATM | ATM germline |
| B8456 | 63 | Prostate adenocarcinoma Gs 7 (3 + 4) | NA | 2011 | No metastasis | None | none | No pathological mutations were detected |
| B778 | 80 | Prostate adenocarcinoma Gs 7 (3 + 4) | NA | 2011 | Biochemical recurrence | Rescue radiation therapy | ATM, FOXA1 | KMT2D germline |
| B8135 | 83 | Prostate adenocarcinoma Gs 7 (3 + 4) | NA | 2011 | No metastasis | None | PTEN, ATM | ATM germline |
| B7970 | 81 | Prostate adenocarcinoma Gs 7 (3 + 4) | Undetectable | 2011 | No metastasis | None | CDK12, AR, FOXA1 | Very low mutation frequency |
| B8234 | 74 | Prostate adenocarcinoma Gs 7 (3 + 4) | Undetectable | 2011 | Biochemical recurrence | Rescue radiation therapy | KMT2D, MED12, FOXA1 | None |
| B6286 | 73 | Prostate adenocarcinoma Gs 7 (3 + 4) | Undetectable | 2011 | Biochemical recurrence | Rescue radiation therapy | ATM, SPOP, FOXA1 | SPOP (hot spot) |
| B6547 | 78 | Prostate adenocarcinoma Gs 7 (3 + 4) | Undetectable | 2011 | Biochemical recurrence | Rescue radiation therapy | CHD1, ZFHX3, TP53, SPOP, FOXA1 | SPOP (hot spot) |
| B6607 | 78 | Prostate adenocarcinoma Gs 7 (3 + 4) | Undetectable | 2011 | Biochemical recurrence | Rescue radiation therapy | APC | Very low mutation frequency |
| B6055 | 69 | Prostate adenocarcinoma Gs 7 (3 + 4) | NA | 2011 | No follow up (alive) | Unavailable data | KMT2D, FOXA1 | None |
| B6395 | 79 | Prostate adenocarcinoma Gs 7 (3 + 4) | Undetectable | 2011 | No metastasis | None | KMT2D, FOXA1 | KMT2D germline |
| B6820 | 75 | Prostate adenocarcinoma Gs 7 (3 + 4) | NA | 2011 | No follow up (alive) | Unavailable data | PI3KCA, ZFHX3, PTEN | None |
| B6224 | 72 | Prostate adenocarcinoma Gs 8 (3 + 5), basocellular carcinoma and squamous cell carcinoma (skin) | NA | 2011 | Lymph node metastasis; total androgen blockade | LHRH analog | APC, KMT2D, ATM, FOXA1 | None |
| B8118 | 77 | Prostate adenocarcinoma Gs 8 (4 + 4) | NA | 2011 | Follow up until 2013 | Postsurgical radiotherapy and hormone therapy | MED12, FOXA1, RB1 | None |
| B8519 | 79 | Prostate adenocarcinoma Gs 8 (4 + 4) | NA | 2010 | No follow up (alive) | Unavailable data | COL5A1, ATM, KMT2D, FOXA1 | None |
| B5172 | 65 | Prostate adenocarcinoma Gs 8 (3 + 5) | NA | 2012 | No metastasis | Postsurgical radiotherapy and hormone therapy | ATM | Very low mutation frequency |
| B1658 | 77 | Prostate adenocarcinoma Gs 8 (3 + 5)and basocellular carcinoma | NA | 2013 | Biochemical recurrence | Rescue radiation therapy | ATM, SPOP | SPOP (hot spot) |
| B2325 | 71 | Prostate adenocarcinoma Gs 8 (4 + 4) | NA | 2013 | Lymph node metastasis | Unavailable data | ATM, SPOP, KTM2D, AR | SPOP (hot spot) |
| B779 | 77 | Prostate adenocarcinoma Gs 8 (5 + 3) | NA | 2013 | Bone metastasis; total androgen blockade | Vertebral radiotherapy | ATM | ATM germline |
| B2471 | 70 | Prostate adenocarcinoma Gs 8 (5 + 3) | NA | 2013 | Biochemical recurrence | Rescue radiation therapy | ATM, SPOP | ATM germline, SPOP (hot spot) |
| B3064 | 77 | Prostate adenocarcinoma Gs 8 (3 + 5) and squamous cell carcinoma (palate) | NA | 2013 | No metastasis | Postsurgical radiotherapy | FOXA1 | none |
| B6007 | 61 | Prostate adenocarcinoma Gs 8 (4 + 4) and b cell lymphoma cutaneous | NA | 2013 | No metastasis; total androgen blockade | Postsurgical radiotherapy | APC, ATM, KMT2D | Very low mutation frequency |
| B4441 | Deceased | Prostate adenocarcinoma Gs 8 (4 + 4) | NA | No surgery | Bone and visceral metastases | Bone radiotherapy, hormone therapy, Cabazitaxel | ATM, TP53 | TP53 germline |
| B435 | Deceased | Prostate adenocarcinoma Gs (5 + 4), urothelial carcinoma and squamous cell carcinoma (lung) | NA | 2013 | Lung cancer relapse | Chemotherapy for lung cancer | KMT2D | KMT2D germline |
| B2777 | 62 | Prostate adenocarcinoma Gs 8 (4 + 5) | NA | 2013 | Lymph node metastasis | Hormone therapy, radiotherapy | TP53 | None |
| B47 | Deceased | Prostate adenocarcinoma Gs (4 + 5), squamous cell carcinoma (larynx) and acinar lung adenocarcinoma | NA | 2014 | Lymph node metastasis | Hormone therapy, radiotherapy | TP53, CDK12 | CDK12 germline |
Note: Patient features, outcome, and putative causing‐disease mutated genes are indicated.
Abbreviations: NA, not available; PSA, prostate‐specific antigen.
