Abstract
Serological assays capable of measuring antibody responses induced by previous infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been critical tools in the response to the COVID-19 pandemic. In this study, we use bead-based multiplex assays to measure IgG and IgA antibodies and IgG avidity to five SARS-CoV-2 antigens (Spike (S), receptor-binding domain (RBD), Nucleocapsid (N), S subunit 2, and Membrane-Envelope fusion (ME)). These assays were performed in several cohorts of healthcare workers and nursing home residents, who were followed for up to eleven months after SARS-CoV-2 infection or up to six months after vaccination. Our results show distinct kinetic patterns of antibody quantity (IgG and IgA) and avidity. While IgG and IgA antibody levels waned over time, with IgA antibody levels waning more rapidly, avidity increased with time after infection or vaccination. These contrasting kinetic patterns allow for the estimation of time since previous SARS-CoV-2 infection. Including avidity measurements in addition to antibody levels in a classification algorithm for estimating time since infection led to a substantial improvement in accuracy, from 62% to 78%. The inclusion of antibody avidity in panels of serological assays can yield valuable information for improving serosurveillance during SARS-CoV-2 epidemics.
Keywords: SARS-CoV-2, serology, multiplex, antibody, avidity, kinetics, time since infection
1. Introduction
Severe acute respiratory syndrome coronavirus 2, or SARS-CoV-2, emerged as a zoonotic virus and was identified as the causative agent of COVID-19 in December 2019. SARS-CoV-2 is a Betacoronavirus belonging to the Sarbecovirus subgenus, like SARS-CoV. Coronaviruses have a positive-sense RNA genome of 26–32 kilobases. This genome encodes four structural proteins: Spike (S), Nucleocapsid (N), Envelope (E) and Membrane (M). The most important for protective immunity is the glycoprotein Spike, which forms a trimeric structure on the virus surface and comprises two subunits. Spike subunit 1 (S1) contains the receptor-binding domain (RBD) responsible for binding to the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell, while Spike subunit 2 (S2) permits the fusion of the viral and cellular membranes. Nucleocapsid plays an important role in transcription enhancement and viral assembly.
The kinetics of the SARS-CoV-2 antibody response following infection or vaccination have been analyzed in detail, with numerous studies demonstrating that specific immunoglobulin antibodies (IgG, IgA and IgM) to SARS-CoV-2 antigens develop between 6–15 days following symptom onset or vaccination. Following an initial period of boosting, antibody levels wane rapidly within the first 3–6 months, followed by a transition to a more slowly waning phase [1,2,3]. The different phases in the kinetics of the antibody response can be explained by a balance between populations of antibody-secreting plasma B cells with a short half-life (predominantly in the spleen) and a long half-life (located in the bone marrow). Over time, memory B cells increasingly differentiate into long-lived plasma cells present in the bone marrow, leading to a more mature antibody response [4]. Long-term follow-up of individuals infected with SARS-CoV-1 has shown that antibodies remain detectable six years after infection but continue to decrease [5].
Affinity maturation is the biological mechanism by which activated B cells undergo rounds of somatic hypermutations in immunoglobin genes, followed by an iterative clonal selection in germinal centers, resulting in the production of antibodies with greater affinities to the antigens over time [6,7]. Structural changes consist of slight amino acid mutations in the variable domains of antibodies, which improves the conformational fit of antibodies into their binding sites, therefore increasing the stability of the immune complexes. Antibody avidity, or functional affinity, measures the total strength of all of the non-covalent interactions between an antibody and its target antigen and can be extended to the total antigen-binding force of antibodies specific to a given antigen in sera. Avidity depends on three parameters: firstly, the binding affinity of the complex of antibodies and the antigen via a non-covalent interaction; secondly, the valency of the antibody; and thirdly, the structural arrangement of the antibody and antigen in the complex. While antibodies with low avidity are produced during the primary response, the progressive increase in the avidity of antibodies over time hence constitutes a useful marker of the maturation of the immune response and could help in providing estimates of time since infection.
In addition to providing insight into the immunology of SARS-CoV-2 infection, the measurement of antibody responses can provide valuable epidemiological information through the implementation of seroprevalence surveys [8]. In the case of SARS-CoV-2, the majority of seroprevalence studies involve the measurement of anti-N or anti-S IgG responses using immunoassays such as enzyme-linked immunosorbent assays (ELISA). In 2020, before the widespread roll-out of vaccines to prevent COVID-19, serological tests based on anti-N or anti-S IgG were demonstrated to have high sensitivity and high specificity for identifying individuals previously infected with SARS-CoV-2 [9]. However, the waning of antibodies was associated with substantial reductions in diagnostic sensitivity over time. The roll-out of COVID-19 vaccines has altered the role of seroprevalence surveys, as assays based on Spike proteins now measure a combination of naturally acquired and vaccine-induced immunity. Measurement of anti-N IgG can be used to distinguish naturally acquired from vaccine-induced immunity; however, this is complicated by the short duration of anti-N IgG antibodies [3], resulting in varying durations of seropositivity following infection [10].
In contrast to monoplex assays such as ELISA, multiplex serological assays can simultaneously measure antibodies to multiple antigens, allowing for more epidemiological information to be obtained from a single test. It has been demonstrated that multiplex assays can have higher accuracy than monoplex assays [11,12]; can distinguish vaccinated from naturally infected individuals [13]; can provide estimates of the time since previous infection [3]; and can simultaneously measure immunity to seasonal coronaviruses [14].
