Summary
Checkpoint inhibitors (CPIs) are routinely employed in relapsed/refractory classical Hodgkin lymphoma. Nonetheless, persistent long‐term responses are uncommon, and one‐third of patients are refractory. Several reports have suggested that treatment with CPIs may re‐sensitize patients to chemotherapy, however there is no consensus on the optimal chemotherapy regimen and subsequent consolidation strategy. In this retrospective study we analysed the response to rechallenge with chemotherapy after CPI failure. Furthermore, we exploratively characterized the clonal evolution profile of a small sample of patients (n = 5) by employing the CALDER approach. Among the 28 patients included in the study, 17 (71%) were primary refractory and 26 (92%) were refractory to the last chemotherapy prior to CPIs. Following rechallenge with chemotherapy, response was recorded in 23 (82%) patients experiencing complete remission and 3 (11%) patients experiencing partial remission. The tumour evolution of the patients inferred by CALDER seemingly occurred prior to the first cycle of therapy and was characterized either by linear or branching evolution patterns. Twenty‐five patients proceeded to allogeneic stem cell transplantation. At a median follow‐up of 21 months, median PFS and OS were not reached. In conclusion, patients who fail CPIs can be effectively rescued by salvage chemotherapy and bridged to allo‐SCT/auto‐SCT.
INTRODUCTION
Classical Hodgkin Lymphoma (cHL) is one of the most common malignancies in adolescents and young adults. Fortunately, most cases are cured by standard chemo‐radiotherapy. However, patients who relapse after standard regimens or are primary refractory have a poorer outcome. In this scenario, checkpoint inhibitors (CPIs) have shown impressive efficacy, producing significant remission rates with manageable toxicities, and have been approved for clinical use. 1 , 2 However, long‐term disease control is uncommon, and most patients ultimately become resistant to CPIs and experience disease progression, with a median overall survival (OS) of around one year upon treatment discontinuation. 3 , 4 Such patients, together with those who do not respond to checkpoint inhibition, represent an unmet medical need. Though promising therapies directed against novel immunological targets are currently under evaluation in clinical trials, 5 , 6 , 7 no treatment alternatives have yet been validated.
In this setting, physicians have recently reported a considerable efficacy of conventional chemotherapy in a small series of solid tumours progressing after PD‐1 inhibitors. 8 , 9 Accordingly, such results were confirmed in relapsed/refractory classical Hodgkin Lymphoma (r/r cHL) in two small French and Italian cohorts 10 , 11 and in a subsequent larger multicentric analysis. 12
We here retrospectively analysed the outcomes of a multi‐centric cohort of chemo‐refractory patients. These patients failed to respond to PD‐1 inhibitors and were subsequently administered salvage chemotherapy and addressed to allogeneic stem cell transplantation (allo‐SCT), in the attempt to identify the optimal therapeutic strategy. Additionally, we performed an exploratory analysis aimed at characterizing the clonal evolution profile of a small sample of patients, to investigate whether subclonal modifications and/or suppression could determine response to chemotherapy.
PATIENTS AND METHODS
Patients
A total of twenty‐eight consecutive patients affected by cHL (age ≥ 18 years) who were refractory (n = 10, 36%) to PD‐1 inhibitors or experienced disease progression after initial response (n = 18, 64%), from March 2017 to November 2020, were included in the study (Table 1). Patients were treated in three different hospitals: Humanitas Research Hospital (n = 17), Istituto Nazionale dei Tumori di Milano (n = 8), Istituto Europeo di Oncologia (n = 3). All the included centers shared similar institutional guidelines and, as such, addressed all patients failing PD‐1 inhibitors to salvage chemotherapy. Sixteen patients (57%) received Nivolumab within the CA209‐205 phase II clinical trial at a fixed dose of 3 mg/kg every 14 days, one patient (4%) received Nivolumab within an extended access program for patients relapsing after allo‐SCT at a dose of 3 mg/kg once a month, eight patients (28%) received EMA‐approved Nivolumab at a flat dose of 240 mg every 14 days, and three patients (11%) received EMA‐approved Pembrolizumab at a flat dose of 200 mg every 21 days. Eligibility criteria included: (i) diagnosis of cHL; (ii) age ≥18 years; (iii) proven disease progression by PET‐CT after therapy with a PD‐1 inhibitor and (iv) Eastern Cooperative Oncology Group performance status (0–1). Patients who exclusively received radiation therapy after disease progression were excluded from the study. Prior allogeneic stem cell transplantation was not considered an exclusion criterion. This study was approved by the institutional Ethical Committee of all participating centers, and all patients provided written informed consent in accordance with the Declaration of Helsinki.
TABLE 1.
Main patients' characteristics
|
All n = 28 |
|
|---|---|
| Gender | |
| Male | 21 (75%) |
| Female | 7 (25%) |
| Median age (range) | 29 (19–71) |
| Stage prior to anti‐PD1 therapy | |
| I–II | 6 (21%) |
| III–IV | 22 (79%) |
| Extranodal disease prior to anti‐PD1 therapy | 21 (75%) |
| B symptoms prior to anti‐PD1 therapy | 10 (36%) |
| Bulky disease prior to anti‐PD1 therapy | 6 (21%) |
| Response to anti PD‐1 therapy | |
| Refractory | 10 (36%) |
| Responsive | 18 (64%) |
| Median number of anti PD‐1 cycles (range) | 13 (3–72) |
| Median duration of anti PD‐1 therapy (mos) | 6 (2–34) |
| Median number of prior therapies (range) | 4 (2–11) |
| Prior BV | 28 (100%) |
| Prior ASCT | 18 (64%) |
| Prior RT | 14 (50%) |
| Response to last chemotherapy prior to anti‐PD1 therapy | |
| Refractory | 26 (92%) |
| Responsive | 2 (8%) |
Study design
The primary objective of this retrospective multi‐centric study was to assess the effectiveness of salvage chemotherapy after failure of PD‐1 inhibitors. The primary endpoint of the study was overall response rate. All patients received various chemotherapy regimens, some of which included agents that had been administered prior to CPIs. Disease response was evaluated by PET‐CT according to the Lugano classification 13 at the end of the regimen; a PET/CT scored as 4 or 5 using the Deauville 5‐point scale was considered positive; when clinically indicated, response assessment was anticipated. Visual analysis was independently performed by two experienced imagers blinded to patients’ characteristics. If an objective response was reached, eligible patients were addressed to allogeneic stem cell transplantation (allo‐SCT). Secondary objectives included overall survival (OS) and progression‐free survival (PFS).
