Abstract
In many bacteria, cell division begins before the sister chromosomes are fully segregated. Specific DNA translocases ensure that the chromosome is removed from the closing septum, such as the transmembrane protein FtsK in Escherichia coli. Bacillus subtilis contains two FtsK homologues, SpoIIIE and SftA. SftA is active during vegetative growth whereas SpoIIIE is primarily active during sporulation and pumps the chromosome into the spore compartment. FtsK and SpoIIIE contain several transmembrane helices, however, SftA is assumed to be a cytoplasmic protein. It is unknown how SftA is recruited to the cell division site. Here we show that SftA is a peripheral membrane protein, containing an N‐terminal amphipathic helix that reversibly anchors the protein to the cell membrane. Using a yeast two‐hybrid screen we found that SftA interacts with the conserved cell division protein SepF. Based on extensive genetic analyses and previous data we propose that the septal localization of SftA depends on either SepF or the cell division protein FtsA. Since SftA seems to interfere with the activity of SepF, and since inactivation of SepF mitigates the sensitivity of a ∆sftA mutant for ciprofloxacin, we speculate that SftA might delay septum synthesis when chromosomal DNA is in the vicinity.
Keywords: cell division, DNA translocase, FtsA, FtsZ, SepF, SftA
Cell division can trap DNA when sister chromosomes are not yet fully segregated. To prevent this, certain cell division proteins can pull the DNA away from the septum. In Gram‐positive bacteria, SftA is such a protein. Here, we show that this protein interacts with the conserved cell division SepF and uses an amphipathic helix to bind to the cell membrane.
1. INTRODUCTION
The key bacterial cell division protein, FtsZ, polymerizes into a ring‐like structure at the midcell, the so‐called Z‐ring. In many bacteria assembly of the Z‐ring commences before the newly formed sister chromosomes are fully segregated (Blaauwen et al., 1999). Specialized DNA translocases bound to the Z‐ring ensure that chromosomal DNA is not trapped by the closing division septum. This is particularly important when the circular chromosomes remain interlinked due to concatenation or homologous recombination events (Crozat et al., 2015; Kaimer & Graumann, 2011). The best‐studied DNA translocase is FtsK of Escherichia coli. FtsK is a multifunctional and multidomain protein present in a wide range of bacteria and archaea (Bigot et al., 2007; Constantinesco et al., 2004; Manzan et al., 2004). Its N‐terminus is required for recruitment to the cell division site and contains 5 transmembrane helices, while the C‐terminus contains the motor domain that supports chromosome translocation (Bigot et al., 2007). In addition, FtsK activates site‐specific recombination by XerCD at dif sites, resolving the two sister chromosomes after replication (Aussel et al., 2002). The motor domain of FtsK is subdivided into an α‐, β‐, and γ‐subdomain, whereby the α‐ and β‐ domains form a double‐stranded DNA translocase, and the γ‐subdomain scans the DNA for specific KOPS (FtsK orienting polar sequences) (Crozat et al., 2015). The ter oriented KOPS motifs assure that FtsK moves toward the dif site so that the γ‐subdomain can interact with XerD, stimulating decatenation of the chromosome dimers (Aussel et al., 2002; Bigot et al., 2005; Grainge et al., 2007; Sherratt et al., 1995; Yates et al., 2006).
In addition to its role as DNA translocase, FtsK is also essential for cell division in E. coli and is targeted to the Z‐ring by interacting with the FtsZ membrane anchors ZipA and FtsA (Pichoff & Lutkenhaus, 2002). FtsK itself is required for the recruitment of FtsQ, FtsL, and FtsI, which are part of the late cell division complex responsible for septum synthesis (Chen & Beckwith, 2001; Wang & Lutkenhaus, 1998; Yu et al., 1998).
The Gram‐positive model bacterium Bacillus subtilis has two FtsK homologues, SpoIIIE and SftA (Figure 1). SpoIIIE is comparable to FtsK, however, SftA lacks clear transmembrane domains. Only the C‐terminal domain, encompassing the ATP‐ and DNA‐binding motifs, share homology with FtsK (Kaimer et al., 2009). In contrast to the E. coli situation, SpoIIIE and SftA are not required for normal cell division. SpoIIIE is primarily active during sporulation and is crucial for pumping one sister chromosome into the forespore compartment (Bath et al., 2000; Kaimer et al., 2009; Wu & Errington, 1994). In addition, SpoIIIE is recruited to the nascent septum when DNA is entrapped after the division has been completed (Biller & Burkholder, 2009; Kaimer et al., 2009; Sharpe & Errington, 1995). SftA functions during vegetative growth and inactivation of SftA results in slightly elongated cells and septum‐entrapped DNA in 1.3% of the cells (Kaimer et al., 2009). It is unknown how SftA is recruited to the cell division site. B. subtilis contains FtsA but not an E. coli ZipA homologue. Deleting FtsA reduces the efficiency of SftA recruitment somewhat but does not prevent it (Najjar et al., 2018). Earlier work suggested that the presence of late cell division proteins, including the transpeptidase Pbp2B, is important (Biller & Burkholder, 2009), but this was called into question in another study (Kaimer et al., 2009).
FIGURE 1.
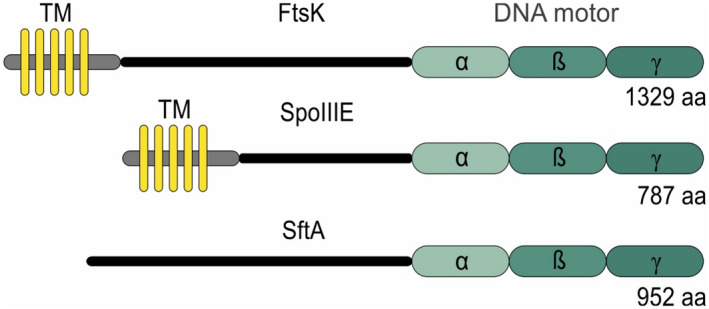
Homology between FtsK, SpoIIIE and SftA. FtsK, SpoIIIE and SftA share the conserved DNA motor subunit consisting of an ATP binding (α), ATP hydrolyzing (β) and DNA binding domain (γ). FtsK and SpoIIIE contain five transmembrane helices (TM) at their N‐terminus. Amino acid (aa) lengths of the proteins are indicated
Gram‐positives, cyanobacteria, and some archaea use the protein SepF to attach FtsZ to the cell membrane. B. subtilis possesses both SepF and FtsA. Deleting either sepF or ftsA does not block cell division, but deleting both genes is lethal (Duman et al., 2013; Feucht et al., 2001). FtsA and SepF bind directly to FtsZ and both contain an amphipathic α‐helix that functions as a reversible membrane anchor (Duman et al., 2013; Pichoff & Lutkenhaus, 2005). SepF polymerizes into large curved structures with a diameter close to 50 nm and this structural characteristic is believed to confine the spread of FtsZ polymers, thus defining the synthesis and thickness of the division septum (Wenzel et al., 2021). In a genome‐wide search for SepF interacting proteins, we found a potential interaction between SepF and SftA. To examine whether this interaction is relevant, we performed extensive mutagenesis and cell biology study. This showed that SepF supports the recruitment of SftA to the Z‐ring. In addition, we found that SftA uses an N‐terminal amphipathic helix to form a dynamic association with the cell membrane. Based on this and other data we propose that SftA can use both SepF and FtsA for its association with the Z‐ring, and we provide some insights into how the interaction with SepF might be related to the activity of SftA.
2. RESULTS
2.1. Yeast two‐hybrid screen
Because of the important role of SepF in cell division, we wondered whether the protein might interact with other proteins than FtsZ. To examine this, we performed a yeast two‐hybrid assay and screened a B. subtilis genomic library for potential interaction partners (Marchadier et al., 2011; Noirot‐Gros et al., 2002). Full‐length SepF fused to the Gal4p DNA binding domain was used as bait. After screening approximately 1 × 108 yeast mating events, several positive clones were found (Figure 2a). Subsequent sequencing showed that the prey plasmids contained fragments of SepF, SftA, the two‐component sensor kinase PsdS, and the putative transcriptional regulator YdeL. Self‐interaction of SepF has been well documented (Duman et al., 2013). The presence of a SftA fragment in the positive clones was interesting since the protein, like SepF, binds specifically to the Z‐ring (Figure 2b). To confirm the interaction between SftA and SepF, the full genes and different fragments were tested in the yeast two‐hybrid assay (Figure 2a, lower panel). The SftA domain responsible for interaction with SepF appeared to reside in the N‐terminal part of the protein, between amino acids 30 and 75 (see Figure S1 for secondary structure prediction of SftA). The SepF region responsible for SftA interaction appeared to be located between amino acids 65–151. This conserved region forms a dimer and is responsible for the polymerization of SepF and FtsZ binding (Duman et al., 2013). We also included FtsZ in this analysis, but no interaction between this protein and SftA was detected, whereas SepF showed a weak interaction with FtsZ, as has been shown in previous yeast two‐hybrid studies (Hamoen et al., 2006; Ishikawa et al., 2006).