The majority of public health applications of serological assays involve the measurement of IgG or IgM antibodies. However, due to the distinct kinetic profiles of antibody responses, there are some notable examples where measuring avidity provides important additional clinical or epidemiological information. Avidity assays can be used to distinguish recent HIV infections from older infections [15], to diagnose cytomegalovirus or rubella viruses during pregnancy [16,17] and to identify cases of measles infection following vaccine failure [18,19]. In this multicentric study, we performed a detailed analysis of the kinetics of the antibody avidity response following infection or vaccination and demonstrated how antibody avidity can be used to improve the estimation of the time since previous SARS-CoV-2 infection.
2. Materials and Methods
2.1. Samples
The samples used in this study are summarized in Table 1. A panel of 522 positive serum samples from 174 healthcare workers with a proven history of SARS-CoV-2 infection in Strasbourg hospitals were collected, with up to three samples per individual collected over a period of nine months following symptom onset [20]. A second panel of samples was collected from healthcare workers in Institut Mutualiste Montsouris (IMM), a Parisian hospital. Of 784 healthcare workers sampled in April 2020, 32 were identified as having previous SARS-CoV-2 infection. For 29/32 of these healthcare workers, an additional sample was collected in February 2021. The longest duration of follow-up after infection in this study was 11 months. The 752 healthcare workers from IMM who were not infected before April 2020 formed the negative group for our study. A pool of serum from 27 PCR-positive healthcare workers from IMM was used for assay calibration.
Table 1.
Panels of samples included in the study.
| Natural Infection | Vaccination | |||||
|---|---|---|---|---|---|---|
| Strasbourg HCWs | Paris HCWs; Infected |
Paris HCWs; Uninfected | Orléans HCWs | Dublin CHR; No Past Infection |
Dublin CHR; Past Infection | |
| Participants | 174 | 32 | 752 | 16 | 47 | 39 |
| Samples | 522 | 64 | 752 | 120 | 126 | 106 |
| Female | 139 | 19 | 543 | 5 | 33 | 23 |
| Male | 35 | 13 | 209 | 11 | 14 | 16 |
| Age | 43 (25–73) | 37 (24–63) | 41 (19–72) | 59 (35–74) | 83 (53–98) | 83 (55–100) |
| Maximum days post-symptom onset | 219 (161–284) | 304 (285–336) | NA | NA | NA | 265 (224–298) * |
| Days post-vaccination | NA | NA | NA | 154 (151–168) | 206 (201–210) | 206 (201–210) |
Abbreviations: HCW, healthcare worker. CHR, care home resident. * Before first sampling.
In a study of vaccine-induced immune responses, 86 residents of nursing homes in Dublin, Ireland, were followed before and after receiving two doses of the Pfizer BNT162b2 vaccine [21]. Samples were collected at baseline and 5 weeks and 6 months after the second dose. Thirty-nine individuals had prior SARS-CoV-2 infection, and forty-seven were SARS-CoV-2 naïve. Another prospective, longitudinal cohort was established in the French city of Orléans to study immunity to SARS-CoV-2 following infection or vaccination. Sixteen individuals who received the Pfizer two-dose vaccine regimen were followed for up to 168 days.
2.2. Serological Assay
A previously described 9-plex bead-based assay was used for simultaneous detection of antibodies to 5 SARS-CoV-2 antigens and 4 seasonal coronaviruses (spike proteins of NL63, 229E, HKU1 and OC43) in 1 μL serum or plasma samples [3]. SARS-CoV-2 antigens of the ancestral lineage were from Spike (whole trimeric Spike, its RBD and S2), Nucleocapsid protein and Membrane-Envelope fusion protein (ME). ME and S2 antigens were purchased from Native Antigen (Oxford, United Kingdom), and all other antigens were produced as recombinant proteins at Institut Pasteur. The mass of proteins coupled to beads was optimized to generate a log-linear standard curve with a pool of 27 positive sera prepared from patients with reverse-transcription quantitative PCR–confirmed SARS-CoV-2. We measured the levels of immunoglobulin G (IgG) and immunoglobulin A (IgA) of each sample in two separate assays. Plates were read using a Luminex MAGPIX® system, and the median fluorescence intensity (MFI) was used for analysis. A 5-parameter logistic curve was used to convert MFI to relative antibody units relative to the standard curve generated on the same plate to account for inter-assay variation.
2.3. Multiplex Avidity Assay
To study avidity, we used a single concentration of a chaotropic agent to destabilize antibodies bound to antigens and measured the proportion of antibodies that remained bound after treatment in order to obtain a relative measure of the total binding strength of antibodies. The protocol was optimized by comparing the effects and dynamic range of three chaotropic agents (urea, guanidine hydrochloride and ammonium thiocyanate) on avidity measurements and testing their side effects on target antigens coupled to beads, incubation times and concentrations. Following assay optimization, a routine avidity assay was performed by treating serum samples at a single concentration of urea at 6M. The protocol for the avidity assay was similar to that of the serological assay with the inclusion of an additional step. After the incubation of beads with serum samples, the bead–Ab complexes were washed with assay buffer PBT (PBS-Tween20 0.05%-BSA 1%) and then incubated for 5 min with 100 μL of urea 6M diluted in water or water alone as a control. After 3 washing steps, 100 μL of anti-IgG secondary antibody conjugated to R-phycoerythrin (Jackson Immunoresearch) diluted at 1/100 was added for 15 min. After 3 final washes to remove unbound secondary antibodies, plates were read using a Luminex® MAGPIX® system, and the median fluorescence intensity (MFI) was used for analysis. Avidity was only assayed for IgG.