Circulating tumour DNA (ctDNA) genotyping
The CAPP‐seq strategy (Cancer Personalized Profiling by deep Sequencing), an ultrasensitive capture‐based targeted sequencing, was used to genotype ctDNA of 5 patients treated with BeGEV (Bendamustine, Gemcitabine, Vinorelbine).
Blood samples as source of plasma and peripheral blood mononuclear cells (PBMCs) were collected prior to chemotherapy initiation, at the time of interim PET and at the end of treatment (this time point is not present for 3 of the 5 patients). We analysed cfDNA (cell‐free DNA) extracted from plasma and paired DNA from PBMCs, as source of germline DNA (gDNA) to filter out polymorphism and sequencing errors.
A targeted resequencing panel optimized to include the coding exons and splice sites of 133 genes (320 Kb) that are recurrently mutated in B‐cell lymphomas was used. Sequencing was performed using the Nextseq 550 platform (Illumina) to obtain a depth of coverage >2000x in >80% of the target region in all samples. A robust and previously validated bioinformatics pipeline was used for somatic analysis. 14
Additional methodological details can be found in the Supplementary Materials.
Reconstruction of longitudinal cancer evolution models via CALDER
We employed the Cancer Analysis of Longitudinal Data through Evolutionary Reconstruction (CALDER) approach, 15 a computational method for the inference of phylogenetic tumour trees from longitudinal bulk sequencing data, to reconstruct the evolution models of 5 treated R/R cHL patients. Notice that approaches with similar goals have been recently proposed to exploit single‐cell sequencing data. 16
In brief, CALDER takes as input the reference and alternative read count of a list of selected genomic variants, detected in samples collected from single patients at subsequent time points.
CALDER solves a Non‐negative Matrix Factorization (NMF) problem and returns both the longitudinal phylogenetic tree describing the accumulation of genomic variants and the clonal prevalence variation at each time point, by maximizing the likelihood with respect to the input data. Every dataset includes the reference and alternative reads count for a panel of genomic variants, for each patient and each time point, as obtained via somatic analysis from ctDNA sequencing, as previously described. Furthermore, to reduce the number of features and the impact of noise, we here clustered all the variants occurring on the same gene, by summing their reference and alternative read counts. As a result, we here consider the counts of all variants hitting a specific gene in that sample. CALDER was run with default parameters and with the Gurobi 8.0.1 optimizer(http://www.gurobi.com). After the inference, we used the CALDER output as input for TimeScape 17 in order to obtain a user‐friendly visualization of the clonal evolution model of every single tumour under analysis. The outcoming clonal evolution models are presented in Figure 3 of the main text.
FIGURE 3.
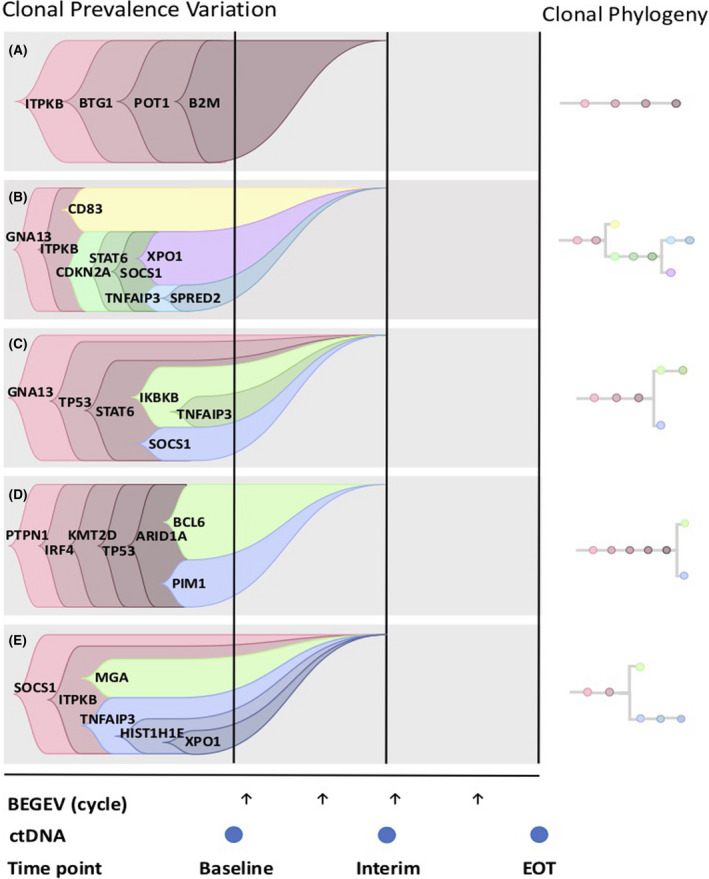
Longitudinal cancer evolution model returned by CALDER from the VAF profiles of 5 consecutive R/R cHL patients failing CPIs and subsequently addressed to BeGEV chemotherapy. Arrows represent BeGEV cycles, while dots indicate plasma collection for ctDNA profiling. Of note, patients C, D and E (0008, 0024 and 0113) do not have a plasma sample at end of treatment (EOT)
Statistical analysis
Analysis was performed using SPSS version 21.0.0 and R version 3.6.1. Categorical variables were expressed as proportions with the respective 95% confidence intervals, and continuous variables were expressed as the medians with the respective range. Fisher’s exact test was used for comparisons between categorical variables. The Kaplan–Meier method was used for OS and PFS analyses, which were calculated starting from the date of initiation of chemotherapy after CPI failure.
