Abstract
Gut microbiota is involved in immune modulation and immune checkpoint inhibitors (ICIs) efficacy. Single‐arm phase II CAVE‐mCRC and CAVE‐LUNG clinical trials investigated cetuximab + avelumab combination in RAS wild‐type (WT) metastatic colorectal cancer (mCRC) and chemo‐refractory nonsmall cell lung cancer (NSCLC) patients, respectively. A comprehensive gut microbiota genetic analysis was done in basal fecal samples of 14 patients from CAVE‐mCRC trial with circulating tumor DNA (ctDNA) RAS/BRAF WT and microsatellite stable (MSS) disease. Results were validated in a cohort of 10 patients from CAVE‐Lung trial. 16S rRNA sequencing revealed 23 027 bacteria species in basal fecal samples of 14 patients from CAVE‐mCRC trial. In five long‐term responding patients (progression‐free survival [PFS], 9‐24 months) significant increases in two butyrate‐producing bacteria, Agathobacter M104/1 (P = .018) and Blautia SR1/5 (P = .023) were found compared to nine patients with shorter PFS (2‐6 months). A significantly better PFS was also observed according to the presence or absence of these species in basal fecal samples. For Agathobacter M104/1, median PFS (mPFS) was 13.5 months (95% confidence interval [CI], 6.5‐20.5 months) vs 4.6 months (95% CI, 1.8‐7.4 months); P = .006. For Blautia SR1/5, mPFS was 5.9 months (95% CI, 2.2‐9.7 months) vs 3.6 months (95% CI, 3.3‐4.0 months); P = .021. Similarly, in CAVE‐Lung validation cohort, Agathobacter M104/1 and Blautia SR1/5 expression were associated with PFS according to their presence or absence in basal fecal samples. Agathobacter and Blautia species could be potential biomarkers of outcome in mCRC, and NSCLC patients treated with cetuximab + avelumab. These findings deserve further investigation.
Keywords: avelumab, cetuximab, gut microbiota, mCRC, NSCLC
What's new?
The gut microbiota has been proposed as a relevant player in cancer development as well as a potential modulator of sensitivity to immunotherapy. Here, the authors performed an extensive analysis of pretreatment fecal microbiota species in patients with metastatic colorectal cancer and nonsmall cell lung cancer treated with cetuximab plus avelumab in the CAVE‐mCRC and CAVE‐lung trials, respectively. For the first time, they demonstrate that two gut bacteria species are associated with longer progression‐free survival. The two butyrate‐producing bacteria could become potential biomarkers for cetuximab plus avelumab antitumor activity in chemo‐refractory colorectal cancer and nonsmall cell lung cancer patients.
Abbreviations
- ADCC
antibody‐dependent cell‐mediated cytotoxicity
- CAVE
cetuximab avelumab
- CI
confidence interval
- CR
complete response
- CRC
colorectal cancer
- CTLA‐4
cytotoxic T‐lymphocyte‐associated antigen
- ctDNA
circulating tumor DNA
- dMMR
deficient mismatch repair
- EGFR
epidermal growth factor receptor
- ICI
immune checkpoint inhibitor
- mAb
monoclonal antibody
- mCRC
metastatic colorectal cancer
- mOS
median overall survival
- mPFS
median PFS
- MSI‐H
microsatellite‐high instability
- MSS
microsatellite stability
- NK
Natural Killer
- NSCLC
nonsmall cell lung cancer
- OS
overall survival
- PD‐1
programmed cell death 1
- PD‐L1
programmed cell death ligand 1
- PFS
progression‐free survival
- PR
partial response
- WT
wild‐type
1. INTRODUCTION
In the last decade, immunotherapy has determined a revolution in the therapeutic approach for various tumors, such as malignant melanoma and lung cancer. 1 , 2 Therapeutic blockade of immune checkpoint programmed cell death 1 (PD‐1), of programmed cell death ligand 1 (PD‐L1) and/or of cytotoxic T‐lymphocyte‐associated antigen (CTLA‐4) has determined significant improvements in patient survival. 3 Unfortunately, the scenario for colorectal cancer (CRC) is completely different. Immune checkpoint inhibitors (ICI) have clinically relevant efficacy only in 3% to 6% of metastatic CRC (mCRC) patients, that have deficient mismatch repair (dMMR) tumors with DNA microsatellite‐high instability (MSI‐H) and that are characterized by a high number of mutations. 4 In mCRC patients with microsatellite stable (MSS) tumors, ICIs are not effective. Therefore, novel immunotherapy strategies are urgently needed for the majority of patients with mCRC. With the advancement of molecular techniques, next‐generation sequencing (NGS) analyses focused on novel approaches for the characterization and identification of microbial communities, which play an important role in different diseases and cancers such as colon cancer, cervical cancer, prostate cancer, lung cancer and gallbladder cancer. 5 , 6 , 7 , 8 , 9 , 10
