Abstract
Objective
Levetiracetam (LEV) is an effective antiseizure medicine, but 10%–20% of people treated with LEV report psychiatric side‐effects, and up to 1% may have psychotic episodes. Pharmacogenomic predictors of these adverse drug reactions (ADRs) have yet to be identified. We sought to determine the contribution of both common and rare genetic variation to psychiatric and behavioral ADRs associated with LEV.
Methods
This case‐control study compared cases of LEV‐associated behavioral disorder (n = 149) or psychotic reaction (n = 37) to LEV‐exposed people with no history of psychiatric ADRs (n = 920). All samples were of European ancestry. We performed genome‐wide association study (GWAS) analysis comparing those with LEV ADRs to controls. We estimated the polygenic risk scores (PRS) for schizophrenia and compared cases with LEV‐associated psychotic reaction to controls. Rare variant burden analysis was performed using exome sequence data of cases with psychotic reactions (n = 18) and controls (n = 122).
Results
Univariate GWAS found no significant associations with either LEV‐associated behavioural disorder or LEV‐psychotic reaction. PRS analysis showed that cases of LEV‐associated psychotic reaction had an increased PRS for schizophrenia relative to contr ols (p = .0097, estimate = .4886). The rare‐variant analysis found no evidence of an increased burden of rare genetic variants in people who had experienced LEV‐associated psychotic reaction relative to controls.
Significance
The polygenic burden for schizophrenia is a risk factor for LEV‐associated psychotic reaction. To assess the clinical utility of PRS as a predictor, it should be tested in an independent and ideally prospective cohort. Larger sample sizes are required for the identification of significant univariate common genetic signals or rare genetic signals associated with psychiatric LEV ADRs.
Keywords: adverse drug reactions, levetiracetam, pharmacogenomics, polygenic risk scoring, psychosis
Key points.
Polygenic risk scores for schizophrenia are predictive of levetiracetam (LEV)–induced psychotic reactions
GWAS analysis reveals no clinically actionable common genetic variants for LEV behavioral or psychiatric adverse drug reactions
Exome analysis identified no burden of rare variants in schizophrenia‐associated genes, or LEV‐target SV2A associated with LEV‐induced psychotic reaction
1. INTRODUCTION
Levetiracetam (LEV) is an effective antiseizure medicine (ASM), first licensed to treat epilepsy in 1999. Upon binding to its target, the synaptic vesicle protein SV2A, seizure activity is suppressed by LEV, which putatively modulates exocytosis from synaptic vesicles, thereby inhibiting presynaptic neurotransmitter release. 1 , 2 As a first‐prescription monotherapy, LEV can provide seizure freedom in over 50% of people with epilepsy. 3 , 4 Adjunctive LEV treatment stopped focal and generalized seizures, which were previously drug resistant. 5 , 6 , 7 LEV is commonly used for both monotherapy and polytherapy to treat a broad spectrum of seizure types. 8 , 9
Adverse drug reactions (ADRs) are associated with LEV treatment. An estimated 18% of people with epilepsy treated with LEV will experience some neuropsychiatric response, resulting in dosage lowering or, more frequently, cessation of treatment. 10 LEV‐associated ADRs cover many phenotypes, including behavioral disorders such as irritability and personality change and affective disorders such as depression and suicidal ideation. Furthermore, ~1% of people exposed to LEV will experience drug‐induced psychotic reactions, a significantly higher rate than associated with other ASMs. 10 , 11 As a group, psychiatric and behavioral side effects have the highest economic burden of all ASM‐related ADRs. 12
Previous pharmacogenomic research into ASM‐associated ADRs, primarily focused on univariate analyses, has identified several clinically important predictors of clinical relevance. For example, human leukocyte antigen (HLA) region alleles HLA‐B*15:02 and HLA‐A*31:01, as well as the cytochrome P450 (CYP)2C9 *3 allele are strong predictors of aromatic ASM‐induced severe cutaneous adverse reactions. 13 , 14 , 15 , 16 A previous effort, focused on a limited number of single nucleotide polymorphisms (SNPs), reported a correlation between LEV‐induced psychiatric ADRs and genetic variation linked to dopaminergic activity. 17 To date, there has been no genomic investigation of LEV psychiatric ADRs.