RESULTS
Patients’ characteristics
The patients’ characteristics are summarized in Table 1. From March 2017 to November 2020, 28 patients with relapsed/refractory cHL (r/r cHL) were included in the study. Median age at diagnosis was 29 years (range, 19–71). Most patients had advanced stage (III/IV) cHL (70%) and had received a median of 4 (range, 2–11) therapies prior to CPI, including Brentuximab Vedotin (BV, 100%), autologous stem cell transplantation (ASCT, 64%) and radiation therapy (50%). One patient had relapsed after allo‐SCT. Almost all patients (n = 26, 92%) were refractory to the last therapy prior to CPIs; 21 (75%) patients presented with extranodal involvement and 10 (36%) had B symptoms. Anti‐PD1 therapy was administered for a median of 13 cycles (range, 3–72), for a median duration of 6 months (range, 2–34). The best overall response (BOR) during CPI treatment was CR in 3 patients (11%; 95% CI 0–22), PR in 15 patients (53%; 95% CI 35–71), SD in 7 patients (25%; 95% CI 9–41) and PD in 3 patients (11%; 95% CI 0–22); however, at end of treatment all patients had PD. Of the 3 patients who achieved CR, one had relapsed after allo‐SCT, one interrupted treatment due to toxicity (grade 3 inflammatory colitis) and relapsed shortly thereafter, and one patient refused allo‐SCT and ultimately progressed at 2 months.
Response to salvage chemotherapy
Disease progression after therapy with PD‐1 inhibitors was assessed by PET‐CT (all patients) and repeat histological evaluation (n = 9). All patients were then addressed to salvage chemotherapy. Median time to chemotherapy was 1.4 months (range 0.3–15.2). Choice of regimen was dependent on both prior administered regimens and center‐specific guidelines/experience. The following chemotherapy regimens were administered: BEGEV (n = 11, 38%), BEACOPP (n = 4, 14%), L‐PAM + ASCT (n = 4, 14%), FEAM +ASCT (n = 2, 7%), Gemcitabine (n = 2, 7%), IGEV (n = 1, 4%), HD‐VP‐16 (n = 1, 4%), BV (n = 1, 4%), Bendamustine (n = 1, 4%), DHAOx (n = 1, 4%). Chemotherapy was well tolerated; one patient developed encephalitis after one cycle of chemotherapy, which was treated with broad spectrum antibiotics and antiviral therapy, with complete resolution. Twenty‐six patients (93%; 95% CI 84–100) were responsive to chemotherapy: 23 patients (82%; 95% CI 68–96) achieved CR and 3 patients (11%; 95% CI 0–22) achieved a partial response. Disease response achieved during treatment with anti‐PD1 inhibitors did not correlate with response to chemotherapy (p = 0.357, Fisher's Exact test, see Figure S1). Interestingly, all patients who received BEGEV chemotherapy after anti‐PD‐1 failure achieved a CR. Furthermore, among such patients, two (18%) had been exposed to BEGEV just prior to PD‐1 inhibition with evidence of progressive disease (PD) at the end of therapy (Figures 1 and 2). Duration of response to post‐CPI chemotherapy was not assessable, as, according to internal policies, all eligible patients were addressed to allo‐SCT, except two patients, one treated with autologous‐SCT and another one, previously treated with allogeneic transplant, who was not transplanted. Therapies prior to and after CPIs with the responses obtained are summarized in Table 2.
FIGURE 1.
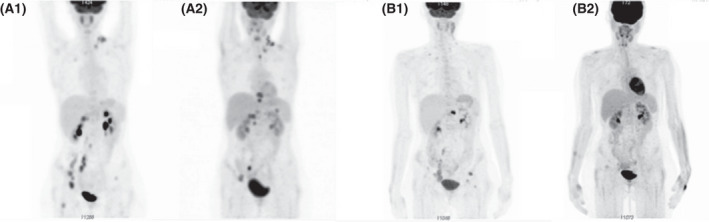
Response to BEGEV: (A) pre‐Nivolumab: persistence of pathological uptake at the left latero‐cervical level, at the hepatic hilum, and in proximity of right iliac vessels. New lesions at the left superior paratracheal level, at the splenic hilum, para‐cavally. Two new bone lesions in two dorsal vertebral bodies; persistence of bone lesions in the left pelvis. Deauville score (DS) 5; (B) post‐Nivolumab. Complete regression of previous areas of uptake. DS3
FIGURE 2.
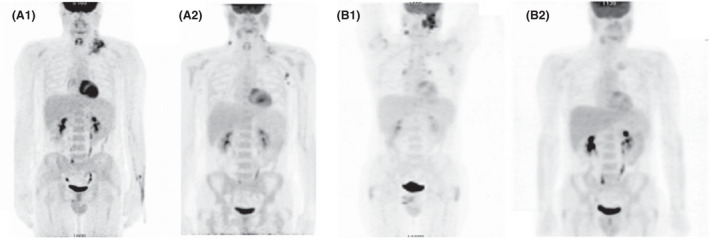
Response to BEGEV: (A) pre‐Nivolumab: appearance of new small adenopathies in the right latero‐cervical area and the left axillary area. DS5; (B) post‐Nivolumab: complete regression of the lesions previously present in the bilateral laterocervical area, subclavicular, mediastinal and pulmonary hilar areas. Complete response to treatment. DS3
TABLE 2.