FIGURE 2.
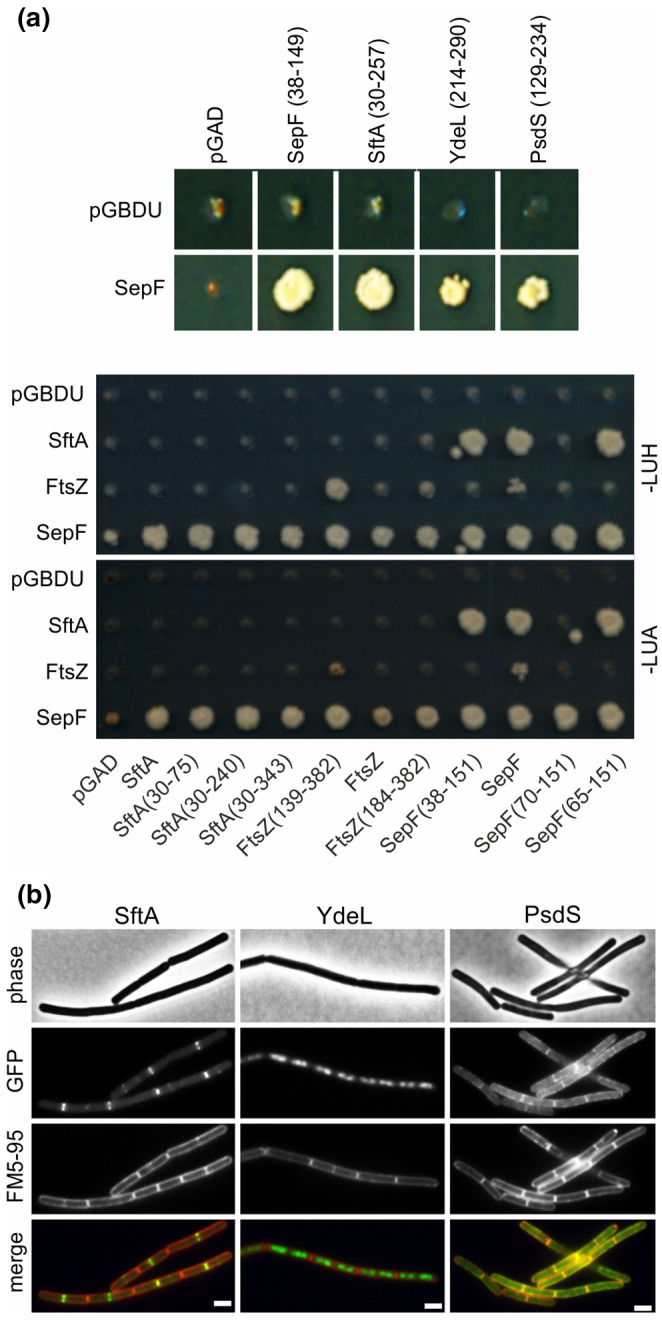
Yeast two‐hybrid screening. (a) Yeast two‐hybrid result showing diploid yeast strains harboring SepF as bait protein (SepF, lower panels) fused to GAL4BD (pGBDU plasmid), and positive clones containing fragments of the indicated genes fused to GAL4AD (pGAD plasmid), selected on synthetic complete medium (SC) (upper panel). Lower panel showing reciprocal yeast‐two hybrid analyses using full genes and gene fragments obtained from previous screens. Diploid yeast colonies expressing SftA, FtsZ, and SepF fused as bait, and prey fusions of SftA, SepF, FtsZ, and fragments of each protein, were selected both on synthetic complete medium lacking leucine, uracil, and adenine (‐LUA) or lacking leucine, uracil, and histidine (‐LUH). Interactions between proteins result in growth. (b) Microscopic images of B. subtilis cells expressing either SftA‐GFP, GFP‐YdeL, or PsdS‐GFP. Membranes were stained with FM5‐95. Scale bars are 2 μm. Strains used: TNVS455, TNVS366, and TNVS810, respectively
To determine whether PsdS and YdeL showed a similar cellular localization pattern as SftA, GFP fusions were constructed and expressed from the ectopic amyE locus. As shown in Figure 2b, YdeL localizes to the nucleoid, which is expected for a transcription factor and is according to what has been shown before for this protein (Meile et al., 2006). The two‐component sensor kinase PsdS contains two transmembrane domains and was indeed membrane‐localized but did not show any clear accumulation at cell division sites (Figure 2b). It seems, therefore, unlikely that SepF forms stable interactions with these two proteins.
2.2. Recruitment of SftA
It is unknown how SftA is recruited to the cell division machinery. The two‐hybrid results suggest that SepF might be involved in this, however, it has been shown that the inactivation of sepF does not affect the localization of SftA (Najjar et al., 2018), and we could confirm this (Figure S2). Other FtsZ binding proteins, including ZapA and EzrA, can also be removed without influencing the recruitment of SftA to the midcell (Figure S2) (Najjar et al., 2018). ZapA links FtsZ polymers together (Gueiros‐Filho & Losick, 2002), whereas EzrA has been shown to suppress the polymerization of FtsZ (Haeusser et al., 2004), but also to support the recruitment of the peptidoglycan transglycosylase/transpeptidase PBP1 to the division sites (Claessen et al., 2008; Cleverley et al., 2014; Levin et al., 1999). It was reported that deletion of ftsA reduces the efficiency of SftA recruitment (Najjar et al., 2018), but on the contrary, a ∆ftsA mutant showed a normal level of SftA‐GFP at cell division sites (Figure S2).
The Z‐ring forms a scaffold for the so‐called late cell division proteins responsible for the synthesis of septal peptidoglycan. These transmembrane proteins include the transpeptidase Pbp2B, transglycosylase FtsW, and three small proteins FtsL, DivIB and DivIC that support the assembly of the late cell division complex (Daniel et al., 2006; Meeske et al., 2016; Scheffers et al., 2004). There is some uncertainty in the literature as to whether the localization of SftA requires the presence of these late cell division proteins or not (Biller & Burkholder, 2009; Kaimer et al., 2009). Since none of the early cell division proteins, aside from FtsZ, appears to be important for SftA recruitment, we checked whether SftA localization might indeed require the late cell division proteins. To this end, the localization of GFP‐tagged SftA in cells depleted for the late cell division Pbp2B was examined. The assembly of late cell division proteins is a highly cooperative process and the absence of Pbp2B abolishes the recruitment of all known late cell division proteins (Daniel et al., 2000; Gamba et al., 2016). SftA‐GFP was ectopically expressed from the amyE locus under the control of the xylose‐inducible Pxyl promoter, and the wild type sftA gene was removed. As a control, we also performed the experiment in cells depleted for FtsZ. Depletion of Pbp2B and FtsZ was achieved by using strains containing either pbp2B or ftsZ under the control of the IPTG‐inducible Pspac promoter. A mCherry‐tagged ZapA fusion was used as a marker for Z‐rings. As shown in Figure S3, the septal localization of SftA‐GFP disappeared when FtsZ was depleted, but not when Pbp2B was depleted, confirming that recruitment of SftA does not rely on the presence of the late cell division proteins, and suggests that the protein in some way interacts with the Z‐ring.
2.3. Membrane targeting domain
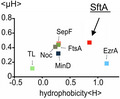
Previous work has shown that the N‐terminal domain of SftA harbors the Z‐ring targeting domain (Najjar et al., 2017). Indeed, when GFP was fused to the first 149 amino acids of SftA and expressed in a ∆sftA background the localization was comparable to that of wild‐type SftA (Figure 3a, middle panel). In fact, it was reported that only the first 67 amino acids of SftA are necessary for septal targeting (Najjar et al., 2017). This region largely overlaps with amino acids 30 to 75 of SftA that can interact with SepF in a yeast two‐hybrid assay (Figure 2a). When we removed amino acids 30 to 75 from the 149 amino acids N‐terminal domain, the binding to cell division sites was completely abolished, and instead, the fluorescence GFP signal became membrane‐localized (Figure 3a, right panel). This was surprising since cellular fractionation studies had shown that most SftA is found in the cytoplasmic fraction, suggesting that SftA is a cytosolic protein (Najjar et al., 2017). Many peripheral membrane proteins, including FtsA and SepF, interact with the cell membrane by inserting an amphipathic α‐helix into the surface region of the lipid bilayer (Duman et al., 2013; Pichoff & Lutkenhaus, 2005). Interestingly, the most N‐terminal predicted alpha‐helix (PSIPRED) (Jones, 1999) of SftA could be an amphipathic helix according to Amphipaseek (Sapay et al., 2006) (Figure 3b,c). A comparison of the mean amphipathic moment (<μH>) and the mean hydrophobicity (<H>) using HeliQuest indicated that the amphipathic helix of SftA is considerably more hydrophobic compared to others (Figure 3d) (Gautier et al., 2008). For comparison, we included the amphipathic helices of FtsA and SepF, those of the FtsZ regulators Noc and MinD, and the weak amphipathic helix of Thermolysin (Adams et al., 2015; Burg et al., 2004; Duman et al., 2013; Hu & Lutkenhaus, 2003; Pichoff & Lutkenhaus, 2005). The N‐terminal transmembrane helix of EzrA was also included (Levin et al., 1999). To confirm that the proposed amphipathic helix can indeed function as a membrane anchor, we fused the first 14 amino acids of SftA to GFP, and this gave a clear fluorescence membrane signal (Figure 3e), suggesting that SftA is a peripheral membrane protein that uses an N‐terminal amphipathic helix for reversible membrane attachment.
FIGURE 3.
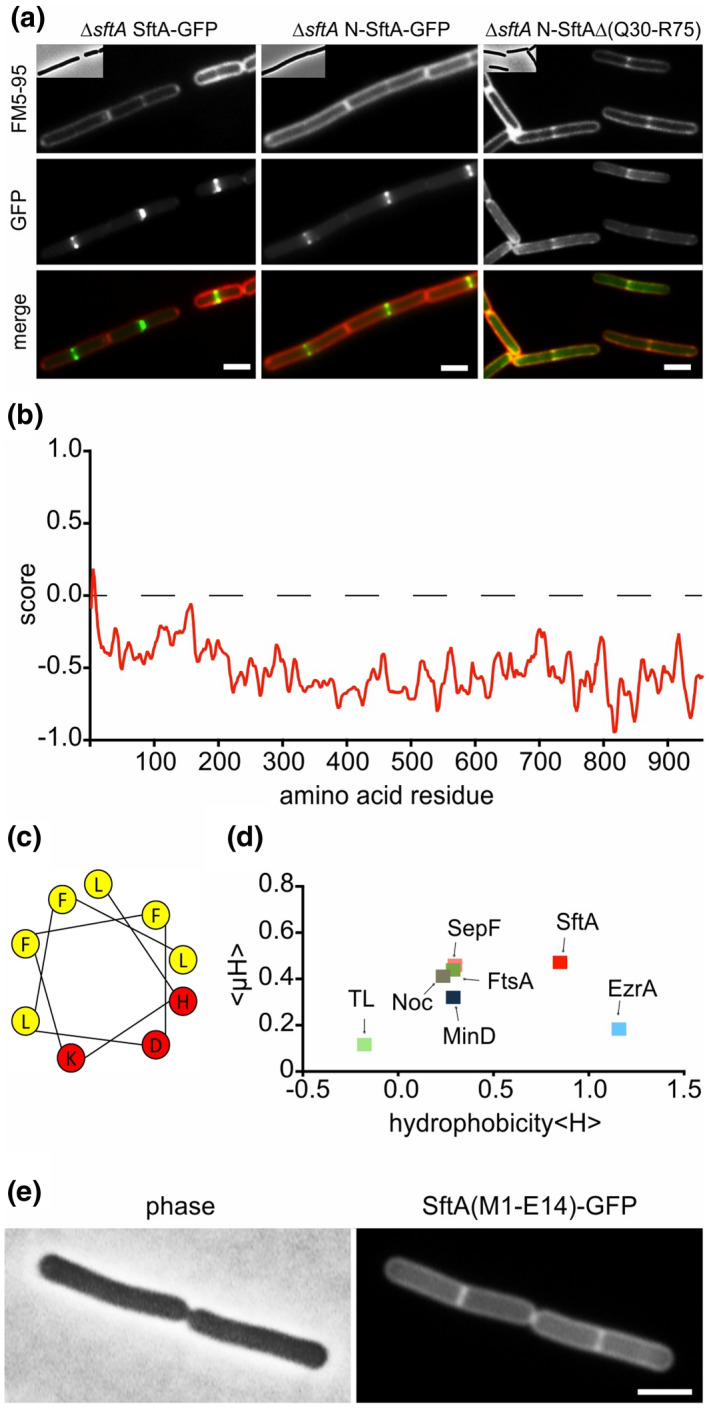
Membrane targeting of SftA. (a) Microscopic images of cells expressing SftA‐GFP, a deletion variant expressing only the N‐terminal 149 amino acids of SftA (N‐SftA‐GFP), and the same fusion construct lacking amino acids Q30 to R75. Membranes were stained with FM5‐95. (b) Amphipaseek plot (Sapay et al., 2006) indicates a possible amphipathic alpha‐helix at the extreme N‐terminus of SftA. (c) Helical wheel projection of this N‐terminal amphipathic helix (amino acids 4–12) with hydrophobic (yellow) and polar (red) amino acids. (d) Hydrophobicity (<H>) of different amphipathic helices plotted against their mean amphipathic moment (<μH>). See main text for details on the different helices. TL = thermolysin. (e) Localization of GFP fused to the N‐terminal amphipathic helix of SftA (amino acids M1‐E14). Expression of GFP fusions was induced with 0.1% xylose. Scale bars are 2 μm. Strains used: (a) TNVS455, TNVS456, TNVS234, and (e) TNV586, respectively
2.4. SepF and FtsA stimulate SftA recruitment
To examine whether the N‐terminal membrane anchor is important for the recruitment of SftA to the Z‐ring, we removed the first 12 amino acids of the SftA‐GFP fusion protein, however, this had no effect on localization (Figure 4). Interestingly, when this fusion protein was expressed in a ∆sepF background, the septal fluorescence signal completely disappeared (Figure 4). This was not the case when either zapA, ezrA, or ftsA were deleted (Figure 4). These results, supported by the yeast 2‐hybrid data, suggest that SepF recruits SftA to the Z‐ring by means of direct interaction.
FIGURE 4.
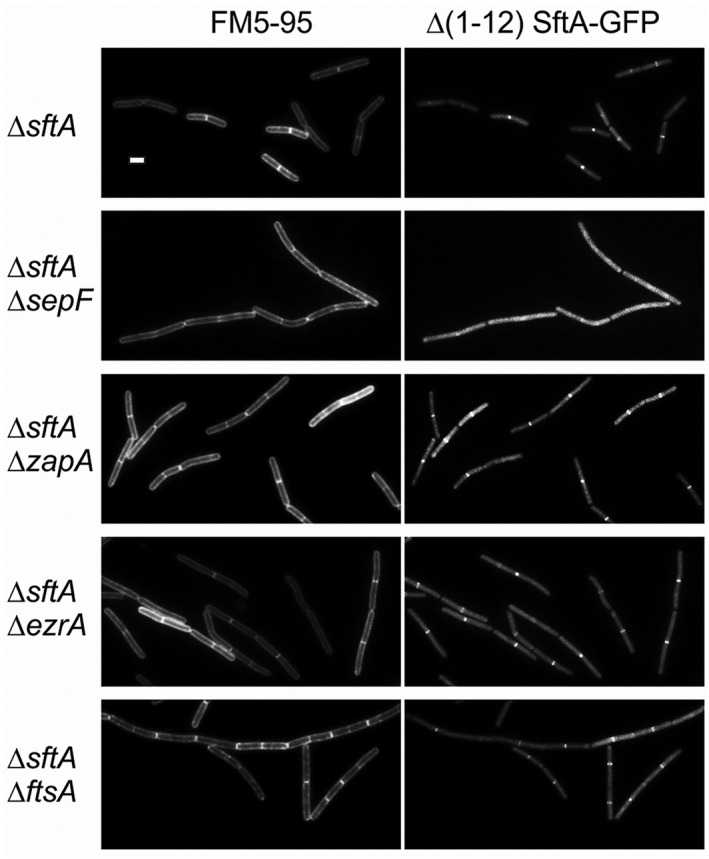
SepF supports SftA localization. Fluorescence microscopy images of different cell division mutants expressing SftA‐GFP, lacking the first 11 amino acids comprising the amphipathic helix membrane anchor. All strains were deleted for the native sftA gene. Left images show the fluorescent membrane stain of FM5‐95. The GFP fusion protein was induced with 0.1% xylose. Scale bars are 2 μm. Strains used: TNVS947, TNVS949, TNVS950, TNVS951, and TNVS952, respectively
Clearly, SepF cannot be the only protein that recruits SftA to the Z‐ring since the absence of SepF does not affect the localization of wild type SftA (Figure S2) (Najjar et al., 2018). It has been reported that FtsA recruits SftA to the cell membrane when both proteins are expressed in a heterologous eukaryotic system, i.e., S2 Schneider cells from Drosophila (Najjar et al., 2018). This makes FtsA an attractive candidate for the alternative protein that links SftA to the Z‐ring. In this case, SftA would contain binding sites for both SepF as well as FtsA, and since the interaction with FtsA requires the first 14 amino acids, it suggests that SftA needs to be attached to the cell membrane to interact with FtsA. Unfortunately, we cannot test this model by removing SepF and FtsA simultaneously since at least one of these proteins is required for FtsZ to polymerise into a Z‐ring (Duman et al., 2013). In an attempt to separate a presumed FtsA binding site from the SepF binding site, we reduced the N‐terminal domain as much as possible and with only 61 amino acids left there was still some fluorescence signal present at midcell (Figure 5, see Figure S4 for large field images showing multiple cells). When this truncation was introduced in a background lacking sepF, the midcell signal was still visible (Figure 5). However, when this fusion was expressed in a ftsA deletion background, no accumulation at cell divisions sites was observed and the fluorescence signal became more cytosolic (Figure 5). This finding supports the notion that FtsA functions as an alternative link between SftA and the Z‐ring.