We have recently reported the results of Cetuximab‐AVElumab‐mCRC (CAVE‐mCRC), a single‐arm, phase II trial, in which the combination of cetuximab, an anti‐epidermal growth factor receptor (EGFR) monoclonal antibody (mAb) plus avelumab, an antiprogrammed death ligand 1 (PD‐L1) mAb, was evaluated as rechallenge strategy in chemo‐refractory RAS wild type (WT) mCRC patients. 11 The rationale for this combination is based on the induction of antibody‐dependent cell‐mediated cytotoxicity (ADCC) by these two IgG isotype mAbs, which could enhance Natural Killer (NK) cell‐mediated antitumor immune response. 12 , 13 This effect may play a major role in their antitumor activity, as suggested by the findings of the CAVE‐Lung trial, in which patients with advanced nonsmall‐cell lung cancer (NSCLC) received the same combination of cetuximab plus avelumab. 14
Seventy‐seven patients with chemo‐refractory RAS WT mCRC were treated in third or further lines of therapy with cetuximab plus avelumab in the CAVE‐mCRC trial. 11 Median overall survival (mOS) was 11.6 months with median progression‐free survival (mPFS) of 3.6 months in the intention to treat patient population. Significantly higher antitumor activity was observed in 48 patients, that, at liquid biopsy plasma analysis of circulating tumor DNA (ctDNA) before treatment, had RAS/BRAF WT tumors (mPFS, 4.1 months; PFS of 6 months of more in 41% of patients; mOS, 17.8 months), providing evidence that cetuximab plus avelumab could be a clinically relevant rechallenge approach for mCRC patients with chemo‐refractory, MSS tumor and plasma ctDNA RAS/BRAF WT.
Potential predictive biomarkers for immunotherapy efficacy are unknown for MSS mCRC. 15 In the recent past, the gut microbiota has been proposed as a relevant player in cancer development and progression as well as a potential modulator of host immune responses and of sensitivity to ICIs. 16 In particular, butyrate‐producing gut bacteria may play a positive role in blocking inflammation and modulating both innate and adaptative immunity. 17
Here, we report an exploratory analysis of basal pretreatment fecal microbiota species in patients from CAVE‐mCRC trial with the aim of identifying gut bacteria, which could be correlated with antitumor activity of avelumab plus cetuximab. To further evaluate and to validate the potential role of intestinal microbiota, we extended this analysis to a subgroup of patients from CAVE‐Lung trial.
2. METHODS
2.1. Study design and patient population
CAVE‐mCRC trial was a nonprofit academic, single‐arm phase II study. 11 Patients had histologically confirmed mCRC with RAS (NRAS and KRAS, exon 2, 3 and 4) WT tumors; obtained a complete (CR) or partial response (PR) during first‐line treatment with an anti‐EGFR‐based regimen and, upon progression, received at least one subsequent line of therapy with an interval of more than 4 months from the last dose of the anti‐EGFR drug. Additional inclusion and exclusion criteria are described in the full protocol available online. 11 CAVE‐Lung trial was a nonprofit academic, single‐arm proof‐of‐concept clinical and translational study. Patients were enrolled with histologically confirmed Stage IIIb/IV or recurrent NSCLC and treated as second‐ or third‐line therapy. Additional inclusion and exclusion criteria are described in the full protocol available online. 14 CAVE‐mCRC (clinical trial registration: NCT04561336); CAVE‐LUNG (clinical trial registration: Eudract‐2017‐004195‐58).
No patients received antibiotic therapy before baseline stool sampling in both trials. PFS and overall survival (OS) were calculated at the follow‐up landmark of August 31, 2021, for both trials. One patient in the CAVE‐Lung trial was still on treatment at the time of the analysis.