Summary of chemotherapy regimens administered before and after treatment with PD‐1 inhibitors
| Patient | Chemo‐refractory a | Last chemo before anti‐PD1 | Disease response | PD‐1 inhibitors (no. of cycles) | Chemo post anti‐PD1 | No. of cycles | Disease response |
|---|---|---|---|---|---|---|---|
| 1 | NO | BRENTUXIMAB | PD | 10 | BEACOPP | 2 | CR |
| 2 | YES | BENDAMUSTINE | PD | 24 | BEACOPP | 2 | CR |
| 3 | YES | BRENTUXIMAB+BENDA | PD | 32 | BEACOPP | 2 | PR |
| 4 | YES | FEAM + ASCT | PD | 10 | BEACOPP+RT | 4 | SD |
| 5 | YES | FEAM + ASCT | SD | 41 | BEGEV | 4 | CR |
| 6 | NO | BRENTUXIMAB | PD | 53 | IGEV | 2 | CR |
| 7 | YES | FEAM + ASCT | SD | 55 | BEGEV | 4 | CR |
| 8 | YES | BRENTUXIMAB | PD | 37 | BEGEV | 4 | CR |
| 9 | YES | RMACOBP | PD | 15 | BRENTUXIMAB | 5 | CR |
| 10 | YES | DHAP | PD | 23 | L‐PAM + ASCT | / | CR |
| 11 | YES | BRENTUXIMAB | PD | 74 | BEGEV | 1 | CR |
| 12 | YES | BEGEV | PD | 12 | BEGEV | 2 | CR |
| 13 | YES | BRENTUXIMAB | PD | 72 | BEGEV | 4 | CR |
| 14 | YES | BEGEV | PD | 12 | BEGEV | 1 | CR |
| 15 | YES | GEM + VNR | PD | 31 | BEGEV | 7 | CR |
| 16 | YES | GVD | PR | 8 | BEGEV | 4 | CR |
| 17 | NA | BRENTUXIMAB | PD | 3 | GEM | 4 | PD |
| 18 | YES | BRENTUXIMAB | PD | 17 | HD‐CTX + FEAM + ASCT | / | CR |
| 19 | YES | GDP | PD | 12 | FEAM + ASCT | / | CR |
| 20 | YES | FEAM + ASCT | PD | 12 | HD‐VP‐16 | / | PR |
| 21 | YES | BRENTUXIMAB | PD | 10 | GEM | 2 | PR |
| 22 | YES | BRENTUXIMAB | PD | 8 | L‐PAM+ ASCT | / | CR |
| 23 | YES | BRENTUXIMAB | PD | 10 | L‐PAM+ ASCT | / | CR |
| 24 | YES | BRENTUXIMAB + BENDA | PD | 7 | L‐PAM+ ASCT+RT | / | CR |
| 25 | YES | BRENTUXIMAB | PD | 10 | BENDA | 3 | CR |
| 26 | YES | BRENTUXIMAB | SD | 29 | BEGEV | 2 | CR |
| 27 | YES | BRENTUXIMAB | PD | 13 | BEGEV | 3 | CR |
| 28 | YES | CTX | PD | 12 | OxDHA + RT | 1 | CR |
BENDA, bendamustine; ASCT, autologous stem cell transplantation; CTX, cyclophosphamide; RT, radiation therapy; HD, high‐dose.
Refractory to 1st line therapy.
ctDNA dynamics during treatment course and reconstruction of clonal phylogeny
We followed serial plasma samples during therapy with PD‐1 inhibitors and subsequent salvage chemotherapy with BeGEV in a small cohort of 5 patients. Serial measurements of ctDNA were normalized to pretreatment levels and expressed as base 10 log hGE/ml.
After 2 cycles of salvage chemotherapy with BeGEV all patients displayed a drop in ctDNA, which is consistent with complete metabolic response assessed by interim PET‐CT (figure not shown). These patients maintained the response, were addressed to allo‐SCT and are currently disease free, supporting previous reports that ctDNA monitoring may be a useful tool in predicting response to chemotherapy also in the salvage setting. 18
Additionally, we applied CALDER 15 to the Variant Allele Frequency profile of a panel of selected genomic variants, computed via variant calling (somatic analysis) from ctDNA sequencing data, for each longitudinal sample of the patients. Variants were aggregated at the gene level, i.e., the read counts of all variants detected on a specific gene were added cumulatively, to reduce the impact of noisy measurements and limit the number of significant variables, as proposed for instance in 19 (See Methods section).
In Figure 3 one can see the longitudinal cancer evolution models returned by CALDER and visualized via TimeScape, 17 in which distinct clones are characterized by different sets of accumulating genomic alterations. Interestingly, the tumour evolution of all patients seems to occur prior to the first cycle of therapy and is characterized either by linear (0003) or branching evolution patterns (0005, 0008, 0024 and 0113), involving gene alterations in temporally ordered accumulation paths. In this respect, we note that the specific temporal orderings among genomic alterations should be considered with a certain caution. Even though the models returned in Figure 3 represent the optimal solutions returned by CALDER, equivalent and slightly different orderings might be possible (as for any deconvolution method), especially due the data‐specific noise levels that are inherently present in the data.
Finally, for all patients, all (sub)clones seem to be depleted before the first ctDNA assessment (interim timepoint) and are not detected thereafter. This notable result shows that ctDNA monitoring can provide an effective tool to support clinicians for the definition (or discontinuation) of cancer therapies.