3.5. Germline mutations and cancer familiarity
We detected different germline variants with likely pathological significance and possible hereditary predisposing‐cancer syndrome in our PCa cohort. In particular, these germline mutations were observed in 10 patients (about 20%) and hit several genes including ATM, KMT2D, TP53, and CDK12. Many germline mutations were found in cases with metastasis and high Gleason score. In fact, of the 10 patients with germline variants, two had a Gleason score 9, three 8, four 7, and only one subject 6. The germline variants R3008H and R805X in ATM as well as the substitution P1275L in CDK12 correlated with cancer familiarity. In particular, we found that the mother of the case carrying the R3008H substitution suffered for breast cancer, while the patient carrying the truncating mutation R805X showed a severe cancer familiarity. His father suffered for gastric carcinoma, while his mother was diagnosed with lung cancer. In addition, two brothers died for lung carcinoma and a sister was deceased for blood cancer (Figure 5). Finally, the mother of the case with the P1275L substitution in CDK12 suffered for breast cancer. No hereditary cancer predisposition linked to the germline mutations K1992T, G2023R, and L2492R in ATM as well as R466C, R5229H, and S5357T in KMT2D were observed (Table 3).
Figure 5.
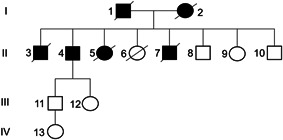
Pedigree of a case with the germline mutation R805X in ATM. Subjects 1 and 2 are deceased for gastric and lung cancer, respectively; Cases 3 and 7 are deceased for lung carcinoma and Subject 5 is dead for a hematological disease. The proband (Case 4) is alive and he suffered from prostate cancer, cholangiocarcinoma, melanoma, and two lung cancers.
Table 3.
Patients carrying mutations linked to hereditary predisposing syndrome
| Sample ID | Gene | Mutation | Frequency (%) | AA change | dbSNP | Mutation pathogenicity | Outcome | Familiarity | References |
|---|---|---|---|---|---|---|---|---|---|
| B8752 | ATM | G6067A | 48.5 | G2023R | rs11212587 | Uncertain | Alive (no metastasis) | None | Tsaousis et al. (2019) |
| B8135 | ATM | T7475G | 49.2 | L2492R | rs56399857 | Uncertain | Alive (no metastasis) | None | Tsaousis et al. (2019) |
| B2471 | ATM | G9023A | 49.8 | R3008H | rs587781894 | Pathogenic | Alive (biochemical recurrence) | Mother with breast cancer | Paglia et al. (2010) |
| B992 | ATM | A5975C | 45.9 | K1992T | rs150757822 | Uncertain | ND | ND | Tsaousis et al. (2019) |
| B779 | ATM | C2413T | 48.3 | R805X | none | Pathogenic | Bone metastasis | Mother with lung cancer and father with gastric carcinoma | Karlsson et al. (2021) |
| B6395 | KMT2D | G16070C | 46.1 | S5357T | none | Uncertain | Alive (no metastasis) | None | None |
| B778 | KMT2D | C1396T | 45.9 | R466C | rs201512665 | Uncertain | Alive (biochemical recurrence) | None | None |
| B435 | KMT2D | G15686A | 49.8 | R5229H | rs201628357 | Uncertain | Deceased (lung cancer) | None | None |
| B4441 | TP53 | G800C | 56.1 | R267P | rs587780075 | Likely pathogenic | Deceased (bone metastasis) | ND | Giacomelli et al. (2018) |
| B47 | CDK12 | C3824T | 47.5 | P1275L | rs34070318 | Uncertain | Deceased (lung cancer and laryngeal carcinoma) | Mother with breast cancer | Jiang et al. (2018); Pratz et al. (2016) |
Note: Mutations, patient outcomes, and familiarity are included.
4. DISCUSSION
The most common alteration found in prostate cancer is the fusion between the androgen‐regulated TMPRSS2 gene and ERG oncogene which occurs in approximately 50% of cases (Alvarez‐Cubero et al., 2017). Nevertheless, it has been reported that translocations involving the ETS family members alone are not sufficient to induce prostate neoplastic transformation and additional alterations such as PTEN and TP53 loss of function could affect the clinical subtype of PCa (Shtivelman et al., 2014). Moreover, the fusion TMPRSS2‐ERG was mainly found in early stage of disease (Yamoah et al., 2021). It is mutually exclusive with other alterations including SPOP and CHD1 loss of function, indicating that TMPRSS2‐ERG negative prostate cancers progress by different tumorigenic processes or represent different cellular subtypes (Shtivelman et al., 2014; Yamoah et al., 2021; Zhu et al., 2021). Thus, the use of new powerful technologies in particular NGS could facilitate the discovery of new somatic and germline mutations improving prognosis and therapeutic response (Alvarez‐Cubero et al., 2017).
The analysis of gene variants in our prostate cancer cohort by NGS shows multiple mutations in different genes that may affect signaling pathways involved in prostate carcinogenesis. In particular, we have analyzed the impact of mutations on several biological processes linked to DNA instability and proliferative signals as well as germline variants associated with hereditary cancer syndrome.