2.2. DNA extraction
Fecal samples were collected at baseline before treatment for CAVE‐mCRC and CAVE‐lung trial patients. Fifty milligrams of feces were homogenized in 500 μL of 10 mg/mL lysozyme solution in Tris‐sucrose buffer (50 mM Tris‐HCl, 40 mM EDTA, 0.75 M sucrose, pH 8.0) and incubated for 1 hour at 37°C. The Maxwell 16 DNA Purification Kit (Promega, Madison, WI) was used for the DNA extraction according to the manufacturer's instructions. The DNA concentration was evaluated using the Qubit HS dsDNA fluorescence assay (Life Technologies, Carlsbad, CA).
2.3. Illumina sequencing of the 16S rRNA genes
The V4 region of the bacterial 16S rRNA gene was amplified by using PCR with the primers 520F and 802R. 18 For each sample, DNA amplification was performed in triplicate by using 5 ng of DNA per reaction, following the conditions previously reported. 19 QIAxcel (Qiagen GmbH, Hilden, Germany) capillary electrophoresis was used to check the resulting amplicons. The amplicons were purified by using the MinElute PCR purification kit (Qiagen GmbH, Hilden, Germany). MiSeq System by using the MiSeq Reagent Kit v3 at a 2 × 250 bp read length configuration Paired‐end sequencing (2 × 250 bp) was carried out on an Illumina MiSeq sequencer (Fasteris, Genève, Switzerland). The OBITools softwares (www.prabi.grenoble.fr/trac/OBITools) were used to analyze the DNA sequence, referring each read to its original sample and de‐replicating reads into unique sequences. Singletons and short sequences (<20 nucleotides) were first removed with the PCR/sequencing errors and chimeras eliminated by obigrep/obiclean command use. For taxonomic assignment of sequences, a reference database, using the ecoPCR program, was built. The taxons were assigned using the EcoTag program. The use of bioinformatic filters allowed to further elimination of putative PCR and/or sequencer artifacts. Sequences characterized by 90% identity to the sequences present in GenBank were considered for further analysis.
2.4. Real‐time PCR
To determine the presence of hydrogen consumers and butyrate producers, PCR was performed on fecal DNA. StepOne Plus Real‐Time PCR System (Applied Biosystems, Foster City, CA) was used for qPCR. The amplification reaction was carried out in 20 μL containing 2 μL DNA, 0.4 mM of each forward and reverse primer, and 10 μL of KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems, Wilmington, MA). Plasmids containing amplified fragments of acs, dsrA, mcrA or genomic DNA from reference strains for BcoAT were used to build standard curves. Results were expressed as gene copy numbers per gram wet feces.
2.5. Nucleotide sequence accession
The sequencing coverage and quality statistics for each sample are summarized in Tables S1 and S2.
2.6. qPCR analysis of plasma samples
Plasma specimens from 67 out of 77 patients were collected at baseline and were suitable for ctDNA evaluation of KRAS, NRAS, BRAF and EGFR extracellular domain S492R mutations by using the automated Idylla TM qPCR‐based platform, as previously reported. 11 Results of the analyses were visualized using the online tool Idylla TM Explore (idyllaexplore.biocartis.com, last accessed May 30, 2020). This protocol has been previously validated and is fully described elsewhere. 20
2.7. Statistical analysis
Search for significant binary correlations between different gut microbiota species and PFS was performed using Kendall Tau‐b or Pearson correlation tests, according to not normal and normal distribution, respectively. P values <.05 were considered statistically significant. Given the exploratory nature of this analysis, no correction was performed for multiplicity. PFS curves were calculated using the Kaplan‐Meier method and compared using the Log‐rank test. Analyses were performed using SPSS package version 24.
3. RESULTS
Chemo‐refractory mCRC patients with plasma ctDNA RAS/BRAF WT and with MSS tumor had the highest clinical benefit from cetuximab rechallenge plus avelumab in CAVE‐mCRC trial. 11 Basal pretreatment fecal samples were collected from 14/48 patients with these characteristics (Table S3). All these 14 patients with plasma ctDNA RAS/BRAF WT and with MSS tumor had left‐sided primary CRC (the primary tumor was located in the sigma or in the rectum) and underwent surgery for primary tumor removal before systemic anticancer treatment. Five patients with PFS of more than 9 months (range: 9‐24 months), defined as long‐term responders and nine patients with PFS ranging from 2 to 6 months, defined as responders, were identified (Figure 1; Figure S1 for OS).
FIGURE 1.