Patient outcomes: allogeneic stem cell transplantation
According to institutional policies, all patients who obtained an objective response were addressed to allo‐SCT, if eligible (n = 25). Only one patient received consolidation with autologous stem cell transplantation. The patient who had relapsed after allo‐SCT, continued solely with clinical observation and experienced PD after approximately 5 months. After a median follow up of 21 months from initiation of chemotherapy after CPIs failure, two‐year OS was 80.0% months (95% confidence interval [CI] 71.9–88.1), while two‐year PFS was 70.7% months (95% CI 61.0–80.4) (Figure 4). Of the twenty‐eight patients included in this study, twenty‐one are alive and in CR, two are alive with PD, one died of PD, three died of PD after allo‐SCT and one died of post‐transplant toxicity (interstitial pneumonia) while in CR. Outcomes of the single patients are summarized in the Swimmer Plot (Figure S2).
FIGURE 4.
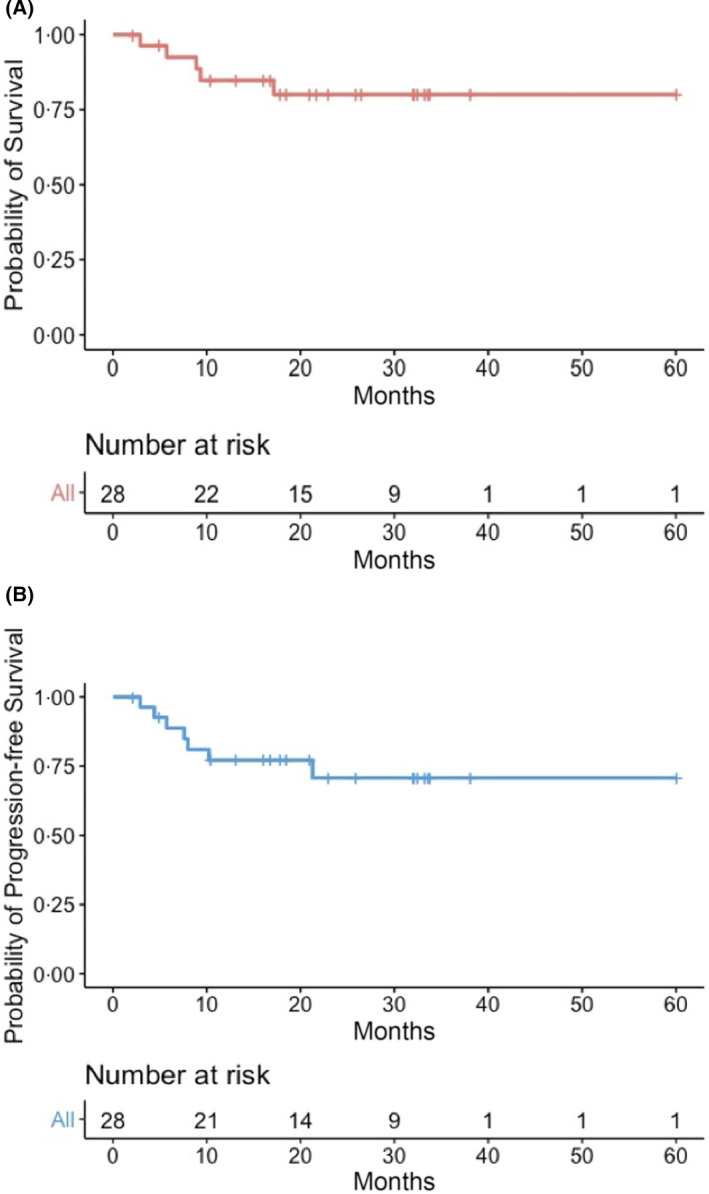
Kaplan‐Meier curves. OS (A) and PFS (B) for the whole population
DISCUSSION
We here demonstrate that administering chemotherapy in patients affected by r/r cHL who failed therapy with CPIs is an effective and feasible option and provides a potential bridge to allogeneic transplantation, thus redefining the prognosis in such patients. In our cohort of heavily pre‐treated chemo‐refractory cHL patients, nearly all patients achieved an objective response; median OS and PFS were not reached after a follow up of 21 months. Our study confirms a previous retrospective report of 17 r/r cHL patients progressing on anti‐PD‐1 therapy, who were administered various chemotherapy regimens with an ORR of 59%. 10 Such results were also recently reported in a larger multicentric analysis including 81 patients, with an ORR of 62% 12 ; in the latter study, however, post‐CPI therapy included also allo‐SCT conditioning and immune‐based therapy. Lastly, an additional monocentric retrospective analysis has also addressed the same issue, with comparable results. 11
Interaction and potential synergistic effects between immunotherapy and chemotherapy have been well established; chemotherapy induces cell death, thereby augmenting tumour antigen exposure to the immune system. Additionally, chemotherapy drugs can alter the tumour microenvironment by decreasing the number of regulatory T cells and myeloid‐derived suppressor cells, 20 , 21 or by downregulating inhibitory checkpoint molecules such as PD‐L1. 22 Such findings have been confirmed in the clinical setting, with combination of chemotherapy and immunotherapy producing enhanced response rates in non‐small cell lung cancer, advanced melanoma as well as cHL. 23 , 24 , 25 Furthermore, recent data have suggested that a sequential strategy of immunotherapy followed by chemotherapy may be equally effective, both in preclinical 26 and clinical settings. 8 , 27 , 28
Our study confirms the significant efficacy of chemotherapy administered after immune‐based therapy, especially considering that most patients were refractory to the last chemotherapy administered prior to CPIs and few of them were re‐treated with drugs that were included in previously ineffective regimens. Due to the retrospective nature of this study, we cannot exclude a selection bias resulting in an over‐estimation of the efficacy data. In particular, we cannot exclude that patients who were addressed to chemotherapy were selected among those with a pre‐existing chemosensitivity. However, prior to CPIs, nearly all patients were overtly refractory to their last chemotherapy. Additionally, in our study, when compared to similar studies, most patients received either high‐dose chemotherapy or a polychemotherapy regimen (n = 23, 82%), which has been suggested to increase the likelihood of response by Casadei et al. (Table 3).