Polygenic risk scoring (PRS) is a method used to assess an individual's cumulative burden of common genetic variants associated with a disease or trait. 18 The predictive potential of PRS in the field of pharmacogenomics has been demonstrated previously. For example, people with bipolar disorder who have a higher PRS for schizophrenia were shown to be less likely to respond to mood‐stabilization treatment with lithium. 19 PRS for non‐melanoma skin cancer has been shown to predict the risk of and time to azathioprine‐associated post‐transplant skin cancer. 20
The role of rare genetic variation in pharmacogenomics is less well assessed. Rare genetic variants in the SLCO1B1 gene seem to influence the clearance of methotrexate, a chemotherapeutic agent. 21 Rare variation in the CYP genes CYP3A4 and CYP2C9 appears to explain the 18.4% and 43.1% spectrum of enzyme activity. 22 Bioinformatic predictions of the contribution of rare variation to drug metabolism suggest that rare variants may account for a substantial proportion of inter‐individual variability of the metabolism of drugs such as warfarin and the statin medication simvastatin. 23
We utilized a variety of approaches to assess the role of genetic variation in psychiatric and behavioral ADRs associated with LEV. First, we applied a univariate genome‐wide association study (GWAS) approach to identify individual common genetic risk loci for LEV‐induced behavioral ADRs or LEV‐associated psychotic reaction. We then applied a polygenic approach, using PRS to test whether a higher polygenic burden for schizophrenia can predict LEV‐associated psychotic reactions. Finally, we performed burden analysis of exome data to identify if rare variants are associated with this clinical condition compared to controls.
2. METHODS
All research participants (or their legal guardians in the case of minors or individuals with intellectual disability) provided written, informed consent. The study was approved by ethics committees at each study site.
2.1. Cohort assembly
Genetic and phenotypic data on cases and controls were obtained from various recruitment sites. All cases and controls were people with epilepsy and a history of treatment with LEV.
EpiPGX Consortium samples were contributed from the following 10 sites:
The Royal College of Surgeons (Dublin, Ireland), Antwerp University Hospital (Belgium), Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) “G. Gaslini” Institute (Genova, Italy), the University of Liverpool (UK), the University of Tubingen (Germany), University Medical Centre, (Utrecht, The Netherlands), UCL Queen Square Institute of Neurology (UK), the University of Glasgow (UK), the University of Bonn (Germany), and the University of Melbourne (Australia).
We obtained additional cases (beyond EpiPGX) from the Beaumont Hospital Epilepsy Biobank (Dublin, Ireland), the Columbia University Medical Center (United States), and the Department of Medicine at the University of Melbourne, Austin Health (Australia).
2.2. Case and control phenotyping
All phenotyping was conducted by the neurology team where the participant was recruited. To meet the criteria of an ADR, each case must have (1) occurred within 6 months of the initiation of LEV treatment, (2) led to withdrawal or dose reduction of LEV, (3) ADR reversed or improved after withdrawal or dose reduction, and (4) ADR not attributed to any other cause by the treating or phenotyping clinician.
We specifically examined two LEV ADR phenotypes:
1: Any LEV‐induced behavioral disorder. Defined as one or more of the following: agitation, aggression, irritability, confusion, or cognitive decline.
2: LEV‐induced psychotic reaction: vivid hallucinations, misidentifications, delusions and or ideas of reference (often of a paranoid or persecutory nature), psychomotor disturbances (excitement or stupor), and an abnormal affect, ranging from intense fear to ecstasy. The sensorium is usually clear, but some degree of clouding of consciousness may be present, although not severe confusion. A psychiatrist must have confirmed the diagnosis. Any cases with a previous history of psychotic illness were excluded.
Controls were LEV‐treated people with epilepsy with no psychiatric side‐effects reported in clinical notes after a minimum of 6 months of treatment.
2.3. Genotyping and quality control
EpiPGX samples were genotyped on various Illumina chips and underwent imputation and quality control (QC) processes, as reported previously. 24
The additional samples from Dublin (Beaumont) and Melbourne were genotyped on the Illumina Global Screening Array chip and imputed on the Sanger imputation server (https://imputation.sanger.ac.uk/) using the Haplotype Reference Consortium release 1.1 panel as a reference. 25 The newly genotyped samples underwent the same QC procedures as the EpiPGX cohort (see Ref. 24), and were then merged with the EpiPGX data set for further analysis.
