Abstract
Norway spruce (Picea abies) is an economically and ecologically important tree species that grows across northern and central Europe. Treating Norway spruce with jasmonate has long‐lasting beneficial effects on tree resistance to damaging pests, such as the European spruce bark beetle Ips typographus and its fungal associates. The (epi)genetic mechanisms involved in such long‐lasting jasmonate induced resistance (IR) have gained much recent interest but remain largely unknown. In this study, we treated 2‐year‐old spruce seedlings with methyl jasmonate (MeJA) and challenged them with the I. typographus vectored necrotrophic fungus Grosmannia penicillata. MeJA treatment reduced the extent of necrotic lesions in the bark 8 weeks after infection and thus elicited long‐term IR against the fungus. The transcriptional response of spruce bark to MeJA treatment was analysed over a 4‐week time course using mRNA‐seq. This analysis provided evidence that MeJA treatment induced a transient upregulation of jasmonic acid, salicylic acid and ethylene biosynthesis genes and downstream signalling genes. Our data also suggests that defence‐related genes are induced while genes related to growth are repressed by methyl jasmonate treatment. These results provide new clues about the potential underpinning mechanisms and costs associated with long‐term MeJA‐IR in Norway spruce.
Keywords: epigenetics, induced resistance, methyl jasmonate, phytohormones, Picea abies, transcriptome
Summary Statment
A bark transcriptome time course shows that MeJA induces both prolonged and transient changes in gene expression. Genes related to the RdDM pathway are repressed, suggesting that epigenetic changes and DNA hypomethylation are important in the establishment and maintenance of MeJA‐IR in Norway spruce.
1. INTRODUCTION
Norway spruce (Picea abies) is an economically and ecologically important tree species that dominates Europe's boreal and sub‐alpine coniferous forests (Caudullo et al., 2016). However, across much of its range, Norway spruce is threatened by the tree‐killing European spruce bark beetle, Ips typographus (Biedermann et al., 2019). The exhaustion of tree defences, and ultimately the success of bark beetle attacks, is thought to be aided by infection of the tree by necrotrophic ophiostomatoid fungi carried by the beetle (Krokene, 2015; Zhao et al., 2019). Common fungal phytopathogens associated with I. typographus include Grosmannia penicillata, Endoconidiophora polonica and Ophiostoma bicolor (Linnakoski et al., 2010; Tanin et al., 2021).
Norway spruce and other members of the Pinaceae family have a comprehensive suite of defences to protect themselves against potentially deadly attackers. In addition to constitutive defences, such as lignified sclerenchyma cells and intracellular deposits of calcium oxalate crystals, these conifers have a range of inducible defences (Franceschi et al., 2005; Krokene, 2015). These include production of traumatic resin ducts (TRDs) filled with a terpene‐rich oleoresin and activation of polyphenolic parenchyma cells rich in phenolic compounds such as stilbenes and flavonoids (Celedon & Bohlmann, 2019; Hammerbacher et al., 2020). Aside from enhanced production of terpenes and phenolics, induction of a hypersensitive cell death response and mobilisation of pathogenesis‐related (PR) proteins are also thought to provide resistance against biotic stress in conifers (Franceschi et al., 2005). Despite this extensive repertoire of inducible defences, I. typographus and its ophiostomatoid fungal associates continue to decimate Norway spruce plantations across central and eastern Europe (Hlásny et al., 2021). Thus, there is an urgent need to understand the defence mechanisms of Norway spruce in more detail and develop novel pest and disease strategies to protect this important tree species.
Exposure of plants to specific environmental stimuli, such as localised pathogen infection, beneficial microbes or specific chemicals, can make them more resistant to subsequent attack (Mauch‐Mani et al., 2017; Pieterse et al., 2014). This phenomenon is known as induced resistance (IR) and provides a feasible route by which particularly young trees in a nursery setting could be protected against pests and diseases. Through direct induction of plant defences, IR can provide immediately, and usually short‐term, protection that lasts for days or a few weeks. More long‐term plant protection by IR may occur through two non‐mutually exclusive mechanisms: (I) prolonged upregulation of inducible defences and (ii) priming of inducible defences (Conrath et al., 2015; De Kesel et al., 2021; Mauch‐Mani et al., 2017; Wilkinson et al., 2019). Prolonged upregulation of inducible defences is where an IR eliciting stimulus (e.g., insect attack) induces defences directly and they remain upregulated for weeks or months. With priming of inducible defences, defences might be transiently activated following an IR eliciting stimulus, but return to basal levels until a subsequent triggering stress activates faster and/or stronger inducible defences. Although long‐term IR may be underpinned by either prolonged upregulation or priming of inducible defences, enhanced plant resistance is often a result of a combination of the two mechanisms (Mageroy, Wilkinson, et al., 2020). Considerable research on the mechanisms involved in IR has been conducted in model angiosperms. For example, increasing evidence suggests that epigenetic mechanisms are involved in the maintenance of particularly longer‐lasting IR (Parker et al., 2022; Wilkinson et al., 2019). DNA methylation, which is partially regulated by RNA‐dependent DNA methylation (RdDM) and DNA demethylases, is one epigenetic mark which has been strongly linked to long‐term IR (Parker et al., 2021). Stress‐induced DNA hypomethylation could be brought about via a repression of machinery involved in RdDM and/or the activation of DNA demethylases.
Application of methyl jasmonate (MeJA) has been shown to induce resistance against I. typographus and its fungal associates in Norway spruce (Erbilgin et al., 2006; Krokene et al., 2008; Mageroy, Christiansen, et al., 2020; Zeneli et al., 2006). MeJA‐IR is linked to both direct and prolonged induction of defences, such as the formation of TRDs and new polyphenolic cells (Celedon & Bohlmann, 2019; Krokene et al., 2008). In addition, MeJA‐IR in Norway spruce has been associated with a priming of inducible defences. Zhao et al. (2011) demonstrated that MeJA treatment alone induces a minor increase in terpene levels in the bark compared to the massive terpene accumulation that occurred following wounding of bark treated with MeJA 4 weeks previously. This accumulation does not appear to be the result of de novo production, as enzymes involved in terpene biosynthesis are generally not primed at a transcriptional level (Mageroy, Wilkinson et al., 2020). Rather, MeJA treatment primed a swathe of PR genes to respond faster and stronger to wounding in mature spruce trees (Mageroy, Wilkinson et al., 2020).
Despite these recent findings, much is still unknown about the molecular mechanisms underpinning the establishment and maintenance of MeJA‐IR in Norway spruce. In this study, we aimed to (i) verify that treating 2‐year old spruce plants with MeJA induces long‐term resistance to necrotrophic ophiostomatoid fungi, (ii) use a detailed mRNA‐seq analysis with multiple time points to obtain a more complete understanding of the transcriptional changes following MeJA treatment, (iii) identify changes in gene expression that could contribute to long‐term MeJA‐IR, and (iv) evaluate costs associated with MeJA‐IR by quantifying growth and resistance of MeJA‐treated spruce plants.
2. METHODS
2.1. Overall experimental design
To address the aims of this study, we set up a series of experiments which are summarised in Figure 1. In all experiments, plants were either treated with MeJA or a water control solution. Subsequently, in the first experiment plants were inoculated with a necrotrophic fungus to verify that MeJA elicited IR. In the second experiment, bark tissue was harvested at various timepoints to establish the impact of MeJA on the bark transcriptome. Finally, two experiments were used to explore the potential costs associated with MeJA‐IR. In one, plants were used for growth and morphology analysis while in the other chlorophyll fluorescence measurements were performed as a rough proxy measure of needle photosynthesis.
Figure 1.
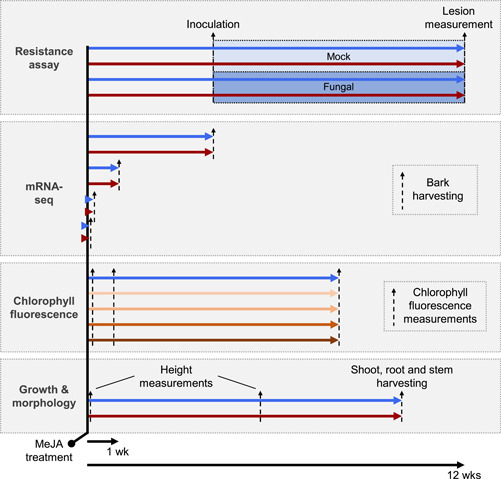
Experimental setup to study establishment and maintenance of methyl jasmonate (MeJA)‐induced resistance in Norway spruce seedlings. Two‐year‐old seedlings were treated with either water (blue lines), 10 mM MeJA (dark red lines) or other concentrations of MeJA (orange lines; details below). For the resistance assay, plants were either inoculated with a necrotrophic fungal pathogen or mock inoculated 4 weeks after treatment. Eight weeks later, necrotic lesions were measured. mRNA‐seq was conducted on mRNA extracted from bark tissue harvested from plants at 3 h, 24 h, 1 week and 4 weeks after MeJA treatment. Chlorophyll fluorescence was measured 1 day, 6 days and 8 weeks after treatment with 0, 5, 25, 50 or 100 mM MeJA. Plant height growth was measured twice over a period of 5.5 weeks. Shoots, roots and stems were then harvested at 10 weeks to explore the impact of MeJA on plant growth and morphology [Color figure can be viewed at wileyonlinelibrary.com]
2.2. Plant materials, growth conditions, and MeJA treatment for pathogen bioassay and mRNA‐Seq
In May 2018, 2‐year‐old Norway spruce seedlings grown in multipot containers (50 cm3 pots with 95 minipots per container, 791 seedlings m−2) were purchased from the nursery Norgesplanter AS. The seedlings, which had overwintered at 7°C with the root plugs wrapped in clingfilm, were transferred to 0.8 L pots (7.5 × 7.5 × 12 cm; Nelson Garden, product no. 5726) containing compost supplemented with mineral fertiliser (Plantasjen, EAN:7058782362802). Seedlings were grown outdoors with an irrigation system providing additional water when required. We performed all manipulations and experimentation from the end of July 2018 onwards, when yearly height and shoot growth had been completed.
On 31 July 2018, half of the spruce seedlings were sprayed with a 10 mM MeJA solution while the remaining half received a 0 mM MeJA control solution. The concentration of MeJA was selected based on a previous study which demonstrated that 10 mM MeJA induced a defence response in 2‐year‐old seedlings and therefore would likely trigger IR (Martin et al., 2002). The MeJA solution consisted of MeJA (Sigma‐Aldrich, 392707) dissolved in tap water and supplemented with 0.1% Tween 20 (Sigma‐Aldrich, P9416) to ensure even coating across all sprayed tissues. The control solution was identical except that it did not contain any MeJA. Using a 1.5 L pressurised spray bottle (Bürkle GmbH, 0309‐0100), each plant was sprayed with a similar volume of solution, which was enough to saturate the entire stem surface. Following spraying, plants of the two pre‐treatments were kept separate for at least 4 h to allow excess solution to evaporate.
2.3. Necrotrophic pathogen (Grosmannia penicillata) bioassay
Grosmannia penicillata isolate 1980‐91/54 (collected: 1980, Akershus) from the Norwegian Institute of Bioeconomy Research (NIBIO) fungal culture collection, was revived from a −150°C culture stock and propagated on malt agar (2% malt extract and 1% agar). Seedlings were inoculated ~4 weeks (29 August 2018) after treatment with water or MeJA (Figure 1). Plants from the control and MeJA treatment groups were inoculated with either sterile malt agar (mock) or G. penicillata (n = 15). The inoculation procedure was identical for all plants regardless of whether they were inoculated with a fungus‐colonised or sterile agar inoculum. First, a wedge‐shaped wound, ~1 cm from top to bottom and half the circumference of the stem in width, was cut in the middle of the first internode by slicing open outer layers of bark with a scalpel. The resulting bark remained attached to the stem and a ~5 mm3 droplet of inoculum was placed behind the flap using a 5 ml needle‐less syringe. To seal the inoculum in place behind the bark flap and to prevent contamination and/or drying out of the wound, Parafilm was wrapped around the stem at the inoculation site. Inoculum was created by homogenising a batch of malt agar, with or without fungus. Inoculated plants were arranged into eight experimental blocks. Seven blocks had two plants of each combination of treatment (water control or MeJA) and inoculation type (mock or fungal), while one block only had one plant per combination. The position of plants was randomised in each block. Approximately 8 weeks after inoculation (25 October 2018), plant resistance was assessed by measuring the lesion length in the inner bark above and below each inoculation site. Parafilm and bark flap were removed, and the inner bark was exposed by removing the outer layers of bark using a scalpel. In both mock and fungal inoculated plants, a distinct area of darkened, necrotic tissue was visible around the wound site (Figure 2a). The axial length of each lesion was measured using calipers (Digital Vernier Caliper, Cocraft).
Figure 2.