2.4. Statistical Analysis
All statistical analyses were performed with R version 4.0.5. Median fluorescence intensities (MFIs) were converted to relative antibody units (RAU) with a 5-parameter logistic curve relative to the standard curve generated on the same plate. The avidity index (AI) was calculated as the MFI of the sample treated with the chaotropic agent divided by the MFI without the chaotropic agent times 100. Linear regression was used to compare the difference between the means of the different groups of time since infection with SARS-CoV-2.
To assess whether avidity is of additional value to antibody measurements in the estimation of time since previous SARS-CoV-2 infection, we predicted time since infection with antibody measurements only and with antibody measurements and avidity estimates. We developed two random forest regression models. For each random forest, the number of trees was set at 1000. For each tree, two-thirds of the observations were used. Predictions were derived from the average of samples’ estimates in the remaining one-third of the samples, the out-of-bag samples. Regressions were built in a step-wise manner. First, the antigen in the regression was selected based on the importance of that antigen, measured by the mean decrease in accuracy on the out-of-bag samples. Subsequently, all other variables were added one by one to the most important antigen in the regression. The antigen associated with the lowest residual sum of squares was kept in the model. This process was repeated until no further decrease in the lowest residual sum of squares was observed. The randomForest package was used to develop and evaluate the random Forest regression models [22].
3. Results
3.1. Kinetics of SARS-CoV-2 Antibody Levels
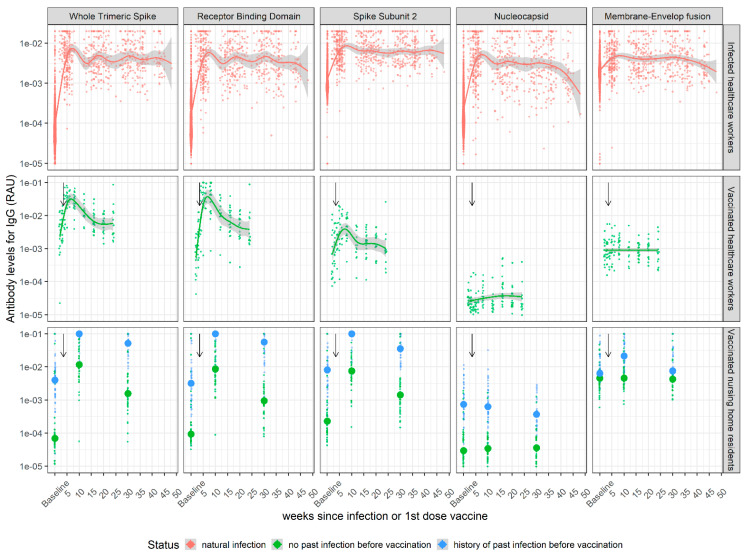
We evaluated SARS-CoV-2-specific antibody responses over time in healthcare workers and care home residents following PCR-confirmed SARS-CoV-2 infection or administration of Pfizer’s BNT162b2 vaccine (Table 1, 308 individuals and 938 samples). IgG antibody kinetics showed a consistent profile, with antibody levels increasing sharply following infection or vaccination, followed by an initial phase of rapid waning over the first 3–6 months and then by a transition to a phase of slower waning (Figure 1). IgA directed against whole Spike and RBD antigens followed a qualitatively similar pattern to IgG kinetics, with the exception that IgA antibodies waned more rapidly than IgG antibodies (Figure S1). Of note, for individuals who were vaccinated and had no history of past infection, no significant antibody signals to Nucleocapsid or Membrane-Envelope antigens were detected, as expected. Compared to nursing home residents with no history of natural infection, residents who were infected before vaccination showed a qualitatively similar kinetic pattern but consistently higher IgG responses to whole Spike, RBD and S2 antigens at all time points.
Figure 1.
IgG antibody kinetics following SARS-CoV-2 infection or vaccination with BNT162b2. IgG antibodies to five SARS-CoV-2 antigens were measured in serum samples using a bead-based multiplex Luminex assay. (First row) Healthcare workers from hospitals in Strasbourg and Paris were followed longitudinally after PCR-confirmed SARS-CoV-2 infection. (Middle row) Healthcare workers from a hospital in Orléans were followed longitudinally after receiving two doses of Pfizer BNT162b2 vaccine. (Bottom row) Residents of a nursing home in Dublin were followed after receiving two doses of Pfizer BNT162b2 vaccine. Individuals with “history of past infection” correspond to individuals with recorded SARS-CoV-2 infection before vaccination and are represented with blue dots. Individuals with no history of past infection are in green. Time is denoted as weeks post-vaccination. Thicker dots represent the median of each group. Black arrows indicate the date of the second vaccine injection.