To ensure European ancestry and genetic homogeneity all samples were merged with the Human Genome Diversity Project (HGDP). 26 Principal component analysis was conducted by thinning for linkage disequilibrium using PLINK 1.9 (‐‐indep‐pairwise 1000, 100, 0.1), and then estimating principal components (PCs). The top two PCs were graphed using R v3.5, 27 and any samples which did not overlay the European HGDP samples on the PCA plot were excluded.
2.4. GWAS
We used the PGA2 software to estimate GWAS power, 28 based on a minimum minor allele frequency of 5% to detect an association to the alpha level of 5 × 10−8 under an additive model. GWAS analyses were carried out using a frequentist association model in SNPTEST, 29 with sex and the top six principal components included as covariates to account for bias and population stratification. The threshold for genome‐wide significance was set at p < 5 × 10−8. We included only autosomal SNPs in our analyses.
2.5. Polygenic risk scoring
Polygenic risk scores for schizophrenia were estimated for all samples with LEV‐induced psychosis, and controls, using PRSice2. 30 GWAS results for schizophrenia were obtained from the Psychiatric Genomics Consortium. 31 All SNPs from the schizophrenia GWAS with p‐values ≤ .5 were included in the PRS analysis. PRS were normalized to mean 0 and SD 1 and then regressed onto LEV psychosis case: control status using R v3.5, with the top six PCs and sex included as covariates.
We used the pROC R package 32 to estimate the area under the receiver‐operating characteristic (ROC) curve of the above PRS model, compared to the null model, and a model ccomprising covariates only (PCs 1–6 and sex).
2.6. Exome sequencing and analysis
Whole‐exome sequencing was conducted at deCODE genetics on the Illumina HiSeq 2500 with the Nextera Rapid Capture Expanded Exome kit (Illumina). Adapter sequences were removed, and the data were put through a Genome Analysis Toolkit (GATK 33 ;) best practices pipeline with the GRCh37 human reference genome 34 for joint calling, recalibration, filtering, and variant annotation. We excluded any variant position with a mean depth of less than 10 in all samples. Only samples with more than 30× mean coverage or more than 70% of the exome intervals covered by at least 20× mean coverage were included for analysis.
We first performed a hypothesis‐free test single‐gene collapsing analysis with the combined and multivariate collapsing (CMC) method with a two‐sided Fisher exact test using rvtests. 35 We then performed gene set collapsing tests with the regression‐based two‐sided SKAT‐O method, 36 testing for a burden of functional variants in genes that had been associated previously with schizophrenia (SLC6A1, SETD1A, RBM12 37 ). PCs 1–6 and sex were included as covariates.
3. RESULTS
3.1. Cohort description
We included 1106 people with epilepsy treated with LEV in our analysis, of which 149 had LEV‐associated behavioral disorder, 37 had LEV‐associated psychotic reaction, and 920 were controls. A full breakdown of case phenotypes and controls is provided in Table 1. Fifty‐four percent of cases in our study were female, compared to 55% of controls. Cases had an average of 46, and controls an average age of 51, with an average age at first seizure of 17 for cases and 21 for controls. Twenty‐seven percent of cases had generalized epilepsy, 59% had focal epilepsy, and 14% had unclassified epilepsy. Twenty‐five percent of controls had generalized epilepsy, 67% had focal epilepsy, and 9% were unclassified. Among cases, the most common EEG finding was generalized spike/wave discharges.
TABLE 1.
Number of post‐QC samples in each phenotypic group
| ADR | Cases | Controls |
|---|---|---|
| LEV behavioral disorder | 149 | 920 |
| LEV psychotic reaction | 37 | 920 |
| LEV psychotic reaction (exome data) | 18 | 122 |
Also shown are the subset (n = 18) of patients with LEV‐psychotic reaction (n = 37) who had exome data available.
Abbreviations: ADR, adverse drug reaction; LEV, levetiracetam; QC, quality control.