TABLE 3.
Summary of studies assessing susceptibility to chemotherapy after checkpoint inhibition in r/r cHL
| Rossi et al. 10 | Carreau et al. 12 | Casadei et al. 11 | Calabretta et al. (this paper) | |
|---|---|---|---|---|
| N of patients | 19 | 81 | 25 | 28 |
| Median age, years | 44 | 39 | 32 | 29 |
| Median N of prior therapies (N, range) | 6 (2–14) | 4 (1–11) | 4 (1–10) | 4 (2–11) |
| Prior auto‐SCT (N, %) | 10 (53%) | 34 (42%) | 11 (44%) | 18 (64%) |
| Number of cycles of anti‐PD1 therapy (median) | 10 | 8 | 14 | 13 |
| Disease status at end of anti‐PD1 therapy (N, %) | ||||
| SD/PD | 17 (89%) | 55 (68%) | 17 (68%) | 28 (100%) |
| PR | 2 (11%) | – | 6 (16%) | – |
| CR | – | 1 (1%) | 2 (8%) | – |
| unknown | – | 25 (31%) | – | – |
| Type of chemotherapy (N, %) | ||||
| Polychemotherapy | 8 (42%) | 30 (37%) | 10 (40%) | 23 (82%) |
| Single agent | 11 (58%) | 22 (27%) | 15 (60%) | 5 (18%) |
| Conditioning regimen a | – | 11 (14%) | – | – |
| Non‐chemotherapy | – | 18 (22%) | – | – |
| Gemcitabine‐containing regimens (N, %) | 4 (21%) | 17 (21%) | 4 (16%) | 13 (45%) |
| ORR (CR), % | 59 (41%) | 62 (42%) | 60 (32%) | 93 (82%) |
| Consolidation with allo‐SCT (N, %) | 3 (16%) | 22 (27%) | 4 (16%) | 25 (89%) |
| Consolidation with auto‐SCT (N, %) | – | 16 (20%) | 4 (16%) | 1 (4%) |
| Median followup (mo) | 12.1 | 18 | 32.4 | 21 |
| Median PFS, mo | 11 | 6.3 | 19.1 | Not reached |
| Median OS, mo | Not reached | 21 | Not reached | Not reached |
Prior to allogeneic stem cell transplantation.
Despite evidence from multiple reports in solid tumours and haematological malignancies that immunotherapy may restore chemosensitivity, there is lack of data on which chemotherapy may produce the best results. It is well established that Gemcitabine displays immunomodulatory effects 29 , 30 , 31 ; indeed, among our small cohort of patients, those treated with Gemcitabine‐containing regimens (n = 13, 45%) were more represented when compared to similar studies (Table 3) and achieved excellent responses. Specifically, the BeGEV regimen, which has been successfully studied as second‐line salvage therapy in cHL, 32 produced CR in all patients. Furthermore, two patients who were refractory to BeGEV before CPIs overcame their chemo‐resistance. Though this finding is limited by the small sample, our data suggest that this regimen may be considered as a primary choice for patients who fail PD‐1 inhibitors.
It is widely accepted that cancer is driven by the accumulation of somatic mutations. In this respect, data‐science methods can exploit the temporal information of longitudinal samples to reconstruct the likely evolutionary history of the tumour, by leveraging accumulating genomic mutations as barcodes to identify putative (sub)clones, as proposed in 15 with bulk sequencing data and, more recently, in 16 with single‐cell sequencing data. Accordingly, the outcoming models allow to highlight the clonal prevalence variation in relation to the distinct therapeutic cycles in any given patient, possibly pinpointing sensitive and resistant subclones. To our knowledge, this is the first report of application of CALDER, a novel algorithm for the reconstruction of clonal phylogenesis, in r/r cHL patients uniformly treated with BeGEV chemotherapy. Surely, the sample is not sufficient to assess a possible predictive value of the inferred models, however the tumour evolution of all patients seems to occur prior to the first cycle of therapy and is characterized either by linear (0003) or branching evolution patterns (0005, 0008, 0024 and 0113), involving genomic alterations in temporally ordered accumulation paths, and is coherent with clinical course. Interestingly, TP53‐mutated subclones were detected in two patients, supporting recent reports associating TP53 gene mutations with refractory cHL. 33 Further, we may speculate that Anti PD‐1 priming followed by chemotherapy could overcome the adverse prognostic influence of TP53 mutations.
Patients who progress after PD‐1 inhibitors have a poor prognosis, with a median OS upon treatment discontinuation of 13.5 months. Although prolongation of CPIs despite PD is associated with clinical benefit and a survival advantage, 3 patients will nonetheless require a new systemic therapy after a median of 8.8 months from initial progression. In this setting, administration of chemotherapy may rescue a substantial number of young patients who can be safely and effectively directed to allo‐SCT. 34 , 35 Indeed, though our case series is limited, we report a 2‐year OS of 80.0% from discontinuation of PD‐1 inhibitors, which is likely affected by the high proportion of patients who underwent allo‐SCT, as compared to similar studies. In our cohort, transplant‐related mortality was low (n = 1), confirming that allo‐SCT in this setting is feasible and can produce long‐lasting disease control. Additionally, we can speculate that patients with chemo‐refractory disease who are unable to undergo auto‐SCT may benefit from exposure to CPIs in terms of re‐sensitization to high‐dose chemotherapy, thus potentially rediscussing the need for consolidation with allo‐SCT. 36
In conclusion, our study confirms previous reports describing the efficacy of chemotherapy when administered after checkpoint inhibition. In this setting, gemcitabine‐containing polychemotherapy regimens may be considered a primary choice for salvage chemotherapy. Further, implementation of ctDNA profiling and bio‐informatic models to current clinical practice, appears to be a promising, but still rather exploratory, approach to monitor disease and potentially predict clinical outcome.