4.1. DNA repair network
Many genes including ATM, CDK12, SPOP, and CHD1 belonging to DNA repair machinery are mutated in PCa and their dysfunction causes genomic instability. Mutations in ATM including the FAT domain were found in PCa indicating that the dysfunction of this kinase may affect the fate of this tumor (Warner et al., 2021). However, these observations are debated since a recent study reports that ATM loss of function is not directly associated with worse outcomes, even if lesions of ATM increase the genomic instability (Neeb et al., 2021). We have detected several mutations of ATM lying in the FAT domain that does not correlate with poor prognosis in our cohort. They are already detected in breast cancer and chronic lymphocytic leukemia (Austen et al., 2007; Bernstein et al., 2010; Podralska et al., 2018) suggesting that these variants might affect cancer development. Outside the FAT domain, we have identified other mutations including the missense variant E365K, processed as uncertain significance, that is very frequent in our cohort, but does not correlate with cancer progression. Conversely, the truncating lesions R805X and L2692X as well as the variant R3008H, defined as pathogenic, are linked to poor prognosis. ATM mutations could alter the DNA damage response (DDR) machinery leading to genomic instability and acquisition of subsequent mutations that could affect prostate carcinogenesis. In different patients with ATM mutations, we have detected lesions in other genes including ZFHX3, FOXA1 and SPOP that are frequently mutated in PCa patients. In particular, the dysfunction of SPOP, that is, another gene implicated in DNA repair is associated with cancer progression (García‐Flores et al., 2014; Ma et al., 2018). The analysis of SPOP variants shows that all pathogenic mutations are localized in a hotspot region within the MATH domain, which is responsible for substrate binding (Ma et al., 2018). Mutations of residues F102, S119, W131, and F133 are already observed in PCa (Barbieri et al., 2012; Boysen et al., 2015; Ma et al., 2018), while the lesion D130fs has never been detected before. The linkage between SPOP mutations and poor prognosis is not well defined, because some authors report that the impairment of SPOP function is associated with less adverse pathologic features and a favorable prognosis (Liu et al., 2018). Our observations indicate that all SPOP pathogenic lesions are associated with patients that have developed biochemical recurrence or lymph node metastasis, but they do not correlate with the most serious cases.
No linkage between CHD1 and CDK12 mutations and cancer progression has been observed in our cohort except for the germline variant P1275L in CDK12 that will be discussed later.
4.2. AR signaling dysfunction
The alteration of androgen receptor‐regulated signaling may affect prostate cancer development and progression. In fact, AR point mutations range from 15% to 30% of patients with metastatic PCa (Fujita & Nonomura, 2019). In our cohort, we have detected the likely pathogenic mutations R727C, M735I, and A736V in AR that are localized in LBD domain. The substitution R727C was also found in patients with 46 XY disorders of sex development (DSD) (Ittiwut et al., 2017), while the other two are new variants. It was reported that the relevance of AR mutations in patients with advanced PCa remains unclear (Eisermann et al., 2013). In this study, we have not found AR mutations associated with poor prognosis. On the other hand, most AR lesions linked to worse outcomes are splice variants (AR‐Vs), which are constitutively activated by the truncation of the COOH‐terminal domain (Antonarakis et al., 2016).
Mutations of FOXA1, a protein that functions as a pioneer factor to facilitate AR transactivation and PCa growth (Zhao et al., 2014), are very frequent in our cohort. FOXA1 is a transcription factor that modulates AR‐driven transcription and mutations strictly affected residues of the Forkhead domain in PCa (Barbieri et al., 2012). Consistently, the most of FOXA1 mutations detected in our cases lie in a hotspot region of the forkhead domain. M253K, C258Y/R, Y259H, and R261C substitutions were already described (Adams et al., 2019; Barbieri et al., 2012; Ritter et al., 2020), but the other lesions found in this domain are novel mutations. Variants of forkhead domain likely cause the alteration of protein function leading to cancer development and progression (Adams et al., 2019). Moreover, mutations in this region promote PCa progression regulating the expression of genes that mediate EMT and metastasis (Gao et al., 2019). Furthermore, it was observed that FOXA1 mutations are associated with a worse clinical outcome (Shah & Brown, 2019). In our cases, most of the mutations found in forkhead domain of FOXA1 are associated with biochemical recurrence.
4.3. Tumor suppressor proteins
Many tumors including prostate cancer rise, develop, and expand due to mutation in tumor suppressor genes including KMT2D, PTEN, RB1, TP53, and ZFHX3. KTM2D is the most mutated gene in our cohort. Eighty‐three mutations were detected in this gene suggesting that the dysfunction of this protein may affect prostate carcinogenesis. In fact, it is emerging that this gene is one of the most frequently mutated in a variety of tumors including PCa (Guo et al., 2013). Moreover, mutations in KMT2D are more frequent in metastatic than in primary tumors (Testa et al., 2019). In contrast to these observations, we report that mutations of KTM2D are prevalent in PCa patients with good outcome. On the other hand, the most of KMT2D mutations found in our cases have a low frequency or are classified as benign except the somatic stop gain E568X that is associated with biochemical recurrence. The germline variants R466C, R5229H, and S5357T will be discussed later.
Most of PTEN and RB1 mutations found in our cohort are variants with uncertain significance and do not correlate with tumor progression. We have also identified the pathogenic truncating lesion C211Lfs in PTEN, but unfortunately, no follow‐up data for the case carrying this variant are available.
Many mutations in ZFHX3, a tumor suppressor gene frequently mutated in prostate cancer (Sun et al., 2005, 2015), were identified. These are mainly clustered in a region lying between the fifth and sixth zinc‐finger domain. It has been reported that the inactivation of ZFHX3 may correlate with tumor aggressiveness, especially in subjects with the deletion of chromosome 16q that contains this gene (Sun et al., 2005). No linkage between ZFHX3 mutations and poor prognosis we have observed, probably because several variants are considered benign or with uncertain significance while those characterized as likely pathogenic have low frequency and could be irrelevant for disease progression. On the contrary, different pathogenic mutations in TP53 correlate with worse outcomes in our PCa cases; in particular, the mutations Y163H, T172Ifs, and R267P were detected in patients with metastasis. The mutation V274A also considered pathogenic is not linked to cancer progression, however, it was predominantly found in breast cancer (Végran et al., 2013). Lesions in TP53 are associated with more aggressive disease not only in PCa but also in many other solid tumors (Mateo et al., 2020; Vodicka et al., 2021) and our data support these observations.