FIGURE 5.
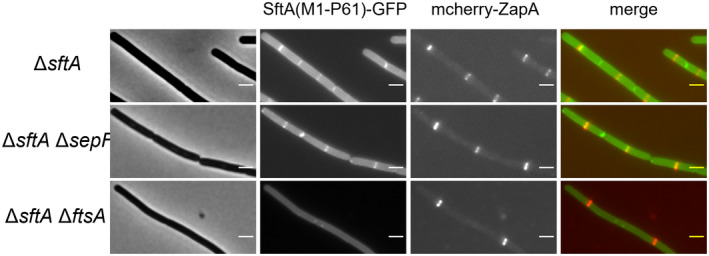
FtsA supports N‐terminal SftA localization. Localization of GFP fused to the first 61 amino acids of SftA in different cell division mutants. All strains lacked wild type sftA. ZapA fused to mCherry (red) was used to indicate the Z‐ring. Expression of the fusion proteins was induced with 0.1% xylose. Scale bars are 2 μm. Large field images showing multiple cells are shown in Figure S4. Strains used, TZH029, TZH119, TZH120, TZH121, respectively
2.5. SftA overproduction affects cell division
The binding of SftA to SepF might affect its activity and thus cell division. To test this, we overproduced SftA in different cell division mutants by introducing an extra copy of sftA expressed from the ectopic amyE locus and driven by the xylose‐inducible Pxyl promoter (Kim et al., 1996; Rygus & Hillen, 1991). Interestingly, overproduction of SftA strongly increased the cell length in a ∆ezrA background and reduced its viability (Figure 6a,b). To examine whether the N‐terminal domain of SftA is responsible for this, we overproduced the first 149 amino acids of SftA in a ∆sftA ∆ezrA double mutant and tested the effect on growth using a spot‐dilution assay. As shown in Figure 6c, overexpression of this domain also strongly impaired viability.
FIGURE 6.
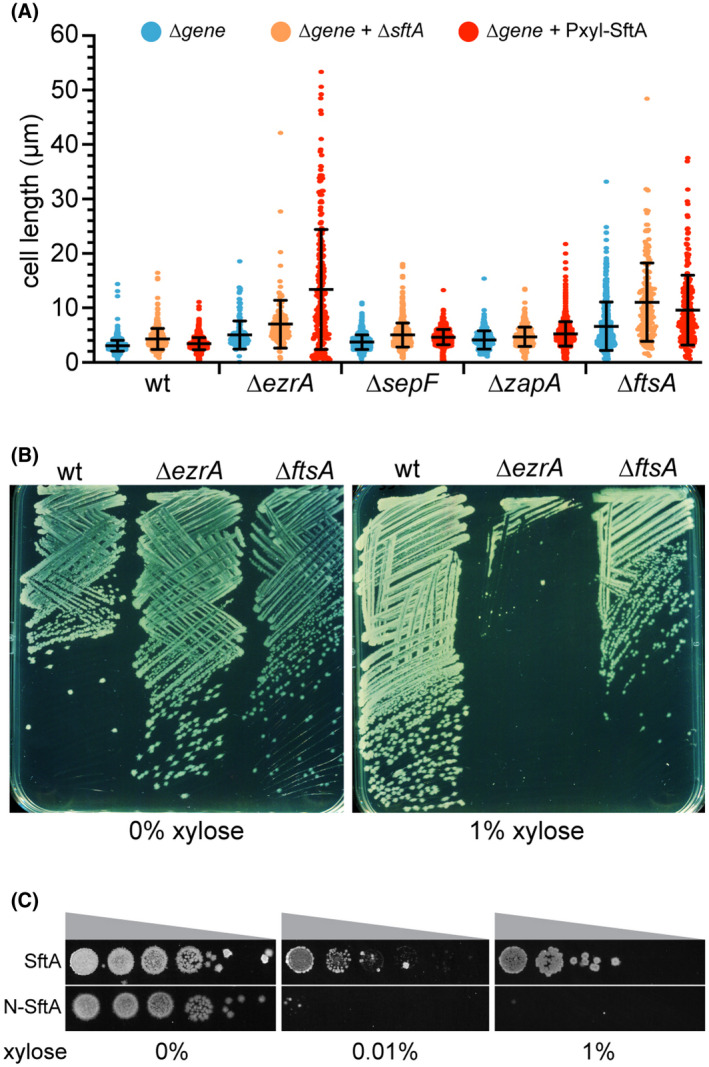
SftA overexpression influences cell division of an ezrA mutant. (a) Scatter plot showing cell lengths of different cell division mutants (blue), combined with either a sftA deletion (orange) or after overexpression of SftA (red). SftA overexpression was achieved by induction with 1% xylose for 1 h. On average 300 cells were analyzed. Means and standard deviations are indicated. (b) Overexpression of SftA in a ∆ezrA mutant affects its viability on the plate. The ∆ezrA and ∆ftsA backgrounds contain an extra xylose‐inducible sftA copy. (c) Dilution spot assay showing the effect on the viability of a ∆ezrA mutant when either full‐length SftA (SftA) or the N‐terminal 149 amino acids (N‐SftA) is induced with increasing xylose concentrations. Strains used in (a): wild type (wt), TNVS083, TNVS390, TNVS158, TNVS171, TNVS591, TNVS159, TNVS152 TNVS605, TNVS193, TNVS197, TNVS606, TNVS281, TNVS869, TNVS622, respectively. Strains used in (b): TNVS390, TNVS591, and TNVS622, respectively. Strains used in (c): TNVS591 and TNVS602, respectively
Previous work has shown that the inactivation of SepF in a ∆ezrA strain is lethal. Under these conditions, the cell division machinery still assembles but somehow fails to initiate septum synthesis (Hamoen et al., 2006). A comparable phenotype was observed when SftA was overexpressed in a ∆ezrA background. As shown in Figure 7, overexpression of SftA blocked cell division (see membrane stain), but the early cell division protein reporters YFP‐FtsA and YFP‐SepF still formed regular bands, indicative of Z‐ring formation. However, the late cell division protein reporter YFP‐Pbp2B only occasionally formed weak fluorescent bands, indicating a problem with the recruitment of late cell division proteins. Finally, we checked whether SftA directly interferes with the polymerization of SepF. SepF polymerizes into large regular ring‐like structures with a diameter of approximately 50 nm. The polymerization of purified SepF into these rings can be observed using negative staining electron microscopy (Gündoğdu et al., 2011) (Figure S5). When purified SftA was added to SepF at a molecular ratio of 1 to 3, rings were no longer observed (Figure S5). These data suggest that the interaction of SftA interferes with the activity of SepF.
FIGURE 7.