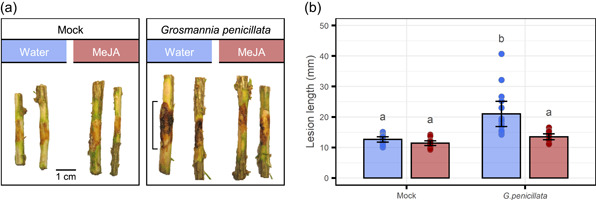
Methyl jasmonate (MeJA) elicits induced resistance against Grosmannia penicillata. Two‐year‐old Norway spruce seedlings were treated with either water (blue) or 10 mM MeJA (red) 4 weeks before inoculation with either a G. penicillata or sterile malt agar inoculum. Symptoms were assessed 8 weeks post inoculation. (a) The first stem internode, with wound and/or fungal induced cell death, from all plants of one representative experimental block. (b) The full axial length of wounds or necrotic lesions (indicated in [a]) were measured on all stems. Results are presented as mean ± 95% confidence interval of the mean. Points show individual replicates (n = 15). Bars with different letters are significantly different (ANOVA followed by Tukey post hoc test, p < 0.05) [Color figure can be viewed at wileyonlinelibrary.com]
2.4. Pathogen bioassay statistical analysis
A three‐factor ANOVA was performed to assess the effect of water/MeJA treatment and inoculation on lesion length, using R v3.6.1 (R Core Team, 2019). To ensure the data conformed to ANOVA's normality assumption, lesion lengths were log‐transformed. The ANOVA included treatment, inoculation, and block as fixed effects, as well as the treatment × inoculation interaction. Due to an imbalance in the number of replicates per block and a significant interaction effect, type III sum of squares were used. A Tukey post‐hoc test with a statistical significance threshold of p < 0.05 was used to evaluate whether mean lesion lengths differed significantly between treatment and inoculation combinations.
2.5. Bark harvesting and RNA extraction
Bark was harvested 3 h, 6 h, 24 h, 72 h, 1 week and 4 weeks after treatment, from the first stem internode of four seedlings per treatment (water and MeJA). Each seedling was treated as a separate sample, therefore each treatment (e.g., MeJA treated) had four replicates for each harvest time. Immediately following harvesting the bark samples were flash frozen in liquid nitrogen and then stored at −80°C. Using a pestle and mortar, all bark samples were later ground to a powder in liquid nitrogen. For the samples collected 4 weeks posttreatment only half of the harvested bark was ground to a powder.
Total RNA was extracted from 30 to 50 mg of bark powder per sample using the MasterPure Complete DNA and RNA Purification Kit (Lucigen, MC85200). To denature ribonucleases and reduce polyphenolic contamination of extracted nucleic acids, 0.5% β‐mercaptoethanol (Sigma‐Aldrich, M3148) and 1% polyvinylpyrrolidone (PVP; Sigma‐Aldrich, P‐5288) were added to the extraction buffer. To reduce carbohydrate contamination, nucleic acids were precipitated using 0.5 volumes of 7.5 M lithium chloride precipitation solution (ThermoFisher Scientific, AM9480), subject to an incubation step at −20°C for 2 h, and centrifuged for 30 min at 16 500 × g and 4°C. Nucleic acids were resuspended in 60 μl of nuclease‐free water.
2.6. RT‐qPCR analysis
The Maxima First Strand cDNA Synthesis Kit for RT‐qPCR with dsDNase (ThermoFisher Scientific, K1671) was used, according to manufacturer's instructions, to remove contaminant genomic DNA and reverse transcribe extracted mRNA to cDNA. For each sample, 200 ng of total RNA was used for cDNA synthesis. qPCR assessment of gene expression was performed with 2‐fold diluted cDNA, SYBR™ Select Master Mix (ThermoFisher Scientific, 4472919) and the Applied Biosystems® ViiA™ 7 Real‐Time PCR System. qPCRs were set up according to manufacturer's recommendations. Primer sequences are shown in Table S1. Ct values, outputted by the ViiA™ 7 system, were adjusted for amplification efficiency calculated using the programme LinRegPCR (Ruijter et al., 2009). Adjusted Ct values were first calibrated to one of the four 3 h water treated samples. Calibrated Ct values were then normalised using the mean of two endogenous reference genes, actin and alpha‐tubulin. Finally, normalised Ct values were expressed relative to the mean of the water treated replicates from the 3 h timepoint. Plots displaying relative expression values were created using the R package ggplot2.
2.7. mRNA library preparation and sequencing
Preliminary RT‐qPCR analysis (Figure S1) suggested that the 3 h, 24 h, 1 week and 4 weeks time points were sufficient to capture the key temporal trends in the transcriptome data. Therefore, the RNA samples from these time points were selected for global transcriptome analysis. Before sequencing, quantity and quality of RNA was assessed using a Nanodrop and Agilent 2100 Bioanalyzer. All samples had an RNA integrity number (RIN) value ≥ 8.5. Library preparation and sequencing of all RNA samples were performed by BGI Tech Solutions (Tai Po, Hong Kong). Across the 32 samples, 1.2 billion 150 bp paired‐end (PE) clean reads were generated in total, with the minimum, maximum and mean number of read pairs per sample being 27.9, 38.1 and 36.3 million, respectively (Data Set S1). BGI removed adaptor sequences, contamination and low‐quality reads. They reported that for all samples, ≥96% of nucleotides had a Phred quality score of >20.
2.8. Read alignment and counting
The Bowtie 2 package v2.3.1 (Langmead & Salzberg, 2012) was used to align reads to the curated Norway spruce reference transcriptome described in Mageroy, Wilkinson, et al., 2020). First, the transcriptome was indexed using the bowtie2‐build function, and then reads were aligned using the bowtie2 function run with the parameter settings: ‘‐‐very‐sensitive' ‘‐q' ‘‐k 10'. With option k being set to 10, up to 10 valid alignments were reported per read pair rather than the default single best alignment. This approach allowed detection and removal of multi‐mapping reads. On average, 22.5% (8.2 million) of the raw read pairs in each sample remained following alignment and subsequent multi‐mapper removal (Data Set S1). The number of uniquely aligned fragments mapping to each gene were counted using the featureCounts function from the Rsubread R package v2.0.1 (Liao et al., 2014, 2019). The following options were specified in the featureCounts function: ‘isGTFAnnotationFile = TRUE', ‘isPairedEnd = TRUE'. The Gene Transfer Format (GTF) reference transcriptome annotation file was created using AUGUSTUS run with the parameter settings: ‘‐‐strand=both', ‘‐‐genemodel=partial', ‘‐ ‐species=arabidopsis' (Stanke et al., 2004).
2.9. Global pattern assessment and differential expression analysis
To assess the global patterns in the data, count tables created by featureCounts were loaded into R and all genes with <100 total read counts across all samples were removed. Remaining read counts were transformed with a variance stabilising transformation (vst) (Anders & Huber, 2010) using the DESeq. 2 vst function run with the following option settings: ‘blind = TRUE', ‘nsub=1000', ‘fitType = "parametric"'. Transformed count data was used for a principal component analysis (PCA), a hierarchical clustering analysis (HCA), and heatmap analysis of sample‐to‐sample distances. The PCA was performed using the DESeq. 2 plotPCA function run with the following options: ‘intgroup = c(“Treatment”, “Time”)', ‘ntop = all genes ≥ 100 read counts', ‘returnData = TRUE'. The outcome of the PCA was displayed using the R package ggplot2 v3.2.1. The R package pheatmap v1.0.12 (Kolde, 2019) was used to cluster samples and create heatmaps displaying sample‐to‐sample distances. Samples were clustered using the complete‐linkage method and the euclidean distances between samples.
Differential expression analysis was conducted in R using the package DESeq. 2 v1.24.0 (Love et al., 2014). Genes showing a significantly altered expression profile across time as a result of MeJA treatment were identified using the DEseq function run with the following parameter settings: ‘test = “LRT”', ‘full = ~ time + treatment:time', ‘reduced = ~ time'. Genes with an adjusted p < 0.001 were selected from the results table created by the DESeq. 2 results function run with the parameter settings: ‘alpha = 0.001', ‘cooksCutoff = T', ‘lfcThreshold = 0' (Benjamini & Hochberg, 1995).
2.10. Hierarchical clustering and expression pattern visualisation
Genes were grouped by expression pattern using agglomerative hierarchical clustering. A correlation‐based distance measure was used to calculate between‐gene dissimilarity based on counts transformed using the DESeq. 2 vst function run with the argument values: ‘blind = FALSE', ‘nsub = 1000', ‘fitType = "parametric"'. A dendrogram was created from the dissimilarity data using Ward's minimum variance clustering method. For visualisation, the dendrogram was displayed together with the heatmap, with each row in the heatmap representing a separate gene with transformed counts displayed as Z‐scores. All aforementioned steps were conducted using the aheatmap function from the NMF R package v0.21.0 (Gaujoux & Seoighe, 2010). All bar charts and line plots showing numbers of differentially expressed genes and gene expression profiles, respectively, were created using the R package ggplot2. Expression profile plots used the same transformed counts as those that were used for the hierarchical clustering.
2.11. Protein signature enrichment analysis
Predicted protein sequences for all genes with ≥100 total read counts across all samples were annotated with protein signatures (e.g., protein domain) using the Pfam database v32.0 (El‐Gebali et al., 2019) and hmmscan from the HMMER package v3.2.1 (Wheeler & Eddy, 2013). These annotated genes provided the background distribution of Pfam protein signatures for enrichment analysis, which was conducted using the enrichment function from the bc3net R package v1.0 (de Matos Simoes & Emmert‐Streib, 2016). For a Pfam protein signature to be classified as enriched in a candidate protein list it had to have an adjusted p ≤ 0.05 (Benjamini & Hochberg, 1995). Fold‐enrichment plots displaying significantly enriched protein signatures were created using ggplot2.
2.12. Gene ontology (GO) enrichment analysis
Enrichment analysis of biological function GO terms was conducted using the enrichment tool of ConGenIE (http://congenie.org/), the online host of information and data relating to the Norway spruce genome project (Nystedt et al., 2013; Sundell et al., 2015). The background for the analysis consisted of all genes in the spruce genome assigned at least one biological function GO term. For a GO term to be reported it had to be assigned to one or more transcript in a cluster and it had to have a false discovery rate (FDR) adjusted p < 0.05.
2.13. Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis
GhostKOALA (Kanehisa et al., 2016) was used, where possible, to annotate and map predicted protein sequences for all differentially expressed genes to KEGG pathways. Of the 6330 differentially expressed genes, 2704 (42.7%) were annotated and mapped (Data Set S2). To compare expression patterns of genes related to defence and growth, we selected genes annotated to KEGG pathways related to terpene biosynthesis [00900 Terpenoid backbone biosynthesis (12 genes); 00909 Sesquiterpenoid and triterpenoid biosynthesis (1 gene); 00904 Diterpenoid biosynthesis (6 genes)] and phenolic biosynthesis [00940 Phenylpropanoid biosynthesis (13 genes); 00945 Stilbenoid, diarylheptanoid and gingerol biosynthesis (4 genes); 00941 Flavonoid biosynthesis (11 genes); 00944 Flavone and flavonol biosynthesis (1 gene)] and genes annotated to KEGG pathways related to growth [04110 Cell cycle (27 genes)] and photosynthesis [00195 Photosynthesis (22 genes); 00196 Photosynthesis—antenna proteins (7 genes)].
2.14. Identification of differentially expressed genes related to defence or epigenetics
Differentially expressed genes were annotated using predicted amino acid sequences (blastp v2.8.1; Altschul et al., 1990, 1997; Camacho et al., 2009) and the Swiss‐Prot database (Bateman, 2019). Genes predicted to encode for hormone biosynthesis enzymes, defence regulators, defence metabolite biosynthesis enzymes or PR proteins were identified by searching the blastp outputs for key terms (e.g., 'WRKY', 'Pathogenesis‐related' and 'anthocyanidin reductase'). Differentially expressed genes predicted to encode for epigenetic regulators were identified using the lists of epigenetic regulator genes provided in Mageroy, Wilkinson, et al. (2020).
2.15. Terpene quantification
Terpenes were extracted and quantified according to Flø et al. (2018) from bark samples collected 3 h, 24 h, 1 week and 4 weeks after treatment with water or MeJA. Briefly, ground bark tissue was extracted overnight with hexane containing 10 μg/ml pentadecane as an internal standard. The supernatant was removed for analysis by GC‐MS using a model 6890 N gas chromatograph (Agilent Technologies) connected to a model 5973 mass spectrometer (Agilent Technologies) fitted with an autosampler. Remaining bark tissue was dried at room temperature for 3 days and weighed to determine dry weight. Terpenes were identified using a library of standards. Terpene concentrations were determined by normalising to the internal standard and correcting for dry weight. As the terpene data were non‐parametric and could not be normalised by transforming the data, Aligned Ranks Transformation ANOVA was used to determine significant differences between treatments over time.
2.16. Plant materials, growth conditions, and treatments for chlorophyll fluorescence measurements
Thirty containerised 2‐year‐old Norway spruce seedlings from a single full‐sib family were potted in 8 cm pots on 5–6 June 2001 and placed in a greenhouse. When all plants had broken bud and started shoot elongation (21 June), they were treated with 0, 5, 25, 50 or 100 mM MeJA in water (n = 6 plants per treatment). All solutions were supplemented with 0.1% Tween 20. Chlorophyll fluorescence was measured on three occasions (1 day, 6 days, 8 weeks after MeJA treatment) using a PAM‐2000 fluorometer (Walz, Effeltrich, Germany) employing the saturation pulse method. Needles were dark‐adapted for at least 45 min before fluorescence measurements and the actinic photosynthetic photon flux density used was 250 µmol m−2 s−1. Maximal PSII fluorescence (Fv/Fm) was measured after 3.5 h in darkness using a white saturating pulse (750 µmol m−2 s−1) lasting 0.8 s. The distance to needles was adjusted and fixed before fluorescence measurements were made to avoid either signal overload or weak signals during data collection. Chlorophyll fluorescence data were analysed with a one‐way ANOVA and a Tukey's HSD test. Statistics were performed in R v3.5.1 (R Core Team, 2019).
2.17. Plant materials, growth conditions, and treatments for growth measurements and anatomical analysis
In May 2017, 2‐year‐old Norway spruce seedlings grown in multipot containers (75 cm3 pots with 60 minipots per container, 500 seedlings per m2) were purchased from the nursery Telemark Skogplanter AS. Seedlings had previously been treated with the insecticide Merit Forest (active ingredient: imidacloprid) in the nursery. The seedlings, which had overwintered at 7°C with the root plugs wrapped in clingfilm, were transferred to single pots (9 × 9 × 10 cm; Nelson Garden, Product No. 5726) containing compost supplemented with mineral fertiliser (Hagebutikken, Product: 30632748). Seedlings were placed on 12 watering trays with 15 pots per tray. The trays were placed in a growth room with constant temperature and humidity (20°C, 60% relative humidity) and 20 h light. Each plant was watered with 100–200 ml of water three times per week and trays were rotated within the growth room to ensure equal lighting for the whole research period. On 31 May 2017, the seedlings were sprayed with ~30 ml of 0 or 10 mM MeJA solutions. The 10 mM MeJA solution consisted of MeJA dissolved in tap water and supplemented with 0.1% Tween 20 to ensure even coating across all sprayed tissues. The control solution was identical except that it did not contain any MeJA.
2.18. Growth measurements
Seedling growth was assessed by measuring plant height twice: on the day of MeJA treatment (31 May 2017) and 5 ½ weeks later (9 July 2017). Plants were harvested for shoot, root, and stem analyses ~10 weeks after MeJA treatment (10 August 2017). The 10 uppermost current‐year lateral shoots on each plant were cut off at their base, placed in paper bags, and stored at room temperature for up to 5 months. Dead shoots without needles were not harvested. Roots were harvested by dividing the soil volume in half. The soil from one half was washed away and all new root growth outside the original root plug was cut, put in water and stored for up to 2 days at approx. 4°C. Washed roots were placed in a water bath and scanned using an Epson Expression 11000XL (Epson America Inc.). Root lengths and diameters were analysed from root images using WinRhizo v2013a (Régent Instruments Inc.). Root lengths were binned into the following diameter classes: <0.39; 0.4–0.79; 0.8–1.19; 1.2–1.59; 1.6–1.99; ≥2 mm. Roots were then placed in paper bags. To determine shoot and root dry weight, lateral shoots and roots in paper bags were dried for 24 h in a drying cabinet set at 65°C and weighed with a Mettler‐Toledo PB602 balance (accuracy to 0.01 g; Mettler‐Toledo GmbH). For growth measurements, two‐sided t‐tests were performed. Root length data were normalised by log transformation and p‐values were adjusted to account for multiple testing using the Benjamini and Hochberg correction. Statistics were performed in R v3.5.1 (R Core Team, 2019).
2.19. Anatomical analysis of stems
A 1‐cm stem section was taken ~10 weeks after MeJA treatment (10 August 2017) from the lower stem of each plant. Thin stem cross‐sections were cut using razor blades, placed on glass slides, and analysed using a light microscope (Leica 020‐525.732, Leica Microsystems Wetzlar GmbH) equipped with the Leica Application Suite software v4.0 (Leica Microsystems Limited). A randomly selected 90‐degree sector of each cross‐section was magnified 2.5× and inside this sector, the following anatomical parameters were measured: the proportion of TRDs across the tangential width of the 2017 annual sapwood growth, the radial thickness of the 2017 annual sapwood growth, and the radial thickness of the secondary phloem (the organised part of the inner bark). These anatomical parameters are illustrated in Figure S2. Two‐sided t‐tests were performed to assess the effect of MeJA treatment on anatomical parameters. Statistics were performed in R v3.5.1 (R Core Team, 2019).
3. RESULTS
3.1. MeJA elicits IR against a necrotrophic pathogen in spruce seedlings
To confirm the protective effect of the MeJA treatment, water (control) and 10 mM MeJA‐treated spruce plants were challenged with the necrotrophic fungal pathogen G. penicillata or a mock agar control (Figure 1). Grosmannia penicillata was chosen for the experiment as it causes substantial necrosis when inoculated in the phloem of unprotected spruce trees (Zhao et al., 2019). Lesion lengths in plants treated with MeJA before fungal inoculation were similar to those of the mock inoculated control plants, while water treated and fungal inoculated plants had significantly larger lesions (Figure 2). These results provide evidence of MeJA‐IR and confirm the suitability of the experimental setup we used to investigate the mechanisms behind MeJA‐IR in spruce.
3.2. Profiling the transcriptional response to MeJA treatment
To assess the plant transcriptional response to resistance‐inducing MeJA treatment, mRNA‐seq was performed on bark tissue harvested 3 h, 24 h, 1 week and 4 weeks after treatment with water or MeJA (Figure 1). Principal component analysis (PCA) and a hierarchical clustering analysis (HCA) were performed to confirm that replicates behaved consistently and to check the global patterns in the data. These analyses suggested that MeJA treatment induced a transient shift in the bark transcriptome (Figure 3). While the 3 h samples were somewhat similar between treatments, by 24 h after treatment plants treated with MeJA were distinct from the water controls. The separation between the two treatments was reduced at 1 week and by 4 weeks after treatment the MeJA samples were very similar to the water controls (Figure 3). This was evidenced by the 4 weeks samples not clustering perfectly by treatment (Figure 3b). Apart from the 4 weeks time‐point, individual replicates of all other treatments clustered together, further confirming the quality of the data set.
Figure 3.
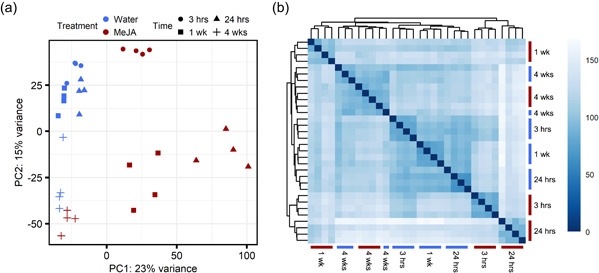
Treatment with methyl jasmonate (MeJA) induces a transient shift in the transcriptome of Norway spruce bark. Principle component analysis (PCA) (a) and hierarchical clustering analysis (HCA) (b) plots displaying how treatment of 2‐year‐old spruce seedlings with water (blue) or 10 mM MeJA (red) impacts on the bark transcriptome over the subsequent 4 weeks. All genes with a total mRNA‐seq read count of ≥100 across the 32 samples (n = 4 per treatment and time point) were included in the analyses. Both the PCA and the HCA utilised counts normalised with a variance‐stabilising transformation. Samples in the HCA were clustered using the Euclidean distances between samples (darker blue for lower distances) and the complete‐linkage method [Color figure can be viewed at wileyonlinelibrary.com]
Having shown that MeJA induced a transient shift in the global transcriptome (Figure 3), next we identified individual genes that showed a significantly (FDR‐adjusted p‐value (p.adj) < 0.001) altered expression profile across time after MeJA treatment. In total, 6330 differentially expressed genes were identified (Data Set S2). Based on expression pattern, these genes separated into 13 clusters (Figure 4a). While each cluster had a subtly different expression profile, there were four main expression patterns (Figure 4; Data Set S2):
-
Pattern 1:
genes transiently upregulated in response to MeJA treatment, before returning to a basal expression level (i.e., the expression level in the water controls; cluster 2, dark pink; cluster 5, purple), or a near basal expression level (cluster 1, yellow; cluster 4, orange; cluster 13, sandy brown).
-
Pattern 2:
genes initially upregulated in response to MeJA treatment, then returning to a basal level before exhibiting slightly repressed expression long‐term (cluster 3, dark green; cluster 9, turquoise).
-
Pattern 3:
genes repressed in response to MeJA treatment, before returning to a basal (cluster 8, pale pink; cluster 12, brown) or near basal expression level (cluster 6, dark grey; cluster 7, fluorescent green; cluster 11, light blue) 4 weeks after treatment.
-
Pattern 4:
genes first repressed in response to MeJA treatment, before climbing back to and above basal expression levels (cluster 10, red).
Figure 4.
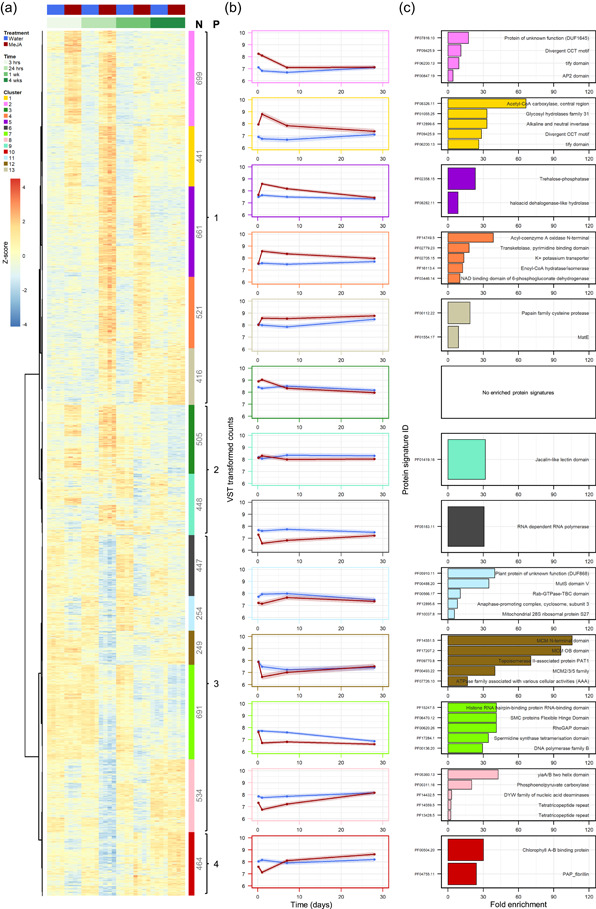
Response of the Norway spruce bark transcriptome to methyl jasmonate (MeJA) treatment. Expression profiles and functional characterisation of 6330 genes showing a significantly (adjusted p < 0.001) altered expression pattern, over a 28‐day period, between bark tissues of 2‐year‐old spruce seedlings treated with water (control, blue) or 10 mM MeJA (red). (a) Differentially expressed genes were grouped into 13 clusters (indicated by the coloured boxes with associated gene numbers [N]) using Spearman distances and Ward's method. The 13 clusters were assigned to one of four patterns (P) based on their general expression profile in MeJA relative to water‐treated plants: 1—Upregulated; 2—Upregulated, then downregulated; 3—Downregulated; 4—Downregulated, then upregulated. Read counts are displayed as per gene z‐scores normalised for library size and transformed using the DESeq. 2 function vst. (b) Per‐cluster mean expression profiles with 95% confidence intervals. Variance‐stabilising transformation (VST) transformed counts are approximately on a log2 scale. (c) Significantly overrepresented protein signatures (adjusted p ≤ 0.05). If a cluster had more than five significantly enriched protein signatures, only the five with the highest fold enrichment are displayed. See Data Set S3 for the full list of enriched protein signatures [Color figure can be viewed at wileyonlinelibrary.com]
To assess whether there were particular functional annotations associated with each of the expression patterns, we performed Protein family (Pfam) domain and GO term enrichment analysis (Figure 4c; Data Sets S3 and S4). Annotations related to ‘response to biotic stress' were overrepresented in the pattern 1‐clusters (Figure 4c; Data Sets S3 and S4). This was exemplified by clusters 1 and 2. Enriched GO terms associated with one or both clusters included ‘regulation of innate immune response’ (GO:0045088), ‘regulation of defence response to fungus’ (GO:1900150), ‘response to chitin’ (GO:0010200), ‘regulation of plant‐type hypersensitive response’ (GO:0010363) and ‘response to wounding’ (GO:0009611; Data Set S4). Defensive hormone‐related GO terms were also enriched in pattern 1‐clusters, including: ‘salicylic acid‐mediated signalling pathway’ (GO:0009863), ‘jasmonic acid‐mediated signalling pathway’ (GO:0009867) and ‘jasmonic acid biosynthetic process’ (GO:0009695; Data Set S4). Additionally, clusters 1 and 2 contain genes encoding for JAZMONATE ZIM DOMAIN (JAZ) proteins, which are key regulators of the response to JA (Howe et al., 2018). This was evidenced by the enrichment of JAZ proteins associated domains ‘tify domain’ (PF06200.13) and ‘divergent CCT motif’ (PF09425.9; renamed ‘Jas motif’) (Figure 4c; Data Set S3). Notably, these two pattern 1‐clusters also contained ethylene (ET) and abscisic acid (ABA) responsive genes, as evidence by the enriched GO terms ‘response to ethylene stimulus’ (GO:0009723) and ‘response to abscisic acid stimulus’ (GO:0009737; Data Set S4). In addition, related to defence, pattern 1‐clusters 1 and 4 had enriched terms associated with terpenoid production, such as ‘terpene synthase family, metal binding domain’ (PF03936.15) and ‘terpenoid biosynthetic process’ (GO:0016114), respectively (Data Sets S3 and S4). Interestingly, genes involved in terpenoid biosynthesis were also overrepresented in pattern 3 and 4 clusters, which were repressed in response to MeJA (Data Set S4).
Other enriched Pfam domains and GO terms associated with the clusters displaying MeJA‐induced repression were generally related to processes involved in primary cell functioning and metabolism, such as cell division, the Krebs cycle, photosynthesis, and transcription. For example, pattern 3‐cluster 7 was enriched for terms such as ‘DNA polymerase family B’ (PF00136.20), ‘regulation of cell cycle phase transition’ (GO:1901987), ‘DNA replication’ (GO:0006260) and ‘gene silencing’ (GO:0016458; Data Sets S3 and S4). Furthermore, ‘phosphoenolpyruvate carboxylase’ (PF00311.16) and ‘photosystem II assembly’ (GO:0010207) were enriched among the genes of cluster 8. Photosynthesis‐related terms such as ‘photosynthesis, light harvesting’ (GO:0009765) and ‘chlorophyll biosynthetic process’ (GO:0015995) were also enriched for pattern 4‐cluster 10 (Figure 4c; Data Sets S3 and S4).
3.3. Biosynthesis of defence hormones in response to MeJA treatment
Based on the results of the protein signature and GO term enrichment analysis and the known role of JA and SA in regulation of defences against biotic stress, we performed a more detailed analysis of the response to MeJA of genes involved in JA and SA biosynthesis and downstream signalling genes. We searched the list of 6330 genes that were differentially expressed after MeJA treatment for genes annotated as encoding JA or SA biosynthesis enzymes, receptors or related transcriptional regulators. Genes predicted to encode enzymes involved in all steps of the conversion of galactolipids to the bioactive form of JA, (+)−7‐iso‐jasmonoyl‐l‐isoleucine, were generally upregulated following MeJA treatment (pattern 1‐clusters; Figures 4 and 5; Data Set S5).
Figure 5.
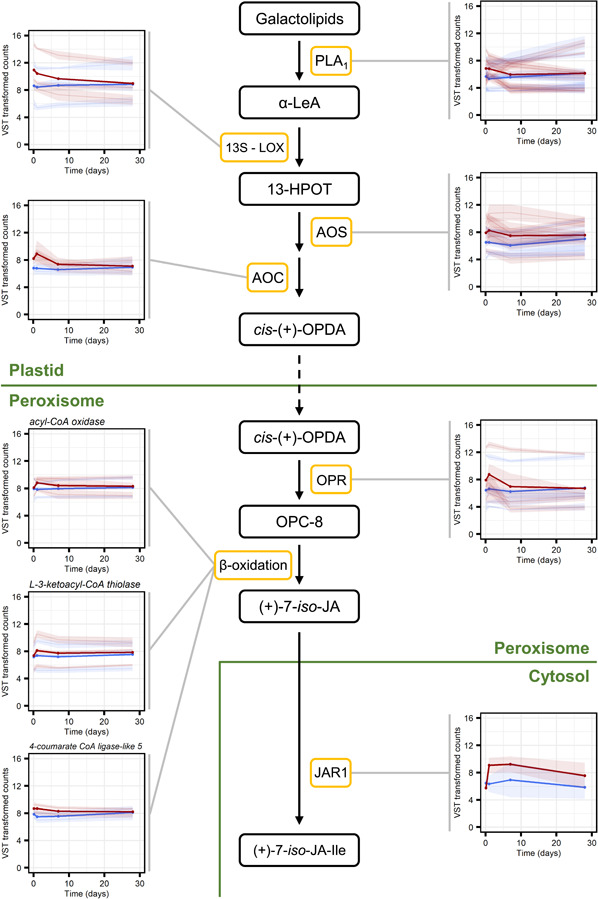
Jasmonic acid (JA) biosynthesis genes are upregulated, either transiently or for at least 4 weeks, in Norway spruce bark in response to methyl jasmonate (MeJA) treatment. Expression profiles of genes displaying a significantly (adjusted p < 0.001) altered expression pattern across time as a result of MeJA treatment and which were annotated as encoding for enzymes (yellow boxes) involved in the (+)−7‐iso‐JA‐Ile biosynthesis pathway. In the plots, faint lines indicate the mean expression profiles, with 95% confidence intervals, of individual transcripts and the thicker lines depict the mean per enzyme category profile, for water controls (blue) and MeJA treatments (red). Read counts were normalised using the variance stabilising transformation (vst) in DEseq. 2. The JA biosynthesis pathway is based on knowledge from angiosperms such as Arabidopsis thaliana and Solanum lycopersicum and was adapted from Wasternack and Hause (2013) and Wasternack and Song (2017). Compound abbreviations: α‐LeA, α‐linolenic acid; 13‐HPOT, (13S)‐hydroperoxyoctadecatrienoic acid; cis‐(+)‐OPDA, cis‐(+)−12‐oxophytodienoic acid; OPC‐8, 3‐oxo‐2‐(2‐pentenyl)‐cyclopentane‐1‐octanoic acid; (+)−7‐iso‐JA, (+)−7‐iso‐jasmonic acid; (+)−7‐iso‐JA‐Ile, (+)−7‐iso‐jasmonoyl‐l‐isoleucine. Enzyme abbreviations: 13S‐LOX, 13S‐lipoxygen‐ase; AOC, allene oxide cyclase; AOS, allene oxide synthase; JAR1, JA‐amino acid synthetase; OPR, OPDA reductase; PLA1, phospholipase A1. [Color figure can be viewed at wileyonlinelibrary.com]
Interestingly, considering the antagonism between JA and SA in some angiosperms (Thaler et al., 2012), genes predicted to encode enzymes involved in the biosynthesis of the precursors of SA, such a phenylalanine ammonia‐lyase (PAL), were also predominantly found in the pattern 1‐clusters (Figures 4 and 6; Data Set S5). The same was true for the downstream signalling pathways. Homologues of master regulators that control SA‐ and JA‐dependent defence responses in Arabidopsis were, generally, also found in the pattern 1‐clusters (Figures 4 and 7; Data Set S5).
Figure 6.
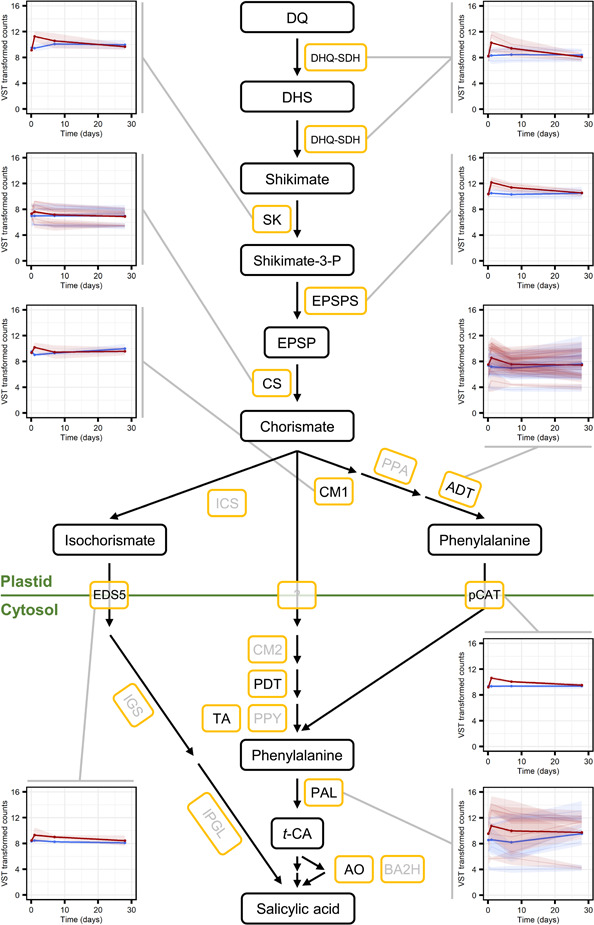
Genes involved in the biosynthesis of salicylic acid (SA) are transiently upregulated in Norway spruce bark in response to methyl jasmonate (MeJA) treatment. Expression profiles of genes displaying a significantly (adjusted p < 0.001) altered expression pattern across time as a result of MeJA treatment and which were annotated as encoding for enzymes and transporters involved in the biosynthesis of SA (yellow boxes). In the plots, faint lines indicate the mean expression profiles, with 95% confidence intervals, of individual transcripts and the thicker lines depict the mean per protein category profile, for water (blue) and MeJA (red) treatments. Read counts were normalised using the variance stabilising transformation (vst) in DEseq. 2. The pathway is based on what is known about the biosynthesis of SA in angiosperms. Enzymes in grey are involved in the biosynthesis pathway but no differentially expressed genes were annotated to encode for them. Due to space limitations not all enzymes have associated expression profiles, see Data Set S5 for the full list of differentially expressed biosynthesis genes. Compound abbreviations: DQ, 3‐dehydroquinate; DHS, 3‐dehydroshikimate; Shikimate‐3‐P, shikimate 3‐phosphate; EPSP; 5‐enolpyruvoyl‐shikimate 3‐phosphate; t‐CA, trans‐cinnamic acid. Enzyme/transporter abbreviations: ADT/PDT, arogenate dehydratase/prephenate dehydratase (annotated as being encoded for by the same genes); AO, aldehyde oxidase; BA2H, benzoic acid 2‐hydroxylase; CM1, chorismate mutase 1; CM2, chorismate mutase 2; CS, chorismate synthase; DHQ‐SDH, dehydroquinate‐shikimate dehydrogenase; EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5; EPSPS, 5‐enolpyruvylshikimate‐3‐phosphate synthase; ICS, isochorismate synthase; IGS, isochorismoyl‐glutamate synthase; IPGL, IC‐9‐Glu pyruvoyl‐glutamate lyase; PAL, phenylalanine ammonia‐lyase; pCAT, plastidial cationic amino‐acid transporter; PPA, prephenate aminotransferase; PPY, phenylpyruvate aminotransferase; SK, shikimate kinase; TAT, tyrosine aminotransferase [Color figure can be viewed at wileyonlinelibrary.com]
Figure 7.
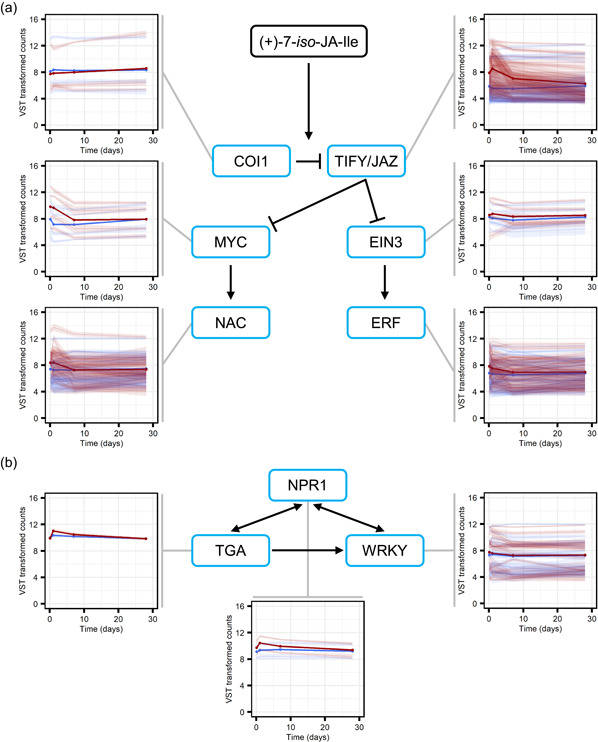
Methyl jasmonate (MeJA) treatment induces a rapid and transient upregulation of regulators of jasmonic and salicylic acid (JA and SA) dependent defences in Norway spruce bark. Expression profiles of genes displaying a significantly (adjusted p < 0.001) altered expression pattern across time as a result of MeJA treatment and which were annotated as encoding for proteins involved in regulation of JA (a) and SA (b) dependent defences. In the plots, faint lines indicate the mean expression profiles, with 95% confidence intervals, of individual transcripts and the thicker lines depict the mean per protein category profile, for water (blue) and MeJA (red) treatments. Read counts were normalised using the variance stabilising transformation (vst) in DEseq. 2. The signalling pathways are based on what is known in Arabidopsis thaliana. Compound abbreviations: (+)−7‐iso‐JA‐Ile, (+)−7‐iso‐jasmonoyl‐l‐isoleucine. Protein abbreviations: COI1, CORONATINE INSENSITIVE1; EIN3, ETHYLENE INSENSITIVE 3; ERF, ETHYLENE RESPONSIVE FACTOR; JAZ, JASMONATE‐ZIM‐DOMAIN PROTEIN; NAC, Petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2; NPR1, NONEXPRESSER OF PR GENES 1; TGA, TGACG motif‐binding [Color figure can be viewed at wileyonlinelibrary.com]
Previous work in conifers has demonstrated that ET accumulates in response to MeJA treatment and in turn is an important signalling compound for the induction of defences by MeJA (Hudgins & Franceschi, 2004). Thus, we also explored in more detail the expression of ET biosynthesis genes. ET is synthesised from the amino acid methionine in a three‐step process involving the enzymes S‐adenosyl‐l‐methionine (SAM) synthetase, 1‐aminocyclopropane‐1‐carboxylic acid (ACC) synthase (ACS), and ACC oxidase (ACO) (Lin et al., 2009). All differentially expressed ET biosynthesis genes were in pattern 1‐clusters, that is, they were transiently upregulated in response to MeJA treatment, before returning to a basal or near basal expression level (Figure 4; Data Set S4). Two genes predicted to encode for SAM synthases were in cluster 1 and one ACO gene was in cluster 5 (Figure 4; Data Set S5). Notably, no ACS genes were differentially expressed in response to MeJA treatment.
3.4. Spruce defence genes are differentially expressed in response to MeJA treatment
The upregulation of genes involved in defence hormone biosynthesis and downstream signalling genes suggested that MeJA treatment directly induced spruce defences. Evidence in support of this conclusion is that genes involved in the biosynthesis of terpenes, the major component of oleoresin, were differentially expressed in response to MeJA treatment (Figure 4; Data Sets S3 and S4). However, while the amounts of some individual terpenes changed, the overall terpene concentration did not increase after MeJA treatment, which is the same as what has previously been observed (Table S2; Mageroy, Wilkinson, et al., 2020; Zhao et al., 2011)
Phenolic compounds also are an important chemical defence in spruce (Krokene, 2015). As mentioned above, MeJA treatment was found to transiently upregulate genes annotated to encode for PAL, which catalysers the first step of the general phenylpropanoid pathway and contributes to biosynthesis of SA and defensive phenolics (Figure 6; Data Set S5). To focus on phenylpropanoid pathway genes associated with MeJA‐IR, we searched the list of differentially expressed genes for genes predicted to encode biosynthesis enzymes for defensive phenolics involved in spruce defence against ophiostomatoid fungal pathogens (Hammerbacher et al., 2011, 2013, 2018, 2019). Among the genes differentially expressed by MeJA treatment we identified genes predicted to encode chalcone synthase, chalcone isomerase, flavanone‐3‐hydroxylase, flavonol‐3′‐hydroxylase, flavonol‐3′5′‐hydroxylase, dihydroflavonol 4‐reductase, anthocyanidin synthase, anthocyanidin reductase and leucoanthocyanidin reductase (Data Set S2). Generally, these biosynthesis genes were upregulated in response to MeJA treatment (pattern 1‐clusters; Figure 4; Data Set S2). Notably, these genes are all involved in the production of flavonoids and none were annotated as encoding for stilbene synthases. Overall, the transcriptome analysis suggested that the biosynthesis of defence‐related phenylpropanoids was upregulated by MeJA.
In addition to terpene‐ and phenolic‐based defences, PR proteins are also thought to be an important component of spruce defence against biotic stress (Franceschi et al., 2005). Hypothetically, MeJA treatment could trigger an upregulation of PR genes and a long‐term accumulation of PR proteins, which could provide enhanced resistance to subsequent attack. To assess this possibility, we searched the list of differentially expressed genes for genes annotated as PR proteins. Interestingly, there was not a consensus pattern (Figure S3) and genes predicted to encode for PR proteins were found in clusters belonging to all four expression patterns (Figure 4; Data Set S5). Thus, although some PR proteins may accumulate in response to MeJA treatment, based on gene expression data alone, this is not the general pattern.
3.5. MeJA treatment induces differential expression of epigenetic regulators
Increasing evidence suggests that epigenetic mechanisms are involved in the immunological memory of plants and in maintenance of IR (Parker et al., 2022; Wilkinson et al., 2019). Interestingly, our GO term enrichment analysis suggested that many genes encoding for epigenetic regulators were differentially expressed following MeJA treatment. This was exemplified by the cluster 7 MeJA‐repressed genes (Figure 4). Overrepresented GO terms in this cluster included 'DNA methylation or demethylation' (GO:0044728), 'chromatin silencing' (GO:0006342) and 'histone modification' (GO:0016570; Data Set S4).
To further investigate the specific epigenetic regulators that were differentially expressed following MeJA treatment we checked our list of 6330 differentially expressed genes against a list of epigenetic regulators in Norway spruce compiled by (Mageroy, Wilkinson, et al., 2020). Many of these epigenetic regulators were differentially expressed in response to MeJA treatment (Figure 8a). They followed several expression patterns and fell into all the 13 clusters shown in Figure 4 (Data Sets S2 and S6). MeJA induced a transient upregulation of most histone acetyltransferase genes, whereas, regulators of DNA methylation were repressed following MeJA treatment (Figure 8b), in particular genes related to RNA‐directed DNA methylation (RdDM; Figure 8b; Data Set S6). These genes included DNA‐directed RNA polymerase V subunit 1 (NRPE1), the largest subunit of Pol IV NUCLEAR RNA POLYMERASE D 1 (NRPD1), and RNA‐DEPENDENT RNA POLYMERASE 2 (RDR2). Additionally, two of the differentially expressed DNA methyltransferases were predicted to encode for DNA (cytosine‐5)‐methyltransferase (DRMs; Figure 8b; Data Set S6). In Arabidopsis, DRMs are guided by small RNAs (sRNAs), derived from Pol IV or Pol II produced transcripts, to specific genome locations where they establish DNA methylation (Zhang et al., 2018). Over 60% of the differentially expressed RdDM‐related genes we detected in Norway spruce were in pattern 3‐clusters, with the mean pattern suggesting that RdDM began to be repressed between 3 and 24 h after MeJA treatment (Figure 8b; Data Set S6). Additionally, one differentially expressed DNA demethylase (MA_111413g0010) was identified. This transcript was initially marginally upregulated before being downregulated to below basal expression levels (Figure 8b).
Figure 8.
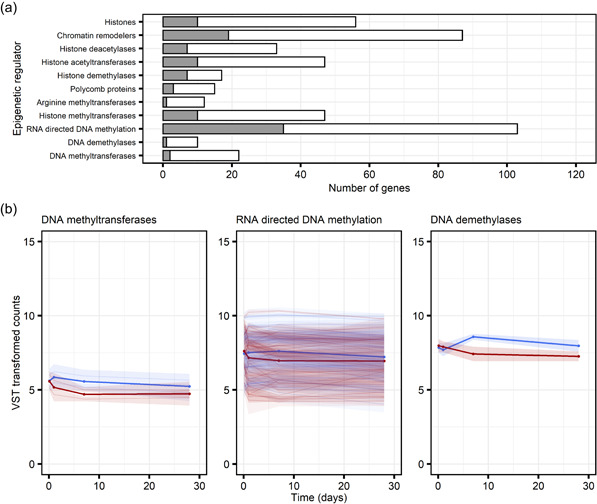
Methyl jasmonate (MeJA) treatment alters the expression of regulators of multiple epigenetic modifications in Norway spruce bark, including DNA methylation. (a) Bars show the number of annotated genes in different epigenetic regulator categories. Genes showing a significantly (adjusted p < 0.001) altered expression pattern across time as a result of treatment are indicated in grey. (b) The individual transcripts (faint lines with 95% confidence intervals) and category means (solid lines) for differentially expressed regulators of DNA methylation homoeostasis in water (blue) and MeJA (red) treated bark (see Data Set S6). Read counts were normalised using the variance stabilising transformation (VST) in DEseq. 2. Epigenetic regulator categories are taken from Mageroy, Wilkinson, et al. (2020) [Color figure can be viewed at wileyonlinelibrary.com]
3.6. Evaluating costs of MeJA treatment
Trade‐offs between growth and defence exist in plants due to the need to allocate limited resources optimally (Züst & Agrawal, 2017). Thus, while MeJA treatment promotes resistance against necrotrophic plant pathogens, it may also incur some costs. To compare expression of genes related to growth and defence, we mapped differentially expressed genes to pathways pertaining to growth or defence using KEGG pathway analysis. This analysis showed that genes in pathways associated with the biosynthesis of important spruce defence compounds were skewed towards MeJA‐induced expression patterns 1 and 2, while genes in pathways associated with cell cycle and photosynthesis were skewed towards MeJA‐repressed expression patterns 3 and 4 (Figure S4; Data Set S2; see Figure 3 for the expression patterns).
To further evaluate potential costs associated with MeJA‐IR, we determined how MeJA treatment affected above‐ and below‐ground growth. MeJA treatment reduced apical leader growth and shoot dry weight (Figure 9a,b). Annual sapwood growth was also reduced by MeJA treatment, whereas the secondary phloem was enlarged (Figure 9c‐d) (Franceschi et al., 2002). In contrast, MeJA treatment had no effect on below ground, with similar root dry weight and length in water and MeJA treated plants (Figure 9f–g). We also measured the effects of a range of MeJA concentrations on chlorophyll fluorescence at 1 day, 6 days, and 8 weeks after treatment. Only a high concentration of 100 mM MeJA induced a transient reduction in chlorophyll fluorescence (Figure S5).
Figure 9.
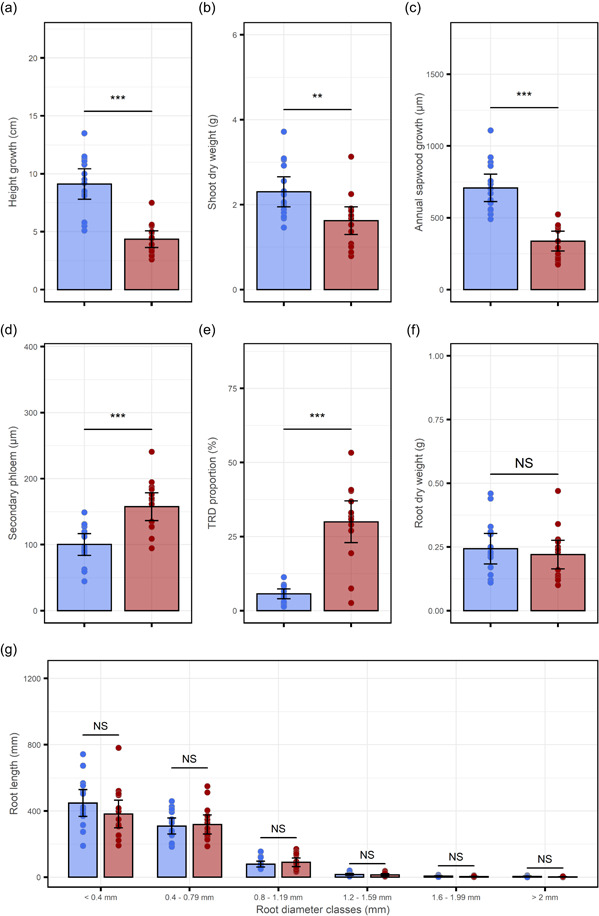
Effect of methyl jasmonate (MeJA) treatment on growth and traumatic resin duct development. 2‐year‐old Norway spruce plants were sprayed with water (blue) or 10 mM MeJA (red). (a) Plant height was measured twice over a period of 5½ weeks after MeJA treatment. Shoots, roots and stem cross‐sections for (b–g) were harvested 10 weeks after treatment. (b) Dry weight of the 10 uppermost current‐year lateral shoots on each plant. (c, d) Radial thickness (i.e., width) of the current‐year annual sapwood growth (c) and secondary phloem (d) on stem cross‐sections. (e) Proportion of traumatic resin ducts (TRD) across a quarter of the tangential width of the current‐year annual sapwood growth. (f) Root dry weight. (g) Total root length for different root diameter classes. All results are presented as mean ± 95% confidence interval of the mean. Points show individual data points (n = 15). Asterisks indicate significant differences (Two‐sided t‐test, n.s, not significant; ***p < 0.001, **p < 0.01) [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
The plant hormone MeJA has previously been demonstrated to elicit IR against pests and pathogens in both young and mature Norway spruce trees (Erbilgin et al., 2006; Mageroy, Christiansen, et al., 2020; Puentes et al., 2021; Zeneli et al., 2006). In this study, we confirmed that MeJA elicits IR by demonstrating that MeJA treatment of 2‐year‐old seedlings increases resistance to infection by the bark beetle‐vectored fungal pathogen G. penicillata 4 weeks later (Figure 2). Subsequently, we explored the molecular mechanisms behind the establishment and maintenance of MeJA‐IR. This analysis provided an understanding of the general transcriptional responses to MeJA in the bark of Norway spruce seedlings. Furthermore, our results suggested that there are costs of MeJA‐IR mainly in the form of reduced above‐ground growth.
4.1. Transcriptional response of Norway spruce bark to MeJA
The transcriptional response to jasmonate treatment has been explored in multiple previous studies on several plant species (e.g., Benevenuto et al., 2019; Hickman et al., 2017; Liu et al., 2017). Although these previous analyses were generally conducted over much shorter timeframes than our study (hours and days rather than hours, days and weeks) and used leaf rather than woody stem tissues, their findings have many similarities to those of our study. In Arabidopsis, the transcriptional response to MeJA has been explored in detail. For instance, Hickman et al. (2017) performed mRNA‐seq on samples harvested at 14 time points over a 16‐h period after MeJA treatment. This high‐resolution time course analysis suggests that in Arabidopsis there is a rapid pulse of differential expression, with many genes showing peak change within 3 h after treatment. While we similarly observed a pulse of transcriptomic change, the peak up‐ or downregulation of most genes appeared to occur many hours later in Norway spruce than in Arabidopsis (Figure 4). One possible explanation is that it may take longer for MeJA to enter and move through bark than leaf tissue. If so, there would be a longer delay between application of MeJA and MeJA entering spruce cells, where it is converted to the bioactive form of JA, JA‐isoleucine, which then stimulates changes in transcription. This hypothesis could be tested using a cell type‐specific metabolomics and transcriptomics approach, the latter as demonstrated by Celedon et al. (2017).
Another similar transcriptomic study in whitebark pine (Pinus albicaulis) seedlings demonstrated that MeJA treatment of needles resulted in a transient upregulation of numerous genes related to hormone signalling, including hormone biosynthesis genes, hormone‐related transcriptional regulators (e.g., homologues of MYC2 and ethylene responsive TFs), and phenylpropanoid and terpenoid biosynthesis genes (Liu et al., 2017). We found similar genes upregulated in our study. However, unlike whitebark pine needles, we did not observe upregulation of the ethylene biosynthesis gene ACC synthase in spruce bark (Data Set S5). A similar MeJA response in whitebark pine needles and Norway spruce bark was the downregulation of multiple genes involved in photosynthesis (Liu et al., 2017). At least in the first 24 h after MeJA treatment, we found repression of genes involved in photosynthesis (Figure 4; Data Sets S2–S4). Although bark tissue, particularly of mature trees, appears brown and lacking in chlorophyll, stem photosynthesis does occur in conifers including Norway spruce (Tarvainen et al., 2018). Thus, some transcriptional responses to MeJA seem to be conserved across angiosperms and gymnosperms and across multiple tissue types.
4.2. Hormonal regulation of MeJA‐Induced defences
Many genes that were upregulated after MeJA treatment were annotated as being involved in defence regulation (Figure 4; Data Sets S2–S4). More specifically, many of the upregulated genes were involved in biosynthesis and signalling of the major defence regulatory hormones JA and SA, along with the supporting hormone ET (Figures 4, 5, 6, 7; Data Sets S2–S5). MeJA treatment has previously been demonstrated to induce an accumulation of JA and SA in the bark of 6–7‐year‐old Norway spruce saplings (Schmidt et al., 2011). Furthermore, a study in 4‐year‐old saplings of Douglas fir (P. menziesii) demonstrated that MeJA induced ET accumulation (Hudgins & Franceschi, 2004). Thus, it is unsurprising that genes encoding many of the enzymes required to produce JA, SA and ET hormones were transiently upregulated following MeJA treatment in our study (Figures 5, 6, 7).
In addition to hormone biosynthesis genes, we observed upregulation of homologues of MYC, EIN3/EIL1 and NPR1, major regulators of JA, JA/ET and SA dependent signalling pathways and defence responses. Studies in Arabidopsis and tomato have demonstrated that JA activates MYC and EIN3/EIL1 by targeting JAZ repressors for degradation (Chang et al., 2013; Fernández‐Calvo et al., 2011). As reported in other species (e.g., Hickman et al., 2017; Sun et al., 2017; Wang et al., 2020), we found that numerous genes encoding for JAZ proteins were upregulated by MeJA treatment. This upregulation likely acts to prevent over‐activation of JA‐dependent responses.
Other TF families that were upregulated in response to MeJA treatment included the NAC (CUP‐SHAPED COTYLEDON/NO APICAL MERISTEM/ATAF), myb domain protein (MYB), ethylene response factor (ERF), and WRKY families. In Arabidopsis and other well‐studied angiosperm species, members of these TF families can play an intermediate role in JA, JA/ET and SA signalling pathways and are important regulators of defence responses (Erb & Reymond, 2019; Yuan et al., 2019). For instance, MYB TFs are known to be key regulators of phenylpropanoid biosynthesis in many species (Liu et al., 2015). We also found NACs, MYBs, ERFs and WRKYs that were downregulated by MeJA treatment. This could be because they are negative regulators of defences or, alternatively, they regulate other pathways and processes which are repressed by MeJA treatment (e.g., growth and development). Additionally, these TFs do not always act below the master regulators (e.g., MYC2) in hormone signalling pathways. For example, there is evidence that specific MYBs can interact directly with JAZ repressors (Qi et al., 2011). Future studies using RNA interference (RNAi) to repress MeJA‐inducible TFs would be helpful for assessing the role of TFs in the regulation of defence responses in Norway spruce (e.g., PR gene expression, terpene and phenolic accumulation).
4.3. Effects of MeJA treatment on spruce defences
IR can be underpinned by a direct and prolonged upregulation of defences induced by an IR‐eliciting stimulus (Wilkinson et al., 2019). In spruce, MeJA treatment has previously been shown to directly induce a range of defences. For example, Zulak et al. (2009) demonstrated that in 2‐year‐old Norway spruce seedlings MeJA treatment induced terpene biosynthesis and the subsequent accumulation of terpenes in bark tissue. However, this induction was transient and only a few specific terpenes remained at high levels 1–2 months after treatment. In our study, we found that genes related to terpene biosynthesis exhibited both upregulated and downregulated express patterns (Figure 4; Data Set S2). The lack of a consistent upregulation of terpene biosynthesis in Norway spruce following MeJA treatment is mirrored by the fact that we did not see a significant increase in the total terpene concentration in MeJA treated bark (Table S2). While these results appear to be at odds with the accumulation of terpenes reported by Zulak et al. (2009), they are in line with our previous studies that found that terpenes were not significantly accumulated in intact, MeJA treated bark, but were induce by subsequent wounding 4‐weeks later (Mageroy, Wilkinson, et al., 2020; Zhao et al., 2011). Interestingly, this primed accumulation of terpenes is unlikely to be explained by a primed gene induction, as terpene biosynthesis genes are not induced by wounding of MeJA treated bark (Mageroy, Wilkinson, et al., 2020). Conceivably wounding of MeJA treated bark releases local resin pools and induces a flow of resin from surrounding bark through axial and radial resin ducts (Krokene, 2015; Nagy et al., 2000). Genotypic differences between trees used in various MeJA‐IR Norway spruce studies could also be an explanation for the dissimilarities in terpene accumulation. Future work should explore whether some genotypes rely more on a prolonged accumulation of terpenes following an IR‐stimuli, while others rely more on a primed accumulation following subsequent challenge.
Multiple studies have shown that MeJA treatment induces the swelling of polyphenolic cells and an accumulation of phenolic compounds in Norway spruce bark (Franceschi et al., 2002; Krokene et al., 2008; Li et al., 2012). Over the past decade, Hammerbacher and colleagues described the biosynthesis of specific polyphenolic compounds in spruce bark (e.g., stilbenes and flavonoids) that provide resistance to ophiostomatoid fungi (Hammerbacher et al., 2011, 2018, 2019). In this study, we observed an upregulation of genes predicted to encode for enzymes involved in flavonoid biosynthesis following MeJA treatment (Data Set S2). The upstream general phenylpropanoid pathway, which provides the precursor metabolites for flavonoid biosynthesis, was also transiently upregulated. Thus, although previous studies have mostly worked with Endoconidiophora polonica, the model ophiostomatoid species in Norway spruce, an accumulation of phenolic compounds derived from the phenylpropanoid pathway also seems to be key to the MeJA‐IR against G. penicillata we observed in this study. A holistic study of phenolic biosynthesis that profiles metabolites, protein levels and enzyme activities should be conducted, like the study Zulak et al. (2009) did on terpenes.
PR proteins are another important defence against pathogens in plants, but have been less extensively studied in conifer than terpenes and phenolics. Numerous genes predicted to encode for PR proteins were differentially expressed in Norway spruce bark following MeJA treatment. While we did not find a consensus expression pattern exhibited by PR genes (Figure S3), there were some interesting results. For instance, some PR genes were transiently induced following MeJA treatment, while others were mildly prolonged upregulated (Figure S3; Data Set S5). This inducibility of PR genes in response to jasmonate is consistent with previous work in conifers (Davis et al., 2002; Pervieux et al., 2004). Additionally, we have previously demonstrated that in mature spruce trees treated with MeJA, numerous PR genes were primed for an augmented induction in response to subsequent wounding (Mageroy, Wilkinson, et al., 2020). Our new data suggest that MeJA‐IR is underpinned by a mixture of prolonged induction and priming of PR genes.
4.4. Role of epigenetic mechanisms in maintaining of MeJA‐IR
MeJA‐IR in Norway spruce seems to be underpinned by both a prolonged upregulation and priming of PR genes and other defence‐related genes. Long‐term changes in basal gene expression and/or gene responsiveness are likely underpinned by epigenetic mechanisms (Wilkinson et al., 2019). For instance, histone tail modifications leading to chromatin decompaction and enhanced TF binding to promoter regions have been linked with defence gene priming in Arabidopsis (Baum et al., 2019). Furthermore, changes in DNA methylation has been linked to both a prolonged upregulation and priming of genes (Cambiagno et al., 2018; Furci et al., 2019; López Sánchez et al., 2016). We found that in response to MeJA treatment, many genes predicted to encode for proteins involved in histone tail modifications, changes in DNA methylation and chromatin remodelling were differentially expressed. These genes included RdDM pathway genes (e.g., DRM2, NRPE1, RDR6) which were repressed following MeJA treatment. In Arabidopsis, RdDM pathways establish de novo DNA methylation and maintain DNA methylation at CHH context sites (Zhang et al., 2018). Arabidopsis and rice (Oryza sativa) mutants with defects in RdDM genes are hypomethylated, particularly in CHH contexts (Stroud et al., 2013; Tan et al., 2016). The canonical RdDM pathway involves 24 nucleotide (nt) small RNAs (sRNAs) and long noncoding scaffold RNAs that help guide the methylation machinery, such as Argonaute 4 (AGO4), to specific loci (Zhang et al., 2018). However, it is has been reported in Arabidopsis that 21/22 nt sRNAs can also guide DNA methylation via a noncanonical RdDM pathway which involves AGO6 (Cuerda‐Gil & Slotkin, 2016). Interestingly, the expression of 24 nt sRNAs is very low in Norway spruce vegetative tissues whereas 21/22 nt sRNAs are common (Nystedt et al., 2013; Wilkinson et al., 2021). We may thus speculate that noncanonical RdDM pathway with 21/22 nt guide sRNAs is the canonical pathway in vegetative Norway spruce tissues (Wilkinson et al., 2021). In addition to RdDM genes, we also found that MeJA treatment repressed genes encoding for DNA glycosylases, enzymes which catalyse the first step in active in removal of DNA methylation (Zhang et al., 2018), further supporting our hypothesis that MeJA treatment triggers a loss of DNA methylation in Norway spruce. Repression of DNA glycosylases occurred later than the RdDM pathway repression suggesting the presence of a RdDM‐DNA glycosylase feedback phenomenon like that previously observed in Arabidopsis (Lei et al., 2015). Taken together, our data suggest that RdDM and other DNA methylation pathways described in angiosperms also function in Norway spruce and that MeJA treatment induces DNA hypomethylation. Techniques such as FAIRE (formaldehyde‐assisted isolation of regulatory elements) sequencing (Baum et al., 2019) should be used in the future to determine what changes MeJA induces to the epigenetic landscape in spruce bark and how these changes contribute to shifts in gene expression associated with MeJA‐IR.
4.5. Costs of long‐term MeJA‐IR
It has been suggested that Norway spruce may increase survival during periods of stress by allocating carbon to secondary metabolites over growth and respiration (Huang et al., 2019). In our transcriptome analysis, genes related to photosynthesis were enriched in expression pattern 4 gene clusters (Figures 4 and S4; Data Sets S3 and S4). This suggests that while the maintenance and functioning of photosynthesis may initially be repressed by MeJA, in the longer term it may be upregulated to higher than basal levels. However, based on our chlorophyll fluorescence data, it seems that treatment of bark with 10 mM MeJA does not influence the level of photosynthesis in the needles (Figure S5). We did not evaluate changes in bark photosynthesis, which may contribute significantly to carbon assimilation in young trees (Wittmann & Pfanz, 2007) and cannot rule out that photosynthesis in bark was more compromised by MeJA treatment than in needles.
We also observed repression of cell cycle‐related genes by MeJA treatment and this was associated with clear negative effects on shoot and sapwood growth (Figures 4, 9 and S4; Data Sets S3 and S4). The growth reduction could be due to the allocation of carbon into the formation of MeJA‐induced TRDs (Figure 9e), which are filled with metabolically costly defence‐related terpenes (Gershenzon, 1994), or the swelling of phenolic rich polyphenolic cells, (which could explain the increased width of the secondary phloem observed in MeJA treated trees (Figure 9d). TRD formation and activation of polyphenolic cells are likely important for long‐term MeJA‐IR in Norway spruce and suggest that enhanced resistance to biotic stress may come at the expense of growth. Whether this has an impact on long‐term fitness and reproduction of MeJA‐treated trees is difficult to assess in a tree species that does not reproduce until it is 25 years old. But, as discussed by Mageroy, Christiansen, et al. (2020), when the alternative is death from bark beetle attack investment in defence may be adaptive for a long‐lived plant species.
5. CONCLUSIONS
This detailed study of transcriptional changes in Norway spruce bark after MeJA treatment showed that changes in gene expression can be detected as early as 3 h after treatment. Expression of some transcripts returned to basal levels by 1 week, while others remained induced for at least 4 weeks after treatment. MeJA‐induced transcript clusters were strongly associated with defence responses to biotic stress, whereas many MeJA‐repressed gene clusters were linked with primary cell functioning and metabolism. Additionally, we found that genes encoding machinery related to the RdDM pathway were repressed, suggesting that epigenetic changes and such as DNA hypomethylation are important in the establishment and maintenance of MeJA‐IR in Norway spruce.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
ACKNOWLEDGEMENTS
We thank the technical staff at NIBIO for their help with experiments, particularly Hans Ragnar Norli for his assistance with terpene analysis. Bioinformatics were performed on the Abel Cluster, owned by the University of Oslo and Uninett/Sigma2, and operated by the Department for Research Computing at USIT, the University of Oslo IT‐department. http://www.hpc.uio.no/(nn9433k). The work presented in this publication was supported by Research Council of Norway grants (249920 and 249958) to Paal Krokene and Melissa H. Mageroy, respectively. Funds were also provided from Borregaard Forskningfond for the master thesis of TOS.
Wilkinson, S.W. , Dalen, L.S. , Skrautvol, T.O. , Ton, J. , Krokene, P. & Mageroy, M.H. (2022) Transcriptomic changes during the establishment of long‐term methyl jasmonate‐induced resistance in Norway spruce. Plant, Cell & Environment, 45, 1891–1913. 10.1111/pce.14320
DATA AVAILABILITY STATEMENT
Raw reads from mRNA sequencing have been submitted to NCBI. BioProject accession: PRJNA768356.
REFERENCES
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. & Lipman, D.J. (1990) Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. Available from: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. et al. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402. Available from: 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. & Huber, W. (2010) Differential expression analysis for sequence count data. Genome Biology, 11, 1–12. Available from: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A. (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research, 47, D506–D515. Available from: 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, S. , Reimer‐Michalski, E.M. , Bolger, A. , Mantai, A.J. , Benes, V. , Usadel, B. & Conrath, U. (2019) Isolation of open chromatin identifies regulators of systemic acquired resistance. Plant Physiology, 181, 673–833. Available from: 10.1104/pp.19.00673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevenuto, R.F. , Seldal, T. , Hegland, S.J. , Rodriguez‐Saona, C. , Kawash, J. & Polashock, J. (2019) Transcriptional profiling of methyl jasmonate‐induced defense responses in bilberry (Vaccinium myrtillus L.). BMC Plant Biology, 19, 1–18. Available from: 10.1186/s12870-019-1650-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. & Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B, 57, 289–300. Available from: 10.2307/2346101 [DOI] [Google Scholar]
- Biedermann, P. , Müller, J. , Grégoire, J.C. , Gruppe, A. , Hagge, J. , Hammerbacher, A. et al. (2019) Bark beetle population dynamics in the anthropocene: challenges and solutions. Trends in Ecology and Evolution (Personal Edition), 34, 914–924. Available from: 10.1016/j.tree.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Camacho, C. , Coulouris, G. , Avagyan, V. , Ma, N. , Papadopoulos, J. , Bealer, K. et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics, 10, 421. Available from: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambiagno, D.A. , Nota, F. , Zavallo, D. , Rius, S. , Casati, P. , Asurmendi, S. et al. (2018) Immune receptor genes and pericentromeric transposons as targets of common epigenetic regulatory elements. The Plant Journal, 96, 1178–1190. Available from: 10.1111/tpj.14098 [DOI] [PubMed] [Google Scholar]
- Caudullo, G. , Tinner, W. & Rigo, D. et al. (2016) Picea abies in Europe: distribution, habitat, usage and threats G. In: San‐Miguel‐Ayanz, J , Rigo, D , Caudullo, G , Houston, Durrant, T & Mauri, A (Eds.) European atlas of forest tree species. Publication Office of the European Union, pp. 114–116. [Google Scholar]
- Celedon, J.M. & Bohlmann, J. (2019) Oleoresin defenses in conifers: chemical diversity, terpene synthases and limitations of oleoresin defense under climate change. New Phytologist, 224, 1444–1463. Available from: 10.1111/nph.15984 [DOI] [PubMed] [Google Scholar]
- Celedon, J.M. , Yuen, M.M.S. , Chiang, A. , Henderson, H. , Reid, K.E. & Bohlmann, J. (2017) Cell‐type‐ and tissue‐specific transcriptomes of the white spruce (Picea glauca) bark unmask fine‐scale spatial patterns of constitutive and induced conifer defense. The Plant Journal, 92, 710–726. Available from: 10.1111/tpj.13673 [DOI] [PubMed] [Google Scholar]
- Chang, K.N. , Zhong, S. , Weirauch, M.T. , Hon, G. , Pelizzola, M. , Li, H. et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross‐regulation in Arabidopsis. eLife, 2, 2013. Available from: 10.7554/eLife.00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath, U. , Beckers, G.J.M.M. , Langenbach, C.J.G.G. & Jaskiewicz, M.R. (2015) Priming for enhanced defense. Annual Review of Phytopathology, 53, 97–119. Available from: 10.1146/annurev-phyto-080614-120132 [DOI] [PubMed] [Google Scholar]
- Cuerda‐Gil, D. & Slotkin, R.K. (2016) Non‐canonical RNA‐directed DNA methylation. Nature Plants, 2, 16163. Available from: 10.1038/nplants.2016.163 [DOI] [PubMed] [Google Scholar]
- Davis, J.M. , Wu, H. , Cooke, J.E.K. , Reed, J.M. , Luce, K.S. & Michler, C.H. (2002) Pathogen challenge, salicylic acid, and jasmonic acid regulate expression of chitinase gene homologs in pine. Molecular Plant‐Microbe Interactions, 15, 380–387. Available from: 10.1094/MPMI.2002.15.4.380 [DOI] [PubMed] [Google Scholar]
- De Kesel, J. , Conrath, U. , Flors, V. , Luna, E. , Mageroy, M.H. , Mauch‐Mani, B. et al. (2021) The induced resistance lexicon: Do's and Don'ts. Trends in Plant Science, 26, 685–691. Available from: 10.1016/j.tplants.2021.01.001 [DOI] [PubMed] [Google Scholar]
- de Matos Simoes, R. & Emmert‐Streib, F. (2016). bc3net: Gene regulatory network inference with Bc3net. Available from: https://cran.r-project.org/package=bc3net.
- El‐Gebali, S. , Mistry, J. , Bateman, A. , Eddy, S.R. , Luciani, A. , Potter, S.C. et al. (2019) The Pfam protein families database in 2019. Nucleic Acids Research, 47, D427–D432. Available from: 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb, M. & Reymond, P. (2019) Molecular interactions between plants and insect herbivores. Annual Review of Plant Biology, 70, 527–557. Available from: 10.1146/annurev-arplant-050718-095910 [DOI] [PubMed] [Google Scholar]
- Erbilgin, N. , Krokene, P. , Christiansen, E. , Zeneli, G. & Gershenzon, J. (2006) Exogenous application of methyl jasmonate elicits defenses in Norway spruce (Picea abies) and reduces host colonization by the bark beetle Ips typographus . Oecologia, 148, 426–436. Available from: 10.1007/s00442-006-0394-3 [DOI] [PubMed] [Google Scholar]
- Fernández‐Calvo, P. , Chini, A. , Fernández‐Barbero, G. , Chico, J.M. , Gimenez‐Ibanez, S. , Geerinck, J. et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell, 23, 701–715. Available from: 10.1105/tpc.110.080788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flø, D. , Norli, H.R. , Økland, B. & Krokene, P. (2018) Successful reproduction and pheromone production by the spruce bark beetle in evolutionary naïve spruce hosts with familiar terpenoid defences. Agricultural and Forest Entomology, 20, 476–486. Available from: 10.1111/afe.12280 [DOI] [Google Scholar]
- Franceschi, V.R. , Krekling, T. & Christiansen, E. (2002) Application of methyl jasmonate on Picea abies (Pinaceae) stems induces defense‐related responses in phloem and xylem. American Journal of Botany, 89, 578–586. Available from: 10.3732/ajb.89.4.578 [DOI] [PubMed] [Google Scholar]
- Franceschi, V.R. , Krokene, P. , Christiansen, E. & Krekling, T. (2005) Anatomical and chemical defenses of conifer bark against bark beetles and other pests. New Phytologist, 167, 353–376. Available from: 10.1111/j.1469-8137.2005.01436.x [DOI] [PubMed] [Google Scholar]
- Furci, L. , Jain, R. , Stassen, J. , Berkowitz, O. , Whelan, J. , Roquis, D. et al. (2019) Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis. eLife, 8, 1–23. Available from: 10.7554/elife.40655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux, R. & Seoighe, C. (2010) A flexible R package for nonnegative matrix factorization. BMC Bioinformatics, 11, 367. Available from: 10.1186/1471-2105-11-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon, J. (1994) Metabolic costs of terpenoid accumulation in higher plants. Journal of Chemical Ecology, 20, 1281–1328. Available from: 10.1007/BF02059810 [DOI] [PubMed] [Google Scholar]
- Hammerbacher, A. , Kandasamy, D. , Ullah, C. , Schmidt, A. , Wright, L.P. & Gershenzon, J. (2019) Flavanone‐3‐Hydroxylase plays an important role in the biosynthesis of spruce phenolic defenses against bark beetles and their fungal associates. Frontiers of Plant Science, 10, 208. Available from: 10.3389/fpls.2019.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerbacher, A. , Raguschke, B. , Wright, L.P. & Gershenzon, J. (2018) Gallocatechin biosynthesis via a flavonoid 3′,5′‐hydroxylase is a defense response in Norway spruce against infection by the bark beetle‐associated sap‐staining fungus Endoconidiophora polonica . Phytochemistry, 148, 78–86. Available from: 10.1016/j.phytochem.2018.01.017 [DOI] [PubMed] [Google Scholar]
- Hammerbacher, A. , Ralph, S.G. , Bohlmann, J. , Fenning, T.M. , Gershenzon, J. & Schmidt, A. (2011) Biosynthesis of the major tetrahydroxystilbenes in spruce, astringin and isorhapontin, proceeds via resveratrol and is enhanced by fungal infection. Plant Physiology, 157, 876–890. Available from: 10.1104/pp.111.181420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerbacher, A. , Schmidt, A. , Wadke, N. , Wright, L.P. , Schneider, B. , Bohlmann, J. et al. (2013) A common fungal associate of the spruce bark beetle metabolizes the stilbene defenses of Norway spruce. Plant Physiology, 162, 1324–1336. Available from: 10.1104/pp.113.218610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerbacher, A. , Wright, L.P. & Gershenzon, J. (2020) Spruce phenolics: biosynthesis and ecological functions. In: Porth, I. & De la Torre, A. (Eds.) The Spruce Genome. Compendium of Plant Genomes. Springer, pp. 193–214. 10.1007/978-3-030-21001-4_12 [DOI] [Google Scholar]
- Hickman, R. , Van Verk, M.C. , Van Dijken, A.J.H. , Mendes, M.P. , Vroegop‐Vos, I.A. , Caarls, L. et al. (2017) Architecture and dynamics of the jasmonic acid gene regulatory network. The Plant Cell, 29, 2086–2105. Available from: 10.1105/tpc.16.00958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlásny, T. , König, L. , Krokene, P. , Lindner, M. , Montagné‐Huck, C. & Müller, J. et al. (2021) Bark beetle outbreaks in Europe: state of knowledge and ways forward for management. Current Forestry Reports, 7, 138–165. Available from: 10.1007/S40725-021-00142-X [DOI] [Google Scholar]
- Howe, G.A. , Major, I.T. & Koo, A.J. (2018) Modularity in jasmonate signaling for multistress resilience. Annual Review of Plant Biology, 69, 387–415. Available from: 10.1146/annurev-arplant-042817-040047 [DOI] [PubMed] [Google Scholar]
- Huang, J. , Hammerbacher, A. , Weinhold, A. , Reichelt, M. , Gleixner, G. , Behrendt, T. et al. (2019) Eyes on the future—evidence for trade‐offs between growth, storage and defense in Norway spruce. New Phytologist, 222, 144–158. Available from: 10.1111/nph.15522 [DOI] [PubMed] [Google Scholar]
- Hudgins, J.W. & Franceschi, V.R. (2004) Methyl jasmonate‐induced ethylene production is responsible for conifer phloem defense responses and reprogramming of stem cambial zone for traumatic resin duct formation. Plant Physiology, 135, 2134–2149. Available from: 10.1104/pp.103.037929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , Sato, Y. & Morishima, K. (2016) BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. 428, 726–731. Available from: 10.1016/J.JMB.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Kolde, R. (2019) pheatmap: Prettyheatmaps. Available from: https://cran.r-project.org/package=pheatmap.
- Krokene, P. (2015) Conifer defense and resistance to bark beetles. In: Vega, F.E. & Hofstetter, R.W. (Eds.) Bark beetles: biology and ecology of native and invasive species. Elsevier, pp. 177–207. 10.1016/B978-0-12-417156-5.00005-8 [DOI] [Google Scholar]
- Krokene, P. , Nagy, N.E. & Solheim, H. (2008) Methyl jasmonate and oxalic acid treatment of Norway spruce: anatomically based defense responses and increased resistance against fungal infection. Tree Physiology, 28, 29–35. Available from: 10.1093/treephys/28.1.29 [DOI] [PubMed] [Google Scholar]
- Langmead, B. & Salzberg, S.L. (2012) Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9, 357–359. Available from: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, M. , Zhang, H. , Julian, R. , Tang, K. , Xie, S. & Zhu, J.K. (2015) Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 112, 3553–3557. Available from: 10.1073/pnas.1502279112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.H. , Nagy, N.E. , Hammerbacher, A. , Krokene, P. , Niu, X.M. , Gershenzon, J. et al. (2012) Localization of phenolics in phloem parenchyma cells of norway spruce (Picea abies). ChemBioChem, 13, 2707–2713. Available from: 10.1002/cbic.201200547 [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G.K. & Shi, W. (2014) FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. Available from: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G.K. & Shi, W. (2019) The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Research, 47, e47. Available from: 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z. , Zhong, S. & Grierson, D. (2009) Recent advances in ethylene research. Journal of Experimental Botany, 60, 3311–3336. Available from: 10.1093/JXB/ERP204 [DOI] [PubMed] [Google Scholar]
- Linnakoski, R. , de Beer, Z.W. , Ahtiainen, J. , Sidorov, E. , Niemelä, P. & Pappinen, A. et al. (2010) Ophiostoma spp. associated with pine and spruce‐infesting bark beetles in Finland and Russia. Persoonia: Molecular Phylogeny and Evolution of Fungi, 25, 72–93. Available from: 10.3767/003158510X550845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.J. , Osbourn, A. & Ma, P. (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Molecular Plant, 8, 689–708. Available from: 10.1016/j.molp.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Liu, J.J. , Williams, H. , Li, X.R. , Schoettle, A.W. , Sniezko, R.A. , Murray, M. et al. (2017) Profiling methyl jasmonate‐responsive transcriptome for understanding induced systemic resistance in whitebark pine (Pinus albicaulis). Plant Molecular Biology, 95, 359–374. Available from: 10.1007/s11103-017-0655-z [DOI] [PubMed] [Google Scholar]
- López Sánchez, A. , Stassen, J.H.M. , Furci, L. , Smith, L.M. & Ton, J. (2016) The role of DNA (de)methylation in immune responsiveness of Arabidopsis. The Plant Journal, 88, 361–374. Available from: 10.1111/tpj.13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. & Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq. 2. Genome Biology, 15, 550. Available from: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mageroy, M.H. , Christiansen, E. , Långström, B. , Borg‐Karlson, A.‐K. , Solheim, H. , Björklund, N. et al. (2020) Priming of inducible defenses protects Norway spruce against tree‐killing bark beetles. Plant, Cell and Environment, 43, 420–430. Available from: 10.1111/pce.13661 [DOI] [PubMed] [Google Scholar]
- Mageroy, M.H. , Wilkinson, S.W. , Tengs, T. , Cross, H. , Almvik, M. , Pétriacq, P. et al. (2020) Molecular underpinnings of methyl jasmonate‐induced resistance in Norway spruce. Plant, Cell and Environment, 43, 1827–1843. Available from: 10.1111/pce.13774 [DOI] [PubMed] [Google Scholar]
- Martin, D. , Tholl, D. , Gershenzon, J. & Bohlmann, J. (2002) Methyl jasmonate induces traumatic resin ducts, terpenoid resin biosynthesis, and terpenoid accumulation in developing xylem of Norway spruce stems. Plant Physiology, 129, 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch‐Mani, B. , Baccelli, I. , Luna, E. & Flors, V. (2017) Defense priming: an adaptive part of induced resistance. Annual Review of Plant Biology, 68, 485–512. Available from: 10.1146/annurev-arplant-042916-041132 [DOI] [PubMed] [Google Scholar]
- Nagy, N.E. , Franceschi, V.R. , Solheim, H. , Krekling, T. & Christiansen, E. (2000) Wound‐induced traumatic resin duct development in stems of Norway spruce (Pinaceae): anatomy and cytochemical traits. American Journal of Botany, 87, 302–313. Available from: 10.2307/2656626 [DOI] [PubMed] [Google Scholar]
- Nystedt, B. , Street, N.R. , Wetterbom, A. , Zuccolo, A. , Lin, Y.C. , Scofield, D.G. et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature, 497, 579–584. Available from: 10.1038/nature12211 [DOI] [PubMed] [Google Scholar]
- Parker, A.H. , Wilkinson, S.W. , Ton, J. , Hannan Parker, A. , Wilkinson, S.W. & Ton, J. (2022) Epigenetics: a catalyst of plant immunity against pathogens. New Phytologist, 233, 66–83. Available from: 10.1111/nph.17699 [DOI] [PubMed] [Google Scholar]
- Pervieux, I. , Bourassa, M. , Laurans, F. , Hamelin, R. & Séguin, A. (2004) A spruce defensin showing strong antifungal activity and increased transcript accumulation after wounding and jasmonate treatments. Physiological and Molecular Plant Pathology, 64, 331–341. Available from: 10.1016/j.pmpp.2004.09.008 [DOI] [Google Scholar]
- Pieterse, C.M.J. , Zamioudis, C. , Berendsen, R.L. , Weller, D.M. , Van Wees, S.C.M. & Bakker, P.A.H.M. (2014) Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology, 52, 347–375. Available from: 10.1146/annurev-phyto-082712-102340 [DOI] [PubMed] [Google Scholar]
- Puentes, A. , Zhao, T. , Lundborg, L. , Björklund, N. & Borg‐Karlson, A.‐K. (2021) Variation in methyl jasmonate‐induced defense among norway spruce clones and trade‐offs in resistance against a fungal and an insect pest. Frontiers of Plant Science, 12, 962. Available from: 10.3389/fpls.2021.678959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, T. , Song, S. , Ren, Q. , Wu, D. , Huang, H. , Chen, Y. et al. (2011) The jasmonate‐ZIM‐domain proteins interact with the WD‐repeat/bHLH/MYB complexes to regulate jasmonate‐mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana . The Plant Cell, 23, 1795–1814. Available from: 10.1105/tpc.111.083261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019). R: A language and environment for statistical computing. Available from: http://www.r-project.org
- Ruijter, J.M. , Ramakers, C. , Hoogaars, W.M. , Karlen, Y. , Bakker, O. & van den Hoff, M.J. et al. (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research, 37(6), e45. Available from: 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A. , Nagel, R. , Krekling, T. , Christiansen, E. , Gershenzon, J. & Krokene, P. (2011) Induction of isoprenyl diphosphate synthases, plant hormones and defense signalling genes correlates with traumatic resin duct formation in Norway spruce (Picea abies). Plant Molecular Biology, 77, 577–590. Available from: 10.1007/s11103-011-9832-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke, M. , Steinkamp, R. , Waack, S. & Morgenstern, B. (2004) AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Research, 32, 309–312. Available from: 10.1093/nar/gkh379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud, H. , Greenberg, M.V.C. , Feng, S. , Bernatavichute, Y.V. & Jacobsen, S.E. (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell, 152, 352–364. Available from: 10.1016/j.cell.2012.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. , Chen, L. , Li, J. , Hu, M. , Ullah, A. , He, X. et al. (2017) The JASMONATE ZIM‐domain gene family mediates JA signaling and stress response in cotton. Plant and Cell Physiology, 58, 2139–2154. Available from: 10.1093/pcp/pcx148 [DOI] [PubMed] [Google Scholar]
- Sundell, D. , Mannapperuma, C. , Netotea, S. , Delhomme, N. , Lin, Y.C. , Sjödin, A. et al. (2015) The plant genome integrative explorer resource: plantGenIE.org. New Phytologist, 208, 1149–1156. Available from: 10.1111/nph.13557 [DOI] [PubMed] [Google Scholar]
- Tan, F. , Zhou, C. , Zhou, Q. , Zhou, S. , Yang, W. , Zhao, Y. et al. (2016) Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiology, 171, 2041–2054. Available from: 10.1104/pp.16.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanin, S.M. , Kandasamy, D. & Krokene, P. (2021) Fungal Interactions and Host Tree Preferences in the Spruce Bark Beetle Ips typographus. Frontiers in Microbiology, 12, 695167. Available from: 10.3389/fmicb.2021.695167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarvainen, L. , Wallin, G. , Lim, H. , Linder, S. , Oren, R. , Ottosson Löfvenius, M. et al. (2018) Photosynthetic refixation varies along the stem and reduces CO2 efflux in mature boreal Pinus sylvestris trees. Tree Physiology, 38, 558–569. Available from: 10.1093/treephys/tpx130 [DOI] [PubMed] [Google Scholar]
- Thaler, J.S. , Humphrey, P.T. & Whiteman, N.K. (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends in Plant Science, 17, 260–270. Available from: 10.1016/j.tplants.2012.02.010 [DOI] [PubMed] [Google Scholar]
- Wang, P. , Yu, S. , Han, X. , Xu, J. , He, Q. , Xu, S. et al. (2020) Identification, molecular characterization and expression of JAZ genes in Lycoris aurea . PLoS One, 15, e0230177. Available from: 10.1371/journal.pone.0230177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. & Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany, 111(6), 1021–1058. 10.1093/aob/mct067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. & Song, S. (2017) Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. Journal of Experimental Botany, 68(6), 1303–1321. 10.1093/jxb/erw443 [DOI] [PubMed] [Google Scholar]
- Wheeler, T.J. & Eddy, S.R. (2013) Nhmmer: DNA homology search with profile HMMs. Bioinformatics, 29, 2487–2489. Available from: 10.1093/bioinformatics/btt403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, S.W. , Magerøy, M.H. , López Sánchez, A. , Smith, L.M. , Furci, L. , Cotton, T.E.A. et al. (2019) Surviving in a hostile world: plant strategies to resist pests and diseases. Annual Review of Phytopathology, 57, 505–529. Available from: 10.1146/annurev-phyto-082718 [DOI] [PubMed] [Google Scholar]
- Wilkinson, S.W. , Vivian‐Smith, A. , Krokene, P. & Mageroy, M.H. (2021) The microRNA response associated with methyl jasmonate‐induced resistance in Norway spruce bark. Plant Gene, 27, 100301. Available from: 10.1016/j.plgene.2021.100301 [DOI] [Google Scholar]
- Wittmann, C. & Pfanz, H. (2007) Temperature dependency of bark photosynthesis in beech (Fagus sylvatica L.) and birch (Betula pendula Roth.) trees. Journal of Experimental Botany, 58, 4293–4306. Available from: 10.1093/JXB/ERM313 [DOI] [PubMed] [Google Scholar]
- Yuan, X. , Wang, H. , Cai, J. , Li, D. & Song, F. (2019) NAC transcription factors in plant immunity. Phytopathology Research, 1, 1–13. Available from: 10.1186/s42483-018-0008-0 [DOI] [Google Scholar]
- Zeneli, G. , Krokene, P. , Christiansen, E. , Krekling, T. & Gershenzon, J. (2006) Methyl jasmonate treatment of mature Norway spruce (Picea abies) trees increases the accumulation of terpenoid resin components and protects against infection by Ceratocystis polonica, a bark beetle‐associated fungus. Tree Physiology, 26, 977–988. Available from: 10.1093/treephys/26.8.977 [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Lang, Z. & Zhu, J.K. (2018) Dynamics and function of DNA methylation in plants. Nature Reviews Molecular Cell Biology, 19, 489–506. Available from: 10.1038/s41580-018-0016-z [DOI] [PubMed] [Google Scholar]
- Zhao, T. , Borg‐Karlson, A.K. , Erbilgin, N. & Krokene, P. (2011) Host resistance elicited by methyl jasmonate reduces emission of aggregation pheromones by the spruce bark beetle, Ips typographus. Oecologia, 167, 691–699. Available from: 10.1007/s00442-011-2017-x [DOI] [PubMed] [Google Scholar]
- Zhao, T. , Ganji, S. , Schiebe, C. , Bohman, B. , Weinstein, P. , Krokene, P. et al. (2019) Convergent evolution of semiochemicals across Kingdoms: bark beetles and their fungal symbionts. The ISME Journal: Multidisciplinary Journal of Microbial Ecology, 13, 1535–1545. Available from: 10.1038/s41396-019-0370-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulak, K.G. , Lippert, D.N. , Kuzyk, M.A. , Domanski, D. , Chou, T. , Borchers, C.H. et al. (2009) Targeted proteomics using selected reaction monitoring reveals the induction of specific terpene synthases in a multi‐level study of methyl jasmonate‐treated Norway spruce (Picea abies). The Plant Journal, 60, 1015–1030. Available from: 10.1111/j.1365-313X.2009.04020.x [DOI] [PubMed] [Google Scholar]
- Züst, T. & Agrawal, A.A. (2017) Trade‐offs between plant growth and defense against insect herbivory: an emerging mechanistic synthesis. Annual Review of Plant Biology, 68, 513–534. Available from: 10.1146/annurev-arplant-042916-040856 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Data Availability Statement
Raw reads from mRNA sequencing have been submitted to NCBI. BioProject accession: PRJNA768356.