3.2. Genome‐wide association analyses of LEV‐associated psychiatric ADRs
We conducted a GWAS of 149 cases with LEV‐associated behavioral disorder vs 920 controls. After quality control, 3.8 million SNPs were included in the association analysis. Our analysis had 80% power to detect a genetic variant with a relative risk of 3.34 or greater. We did not observe any variants that surpassed the significance threshold of 5 × 10−8 (Figure 1A). The variant rs1800497, which had been reported previously to predict LEV‐induced psychiatric ADRs, 17 was found in our LEV‐associated behavioral disorder GWAS to have an uncorrected p‐value of .458, although the phenotype criteria used in this study was not an exact match.
FIGURE 1.

Manhattan plots (left), and quantile‐quantile plots (right) for GWAS of (A) LEV‐induced behavioural disorder and (B) LEV‐induced psychosis. Genomic inflation factors displayed as GIF in the QQ plots. LEV, levetiracetam
We next conducted a GWAS of LEV‐induced psychotic reaction, which included 37 cases and 920 controls across 3.8 million SNPs. We estimated 80% power to detect a variant with a relative risk of 7.22 or greater. No genome‐wide significant signals were observed (Figure 1B).
3.3. Polygenic risk score analysis
We tested the hypothesis that people who experience LEV‐induced psychotic reaction harbor an excess of common variants associated with schizophrenia using PRS analysis (see Methods). We found that the PRS for schizophrenia was significantly higher in LEV‐psychotic reaction cases compared to controls (estimate = .4886, standard error [SE] = .1881, p = .0097). Schizophrenia PRS explained 4% of the phenotypic variance in case‐control status. Generating a ROC curve of LEV‐psychotic reaction case: control status from a model of schizophrenia PRS, PCs 1–6, and sex produced a curve with an area under the curve (AUC; predictive power) of 0.65 (Figure 2). This is greater than the AUCs of the null model (0.50) and a model built on covariates alone (0.57). LEV‐psychosis cases make up 3.87% of our cohort (n cases = 37, n controls = 920; Table 1). If we take only samples in the top 10% of the schizophrenia PRS distribution, we find that LEV‐psychosis cases make up 8.3% of the cohort (n cases = 8, n controls = 88). The bottom 10% of the schizophrenia PRS distribution contains only 1% LEV‐psychosis cases (n cases = 1, n controls = 99).
FIGURE 2.
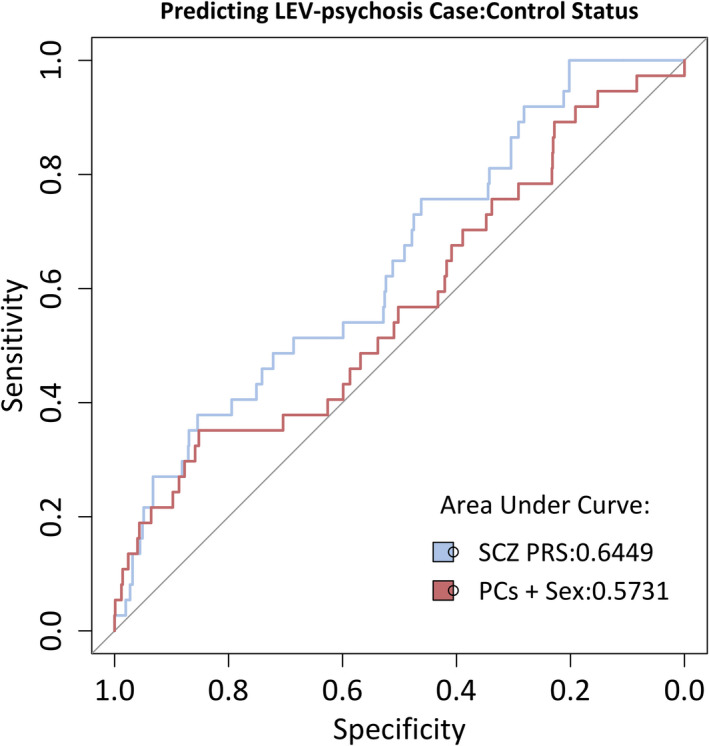
Area under the receiver‐operating characteristic curve for LEV‐psychotic reaction PRS analysis. The red line shows a model built from covariates only (PCs 1–6 + sex), the blue line is a model of both covariates and schizophrenia PRS. LEV, levetiracetam; PRS, polygenic risk score
3.4. Rare variant burden analysis
To test the hypothesis that rare variant burden can contribute to the LEV‐psychotic reaction, we performed rare variant analysis on people with LEV‐induced psychotic reaction. First, all genes were tested individually for the enrichment of variation. After Bonferroni correction for 18 668 protein‐coding genes, no gene reached the threshold of statistical significance (p < 2.67−6). We tested for rare variant burden in genes previously found to harbor rare variants in people with schizophrenia. We found no significant enrichment of rare variation in SLC6A1 (p = .819), SETD1A (p = .030), or RBM12 (p = .220), given a threshold of statistical significance of p < .016. Testing rare‐variant burden in these schizophrenia‐associated genes together as a unit also found no significant enrichment (p = .83). We did not observe a burden of rare variants in the target of LEV SV2A (p = .492).