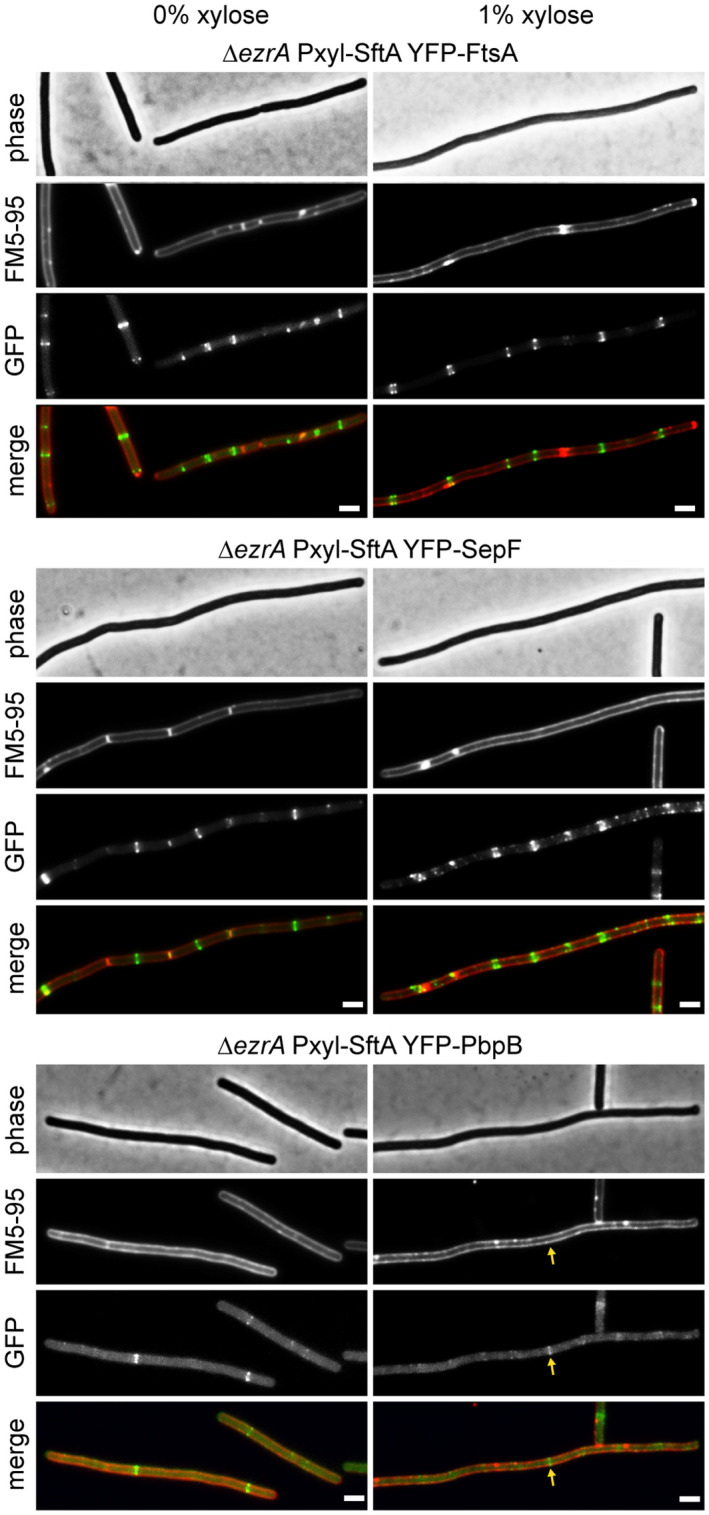
SftA overproduction in ∆ezrA does not affect Z‐ring formation. Fluorescence microscopy images of ∆ezrA cells overexpressing SftA from an ectopic locus by induction with 1% xylose for 1 h. Localization of the early cell division proteins YFP‐FtsA and YFP‐SepF is shown in the upper and middle panel, respectively, and the localization of the late cell division protein YFP‐PbpB is shown in the lower panel. Some elongated cells that were blocked in cell division still formed YFP‐PbpB bands (arrow). Reporter fusions were induced with 0.2 mM IPTG, and membranes were stained with FM5‐95. Scale bars are 2 μm. Strains used: TNVS710, TNVS712, and TNVS713, respectively
2.6. Function of SftA
The absence of SftA leads to bisected nucleoids in 1% to 3% of the cells (Biller & Burkholder, 2009; Kaimer et al., 2009). To examine whether SepF influences the activity of SftA, we measured the fraction of nucleoid bisected cells in a sepF deletion strain using fluorescence microscopy (Figure 8a). In the absence of SepF, approximately a doubling of bisected chromosomes was observed compared to wild type. This number is eightfold higher in a ∆sftA mutant, indicating that the presence of SepF is not required for the activity of SftA. However, the bisection numbers are low and a possible function of SepF might become more apparent when cells are treated with the topoisomerase IV inhibitor ciprofloxacin, which blocks chromosome decatenation (FERNÁNDEZ‐MOREIRA et al., 2000). A spot dilution assay showed that a ∆sftA mutant is much more sensitive to ciprofloxacin than the wild type strain (Figure 8b). Interestingly, this sensitivity was largely alleviated when sepF was also deleted. Cell division is inhibited upon DNA damage and the SOS response protein responsible for this is YneA (Kawai et al., 2003). To examine whether the induction of YneA could be responsible for the observed effects, we repeated the experiment in strains lacking yneA. The absence of this gene did not change the results (Figure 8b). Based on these data, we assume that SepF, at least under certain conditions, is required for the activity of SftA (see Discussion for details).
FIGURE 8.
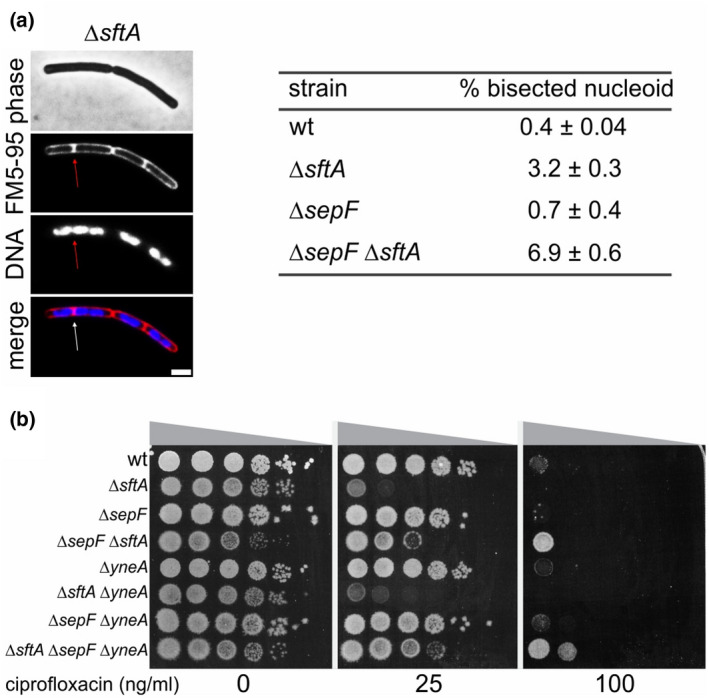
DNA bisection and sensitivity to ciprofloxacin. (a) Chromosome bisection in B. subtilis ∆sftA and ∆sepF mutants. A microscopic example is presented in the left panel. The table indicates the percentage of bisection based on approximately 500–1000 cells from independent biological triplicates. Membrane and DNA were stained with FM5‐95 and DAPI, respectively. The scale bar is 2 μm. Strains used: wild type (wt), TNVS083, TNVS159, and TNVS152, respectively. (b) Spot dilution assay showing the sensitivity of different B. subtilis mutants for the topoisomerase IV inhibitor ciprofloxacin. Strains used: wild type (wt), TNVS083, TNVS089, TNVS152, TNVS159, TNVS371, TNVS373, and TNVS431, respectively
3. DISCUSSION
3.1. SftA localization
In contrast to FtsK and SpoIIIE, SftA does not contain clear transmembrane domains, although based on an extensive Blast study it has been postulated that SftA might contain 3 putative transmembrane domains at its N‐terminus (Crozat et al., 2015). Another experimental study suggested that SftA is a cytoplasmic protein that requires interaction with a membrane protein to bind to the cell membrane (Najjar et al., 2017). Here, we show that SftA binds directly to the cell membrane by means of an N‐terminal amphipathic helix. Membrane targeting amphipathic helices provide a relatively weak and reversible interaction with membranes, which explains why SftA is found both in the cytoplasm as well as in the membrane (Najjar et al., 2017).
In a previous study, it was shown that the absence of FtsA reduces the dwell time of SftA molecules at division sites and that FtsA is able to recruit SftA to the membrane in a heterologous eukaryotic expression system (Drosophila S2 Schneider cells) (Najjar et al., 2018). Here, we have shown that a 61 amino acid long N‐terminal domain of SftA is no longer recruited to cell division sites in a ∆ftsA strain, providing further support for the role of FtsA in the recruitment of SftA. On the contrary, we have shown that SepF also interacts with SftA and that a SftA lacking the N‐terminal amphipathic helix is no longer recruited to the cell division site when SepF is absent. Finally, we have shown that purified SftA inhibited polymerization of SepF. Based on these data, we postulate that both SepF and FtsA can recruit SftA to the Z‐ring, which also explains why deleting either sepF or ftsA does not influence the binding of SftA to the Z‐ring. It cannot be excluded that SftA binds directly to FtsZ, however in our yeast two‐hybrid system we could not find any evidence for this, and thus far extensive FtsZ pulldown experiments have also not been able to identify such a connection (Ishikawa et al., 2006). Moreover, co‐expression of SftA and FtsZ in Drosophila S2 Schneider cells also showed no colocalization of these proteins (Najjar et al., 2018).