3.2. Kinetics of SARS-CoV-2 IgG Antibody Avidity
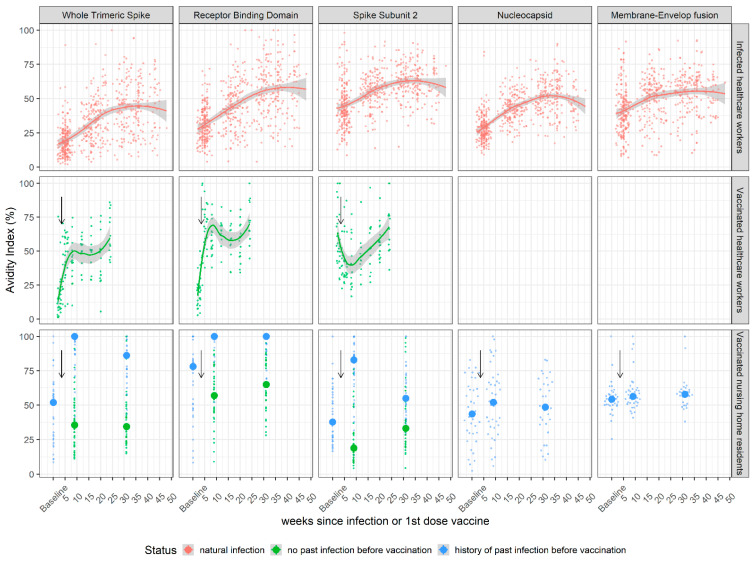
Analysis of the kinetics of the SARS-CoV-2 IgG antibody avidity response revealed a general pattern of increasing avidity over the monitored timeframe for both healthcare workers who were vaccinated or naturally infected (Figure 2, top and middle rows). However, this rise in avidity index showed a slightly different profile in vaccinated or naturally infected individuals for spike-related antigens. The median avidity index (AI) of anti-RBD IgG in vaccinated individuals reached a peak at 60% only 10 weeks after the first vaccine dose, while it took 30 weeks to reach an equivalent median AI in naturally infected individuals. Among elderly individuals, a significantly different kinetic profile was observed according to the history of natural infection preceding vaccination. Individuals who were infected and vaccinated (Figure 2, bottom panel, blue dots) showed a very homogeneous response with a strong increase in AI 10 weeks after vaccination that reached a peak median AI at 100% for whole spike and RBD antigens. In contrast, nursing home residents who were vaccinated with no prior infections (Figure 2, bottom panel, green dots) showed a median AI similar to or slightly lower than vaccinated HCWs (blue dots). Finally, the median AI of anti-N and anti-ME IgG among vaccinated individuals with prior infection (Figure 2, bottom panel, blue dots) remained stable throughout the study period and was therefore not influenced by vaccination.
Figure 2.
Kinetics of IgG avidity following SARS-CoV-2 infection or vaccination with BNT162b2. IgG avidity to five SARS-CoV-2 antigens was measured in serum samples using a bead-based multiplex Luminex assay. (First row) Healthcare workers from hospitals in Strasbourg and Paris were followed longitudinally following PCR-confirmed SARS-CoV-2 infection. (Middle row) Healthcare workers from a hospital in Orléans were followed longitudinally after receiving two doses of Pfizer BNT162b2 vaccine. (Bottom row) Residents of a nursing home in Dublin were followed after receiving two doses of Pfizer BNT162b2 vaccine. Individuals with “history of past infection” correspond to individuals with recorded SARS-CoV-2 infection before vaccination and are represented with blue dots. Individuals with no history of past infection are in green. Time is denoted as weeks post-vaccination. Thicker dots represent the median of each group. Black arrows indicate the date of the second vaccine injection. The avidity indexes of anti-Nucleocapsid and anti-Membrane-Envelope IgG are not shown for unvaccinated individuals, as well as data points for anti-spike (whole spike, RBD and S2) IgG of nursing home residents with no prior history of infection before vaccination.
3.3. Estimation of Time since Infection
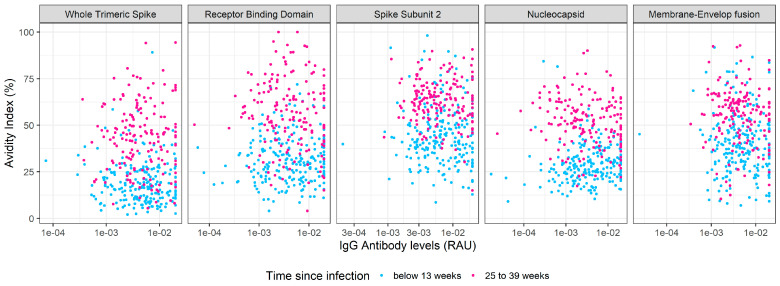
The clear temporal trends in antibody avidity demonstrate that there is a statistical signal for estimating time since previous infection. Figure 3 shows a comparison of anti-SARS-CoV-2 IgG levels and avidity for individuals with recent (less than 3 months) and older (6–9 months ago) naturally acquired SARS-CoV-2 infection. This representation shows that the two populations can be visually distinguished.
Figure 3.
Serological markers of time since infection. Anti-SARS-CoV-2 IgG levels and avidity were measured in samples from individuals with recent (within the previous 3 months, red) and older (6–9 months ago, blue) naturally acquired SARS-CoV-2 infection.