AUTHOR CONTRIBUTIONS
C.C‐S. conceived the project. E.C., A.G., F.R., C.M., M.Ma., S.B., A.d.R., M.Me., and E.D. collected clinical data. M.D.T., performed ctDNA genotyping. D.M. and A.G. performed CALDER analysis. E.C., M.D.T., M.A., D.R., and C.C‐S., analysed and interpreted original data. E.C. performed statistical analysis. E.C., M.D.T., D.M. and A.G. wrote the manuscript. M.Me., S.B., E.D., D.R., P.C., A.S., and C.C‐S. provided critical input and revised the manuscript.
Supporting information
SUPPLEMENTARY MATERIALS
ACKNOWLEDGEMENTS
This work was supported in part by a grant from the Italian Association for Cancer Research (AIRC, grant #20575 to CC‐S).
Calabretta E, Guidetti A, Ricci F, Di Trani M, Monfrini C, Magagnoli M, et al. Chemotherapy after PD‐1 inhibitors in relapsed/refractory Hodgkin lymphoma: Outcomes and clonal evolution dynamics. Br J Haematol. 2022;198:82–92. doi: 10.1111/bjh.18183
REFERENCES
- 1. Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD‐1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2015;372(4):311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen R, Zinzani PL, Fanale MA, Armand P, Johnson NA, Brice P, et al. Phase II study of the efficacy and safety of pembrolizumab for relapsed/refractory classic Hodgkin lymphoma. J Clin Oncol. 2017;35(19):2125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armand P, Engert A, Younes A, Fanale M, Santoro A, Zinzani PL, et al. Nivolumab for relapsed/refractory classic Hodgkin lymphoma after failure of autologous hematopoietic cell transplantation: extended follow‐up of the multicohort single‐arm phase II CheckMate 205 trial. J Clin Oncol. 2018;36(14):1428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen R, Zinzani PL, Lee HJ, Armand P, Johnson NA, Brice P, et al. Pembrolizumab in relapsed or refractory Hodgkin lymphoma: 2‐year follow‐up of KEYNOTE‐087. Blood. 2019;134(14):1144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Phillips TJ, Forero‐Torres A, Sher T, Diefenbach CS, Johnston P, Talpaz M, et al. Phase 1 study of the PI3Kδ inhibitor INCB040093 ± JAK1 inhibitor itacitinib in relapsed/refractory B‐cell lymphoma. Blood. 2018;132(3):293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamadani M, Collins GP, Caimi PF, Samaniego F, Spira A, Davies A, et al. Camidanlumab tesirine in patients with relapsed or refractory lymphoma: a phase 1, open‐label, multicentre, dose‐escalation, dose‐expansion study. Lancet Haematol. 2021;8:e433–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rajendran S, Li Y, Ngoh E, Wong HY, Cheng MS, Wang CI, et al. Development of a bispecific antibody targeting CD30 and CD137 on Hodgkin and reed‐Sternberg cells. Front Oncol. 2019;9:945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park SE, Lee SH, Ahn JS, Ahn MJ, Park K, Sun JM. Increased response rates to salvage chemotherapy administered after PD‐1/PD‐L1 inhibitors in patients with non‐small cell lung cancer. J Thorac Oncol. 2018;13(1):106–11. [DOI] [PubMed] [Google Scholar]
- 9. Dwary AD, Master S, Patel A, Cole C, Mansour R, Mills G, et al. Excellent response to chemotherapy post immunotherapy. Oncotarget. 2017;8(53):91795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rossi C, Gilhodes J, Maerevoet M, Herbaux C, Morschhauser F, Brice P, et al. Efficacy of chemotherapy or chemo‐anti‐PD‐1 combination after failed anti‐PD‐1 therapy for relapsed and refractory Hodgkin lymphoma: a series from Lysa centers. Am J Hematol. 2018;93:1042–9. [DOI] [PubMed] [Google Scholar]
- 11. Casadei B, Argnani L, Morigi A, Lolli G, Broccoli A, Pellegrini C, et al. Effectiveness of chemotherapy after anti‐PD‐1 blockade failure for relapsed and refractory Hodgkin lymphoma. Cancer Med. 2020;9(21):7830–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carreau NA, Pail O, Armand P, Merryman R, Advani RH, Spinner MA, et al. Checkpoint blockade treatment may sensitize Hodgkin lymphoma to subsequent therapy. Oncologist. 2020;25:878–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non‐Hodgkin lymphoma: the Lugano classification. J Clin Oncol. 2014;32(27):3059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rossi D, Condoluci A, Spina V, Gaidano G. Methods for measuring ctDNA in lymphomas. Methods Mol Biol. 2019;1881:253–65. [DOI] [PubMed] [Google Scholar]
- 15. Myers MA, Satas G, Raphael BJ. CALDER: inferring phylogenetic trees from longitudinal tumor samples. Cell Syst. 2019;8(6):514–22.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramazzotti D, Angaroni F, Maspero D, Ascolani G, Castiglioni I, Piazza R, et al. LACE: inference of cancer evolution models from longitudinal single‐cell sequencing data. J Comput Sci. 2022;58:101523. [Google Scholar]
- 17. Smith MA, Nielsen CB, Chan FC, McPherson A, Roth A, Farahani H, et al. E‐scape: interactive visualization of single‐cell phylogenetics and cancer evolution. Nat Methods. 2017;14(6):549–50. [DOI] [PubMed] [Google Scholar]
- 18. Spina V, Bruscaggin A, Cuccaro A, Martini M, Di Trani M, Forestieri G, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131(22):2413–25. [DOI] [PubMed] [Google Scholar]