3.2. Functional homology with FtsK
Based on the homology with E. coli FtsK, it is assumed that SftA functions as a DNA translocase. The protein can bind to DNA (Najjar et al., 2017), but there is no clear evidence that SftA can function as a DNA pump. In fact, in a previous study, it was shown, using an elegant genetic assay, that only SpoIIIE has the ability to translocate chromosomes through the closing septum (Biller & Burkholder, 2009). In addition to its DNA‐translocation activity, FtsK also activates the XerCD recombinases to initiate the resolution of the replicated sister chromosomes at the dif sites (Aussel et al., 2002; Dubarry et al., 2010; Recchia & Sherratt, 1999). Moreover, FtsK stimulates chromosome decatenation by directly activating DNA topoisomerase IV (Bigot & Marians, 2010). A previous study demonstrated that SpoIIIE and SftA are not required for integration of a dif site‐containing plasmid into the B. subtilis chromosome, suggesting that these proteins are not involved in CodV‐ and RipX‐mediated site‐specific recombination (Sciochetti et al., 2001). However, another study found that the nucleoid bisection phenotype of an sftA deletion mutant depends on the presence of the conserved recombination protein RecA, arguing that SftA might promote dimer resolution by RipX and CodV (Biller & Burkholder, 2009). Whether SftA and/or SpoIIIE execute a similar function as FtsK and can activate either the XerCD homologous RipX and CodV and/or topoisomerase remains to be seen (Recchia & Sherratt, 1999).
3.3. Alternative SftA function
We have shown that overexpression of SftA in a ∆ezrA strain blocks cell division, possibly by directly interfering with the activity of SepF. Because of this, and the lack of evidence for DNA‐translocation activity, it might be that one of the main functions of SftA is to delay septum closure by interfering with the activity of SepF, FtsA or another cell division protein, in order to provide sufficient time for the sister chromosomes to fully segregate. The DNA‐binding activity of SftA might regulate this activity by detecting the proximity of chromosomal DNA. In fact, in ∆sftA cells, the formation of a Z‐ring over the nucleoid occurs approximately 1.7‐times more frequent than in wild type cells (Figure S6). This alternative model could explain why deleting sepF in a ∆sftA strain alleviates the detrimental effect of the topoisomerase IV inhibitor ciprofloxacin, since division is delayed when SepF is absent (Hamoen et al., 2006).
4. EXPERIMENTAL PROCEDURES
4.1. General experimental procedures
General growth conditions and construction of B. subtilis strains are described in detail in the Supporting Information. Tables S1, S2, and S3 list the strains, plasmids and primers, respectively, used in this study.
4.2. Yeast‐two‐hybrid assay
To construct the bait vector, sepF was amplified from genomic DNA of B. subtilis and inserted in the ClaI and EcoRI sites in the multiple cloning site of plasmid pGBD‐C1. The prey libraries containing fragments of the complete genome of B. subtilis were constructed as follows. First, the complete chromosome of B. subtilis was partially digested with restriction enzymes AciI, HinPII, MaeII, MspI and TaqI to produce a 5’ CG overhang. Second, the prey vectors pGAD‐C1, C2, and C3 were linearized with ClaI to produce compatible overhangs to the genomic fragments. Then, the linearized vector was treated with Calf Intestine Phosphatase to dephosphorylate the 5′‐end to prevent self‐ligation. Next, the restricted fragments and the linearized plasmid were ligated to produce GAL4 activation‐domain protein fusions. Subsequently, the ligation products were then transformed into E. coli, resulting in 15 different sub‐libraries corresponding to cloning into the pGAD‐C1, ‐C2, and ‐C3. The sub‐libraries were mixed to form three B. subtilis genomic libraries BSL‐C1, ‐C2, and ‐C3. Purified plasmid DNA from each library was used to transform the PJ69‐4a yeast strain. Finally, a minimum of 1.5 × 107 prey‐containing yeast colonies, grown on a synthetic medium complemented with leucine (SC‐L) plates, were combined to form BSL libraries. Aliquots of about 1 × 108 prey‐containing yeast cells were flash‐frozen and stored at −80°C.
To screen for interactions, one thawed aliquot of BSL‐C1, ‐C2, and ‐C3 was mixed with PJ69‐4a containing bait plasmid, plated on a rich medium and incubated for 5 h at 30°C for mating. Cells were then collected, washed, and transferred to synthetic complete medium plates lacking leucine, uracil, histidine (SC‐LUH) and containing 0.5 mM 3‐aminotriazole (3AT). Self‐activating baits were suppressed by the addition of 3AT up to 50 mM to the SC‐LU plates. The plates were incubated for 10–15 days at 30°C, after which parental cells and diploids were selected on SC‐L, SC‐U, and SC‐LU plates. His+ colonies were further transferred to synthetic complete plates without leucine, uracil, and adenine (SC‐LUA) and incubated for 3–5 days at 30°C. The diploid clones containing interaction candidates, thus the His+ and Ade+ colonies, were finally transferred to 96‐well plates and stored at −20°C. To identify the prey DNA inserts, all candidate interactions were PCR amplified, sequenced and aligned to the B. subtilis genome.
4.3. Fluorescence light microscopy and image analysis
For protein localization using fluorescent reporter fusions, strains were grown overnight at 30°C in LB supplemented with 0.1% xylose. Nucleoid bisection frequencies were determined in a rich PAB medium at 37°C based on (Kaimer et al., 2009). In short, overnight cultures were diluted to an optical density of 0.05 in the PAB medium. At an optical density of 0.5, 200 μl of the culture was stained with 2 μg/ml FM5‐95 membrane dye and 1 μg/ml DAPI. 5 μl of the stained culture was spotted onto a thin layer of 1.3% agarose for microscopy. For protein localization and cell length measure studies, LB medium was used. A Nikon Eclipse Ti with CFI Plan Apochromat DM 100x oil objective Intensilight HG 130 W lamp, with a C11440‐22CU Hamamatsu ORCA camera, and NIS elements software was used for image acquisition. For cell length analyses, the ImageJ based program ChainTracer was used (Syvertsson et al., 2016). Images were analyzed using ImageJ (National Institutes of Health) (Schneider et al., 2012).
To determine when the nucleoid is bisected by the membrane, samples were stained with DAPI (to visualize the nucleoid) and FM5‐95 (to visualize the membrane) prior to imaging. ChainTracer was used to align cells for easy visual inspection. A nucleoid was scored bisected when either one of the following criteria was met: (i) a division septum (visualized with the membrane dye FM5‐95) clearly overlapped with DAPI‐stained DNA, (ii) the DAPI‐stained DNA was clearly seen on both sides of the septum (the state when cell division is completed), and (iii) the DAPI signal spanned between the rounded ends of two cells that had begun physical separation.
4.4. SepF ring formation interference
To purify SepF, an overnight culture of E. coli BL21 (DE3) harboring the MBP‐SepF expression plasmid pNC12 (Duman et al., 2013) was diluted in 2 L fresh medium and grown to an optical density of 0.4. Then the culture was induced with a final concentration of 0.5 mM IPTG for 4 h as described in (Duman et al., 2013). The culture was quickly cooled down on slush ice, pelleted at 4000 RPM at 4°C, and washed with ice‐cold phosphor buffer saline containing 1 m PMSF. When necessary, the pellet was flash‐frozen in nitrogen and stored at −80°C. After slowly thawing on ice, the pellet was dissolved in 20 ml buffer AF (50 mM Tris–HCl, pH 7.4, 200 mM KCl, 5 mM EDTA, 0.5 mM DTT) containing 1 cOmplete Mini protease inhibitor tablet (Roche) followed by cell disruption using French Press. The resuspension was then centrifugated at 31 k RCF at 4°C for 1 h to clear the lysate from cell debris. The supernatant was then passed through a 0.2 μm filter and loaded onto a 1 ml MBP‐trap column pre‐equilibrated with buffer AF. The column was washed with buffer AF and subsequently washed with buffer BF (50 mM Tris–HCl, pH 7.4). The MBP‐SepF protein was eluted with 5 ml of buffer BF supplemented with 10 mM maltose. Pooled fractions were subjected to factor Xa protease digestion with 2 mM CaCl2 final concentration overnight at 4°C to separate the MBP and SepF.
The purification of SftA was accomplished by a C‐terminal His6 tag as previously described (Kaimer et al., 2009). To this end, the expression plasmid pET24A was linearized using oligonucleotides TerS373 & TerS374, and the sftA gene was amplified from genomic DNA of B. subtilis 168 using primers TerS377 & TerS378. Gibson assembly of both products resulted in plasmid pTNV082. In our adjusted purification protocol, E. coli BL21 Star (pLys) cells containing plasmid pTNV082 were grown in a 3 L LB medium at 37°C to an OD600 of approximately 0.4, followed by induction with 1 mM IPTG for 3 h. The culture was cooled down on slush ice, harvested by centrifugation, washed once with ice‐cold phosphate buffer saline supplemented with 1 mM PMSF, and after flash freezing in liquid nitrogen stored at −80°C. The pellet was slowly thawed on ice, then resuspended in binding buffer (100 mM Tris–HCl, pH 8.0, 200 mM KCl, 5 mM EDTA, 0.5 mM DTT) with 1 cOmplete Mini protease inhibitor tablet and 30 mM imidazole. Cells were disrupted by French Press and cell debris was pelleted by centrifugation at 31 k RCF and 4°C for 1 h to clear the lysate from cell debris. The supernatant was then passed through a 0.2 μm filter prior to loading on a binding buffer‐equilibrated 1 ml Hi‐trap Talon crude column. The column was then washed with five column volumes of 6% binding buffer containing 30 mM imidazole. To elute SftA, a gradient of 6% to 100% of binding buffer containing 0.5 M imidazole was applied. Fractions containing SftA were pooled and subjected to gel filtration on a Superdex 200 Increase column (GE Healthcare), equilibrated with binding buffer (flow rate 0.2 ml/min). Fractions were collected and analyzed by SDS PAGE. Pure SftA fractions were pooled, and glycerol to a final concentration of 20% was added prior to flash freezing in liquid nitrogen and storage at −80°C.
To test the effect of SftA on SepF ring formation, 2.75 μM MBP‐SepF was digested with Factor Xa protease in SepF binding buffer (20 mM Tris–HCl pH 7.5, 200 mM KCl, 1 mM KCl2, 1 mg/ml BSA, 0.5 mM DTT) as described above to separate SepF from the MBP moiety. During this separation, the freed SepF makes stable SepF rings (Gündoğdu et al., 2011). To test the effect of SftA on SepF rings, 0.75 μM SftA was added to the MBP‐SepF digestion reaction with Factor Xa protease. After the digest, the samples were centrifuged (10 k CFR) to separate MBP from SepF. The pellet was then dissolved in BF buffer without calcium chloride. Protein samples were spotted on glow‐discharged 200 mesh formvar/carbon‐coated copper grids (Agar Scientific) and incubated for 1 min at room temperature. Excess liquid was removed with a paper tissue and samples were negatively stained by adding 100 μl 2% uranyl acetate drop by drop. The excess staining solution was removed with a paper tissue and samples were allowed to air dry. Samples were examined with a Philips CM100 at an electron voltage of 80 kV.
4.5. Spot dilution assays
To test the effect of SftA overexpression and the topoisomerase IV inhibitor ciprofloxacin, overnight cultures were diluted into a fresh 37°C LB medium to an optical density of 0.05 and grown to an optical density of about 0.5. Subsequently, the cultures were serially diluted ten‐fold and 10 μl was spotted onto agar plates containing either xylose or ciprofloxacin and incubated at 37°C.
ETHICAL APPROVAL
This article does not contain any studies involving animals or human participants performed by any of the authors.
AUTHOR CONTRIBUTIONS
Terrens N. V. Saaki, Zihao Teng, Michaela Wenzel performed and Terrens N. V. Saaki, Zihao Teng, Michaela Wenzel, Magali Ventroux, Rut Carballido‐Lόpez, Marie Francoise Noirot‐Gros, Leendert W. Hamoen designed the experiments. Terrens N. V. Saaki, Zihao Teng, Michaela Wenzel, Marie Francoise Noirot‐Gros, and Leendert W. Hamoen wrote the paper.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
We thank the current and previous members of the Bacterial Cell Biology group for useful and constructive discussions. We would like to thank Norbert Vischer for his help with the ImageJ Chain tracer software. This research was funded by NWO STW‐Vici grant 12128, EU ITN grant AMBER, and CSC China Scholarship Council (CSC) fellowship.
Saaki, T. N. , Teng, Z. , Wenzel, M. , Ventroux, M. , Carballido‐Lόpez, R. , Noirot‐Gros, M. F. & Hamoen, L. W. (2022). SepF supports the recruitment of the DNA translocase SftA to the Z‐ring. Molecular Microbiology, 117, 1263–1274. 10.1111/mmi.14906
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Adams, D.W. , Wu, L.J. & Errington, J. (2015) Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. The EMBO Journal, 34, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aussel, L. , Barre, F.‐X. , Aroyo, M. , Stasiak, A. , Stasiak, A.Z. & Sherratt, D. (2002) FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell, 108, 195–205. [DOI] [PubMed] [Google Scholar]
- Bath, J. , Wu, L.J. , Errington, J. & Wang, J.C. (2000) Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell‐prespore division septum. Science, 290, 995–997. [DOI] [PubMed] [Google Scholar]
- Bigot, S. & Marians, K.J. (2010) DNA chirality‐dependent stimulation of topoisomerase IV activity by the C‐terminal AAA+ domain of FtsK. Nucleic Acids Research, 38, 3031–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot, S. , Saleh, O.A. , Lesterlin, C. , Pages, C. , Karoui, M.E. , Dennis, C. et al. (2005) KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. The EMBO Journal, 24, 3770–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot, S. , Sivanathan, V. , Possoz, C. , Barre, F.‐X. & Cornet, F. (2007) FtsK, a literate chromosome segregation machine: FtsK, a literate chromosome segregation machine. Molecular Microbiology, 64, 1434–1441. [DOI] [PubMed] [Google Scholar]
- Biller, S.J. & Burkholder, W.F. (2009) The Bacillus subtilis SftA (YtpS) and SpoIIIE DNA translocases play distinct roles in growing cells to ensure faithful chromosome partitioning. Molecular Microbiology, 74, 790–809. [DOI] [PubMed] [Google Scholar]
- Blaauwen, T.D. , Buddelmeijer, N. , Aarsman, M.E. , Hameete, C.M. & Nanninga, N. (1999) Timing of FtsZ assembly in Escherichia coli . Journal of Bacteriology, 181, 5167–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.C. & Beckwith, J. (2001) FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co‐localization with FtsZ during Escherichia coli cell division. Molecular Microbiology, 42, 395–413. [DOI] [PubMed] [Google Scholar]
- Claessen, D. , Emmins, R. , Hamoen, L.W. , Daniel, R.A. , Errington, J. & Edwards, D.H. (2008) Control of the cell elongation–division cycle by shuttling of PBP1 protein in Bacillus subtilis . Molecular Microbiology, 68, 1029–1046. [DOI] [PubMed] [Google Scholar]
- Cleverley, R.M. , Barrett, J.R. , Baslé, A. , Bui, N.K. , Hewitt, L. , Solovyova, A. et al. (2014) Structure and function of a spectrin‐like regulator of bacterial cytokinesis. Nature Communications, 5, 5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinesco, F. , Forterre, P. , Koonin, E.V. , Aravind, L. & Elie, C. (2004) A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Research, 32, 1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat, E. , Rousseau, P. , Fournes, F. & Cornet, F. (2015) The FtsK family of DNA translocases finds the ends of circles. Journal of Molecular Microbiology and Biotechnology, 24, 396–408. [DOI] [PubMed] [Google Scholar]
- Daniel, R.A. , Harry, E.J. & Errington, J. (2000) Role of penicillin‐binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis . Molecular Microbiology, 35, 299–311. [DOI] [PubMed] [Google Scholar]
- Daniel, R.A. , Noirot‐Gros, M.‐F. , Noirot, P. & Errington, J. (2006) Multiple interactions between the Transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. Journal of Bacteriology, 188, 7396–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry, N. , Possoz, C. & Barre, F. (2010) Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Molecular Microbiology, 78, 1088–1100. [DOI] [PubMed] [Google Scholar]
- Duman, R. , Ishikawa, S. , Celik, I. , Strahl, H. , Ogasawara, N. , Troc, P. et al. (2013) Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proceedings of the National Academy of Sciences of the United States of America, 110, E4601–E4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERNÁNDEZ‐MOREIRA, E. , Balas, D. , González, I. & Campa, A.G.D.L. (2000) Fluoroquinolones inhibit preferentially Streptococcus pneumoniae DNA topoisomerase IV than DNA Gyrase native proteins. Microbial Drug Resistance, 6, 259–267. [DOI] [PubMed] [Google Scholar]
- Feucht, A. , Lucet, I. , Yudkin, M.D. & Errington, J. (2001) Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis . Molecular Microbiology, 40, 115–125. [DOI] [PubMed] [Google Scholar]
- Gamba, P. , Hamoen, L.W. & Daniel, R.A. (2016) Cooperative recruitment of FtsW to the division site of Bacillus subtilis . Frontiers in Microbiology, 7, 1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, R. , Douguet, D. , Antonny, B. & Drin, G. (2008) HELIQUEST: a web server to screen sequences with specific α‐helical properties. Bioinformatics, 24, 2101–2102. [DOI] [PubMed] [Google Scholar]
- Grainge, I. , Bregu, M. , Vazquez, M. , Sivanathan, V. , Ip, S.C. & Sherratt, D.J. (2007) Unlinking chromosome catenanes in vivo by site‐specific recombination. The EMBO Journal, 26, 4228–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros‐Filho, F.J. & Losick, R. (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin‐like protein FtsZ. Genes & Development, 16, 2544–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gündoğdu, M.E. , Kawai, Y. , Pavlendova, N. , Ogasawara, N. , Errington, J. , Scheffers, D. et al. (2011) Large ring polymers align FtsZ polymers for normal septum formation. The EMBO Journal, 30, 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser, D.P. , Schwartz, R.L. , Smith, A.M. , Oates, M.E. & Levin, P.A. (2004) EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Molecular Microbiology, 52, 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen, L.W. , Meile, J. , Jong, W.D. , Noirot, P. & Errington, J. (2006) SepF, a novel FtsZ‐interacting protein required for a late step in cell division. Molecular Microbiology, 59, 989–999. [DOI] [PubMed] [Google Scholar]
- Hu, Z. & Lutkenhaus, J. (2003) A conserved sequence at the C‐terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Molecular Microbiology, 47, 345–355. [DOI] [PubMed] [Google Scholar]
- Ishikawa, S. , Kawai, Y. , Hiramatsu, K. , Kuwano, M. & Ogasawara, N. (2006) A new FtsZ‐interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis . Molecular Microbiology, 60, 1364–1380. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. (1999) Protein secondary structure prediction based on position‐specific scoring matrices1 1Edited by G. Von Heijne. Journal of Molecular Biology, 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Kaimer, C. , González‐Pastor, J.E. & Graumann, P.L. (2009) SpoIIIE and a novel type of DNA translocase, SftA, couple chromosome segregation with cell division in Bacillus subtilis . Molecular Microbiology, 74, 810–825. [DOI] [PubMed] [Google Scholar]
- Kaimer, C. & Graumann, P.L. (2011) Players between the worlds: multifunctional DNA translocases. Current Opinion in Microbiology, 14, 719–725. [DOI] [PubMed] [Google Scholar]
- Kawai, Y. , Moriya, S. & Ogasawara, N. (2003) Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis . Molecular Microbiology, 47, 1113–1122. [DOI] [PubMed] [Google Scholar]
- Kim, L. , Mogk, A. & Schumann, W. (1996) A xylose‐inducible Bacillus subtilis integration vector and its application. Gene, 181, 71–76. [DOI] [PubMed] [Google Scholar]
- Levin, P.A. , Kurtser, I.G. & Grossman, A.D. (1999) Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis . Proceedings of the National Academy of Sciences of the United States of America, 96, 9642–9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzan, A. , Pfeiffer, G. , Hefferin, M.L. , Lang, C.E. , Carney, J.P. & Hopfner, K. (2004) MlaA, a hexameric ATPase linked to the Mre11 complex in archaeal genomes. EMBO Reports, 5, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchadier, E. , Carballido‐López, R. , Brinster, S. , Fabret, C. , Mervelet, P. , Bessières, P. et al. (2011) An expanded protein–protein interaction network in Bacillus subtilis reveals a group of hubs: exploration by an integrative approach. Proteomics, 11, 2981–2991. [DOI] [PubMed] [Google Scholar]
- Meeske, A.J. , Riley, E.P. , Robins, W.P. , Uehara, T. , Mekelanos, J.J. , Kahne, D. et al. (2016) SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature, 537, 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meile, J. , Wu, L.J. , Ehrlich, S.D. , Errington, J. & Noirot, P. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory. Proteomics, 6, 2135–2146. [DOI] [PubMed] [Google Scholar]
- Najjar, N.E. , Andari, J.E. , Kaimer, C. , Fritz, G. , Rösch, T.C. & Graumann, P.L. (2018) Single‐molecule tracking of DNA Translocases in Bacillus subtilis reveals strikingly different dynamics of SftA, SpoIIIE, and FtsA. Applied and Environmental Microbiology, 84, e02610‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar, N.E. , Kaimer, C. , Rösch, T. & Graumann, P.L. (2017) Requirements for Septal localization and chromosome segregation activity of the DNA Translocase SftA from Bacillus subtilis . Journal of Molecular Microbiology and Biotechnology, 27, 29–42. [DOI] [PubMed] [Google Scholar]
- Noirot‐Gros, M.‐F. , Dervyn, E. , Wu, L.J. , Mervelet, P. , Errington, J. , Ehrlich, S.D. et al. (2002) An expanded view of bacterial DNA replication. Proceedings of the National Academy of Sciences of the United States of America, 99, 8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff, S. & Lutkenhaus, J. (2002) Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli . The EMBO Journal, 21, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff, S. & Lutkenhaus, J. (2005) Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA: membrane tethering of Z ring by FtsA. Molecular Microbiology, 55, 1722–1734. [DOI] [PubMed] [Google Scholar]
- Recchia, G.D. & Sherratt, D.J. (1999) Conservation of xer site‐specific recombination genes in bacteria. Molecular Microbiology, 34, 1146–1148. [DOI] [PubMed] [Google Scholar]
- Rygus, T. & Hillen, W. (1991) Inducible high‐level expression of heterologous genes in Bacillus megaterium using the regulatory elements of the xylose‐utilization operon. Applied Microbiology and Biotechnology, 35, 594–599. [DOI] [PubMed] [Google Scholar]
- Sapay, N. , Guermeur, Y. & Deléage, G. (2006) Prediction of amphipathic in‐plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinformatics, 7, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers, D. , Jones, L.J.F. & Errington, J. (2004) Several distinct localization patterns for penicillin‐binding proteins in Bacillus subtilis . Molecular Microbiology, 51, 749–764. [DOI] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. & Eliceiri, K.W. (2012) NIH image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciochetti, S.A. , Piggot, P.J. & Blakely, G.W. (2001) Identification and characterization of the dif site from Bacillus subtilis . Journal of Bacteriology, 183, 1058–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe, M.E. & Errington, J. (1995) Postseptational chromosome partitioning in bacteria. Proceedings of the National Academy of Sciences of the United States of America, 92, 8630–8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt, D.J. , Arciszewska, L.K. , Blakely, G. , Colloms, S. , Grant, K. , Leslie, N. et al. (1995) Site‐specific recombination and circular chromosome segregation. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 347, 37–42. [DOI] [PubMed] [Google Scholar]
- Syvertsson, S. , Vischer, N.O.E. , Gao, Y. & Hamoen, L.W. (2016) When phase contrast fails: ChainTracer and NucTracer, two ImageJ methods for semi‐automated single cell analysis using membrane or DNA staining. PLoS One, 11, e0151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Burg, B. (2004) Thermolysin and related Bacillus metallopeptidases. In: Barrett, A. J. , Rawlings, N. D. & Woessner, J. F. (Eds.) Handbook of Proteolytic Enzymes, 2nd edition. London: Academic Press, pp. 374–387. [Google Scholar]
- Wang, L. & Lutkenhaus, J. (1998) FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Molecular Microbiology, 29, 731–740. [DOI] [PubMed] [Google Scholar]
- Wenzel, M. , Gulsoy, I.N.C. , Gao, Y. , Teng, Z. , Willemse, J. , Middelkamp, M. et al. (2021) Control of septum thickness by the curvature of SepF polymers. Proceedings of the National Academy of Sciences of the United States of America, 118, e2002635118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L.J. & Errington, J. (1994) Bacillus subtilis spoIIIE protein required for DNA segregation during asymmetric cell division. Science, 264, 572–575. [DOI] [PubMed] [Google Scholar]
- Yates, J. , Zhekov, I. , Baker, R. , Eklund, B. , Sherratt, D.J. & Arciszewska, L.K. (2006) Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Molecular Microbiology, 59, 1754–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Tran, A.H. , Sun, Q. & Margolin, W. (1998) Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N‐terminal targeting domain. Journal of Bacteriology, 180, 1296–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