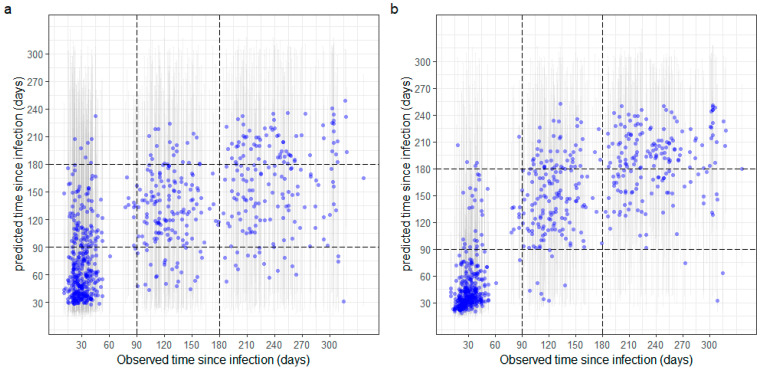
Estimation of time since infection was conducted with two random forest regression models. In the first model, we considered IgG and IgA antibodies. In the second model, we also included avidity measurements. The regression model without avidity estimates showed that RAU to NP IgA provided the highest accuracy, followed by NP IgG and RBD IgA (Figure S2a). The final model predicting time since infection without avidity included the following biomarkers: NP IgA, Spike IgA, NP IgG, S2 IgA, RBD IgA and S2 IgG. With these antigens, the regression model yielded a residual sum of squares of 4788 (Figure S2b). Categorizing each prediction into the categories of 3 months and less, 4 to 6 months, and infections longer than 6 months ago could classify 62.5% of the samples correctly (Figure 4a). Combining antibody levels and avidity led to improved accuracy of time since infection estimates, with the most informative biomarkers being NP avidity followed by Spike avidity. Antigens that reduced the residual sum of squares to 2608 were NP avidity, NP IgA, NP IgG, Spike avidity, RBD avidity and S2 IgA (Figure S2c). The predictions of this regression model are shown in Figure 4b. When we categorized the predictions into the three categories, 78% of the classifications were correct: 90% of samples taken within 3 months after symptom onset were classified correctly, 69% of samples 4 to 6 months were classified correctly, and 63% of samples taken 6 to 9 months after symptom onset were classified correctly.
Figure 4.
Predictions of time since infection without (a) and with (b) avidity measurements. Predictions are derived from random forest regression models, with point estimates in blue and 95% uncertainty intervals as vertical bars. (a) Predictions without avidity measurements from a random forest model with 6 biomarkers: NP IgA, S IgA, NP IgG, S2 IgA, RBD IgA and S2 IgG. (b) Predictions with avidity measurements from a random forest model with 6 biomarkers: NP avidity, NP IgA, NP IgG, S avidity, RBD avidity and S2 avidity.
4. Discussion
COVID-19 remains a critical threat to public health, and a deeper understanding of how antibodies are produced during infection is crucial. As can be seen from our results and others [23,24,25], antibody kinetics differ according to isotype and target antigen. Indeed, IgA wanes more rapidly than IgG antibodies for the studied SARS-CoV-2 antigens. This has potential implications for the duration of protective immunity against SARS-CoV-2 infection. The effect of the antibody level on immunity to SARS-CoV-2 has been well studied, with anti-Spike IgG levels and neutralizing antibody titers shown to be correlated and associated with protection from infection in vaccine studies [26,27,28]. The effect of the quality of the antibody response to SARS-CoV-2 has received comparatively less attention. As avidity measures the overall binding strength between antibodies and antigens, it has been suggested that avidity may be associated with protection against infection and has the potential to complement antibody titer data in the search for biological correlates of clinical protection [29].
The rationale is that for an equivalent number of antibodies, avidity can make a difference in terms of the quality of the response. Similarly, declining levels of antibodies over time could be partly compensated by an increase in avidity, thus maintaining an equivalent or even superior protective efficacy compared to serum with high antibody titers with poor avidity. This phenomenon has been observed in studies of vaccines against other pathogens, such as Plasmodium falciparum malaria [30] or Streptococcus pneumonia [31], where both antibody levels and avidity were shown to be significantly associated with vaccine efficacy.
In accordance with other studies [32], we found a progressive increase in IgG avidities to SARS-CoV-2 antigens in convalescent and vaccinated individuals after exposure. Although both groups were able to reach similar levels, we observed a significant difference in the rate at which this response took place. The median avidity index of vaccinated individuals exceeded 50% three times faster than the avidity index developed by naturally infected individuals (10 vs. 30 weeks). We could observe the same rapid boost in antibody avidity 10 weeks after vaccination in individuals who were previously infected, up to the maximal value of 100%. Other studies have reported a similar net increase in antibody avidity in previously exposed individuals receiving a vaccine booster dose, which was associated with a more efficient binding inhibition of spike to ACE2 in an in vitro competition assay [33]. It would be interesting to investigate whether this steeper increase in avidity associated with vaccination is caused by an acceleration of affinity maturation in germinal centers or simply reflects an ongoing trajectory towards a more complete process of affinity maturation. While our data seem to indicate the stabilization of anti-RBD IgG avidity at the end of follow-up in healthcare workers and care home residents, actually, it would be helpful to extend this follow-up to see how it evolves and correlates with neutralization efficiency.
In addition to being a critical tool for understanding the determinants of clinical protection, serological data can also provide important epidemiological information. Antibody avidity assays can be useful for the diagnosis of recent infections, as previously demonstrated for other pathogens such as measles, CMV or HIV [15,16,18]. The main objective of our study was to see whether avidity data could improve our previous model relying on IgG and IgA levels to spike and nucleocapsid antigens to produce estimates of time since infection based on its distinct kinetic pattern. By using machine learning algorithms to combine the data types, we conclude that antibody avidity is an even more accurate biomarker for identifying recent infection and leads to a substantial improvement to 78% accuracy.
One concern in the current situation is the continuous emergence and spread of SARS-CoV-2 variants characterized by antigenic changes in the Spike protein. These mutations are associated with reduced susceptibility to infection- or vaccine-induced immunity. Impaired binding efficiency of IgG to mutant RBD epitopes and enhanced stability of the ACE2-RBD complex [34] induced by key changes, such as mutation of residue 484, are two complementary mechanisms that would be very interesting to quantify at the epidemiological level with avidity measures in order to better know the determinants of transmission dynamics and the correlates of protection. In particular, it would be useful to study how these differential avidity responses to new variants can impact our prediction model and help to reconstruct successive waves of infection with variants of concerns.
Altogether, we provide further evidence that the integration of the IgG avidity parameter in the serological toolkit combined with epidemiological models can play a useful role in the response to the COVID-19 pandemic.
Acknowledgments
The authors are grateful to healthcare workers and nursing home residents who participated in this study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14071491/s1. Figure S1: IgA antibody kinetics following SARS-CoV-2 infection or vaccination with BNT162b2; Figure S2: Model development of random forest regressions predicting time since SARS-CoV-2 infection without and with avidity estimates.
Author Contributions
L.G., F.D., A.H.D., J.R. and S.P. performed the assays. L.G., S.P. and T.W. analyzed data, and T.W. wrote the algorithm to produce time since infection estimates. A.H.D., D.P., T.B., O.S., T.P., A.V., S.F.-K., I.B., C.R., E.C., M.M., S.P.K. and N.M.B. were involved in study design, patient recruitment, supervision, sample collection and distribution. M.T.W. and S.P. conceptualized and supervised the study. L.G., T.W., S.P. and M.T.W. wrote the initial draft. All authors contributed to the review and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Serum samples were biobanked at the Clinical Investigation and Access to BioResources platform at Institut Pasteur (Paris, France). Samples were obtained from consenting individuals through the CORSER study (ClinicalTrials.gov Identifier: NCT04325646), directed by Institut Pasteur and approved by the Comité de Protection des Personnes Ile de France III (on 19 February 2020), and the French COVID cohort (ClinicalTrials.gov Identifier: NCT04262921), sponsored by Inserm and approved by the Comité de Protection des Personnes Ile de France VI. Samples from healthcare workers in Strasbourg University Hospitals followed longitudinally were collected as part of an ongoing clinical trial (ClinicalTrials.gov Identifier: NCT04441684), which received ethical approval from the Comité de Protection des Personnes Ile de France III. Full ethical approval for the study in nursing home residents in Dublin was granted by the local ethics committee (Reference: 20-NREC-COV-049). The Orléans study was approved by the ILE DE FRANCE IV ethical committee. Blood sampling from vaccinated individuals was conducted as part of a clinical trial (ClinicalTrials.gov: NCT04750720).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data and code used for reproducing the results are available on request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the Fondation pour la Recherche Médicale (CorPopImm to MW), the French Government’s Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (Investissement d’Avenir grant n°ANR-10-LABX-62-IBEID), and INCEPTION programs (Investissement d’Avenir grant ANR-16-CONV-0005). SFK’s laboratory is funded by Strasbourg University Hospitals (SeroCoV-HUS; PRI 7782), the Agence Nationale de la Recherche (ANR-18-CE17–0028), Laboratoire d’Excellence TRANSPLANTEX (ANR-11-LABX-0070_TRANSPLANTEX) and Institut National de la Santé et de la Recherche Médicale (UMR_S 1109). The NH-COVAIR Study was funded by a grant from the Meath Foundation, Tallaght University Hospital. A.H.D. has been awarded the Irish Clinical Academic Training (ICAT) Programme, supported by the Wellcome Trust and the Health Research Board (Grant Number 203930/B/16/Z), the Health Service Executive, National Doctors Training and Planning, and the Health and Social Care, Research and Development Division, Northern Ireland. N.B. is funded under the Science Foundation Ireland Phase 2 COVID-19 Rapid Response Call (20/COV/8487) and the Health Research Board COVID-19 Rapid Response Call (COV19e2020e053).