4.4. Cell growth and invasion
We have analyzed mutations in genes associated with cell proliferation and motility such as COL5A1, PIK3CA, APC, and MED12. Mutations found in PIK3CA, COL5A1, and APC have not a significant impact on patient outcomes in our cohort. Regarding MED12, it was reported that mutations in this gene are frequent in PCa (Barbieri et al., 2012). We have detected variants of MED12 in 7 of 48 patients (14.5%). All pathogenic mutations detected in MED12 lie in the leucine‐serin‐rich domain except the variant A157T, suggesting that this protein region may be involved in the tumorigenesis of PCa. Actually, this domain is strongly conserved and mutations located inside this region are associated with prostate tumor (Barbieri et al., 2012; Kämpjärvi et al., 2016). Interestingly, some studies report that the missense mutation L1224F is a recurrent variant in prostate cancer (Barbieri et al., 2012), while others did not observe this lesion in any of their cases (Stoehr et al., 2013). We have found this mutation solely in one subject with a low tumor stage and without metastasis. Moreover, MED12 mutations found in our cohort do not correlate with cancer progression in most of cases, suggesting that MED12 dysfunction could not be associated with tumor metastasis.
4.5. Germline mutations and cancer familiarity
We have searched germline mutations that could be associated with inherited cancer. Ten variants in heterozygous form also expressed in normal tissue were detected in ATM, KMT2D, TP53, and CDK12. Germline mutations of ATM such as K1992T, G2023R, and L2492R have uncertain significance (Tsaousis et al., 2019); therefore, their role in hereditary cancer is not well defined. We have observed that cases carrying G2023R and L2492R mutations have neither metastasis nor cancer familiarity, while no information on clinical outcome for the patient with the K1992T variant is available. On the contrary, the subject carrying the germline mutation R3008H has developed biochemical recurrence and his mother suffered from breast cancer. Accordingly, this lesion has been already associated with hereditary breast cancer (Paglia et al., 2010), but in PCa it was never found before. Interestingly, one case carrying the truncating variant R805X in ATM has suffered for five different cancers and shows a severe cancer familiarity. In particular, mother and father are deceased for lung and gastric cancer, respectively. Furthermore, four siblings are deceased; two brothers with lung cancer, one sister for leukemia, and the second for a disease not linked to cancer (pedigree of Figure 5). The proband is alive and, in addition to prostate cancer, two lung tumors, one cholangiocarcinoma, and one melanoma were diagnosed. Currently, the truncating variant R805X has been described only in breast cancer, however truncating mutations in ATM such as stop gain or frameshift were also found in familial PCa (Karlsson et al., 2021). In addition, germline mutations of ATM are associated with gastric cancer as well as lung carcinoma (Huang et al., 2015; Parry et al., 2017). Taken together, these observations suggest that the lesion R805X could be associated with a high risk to develop tumors; moreover, ATM pathogenic germline lesions could be considered possible markers for familial cancer.
We have found germline mutations also in KMT2D; the variants R466C, R5259H, and S5357T are classified as uncertain significance and none of these is associated with familial cancer. However, patients carrying the R466C and R5229H substitutions have developed biochemical recurrence and lung cancer, respectively. Consistently, it is known that KMT2D is among the most highly inactivated epigenetic modifiers in lung cancer (Alam et al., 2020). Interestingly, in a subject with advanced PCa and bone metastasis, we have detected the germline mutation R267P in TP53. This variant causes the dysfunction of TP53 protein and was already detected in both liver and lung carcinoma (Giacomelli et al., 2018). Unfortunately, this patient is deceased and information about hereditary cancer predisposition is no longer available. Finally, we identified the germline mutation P1275L of CDK12 in a case deceased for multiple cancers. In addition to PCa, this patient has suffered from lung carcinoma and laryngeal cancer; moreover, his mother is deceased of breast cancer. Importantly, in this patient, the somatic mutation Y163H in TP53 that is associated with lung cancer was also detected (Vega et al., 1997). The germline variant P1275L was observed in myeloproliferative neoplasms and in EGFR‐mutated tumors (Jiang et al., 2018; Pratz et al., 2016), but its role in both prostate and breast cancer should be further investigated.
5. CONCLUSIONS
NGS analysis performed in 48 normal and corresponding prostate cancer tissues has allowed the detection of several lesions in TP53, ATM, FOXA1, and SPOP associated with cancer progression. Moreover, we described first‐time hotspot mutations in ZFHX3 and novel mutations in the hotspot region of FOXA1. Furthermore, this study has led to the identification of different germline mutations, some of which in cases with familial cancer were found.
Our data indicate that mutations detected mainly in ATM and TP53 could be used as biomarkers for poor prognosis in prostate cancer. Moreover, mutations altering pathways involved in prostate carcinogenesis including FOXA1‐, SPOP‐ and ATM‐regulated signals could be useful to discover new therapeutic targets for the treatment of metastatic PCa.
AUTHOR CONTRIBUTIONS
Gianluca Aguiari and Alessandra Mangolini designed the project. Christian Rocca, Carmelo Ippolito, Lucio Dell' Atti, Giovanni Lanza, and Roberta Gafà collected the samples and managed patient follow up. Alessandra Mangolini and Nicoletta Bianchi performed the experiments. Alessandra Mangolini, Cristian Bassi, and Gianluca Aguiari analyzed the data. Paolo Pinton, Massimo Negrini, and Gianluca Aguiari discussed the experiments. Gianluca Aguiari wrote the manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supporting information.
ACKNOWLEDGMENT
This study was supported by University of Ferrara local Funds (FAR 2018‐2020) and Ricerca Finalizzata 2011‐2012 Grant: GR‐2011‐02346964. Open Access Funding provided by Universita degli Studi di Ferrara within the CRUI‐CARE Agreement.
Mangolini, A. , Rocca, C. , Bassi, C. , Ippolito, C. , Negrini, M. , Dell'Atti, L. , Lanza, G. , Gafà, R. , Bianchi, N. , Pinton, P. , & Aguiari, G. (2022). Detection of disease‐causing mutations in prostate cancer by NGS sequencing. Cell Biology International, 46, 1047–1061. 10.1002/cbin.11803
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- Adams, E. J. , Karthaus, W. R. , Hoover, E. , Liu, D. , Gruet, A. , Zhang, Z. , Cho, H. , DiLoreto, R. , Chhangawala, S. , Liu, Y. , Watson, P. A. , Davicioni, E. , Sboner, A. , Barbieri, C. E. , Bose, R. , Leslie, C. S. , & Sawyers, C. L. (2019). FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature, 571, 408–412. 10.1038/s41586-019-1318-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam, H. , Tang, M. , Maitituoheti, M. , Dhar, S. S. , Kumar, M. , Han, C. Y. , Ambati, C. R. , Amin, S. B. , Gu, B. , Chen, T. Y. , Lin, Y. H. , Chen, J. , Muller, F. L. , Putluri, N. , Flores, E. R. , DeMayo, F. J. , Baseler, L. , Rai, K. , & Lee, M. G. (2020). KMT2D Deficiency impairs super‐enhancers to confer a glycolytic vulnerability in lung cancer. Cancer Cell, 37, 599–617. 10.1016/j.ccell.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Cubero, M. J. , Martinez‐Gonzalez, L. J. , Robles‐Fernandez, I. , Martinez‐Herrera, J. , Garcia‐Rodriguez, G. , Pascual‐Geler, M. , Cozar, J. M. , & Lorente, J. A. (2017). Somatic mutations in prostate cancer: Closer to personalized medicine. Molecular Diagnosis & Therapy, 21, 167–178. 10.1007/s40291-016-0248-6 [DOI] [PubMed] [Google Scholar]
- Antonarakis, E. S. , Armstrong, A. J. , Dehm, S. M. , & Luo, J. (2016). Androgen receptor variant‐driven prostate cancer: clinical implications and therapeutic targeting. Prostate Cancer and Prostatic Diseases, 19, 231–241. 10.1038/pcan.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austen, B. , Skowronska, A. , Baker, C. , Powell, J. E. , Gardiner, A. , Oscier, D. , Majid, A. , Dyer, M. , Siebert, R. , Taylor, A. M. , Moss, P. A. , & Stankovic, T. (2007). Mutation status of the residual ATM allele is an important determinant of the cellular response to chemotherapy and survival in patients with chronic lymphocytic leukemia containing an 11q deletion. Journal of Clinical Oncology, 25, 5448–5457. 10.1200/JCO.2007.11.2649 [DOI] [PubMed] [Google Scholar]
- Barbieri, C. E. , Baca, S. C. , Lawrence, M. S. , Demichelis, F. , Blattner, M. , Theurillat, J. P. , White, T. A. , Stojanov, P. , Van Allen, E. , Stransky, N. , Nickerson, E. , Chae, S. S. , Boysen, G. , Auclair, D. , Onofrio, R. C. , Park, K. , Kitabayashi, N. , MacDonald, T. Y. , Sheikh, K. , … Garraway, L. A. (2012). Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature Genetics, 44, 685–689. 10.1038/ng.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, J. L. , Haile, R. W. , Stovall, M. , Boice, J. D., Jr. , Shore, R. E. , Langholz, B. , Thomas, D. C. , Bernstein, L. , Lynch, C. F. , Olsen, J. H. , Malone, K. E. , Mellemkjaer, L. , Borresen‐Dale, A. L. , Rosenstein, B. S. , Teraoka, S. N. , Diep, A. T. , Smith, S. A. , Capanu, M. , Reiner, A. S. , … Concannon, P. (WECARE Study Collaborative Group 2010). Radiation exposure, the ATM Gene, and contralateral breast cancer in the women's environmental cancer and radiation epidemiology study. Journal of the National Cancer Institute, 102, 475–483. 10.1093/jnci/djq055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen, G. , Barbieri, C. E. , Prandi, D. , Blattner, M. , Chae, S. S. , Dahija, A. , Nataraj, S. , Huang, D. , Marotz, C. , Xu, L. , Huang, J. , Lecca, P. , Chhangawala, S. , Liu, D. , Zhou, P. , Sboner, A. , de Bono, J. S. , Demichelis, F. , Houvras, Y. , & Rubin, M. A. (2015). SPOP mutation leads to genomic instability in prostate cancer. eLife, 4, e09207. 10.7554/eLife.09207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejous, C. , & Krishnan, U. M. (2020). Sensors for diagnosis of prostate cancer: Looking beyond the prostate specific antigen. Biosensors and Bioelectronics, 173, 112790. 10.1016/j.bios.2020.112790 [DOI] [PubMed] [Google Scholar]
- Deshpande, A. , Lang, W. , McDowell, T. , Sivakumar, S. , Zhang, J. , Wang, J. , & Scheet, P. (2018). Strategies for identification of somatic variants using the Ion Torrent deep targeted sequencing platform. BMC Bioinformatics, 19, 5. 10.1186/s12859-017-1991-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisermann, K. , Wang, D. , Jing, Y. , Pascal, L. E. , & Wang, Z. (2013). Androgen receptor gene mutation, rearrangement, polymorphism. Translational Andrology and Urology, 2, 137–147. 10.3978/j.issn.2223-4683.2013.09.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, S. , Nelson, P. , & Vasioukhin, V. (2018). Recent advances in prostate cancer research: large‐scale genomic analyses reveal novel driver mutations and DNA repair defects. F1000Research, 2(7), 1173. 10.12688/f1000research.14499.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, K. , & Nonomura, N. (2019). Role of androgen receptor in prostate cancer: A review. The World Journal of Men's Health, 37, 288–295. 10.5534/wjmh.180040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi, J. , Afridi, A. , Vatsia, S. , Joshi, G. , Joshi, G. , Kaplan, S. A. , Smith, N. L. , & Khan, S. A. (2018). The molecular biology of prostate cancer: current understanding and clinical implications. Prostate Cancer and Prostatic Diseases, 1, 22–36. 10.1038/s41391-017-0023-8 [DOI] [PubMed] [Google Scholar]
- Gao, S. , Chen, S. , Han, D. , Barrett, D. , Han, W. , Ahmed, M. , Patalano, S. , Macoska, J. A. , He, H. H. , & Cai, C. (2019). Forkhead domain mutations in FOXA1 drive prostate cancer progression. Cell Research, 29, 770–772. 10.1038/s41422-019-0203-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Flores, M. , Casanova‐Salas, I. , Rubio‐Briones, J. , Calatrava, A. , Domínguez‐Escrig, J. , Rubio, L. , Ramírez‐Backhaus, M. , Fernández‐Serra, A. , García‐Casado, Z. , & López‐Guerrero, J. A. (2014). Clinico‐pathological significance of the molecular alterations of the SPOP gene in prostate cancer. European Journal of Cancer, 50, 2994–3002. 10.1016/j.ejca.2014.08.009 [DOI] [PubMed] [Google Scholar]
- Gasi Tandefelt, D. , Boormans, J. , Hermans, K. , & Trapman, J. (2014). ETS fusion genes in prostate cancer. Endocrine‐related Cancer, 21, R143–R152. 10.1530/ERC-13-0390 [DOI] [PubMed] [Google Scholar]
- Giacomelli, A. O. , Yang, X. , Lintner, R. E. , McFarland, J. M. , Duby, M. , Kim, J. , Howard, T. P. , Takeda, D. Y. , Ly, S. H. , Kim, E. , Gannon, H. S. , Hurhula, B. , Sharpe, T. , Goodale, A. , Fritchman, B. , Steelman, S. , Vazquez, F. , Tsherniak, A. , Aguirre, A. J. , … Hahn, W. C. (2018). Mutational processes shape the landscape of TP53 mutations in human cancer. Nature Genetics, 50, 1381–1387. 10.1038/s41588-018-0204-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, C. , Chen, L. H. , Huang, Y. , Chang, C. C. , Wang, P. , Pirozzi, C. J. , Qin, X. , Bao, X. , Greer, P. K. , McLendon, R. E. , Yan, H. , Keir, S. T. , Bigner, D. D. , & He, Y. (2013). KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget, 4, 2144–2153. 10.18632/oncotarget.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D. S. , Tao, H. Q. , He, X. J. , Long, M. , Yu, S. , Xia, Y. J. , Wei, Z. , Xiong, Z. , Jones, S. , He, Y. , Yan, H. , & Wang, X. (2015). Prevalence of deleterious ATM germline mutations in gastric cancer patients. Oncotarget, 6, 40953–40958. 10.18632/oncotarget.5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittiwut, C. , Pratuangdejkul, J. , Supornsilchai, V. , Muensri, S. , Hiranras, Y. , Sahakitrungruang, T. , Watcharasindhu, S. , Suphapeetiporn, K. , & Shotelersuk, V. (2017). Novel mutations of the SRD5A2 and AR genes in Thai patients with 46, XY disorders of sex development. Journal of Pediatric Endocrinology and Metabolism, 30, 19–26. 10.1515/jpem-2016-0048 [DOI] [PubMed] [Google Scholar]
- Jiang, J. , Protopopov, A. , Sun, R. , Lyle, S. , & Russell, M. (2018). Genomic profiling on an unselected solid tumor population reveals a highly mutated Wnt/β‐catenin pathway associated with oncogenic EGFR mutations. Journal of Personalized Medicine, 8, 13. 10.3390/jpm8020013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpjärvi, K. , Kim, N. H. , Keskitalo, S. , Clark, A. D. , von Nandelstadh, P. , Turunen, M. , Heikkinen, T. , Park, M. J. , Mäkinen, N. , Kivinummi, K. , Lintula, S. , Hotakainen, K. , Nevanlinna, H. , Hokland, P. , Böhling, T. , Bützow, R. , Böhm, J. , Mecklin, J. P. , Järvinen, H. , … Vahteristo, P. (2016). Somatic MED12 mutations in prostate cancer and uterine leiomyomas promote tumorigenesis through distinct mechanisms. Prostate, 76, 22–31. 10.1002/pros.23092 [DOI] [PubMed] [Google Scholar]
- Karlsson, Q. , Brook, M. N. , Dadaev, T. , Wakerell, S. , Saunders, E. J. , Muir, K. , Neal, D. E. , Giles, G. G. , MacInnis, R. J. , Thibodeau, S. N. , McDonnell, S. K. , Cannon‐Albright, L. , Teixeira, M. R. , Paulo, P. , Cardoso, M. , Huff, C. , Li, D. , Yao, Y. , Scheet, P. , … Kote‐Jarai, Z. (2021). Rare germline variants in ATM predispose to prostate cancer: A PRACTICAL Consortium study. Eur Urol Oncol, 4, S2588‐9311 30209–1. 10.1016/j.euo.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Takhar, M. , Alshalalfa, M. , Erho, N. , Shoag, J. , Jenkins, R. B. , Karnes, R. J. , Ross, A. E. , Schaeffer, E. M. , Rubin, M. A. , Trock, B. , Klein, E. A. , Den, R. B. , Tomlins, S. A. , Spratt, D. E. , Davicioni, E. , Sboner, A. , & Barbieri, C. E. (2018). Impact of the SPOP mutant subtype on the interpretation of clinical parameters in prostate cancer. JCO Precision Oncology, 2018, 2018, PO.18.00036. 