- 19. Caravagna G, Graudenzi A, Ramazzotti D, Sanz‐Pamplona R, De Sano L, Mauri G, et al. Algorithmic methods to infer the evolutionary trajectories in cancer progression. Proc Natl Acad Sci U S A. 2016;113(28):E4025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suzuki E, Kapoor V, Jassar AS, Kaiser LR, Albelda SM. Gemcitabine selectively eliminates splenic gr‐1+/CD11b+ myeloid suppressor cells in tumor‐bearing animals and enhances antitumor immune activity. Clin Cancer Res. 2005;11(18):6713–21. [DOI] [PubMed] [Google Scholar]
- 21. Ghiringhelli F, Larmonier N, Schmitt E, Parcellier A, Cathelin D, Garrido C, et al. CD4+CD25+ regulatory T cells suppress tumor immunity but are sensitive to cyclophosphamide which allows immunotherapy of established tumors to be curative. Eur J Immunol. 2004;34(2):336–44. [DOI] [PubMed] [Google Scholar]
- 22. Chen G, Emens LA. Chemoimmunotherapy: reengineering tumor immunity. Cancer Immunol Immunother. 2013;62(2):203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paz‐Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, et al. Pembrolizumab plus chemotherapy for squamous non‐small‐cell lung cancer. N Engl J Med. 2018;379(21):2040–51. [DOI] [PubMed] [Google Scholar]
- 24. Hersh EM, O'Day SJ, Powderly J, Khan KD, Pavlick AC, Cranmer LD, et al. A phase II multicenter study of ipilimumab with or without dacarbazine in chemotherapy‐naïve patients with advanced melanoma. Invest New Drugs. 2011;29(3):489–98. [DOI] [PubMed] [Google Scholar]
- 25. Moskowitz AJ, Shah G, Schöder H, Ganesan N, Drill E, Hancock H, et al. Phase II trial of pembrolizumab plus gemcitabine, vinorelbine, and liposomal doxorubicin as second‐line therapy for relapsed or refractory classical Hodgkin lymphoma. J Clin Oncol. 2021;39(28):3109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fridlender ZG, Sun J, Singhal S, Kapoor V, Cheng G, Suzuki E, et al. Chemotherapy delivered after viral immunogene therapy augments antitumor efficacy via multiple immune‐mediated mechanisms. Mol Ther. 2010;18(11):1947–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Inayama Y, Hamanishi J, Matsumura N, Murakami R, Abiko K, Yamaguchi K, et al. Antitumor effect of nivolumab on subsequent chemotherapy for platinum‐resistant ovarian cancer. Oncologist. 2018;23(11):1382–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saleh K, Daste A, Martin N, Pons‐Tostivint E, Auperin A, Herrera‐Gomez RG, et al. Response to salvage chemotherapy after progression on immune checkpoint inhibitors in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck. Eur J Cancer. 2019;121:123–9. [DOI] [PubMed] [Google Scholar]
- 29. Homma Y, Taniguchi K, Nakazawa M, Matsuyama R, Mori R, Takeda K, et al. Changes in the immune cell population and cell proliferation in peripheral blood after gemcitabine‐based chemotherapy for pancreatic cancer. Clin Transl Oncol. 2014;16(3):330–5. [DOI] [PubMed] [Google Scholar]
- 30. Deshmukh SK, Tyagi N, Khan MA, Srivastava SK, Al‐Ghadhban A, Dugger K, et al. Gemcitabine treatment promotes immunosuppressive microenvironment in pancreatic tumors by supporting the infiltration, growth, and polarization of macrophages. Sci Rep. 2018;8(1):12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nowak AK, Robinson BW, Lake RA. Gemcitabine exerts a selective effect on the humoral immune response: implications for combination chemo‐immunotherapy. Cancer Res. 2002;62(8):2353–8. [PubMed] [Google Scholar]
- 32. Santoro A, Mazza R, Pulsoni A, Re A, Bonfichi M, Zilioli VR, et al. Bendamustine in combination with gemcitabine and vinorelbine is an effective regimen as induction chemotherapy before autologous stem‐cell transplantation for relapsed or refractory Hodgkin lymphoma: final results of a multicenter phase II study. J Clin Oncol. 2016;34(27):3293–9. [DOI] [PubMed] [Google Scholar]
- 33. Mata E, Fernández S, Astudillo A, Fernández R, García‐Cosío M, Sánchez‐Beato M, et al. Genomic analyses of microdissected Hodgkin and reed‐Sternberg cells: mutations in epigenetic regulators and p53 are frequent in refractory classic Hodgkin lymphoma. Blood Cancer J. 2019;9(3):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Merryman RW, Kim HT, Zinzani PL, Carlo‐Stella C, Ansell SM, Perales MA, et al. Safety and efficacy of allogeneic hematopoietic stem cell transplant after PD‐1 blockade in relapsed/refractory lymphoma. Blood. 2017;129(10):1380–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Philippis C, Legrand‐Izadifar F, Bramanti S, Giordano L, Montes de Oca C, Duléry R, et al. Checkpoint inhibition before haploidentical transplantation with posttransplant cyclophosphamide in Hodgkin lymphoma. Blood Adv. 2020;4(7):1242–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Merryman RW, Redd RA, Nishihori T, Chavez J, Nieto Y, Darrah JM, et al. Autologous stem cell transplantation after anti‐PD‐1 therapy for multiply relapsed or refractory Hodgkin lymphoma. Blood Adv. 2021;5(6):1648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIALS