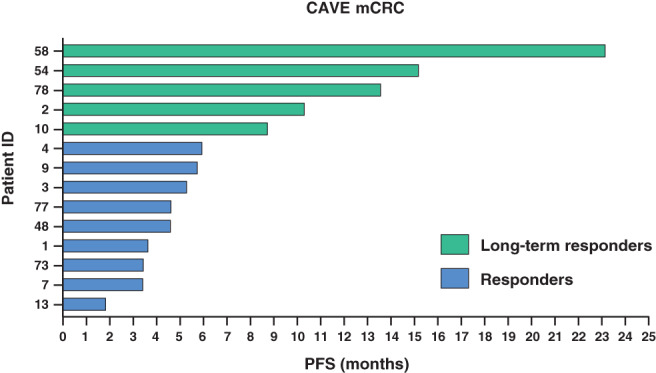
Swimmer plot of progression‐free survival for 14 patients with basal fecal sample in the CAVE‐mCRC trial [Color figure can be viewed at wileyonlinelibrary.com]
Gut microbiota species were genetically evaluated by bacteria 16S rRNA sequencing. This allowed to identify 23 027 species (data not shown). Higher expression of 10 gut microbiota species was associated with better PFS in these patients, whereas opposite correlation with PFS was found for the other five species (Table S4). By performing comparative and quantitative analysis between long‐term responders and responders, two bacteria species, that could be involved in host immune modulation, as butyrate‐producing bacteria (11), were differentially expressed in the basal fecal samples: Bacteria, Firmicutes, Clostridia, Clostridiales, Lachnospiraceae, Agathobacter M104/1 (P = .023) and Bacteria, Firmicutes, Clostridia, Clostridiales, Lachnospiraceae, Blautia SR1/5 (P = .018; Figure 2A,B). To further investigate how these two gut microbiota species could correlate with therapy outcomes, PFS Kaplan‐Meier estimates were calculated according to their presence or absence in the basal fecal samples. Significantly better PFS was observed for those patients with either Agathobacter M104/1 or Blautia SR1/5 positive fecal samples as compared to patients with negative fecal samples (Figure 2C,D). In fact, for Agathobacter M104/1, median PFS (mPFS) was 13.5 months (95% confidence interval [CI], 6.5‐20.5 months) vs 4.6 months (95% CI, 1.8‐7.4 months); P = .006. For Blautia SR1/5, mPFS was 5.9 months (95% CI, 2.2‐9.7 months) vs 3.6 months (95% CI, 3.3‐4.0 months); P = .021.
FIGURE 2.

(A,B) Expression of Aghatobacter M104/1 (A) and Blautia SR1/5 (B) in the basal fecal samples of long responders and responders in CAVE‐mCRC trial. (C,D) Kaplan‐Meier estimates of progression‐free survival according to presence or absence in the basal fecal samples of Aghatobacter M104/1 (C) and Blautia SR1/5 (D) in CAVE‐mCRC trial [Color figure can be viewed at wileyonlinelibrary.com]
To extend and to validate the results observed in CAVE‐mCRC trial, we performed a similar assessment in NSCLC patients, that were treated in CAVE‐Lung trial, in which they received the same combination of cetuximab plus avelumab. In 10 NSCLC patients, whose basal fecal samples were available for gut microbiota analysis (see Table S5 for patient characteristics), two subgroups were identified: four long‐term responders with PFS of more than 8 months (range, 8‐32 months) and six responders with shorter PFS (range, 1.4‐5 months) (R) (Figure 3; see Figure S2 for OS). A similar number of gut microbiota species were identified in the fecal samples of NSCLC patients as compared to mCRC patients (data not shown). Higher expression was associated with better PFS for 92 gut microbiota species, whereas a negative correlation with PFS was observed for nine species (Table S6). Analysis of the basal fecal samples from patients with NSCLC treated in the CAVE‐Lung trial identified for both Agathobacter M104/1 (P = .016) and Blautia SR1/5 (P = .0008) differential expression according to antitumor activity of cetuximab plus avelumab (Figure 4A,B). Kaplan‐Meier estimates for PFS, which were calculated according to the presence or the absence of Agathobacter or of Blautia species in basal pretreatment fecal samples, revealed different PFS for CAVE‐Lung patients. Median PFS was 7.8 months (95% CI, 1‐20 months) for patients with basal fecal samples, that were positive for Agathobacter M104/1, vs 1.4 months (95% CI, 0.6‐2.1 months) for patients with negative samples; P = .002. For Blautia SR1/5, mPFS was 7.8 months (95% CI, 1.2‐14.3 months) for positive cases vs 1.8 months for negative cases (95% CI, 1.7‐1.8 months; P = .018; Figure 4C,D).