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wheatley A.K., Juno J.A., Wang J.J., Selva K.J., Reynaldi A., Tan H.-X., Lee W.S., Wragg K.M., Kelly H.G., Esterbauer R., et al. Evolution of Immune Responses to SARS-CoV-2 in Mild-Moderate COVID-19. Nat. Commun. 2021;12:1162. doi: 10.1038/s41467-021-21444-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallais F., Gantner P., Bruel T., Velay A., Planas D., Wendling M.-J., Bayer S., Solis M., Laugel E., Reix N., et al. Evolution of Antibody Responses up to 13 Months after SARS-CoV-2 Infection and Risk of Reinfection. EBio Med. 2021;71:103561. doi: 10.1016/j.ebiom.2021.103561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelleau S., Woudenberg T., Rosado J., Donnadieu F., Garcia L., Obadia T., Gardais S., Elgharbawy Y., Velay A., Gonzalez M., et al. Kinetics of the Severe Acute Respiratory Syndrome Coronavirus 2 Antibody Response and Serological Estimation of Time Since Infection. J. Infect. Dis. 2021;224:1489–1499. doi: 10.1093/infdis/jiab375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slifka M.K., Antia R., Whitmire J.K., Ahmed R. Humoral Immunity Due to Long-Lived Plasma Cells. Immunity. 1998;8:363–372. doi: 10.1016/S1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- 5.Tang F., Quan Y., Xin Z.-T., Wrammert J., Ma M.-J., Lv H., Wang T.-B., Yang H., Richardus J.H., Liu W., et al. Lack of Peripheral Memory B Cell Responses in Recovered Patients with Severe Acute Respiratory Syndrome: A Six-Year Follow-up Study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 6.Tas J.M.J., Mesin L., Pasqual G., Targ S., Jacobsen J.T., Mano Y.M., Chen C.S., Weill J.-C., Reynaud C.-A., Browne E.P., et al. Visualizing Antibody Affinity Maturation in Germinal Centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shlomchik M.J., Weisel F. Germinal Center Selection and the Development of Memory B and Plasma Cells. Immunol. Rev. 2012;247:52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 8.Bergeri I., Whelan M., Ware H., Subissi L., Nardone A., Lewis H.C., Li Z., Ma X., Valenciano M., Cheng B., et al. Global Epidemiology of SARS-CoV-2 Infection: A Systematic Review and Meta-Analysis of Standardized Population-Based Seroprevalence Studies, Jan 2020-Dec 2021. medRxiv. 2022 doi: 10.1101/2021.12.14.21267791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ainsworth M., Andersson M., Auckland K., Baillie J.K., Barnes E., Beer S., Beveridge A., Bibi S., Blackwell L., Borak M., et al. Performance Characteristics of Five Immunoassays for SARS-CoV-2: A Head-to-Head Benchmark Comparison. Lancet Infect. Dis. 2020;20:1390–1400. doi: 10.1016/S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y., Yang M., Peng Y., Liang Y., Wei J., Xing L., Guo L., Li X., Li J., Wang J., et al. Longitudinal Analysis of Antibody Dynamics in COVID-19 Convalescents Reveals Neutralizing Responses up to 16 Months after Infection. Nat. Microbiol. 2022;7:423–433. doi: 10.1038/s41564-021-01051-2. [DOI] [PubMed] [Google Scholar]
- 11.Drouot L., Hantz S., Jouen F., Velay A., Lamia B., Veber B., Sibilia J., Lotellier M., Candon S., Alain S., et al. Evaluation of Humoral Immunity to SARS-CoV-2: Diagnostic Value of a New Multiplex Addressable Laser Bead Immunoassay. Front. Microbiol. 2020;11:603931. doi: 10.3389/fmicb.2020.603931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarino C., Larson E., Babasyan S., Rollins A., Joshi L.R., Laverack M., Parrilla L., Plocharczyk E., Diel D.G., Wagner B. Development of a Quantitative COVID-19 Multiplex Assay and Its Use for Serological Surveillance in a Low SARS-CoV-2 Incidence Community. PLoS ONE. 2022;17:e0262868. doi: 10.1371/journal.pone.0262868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brochot E., Souplet V., Follet P., Ponthieu P., Olivier C., Even G., Audebert C., Malbec R. A Multiplex Serological Assay for the Characterization of IgG Immune Response to SARS-CoV-2. PLoS ONE. 2022;17:e0262311. doi: 10.1371/journal.pone.0262311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F.F., Liu A., Gibbs E., Tanunliong G., Marquez A.C., Gantt S., Frykman H., Krajden M., Morshed M., Prystajecky N.A., et al. A Novel Multiplex Electrochemiluminescent Immunoassay for Detection and Quantification of Anti-SARS-CoV-2 IgG and Anti-Seasonal Endemic Human Coronavirus IgG. J. Clin. Virol. 2022;146:105050. doi: 10.1016/j.jcv.2021.105050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chawla A., Murphy G., Donnelly C., Booth C.L., Johnson M., Parry J.V., Phillips A., Geretti A.M. Human Immunodeficiency Virus (HIV) Antibody Avidity Testing To Identify Recent Infection in Newly Diagnosed HIV Type 1 (HIV-1)-Seropositive Persons Infected with Diverse HIV-1 Subtypes. J. Clin. Microbiol. 2007;45:415–420. doi: 10.1128/JCM.01879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prince H.E., Lapé-Nixon M. Role of Cytomegalovirus (CMV) IgG Avidity Testing in Diagnosing Primary CMV Infection during Pregnancy. Clin. Vaccine Immunol. 2014;21:1377–1384. doi: 10.1128/CVI.00487-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agbede O.O., Adeyemi O.O., Olatinwo A.W.O. Significance of IgG-Avidity in Antenatal Rubella Diagnosis. J. Fam. Reprod. Health. 2013;7:131–137. [PMC free article] [PubMed] [Google Scholar]