10.1200/PO.18.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. , Chang, K. , Peng, J. , Shi, Q. , Gan, H. , Gao, K. , Feng, K. , Xu, F. , Zhang, H. , Dai, B. , Zhu, Y. , Shi, G. , Shen, Y. , Zhu, Y. , Qin, X. , Li, Y. , Zhang, P. , Ye, D. , & Wang, C. (2018). SPOP promotes ATF2 ubiquitination and degradation to suppress prostate cancer progression. Journal of Experimental & Clinical Cancer Research, 37, 145. 10.1186/s13046-018-0809-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo, J. , Seed, G. , Bertan, C. , Rescigno, P. , Dolling, D. , Figueiredo, I. , Miranda, S. , Nava Rodrigues, D. , Gurel, B. , Clarke, M. , Atkin, M. , Chandler, R. , Messina, C. , Sumanasuriya, S. , Bianchini, D. , Barrero, M. , Petermolo, A. , Zafeiriou, Z. , Fontes, M. , … de Bono, J. S. (2020). Genomics of lethal prostate cancer at diagnosis and castration resistance. Journal of Clinical Investigation, 30, 1743–1751. 10.1172/JCI132031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottet, N. , van den Bergh, R. C. N. , Briers, E. , Van den Broeck, T. , Cumberbatch, M. G. , De Santis, M. , Fanti, S. , Fossati, N. , Gandaglia, G. , Gillessen, S. , Grivas, N. , Grummet, J. , Henry, A. M. , van der Kwast, T. H. , Lam, T. B. , Lardas, M. , Liew, M. , Mason, M. D. , Moris, L. , … Cornford, P. (2021). EAU‐EANM‐ESTRO‐ESUR‐SIOG Guidelines on Prostate Cancer−2020 Update. Part 1: Screening, diagnosis, and local treatment with curative intent. European Urology, 79, S0302‐2838 30769–7. 10.1016/j.eururo.2020.09.042 [DOI] [PubMed] [Google Scholar]
- Neeb, A. , Herranz, N. , Arce‐Gallego, S. , Miranda, S. , Buroni, L. , Yuan, W. , Athie, A. , Casals, T. , Carmichael, J. , Rodrigues, D. N. , Gurel, B. , Rescigno, P. , Rekowski, J. , Welti, J. , Riisnaes, R. , Gil, V. , Ning, J. , Wagner, V. , Casanova‐Salas, I. , … de Bono, J. S. (2021). Advanced prostate cancer with ATM Loss: PARP and ATR Inhibitors. European Urology, 79, 200–211. 10.1016/j.eururo.2020.10.029 [DOI] [PubMed] [Google Scholar]
- Paglia, L. L. , Laugé, A. , Weber, J. , Champ, J. , Cavaciuti, E. , Russo, A. , Viovy, J. L. , & Stoppa‐Lyonnet, D. (2010). ATM germline mutations in women with familial breast cancer and a relative with haematological malignancy. Breast Cancer Research and Treatment, 119, 443–452. 10.1007/s10549-009-0396-z [DOI] [PubMed] [Google Scholar]
- Parry, E. M. , Gable, D. L. , Stanley, S. E. , Khalil, S. E. , Antonescu, V. , Florea, L. , & Armanios, M. (2017). Germline mutations in DNA repair genes in lung adenocarcinoma. Journal of Thoracic Oncology, 12, 1673–1678. 10.1016/j.jtho.2017.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podralska, M. , Ziółkowska‐Suchanek, I. , Żurawek, M. , Dzikiewicz‐Krawczyk, A. , Słomski, R. , Nowak, J. , Stembalska, A. , Pesz, K. , & Mosor, M. (2018). Genetic variants in ATM, H2AFX and MRE11 genes and susceptibility to breast cancer in the polish population. BMC Cancer, 18, 452. 10.1186/s12885-018-4360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratz, K. W. , Koh, B. D. , Patel, A. G. , Flatten, K. S. , Poh, W. , Herman, J. G. , Dilley, R. , Harrell, M. I. , Smith, B. D. , Karp, J. E. , Swisher, E. M. , McDevitt, M. A. , & Kaufmann, S. H. (2016). Poly (ADP‐Ribose) polymerase inhibitor hypersensitivity in aggressive myeloproliferative neoplasms. Clinical Cancer Research, 22, 3894–3902. 10.1158/1078-0432.CCR-15-2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter, M. , Paradiso, V. , Widmer, P. , Garofoli, A. , Quagliata, L. , Eppenberger‐Castori, S. , Soysal, S. D. , Muenst, S. , Ng, C. , Piscuoglio, S. , Weber, W. , & Weber, W. P. (2020). Identification of somatic mutations in thirty‐year‐old serum cell‐free DNA from patients with breast cancer: A feasibility study. Clinical Breast Cancer, 20, 413–421e1. 10.1016/j.clbc.2020.04.005 [DOI] [PubMed] [Google Scholar]
- Robinson, D. , Van Allen, E. M. , Wu, Y. M. , Schultz, N. , Lonigro, R. J. , Mosquera, J. M. , Montgomery, B. , Taplin, M. E. , Pritchard, C. C. , Attard, G. , Beltran, H. , Abida, W. , Bradley, R. K. , Vinson, J. , Cao, X. , Vats, P. , Kunju, L. P. , Hussain, M. , Feng, F. Y. , … Chinnaiyan, A. M. (2015). Integrative clinical genomics of advanced prostate cancer. Cell, 161, 1215–1228. 10.1016/j.cell.2015.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg, J. M. , Hinz, W. , Rearick, T. M. , Schultz, J. , Mileski, W. , Davey, M. , Leamon, J. H. , Johnson, K. , Milgrew, M. J. , Edwards, M. , Hoon, J. , Simons, J. F. , Marran, D. , Myers, J. W. , Davidson, J. F. , Branting, A. , Nobile, J. R. , Puc, B. P. , Light, D. , … Bustillo, J. (2011). An integrated semiconductor device enabling non‐optical genome sequencing. Nature, 475, 348–352. 10.1038/nature10242 [DOI] [PubMed] [Google Scholar]