4. DISCUSSION
Levetiracetam (or LEV) is a highly effective ASM that is associated with behavioral adverse events in a proportion of patients, including affective disorder, aggression, and psychotic reactions. 38 We applied various analytical models to assess the role of genetic variation in LEV behavioral ADRs. We present evidence that the genetic burden for schizophrenia, as quantified by PRS, is a risk factor for LEV‐induced psychotic reactions in people with epilepsy. We found no evidence of rare variant burden in LEV psychosis. From univariate GWAS analysis, we can conclude that there are no common variants with an OR >7.22 associated with LEV‐induced psychotic reaction, or an OR >3.34 associated with an LEV behavioral disorder.
We then constructed a predictive model for LEV‐psychotic reaction using schizophrenia PRS with a predictive power (as measured by AUC/ROC analysis) of 65%. This model explained 4% of the variation in case: control status for LEV psychosis in our cohort. The schizophrenia GWAS used to estimate the PRS explained 7% of the variation in schizophrenia case: control status, representing the upper limit of phenotypic variation that could be explained by a PRS model generated from it. 31 More powerful GWAS of schizophrenia that explains more phenotypic variation may allow more accurate PRS models in the future.
These results raise the possibility of screening people with epilepsy to identify those at risk of developing psychotic reactions as an ADR, before exposure to LEV. Our findings must be validated in an independent sample ideally collected in a prospective study to clarify clinical potential.
The ability to screen for individuals at risk of developing LEV‐induced psychotic reactions could be improved by including known clinical risk factors such as a history of depression or anxiety or a history of recreational drug use. 39 Given that LEV is a commonly prescribed first‐line ASM, 40 and that up to 18% of people prescribed LEV will experience some side effects, 41 identifying those at risk of ADRs would appear clinically attractive.
Our study has limitations. First, we focused on people of European ancestry. Given that PRS effects cannot be assumed to act consistently across ethnic backgrounds, the role of schizophrenia PRS in non‐European LEV‐psychosis cases must be assessed separately. 42 Second, the relatively low number of cases included in our analyses limited our power to detect effects, particularly in the case of the rare variant analysis. Finally, although the potential dose/concentration‐dependent LEV‐induced psychosis could not be explored in this study, clinicians could consider optimizing therapeutic LEV treatment in the patients.
In summary, we showed that polygenic burden for schizophrenia is a risk factor for LEV‐induced psychotic reactions. To assess the clinical utility of this result, it should be tested in an independent and ideally prospective cohort. The following steps would include testing larger cohorts for univariate GWAS signals and further exome analysis in larger samples to assess the rare variant contribution to LEV psychiatric ADRs. Future research could also perform similar genetic analysis on other ASMs that are known to associate with behavioral and/or psychiatric ADRs.
CONFLICT OF INTEREST
The authors have no financial conflicts of interest to disclose. We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
ACKNOWLEDGMENTS
This work was supported by a grant from the European Commission (7th Framework Programme Grant 279062, EpiPGX) and in part by a research grant from Science Foundation Ireland (SFI) under Grant Number 16/RC/3948 and co‐funded under the European Regional Development Fund and by FutureNeuro industry partners. We would like to thank all the people with epilepsy who kindly donated their DNA and associated clinical data to this study. Without their participation the work would not have been possible. Open access funding provided by IReL. [Correction added on 16 May, 2022, after first online publication: IReL funding statement has been added.]
Campbell C, McCormack M, Patel S, Stapleton C, Bobbili D, Krause R, et al; The EpiPGX Consortium . A pharmacogenomic assessment of psychiatric adverse drug reactions to levetiracetam. Epilepsia. 2022;63:1563–1570. 10.1111/epi.17228
REFERENCES
- 1. Stahl SM. Psychopharmacology of anticonvulsants: levetiracetam as a synaptic vesicle protein modulator. J Clin Psychiatry. 2004;65(9):1162–3. [DOI] [PubMed] [Google Scholar]
- 2. Klitgaard H, Verdru P. Levetiracetam: the first SV2A ligand for the treatment of epilepsy. Expert Opin Drug Discov. 2007;2(11):1537–45. [DOI] [PubMed] [Google Scholar]
- 3. Brodie MJ, Perucca E, Ryvlin P, Ben‐Menachem E, Meencke H‐J. Comparison of levetiracetam and controlled‐release carbamazepine in newly diagnosed epilepsy. Neurology. 2007;68(6):402–8. [DOI] [PubMed] [Google Scholar]
- 4. Stephen LJ, Kelly K, Parker P, Brodie MJ. Levetiracetam monotherapy–outcomes from an epilepsy clinic. Seizure. 2011;20(7):554–7. [DOI] [PubMed] [Google Scholar]
- 5. Mohanraj R, Parker PG, Stephen LJ, Brodie MJ. Levetiracetam in refractory epilepsy: a prospective observational study. Seizure. 2005;14(1):23–7. [DOI] [PubMed] [Google Scholar]
- 6. Ben‐Menachem E, Gilland E. Efficacy and tolerability of levetiracetam during 1‐year follow‐up in patients with refractory epilepsy. Seizure. 2003;12(3):131–5. [DOI] [PubMed] [Google Scholar]
- 7. Delanty N, Jones J, Tonner F. Adjunctive levetiracetam in children, adolescents, and adults with primary generalized seizures: open‐label, noncomparative, multicenter, long‐term follow‐up study. Epilepsia. 2012;53(1):111–9. [DOI] [PubMed] [Google Scholar]
- 8. Lee JW, Dworetzky B. Rational polytherapy with antiepileptic drugs. Pharmaceuticals. 2010;3(8):2362–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kikuchi D, Obara T, Kashiwagura S, Arima Y, Hino H, Miura R, et al. Trends in prescription of anti‐seizure medicines for Japanese pediatric outpatients during 2013–2019. Epilepsy Behav Rep. 2021;16:2013–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen B, Choi H, Hirsch LJ, Katz A, Legge A, Buchsbaum R, et al. Psychiatric and behavioral side effects of antiepileptic drugs in adults with epilepsy. Epilepsy Behav. 2017;76:24–31. [DOI] [PubMed] [Google Scholar]
- 11. Chen Z, Lusicic A, O'Brien TJ, Velakoulis D, Adams SJ, Kwan P. Psychotic disorders induced by antiepileptic drugs in people with epilepsy. Brain. 2016;139(10):2668–78. 10.1093/brain/aww196 [DOI] [PubMed] [Google Scholar]
- 12. de Kinderen RJA, Evers SMAA, Rinkens R, Postulart D, Vader CI, Majoie MHJM, et al. Side‐effects of antiepileptic drugs: the economic burden. Seizure. 2014;23(3):184–90. [DOI] [PubMed] [Google Scholar]
- 13. McCormack M, Alfirevic A, Bourgeois S, Farrell JJ, Kasperavičiūtė D, Carrington M, et al. HLA‐A*3101 and carbamazepine‐induced hypersensitivity reactions in Europeans. N Engl J Med. 2011;364(12):1134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mockenhaupt M, Wang C‐W, Hung S‐I, Sekula P, Schmidt AH, Pan R‐Y, et al. HLA‐B*57:01 confers genetic susceptibility to carbamazepine‐induced SJS/TEN in Europeans. Allergy. 2019;74(11):2227–30. [DOI] [PubMed] [Google Scholar]
- 15. Chung W‐H, Hung S‐I, Hong H‐S, Hsih M‐S, Yang L‐C, Ho H‐C, et al. Medical genetics: a marker for Stevens‐Johnson syndrome. Nature. 2004;428:486. [DOI] [PubMed] [Google Scholar]
- 16. Hung C‐C, Lin C‐J, Chen C‐C, Chang C‐J, Liou H‐H. Dosage recommendation of phenytoin for patients with epilepsy with different CYP2C9/CYP2C19 polymorphisms. Ther Drug Monit. 2004;26(5):534–40. [DOI] [PubMed] [Google Scholar]
- 17. Helmstaedter C, Mihov Y, Toliat MR, Thiele H, Nuernberg P, Schoch S, et al. Genetic variation in dopaminergic activity is associated with the risk for psychiatric side effects of levetiracetam. Epilepsia. 2013;54(1):36–44. [DOI] [PubMed] [Google Scholar]
- 18. Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12(1):44. 10.1186/s13073-020-00742-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amare AT, Schubert KO, Hou L, Clark SR, Papiol S, Heilbronner U, et al. Association of polygenic score for schizophrenia and HLA antigen and inflammation genes with response to lithium in bipolar affective disorder: a genome‐wide association study. JAMA Psychiatry. 2018;75(1):65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stapleton CP, Chang B‐L, Keating BJ, Conlon PJ, Cavalleri GL. Polygenic risk score of non‐melanoma skin cancer predicts post‐transplant skin cancer across multiple organ types. Clin Transplant. 2020;34(8):e13904. 10.1111/ctr.13904 [DOI] [PubMed] [Google Scholar]
- 21. Ramsey LB, Bruun GH, Yang W, Treviño LR, Vattathil S, Scheet P, et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012;22(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ingelman‐Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol Sci. 2004;25(4):193–200. [DOI] [PubMed] [Google Scholar]
- 23. Ingelman‐Sundberg M, Mkrtchian S, Zhou Y, Lauschke VM. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum Genomics. 2018;12(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCormack M, Gui H, Ingason A, Speed D, Wright GEB, Zhang EJ, et al. Genetic variation in CFH predicts phenytoin‐induced maculopapular exanthema in European‐descent patients. Neurology. 2018;90(4):e332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cavalli‐Sforza LL. The Human Genome Diversity Project: past, present and future. Nat Rev Genet. 2005;6(4):333–40. 10.1038/nrg1596 [DOI] [PubMed] [Google Scholar]
- 27. R Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Core Team; 2019. https://www.r‐project.org/ [Google Scholar]
- 28. Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case‐control genetic association analyses. BMC Genet. 2008;9:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome‐wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–13. 10.1038/ng2088 [DOI] [PubMed] [Google Scholar]
- 30. Choi SW, O'Reilly PF. PRSice‐2: polygenic risk score software for biobank‐scale data. GigaScience. 2019;8(7):giz082. 10.1093/gigascience/giz082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ripke S, Neale BM, Corvin A, Walters JTR, Farh K‐H, Holmans PA, et al. Biological insights from 108 schizophrenia‐associated genetic loci. Nature. 2014;511(7510):421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J‐C, et al. pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy‐Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43(1110):11.10.1‐11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32(9):1423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare‐variant association testing with application to small‐sample case‐control whole‐exome sequencing studies. Am J Hum Genet. 2012;91(2):224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rees E, Han J, Morgan J, Carrera N, Escott‐Price V, Pocklington AJ, et al. De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nat Neurosci. 2020;23(2):179–84. 10.1038/s41593-019-0565-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Verrotti A, Prezioso G, Di Sabatino F, Franco V, Chiarelli F, Zaccara G. The adverse event profile of levetiracetam: a meta‐analysis on children and adults. Seizure. 2015;31:49–55. [DOI] [PubMed] [Google Scholar]
- 39. Josephson CB, Engbers JDT, Jette N, Patten SB, Singh S, Sajobi TT, et al. Prediction tools for psychiatric adverse effects after levetiracetam prescription. JAMA Neurol. 2019;76(4):440–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nicholas JM, Ridsdale L, Richardson MP, Ashworth M, Gulliford MC. Trends in antiepileptic drug utilisation in UK primary care 1993–2008: cohort study using the General Practice Research Database. Seizure. 2012;21(6):466–70. [DOI] [PubMed] [Google Scholar]
- 41. Chen Z, Brodie MJ, Liew D, Kwan P. Treatment outcomes in patients with newly diagnosed epilepsy treated with established and new antiepileptic drugs: a 30‐year longitudinal cohort study. JAMA Neurol. 2018;75(3):279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100(4):635–49. 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]