- 18.Mercader S., Garcia P., Bellini W.J. Measles Virus IgG Avidity Assay for Use in Classification of Measles Vaccine Failure in Measles Elimination Settings. Clin. Vaccine Immunol. 2012;19:1810–1817. doi: 10.1128/CVI.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sowers S.B., Rota J.S., Hickman C.J., Mercader S., Redd S., McNall R.J., Williams N., McGrew M., Walls M.L., Rota P.A., et al. High Concentrations of Measles Neutralizing Antibodies and High-Avidity Measles IgG Accurately Identify Measles Reinfection Cases. Clin. Vaccine Immunol. 2016;23:707–716. doi: 10.1128/CVI.00268-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grzelak L., Velay A., Madec Y., Gallais F., Staropoli I., Schmidt-Mutter C., Wendling M.-J., Meyer N., Planchais C., Rey D., et al. Sex Differences in the Evolution of Neutralizing Antibodies to Severe Acute Respiratory Syndrome Coronavirus 2. J. Infect. Dis. 2021;224:983–988. doi: 10.1093/infdis/jiab127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dyer A.H., Noonan C., McElheron M., Batten I., Reddy C., Connolly E., Pierpoint R., Murray C., Leonard A., Higgins C., et al. Previous SARS-CoV-2 Infection, Age, and Frailty Are Associated With 6-Month Vaccine-Induced Anti-Spike Antibody Titer in Nursing Home Residents. J. Am. Med. Dir. Assoc. 2022;23:434–439. doi: 10.1016/j.jamda.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liaw A., Wiener M. Classification and Regression by RandomForest. Comput. Sci. 2007;2:18–22. [Google Scholar]
- 23.Koerber N., Priller A., Yazici S., Bauer T., Cheng C.-C., Mijočević H., Wintersteller H., Jeske S., Vogel E., Feuerherd M., et al. Dynamics of Spike-and Nucleocapsid Specific Immunity during Long-Term Follow-up and Vaccination of SARS-CoV-2 Convalescents. Nat. Commun. 2022;13:153. doi: 10.1038/s41467-021-27649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M., Liu J., Lu R., Zhang Y., Du M., Xing M., Wu Z., Kong X., Zhu Y., Zhou X., et al. Longitudinal Immune Profiling Reveals Dominant Epitopes Mediating Long-Term Humoral Immunity in COVID-19-Convalescent Individuals. J. Allergy Clin. Immunol. 2022;149:1225–1241. doi: 10.1016/j.jaci.2022.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurano M., Morita Y., Nakano Y., Yokoyama R., Shimura T., Qian C., Xia F., He F., Zheng L., Ohmiya H., et al. Response Kinetics of Different Classes of Antibodies to SARS-CoV2 Infection in the Japanese Population: The IgA and IgG Titers Increased Earlier than the IgM Titers. Int. Immunopharmacol. 2022;103:108491. doi: 10.1016/j.intimp.2021.108491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of Protection against Symptomatic and Asymptomatic SARS-CoV-2 Infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune Correlates Analysis of the MRNA-1273 COVID-19 Vaccine Efficacy Clinical Trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing Antibody Levels Are Highly Predictive of Immune Protection from Symptomatic SARS-CoV-2 Infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 29.Bauer G. The Potential Significance of High Avidity Immunoglobulin G (IgG) for Protective Immunity towards SARS-CoV-2. Int. J. Infect. Dis. 2021;106:61–64. doi: 10.1016/j.ijid.2021.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobaño C., Sanz H., Sorgho H., Dosoo D., Mpina M., Ubillos I., Aguilar R., Ford T., Díez-Padrisa N., Williams N.A., et al. Concentration and Avidity of Antibodies to Different Circumsporozoite Epitopes Correlate with RTS,S/AS01E Malaria Vaccine Efficacy. Nat. Commun. 2019;10:2174. doi: 10.1038/s41467-019-10195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Usinger W.R., Lucas A.H. Avidity as a Determinant of the Protective Efficacy of Human Antibodies to Pneumococcal Capsular Polysaccharides. Infect. Immun. 1999;67:2366–2370. doi: 10.1128/IAI.67.5.2366-2370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tauzin A., Gendron-Lepage G., Nayrac M., Anand S.P., Bourassa C., Medjahed H., Goyette G., Dubé M., Bazin R., Kaufmann D.E., et al. Evolution of Anti-RBD IgG Avidity Following SARS-CoV-2 Infection. Viruses. 2022;14:532. doi: 10.3390/v14030532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glück V., Tydykov L., Mader A.-L., Warda A.-S., Bertok M., Weidlich T., Gottwald C., Köstler J., Salzberger B., Wagner R., et al. Humoral Immunity in Dually Vaccinated SARS-CoV-2-Naïve Individuals and in Booster-Vaccinated COVID-19-Convalescent Subjects. Infection. 2022 doi: 10.1007/s15010-022-01817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koehler M., Ray A., Moreira R.A., Juniku B., Poma A.B., Alsteens D. Molecular Insights into Receptor Binding Energetics and Neutralization of SARS-CoV-2 Variants. Nat. Commun. 2021;12:6977. doi: 10.1038/s41467-021-27325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and code used for reproducing the results are available on request.