- Shah, N. , & Brown, M. (2019). The sly oncogene: FOXA1 mutations in prostate cancer. Cancer Cell, 36, 119–121. 10.1016/j.ccell.2019.07.005 [DOI] [PubMed] [Google Scholar]
- Shtivelman, E. , Beer, T. M. , & Evans, C. P. (2014). Molecular pathways and targets in prostate cancer. Oncotarget, 5, 7217–7259. 10.18632/oncotarget.2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt, D. E. , Dai, D. L. Y. , Den, R. B. , Troncoso, P. , Yousefi, K. , Ross, A. E. , Schaeffer, E. M. , Haddad, Z. , Davicioni, E. , Mehra, R. , Morgan, T. M. , Rayford, W. , Abdollah, F. , Trabulsi, E. , Achim, M. , Tapia, E. , Guerrero, M. , Karnes, R. J. , Dicker, A. P. , … Davis, J. W. (2018). Performance of a prostate cancer genomic classifier in predicting metastasis in men with prostate‐specific antigen persistence postprostatectomy. European Urology, 74, 107–114. 10.1016/j.eururo.2017.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoehr, R. , Taubert, H. , Gaisa, N. T. , Smeets, D. , Kneitz, B. , Giedl, J. , Ruemmele, P. , Wieland, W. F. , Rau, T. T. , & Hartmann, A. (2013). Lack of evidence for frequent MED12 p.L1224F mutation in prostate tumours from Caucasian patients. Journal of Pathology, 230, 453–456. 10.1002/path.4208 [DOI] [PubMed] [Google Scholar]
- Sun, X. , Frierson, H. F. , Chen, C. , Li, C. , Ran, Q. , Otto, K. B. , Cantarel, B. L. , Vessella, R. L. , Gao, A. C. , Petros, J. , Miura, Y. , Simons, J. W. , & Dong, J. T. (2005). Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nature Genetics, 37, 407–412. 10.1038/ng1528 [DOI] [PubMed] [Google Scholar]
- Sun, X. , Xing, C. , Fu, X. , Li, J. , Zhang, B. , Frierson, H. F., Jr. , & Dong, J. T. (2015). Additive effect of Zfhx3/Atbf1 and Pten deletion on mouse prostatic tumorigenesis. Journal of Genetics and Genomics, 42, 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa, U. , Castelli, G. , & Pelosi, E. (2019). Cellular and molecular mechanisms underlying prostate cancer development: therapeutic implications. Medicines (Basel), 6, 82. 10.3390/medicines6030082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaousis, G. N. , Papadopoulou, E. , Apessos, A. , Agiannitopoulos, K. , Pepe, G. , Kampouri, S. , Diamantopoulos, N. , Floros, T. , Iosifidou, R. , Katopodi, O. , Koumarianou, A. , Markopoulos, C. , Papazisis, K. , Venizelos, V. , Xanthakis, I. , Xepapadakis, G. , Banu, E. , Eniu, D. T. , Negru, S. , … Nasioulas, G. (2019). Analysis of hereditary cancer syndromes by using a panel of genes: novel and multiple pathogenic mutations. BMC Cancer, 19, 535. 10.1186/s12885-019-5756-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega, F. J. , Iniesta, P. , Caldés, T. , Sanchez, A. , López, J. A. , de Juan, C. , Diaz‐Rubio, E. , Torres, A. , Balibrea, J. L. , & Benito, M. (1997). p53 exon 5 mutations as a prognostic indicator of shortened survival in non‐small‐cell lung cancer. British Journal of Cancer, 76, 44–51. 10.1038/bjc.1997.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Végran, F. , Rebucci, M. , Chevrier, S. , Cadouot, M. , Boidot, R. , & Lizard‐Nacol, S. (2013). Only missense mutations affecting the DNA binding domain of p53 influence outcomes in patients with breast carcinoma. PLoS One, 8, e55103. 10.1371/journal.pone.0055103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodicka, P. , Andera, L. , Opattova, A. , & Vodickova, L. (2021). The interactions of DNA repair, telomere homeostasis, and p53 mutational status in solid cancers: risk, prognosis, and prediction. Cancers (Basel), 13, 479. 10.3390/cancers13030479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, E. , Herberts, C. , Fu, S. , Yip, S. , Wong, A. , Wang, G. , Ritch, E. , Murtha, A. J. , Vandekerkhove, G. , Fonseca, N. M. , Angeles, A. , Beigi, A. , Schönlau, E. , Beja, K. , Annala, M. , Khalaf, D. , Chi, K. N. , & Wyatt, A. W. (2021). BRCA2, ATM, and CDK12 defects differentially shape prostate tumor driver genomics and clinical aggression. Clinical Cancer Research, 27, 1650–1662. 10.1158/1078-0432.CCR-20-3708 [DOI] [PubMed] [Google Scholar]
- Yamoah, K. , Lal, P. , Awasthi, S. , Naghavi, A. O. , Rounbehler, R. J. , Gerke, T. , Berglund, A. E. , Pow‐Sang, J. M. , Schaeffer, E. M. , Dhillon, J. , Park, J. Y. , & Rebbeck, T. R. (2021). TMPRSS2‐ERG fusion impacts anterior tumor location in men with prostate cancer. Prostate, 2, 109–117. 10.1002/pros.24086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Tindall, D. J. , & Huang, H. (2014). Modulation of androgen receptor by FOXA1 and FOXO1 factors in prostate cancer. International Journal of Biological Sciences, 10, 614–619. 10.7150/ijbs.8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Wen, J. , Huang, G. , Mittlesteadt, J. , Wen, X. , & Lu, X. (2021). CHD1 and SPOP synergistically protect prostate epithelial cells from DNA damage. Prostate, 81, 81–88. 10.1002/pros.24080 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