FIGURE 3.

Swimmer plot of progression‐free survival for 10 patients with basal fecal sample analysis in the CAVE‐Lung trial [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 4.

(A,B) Expression of Aghatobacter M104/1 (A) and Blautia SR1/5 (B) in in the basal fecal samples of long responders and responders in CAVE‐Lung trial. (C,D) Kaplan‐Meier estimates of progression‐free survival according to the presence or absence in the basal fecal samples of Aghatobacter M104/1 (C) and Blautia SR1/5 (D) in CAVE‐Lung trial [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
A complex relationship takes place between the host and gut microbiota in normal physiology. In several human diseases, gut microbiota could modulate disease development and progression and could potentially interfere with treatment efficacy. 21 It may play a key role also in human cancer, including the ability to modulate host immune response. 22 , 23 In this context, there is increasing evidence for the correlation of gut microbiota with cancer immunotherapy activity and toxicity. 24 , 25 , 26 In preclinical in vivo animal models, antitumor efficacy of CTLA‐4 blockade was related to the presence of selected gut microbiota species, by a mechanism that could involve suppression of CD4+ T cell infiltration in tumor microenvironment. 26 Similar findings were also reported for anti‐PD‐1/PD‐L1 treatment in germ‐free or in antibiotic‐treated mice, in which fecal microbiota transplantation was able to restore ICI efficacy. 27 , 28
Recently, we and others have provided the first clinical evidence of antitumor activity of cetuximab plus avelumab, via combined EGFR and PD‐L1 blockade, in chemo‐refractory metastatic CRC, NSCLC and squamous anal cancer. 11 , 14 , 29 Here we report the results of an exploratory analysis of basal, pretreatment fecal samples from two cohorts of patients, that received cetuximab plus avelumab within CAVE‐mCRC and CAVE‐Lung trials. None of these patients received antibiotic therapy, which could affect gut microbiota biodiversity. Further, for the mCRC cohort, all patients had left‐sided primary tumor, which was surgically removed (in most cases, by left‐hemicolectomy) before first‐line systemic therapy. Two gut microbiota species (Agathobacter M104/1 and Blautia SR1/5) were identified in mCRC patients, that had the best clinical outcome. The presence of either bacteria species correlated with significantly longer PFS. An obvious limitation of our analysis is the low number of patients that could negatively affect the significance of the data, and the high number of statistical tests that were performed, which is associated with an increased risk of false‐positive association of outcome with the presence of some bacteria species. In this respect, the biological plausibility of the association of clinical outcome with two butyrate‐producing intestinal bacteria species is relevant as well as it is important that similar findings were observed in two completely different disease settings; in fact, the same bacteria were found to be correlated with clinical outcomes in the mCRC cohort and in the NSCLC patient validation cohort. 30
Several studies have demonstrated the influence of gut microbiota diversity on immunotherapy efficacy. In this context, antibiotic treatment, by altering intestinal microbiota composition, negatively affected ICI therapy in NSCLC, renal cell carcinoma, and urothelial cancer patients, with significantly shorter PFS and OS in antibiotic‐treated patients. 28 , 31 , 32 Qualitative evaluation of gut microbiota species found significant abundance of classified and unclassified Firmicutes, Akkermansia and Alistipes bacteria genera in anti‐PD‐1 mAb‐responding NSCLC patients. 32 In particular, production of butyrate, a short‐chain fatty acid, by the two Agathobacter and Blautia species, which have been found in the basal fecal samples of long‐term responding patients from CAVE‐mCRC and CAVE‐Lung trials, may in part explain the mechanism(s) by which these bacteria could increase cetuximab plus avelumab antitumor activity. Butyrate is a key modulator of host immune reactivity in both physiologic and pathologic conditions through activation of innate and adaptive immunity. 33 In this respect, it has been recently demonstrated that dietary supplement of pectin, which is metabolized to butyrate by gut microbiota, increased butyrate production and enhanced anti‐PD‐1 mAb efficacy in tumor‐bearing mice, which were humanized with gut microbiota from patients with CRC (26). Increased immunotherapy efficacy was accompanied by T cell infiltration in tumor microenvironment. 34
In conclusion, here we provide clinical evidence that two butyrate‐producing intestinal bacteria (Agathobacter M104/1 and Blautia SR1/5) could favorably modulate host immune response and could be potential biomarkers for cetuximab plus avelumab antitumor activity in metastatic and chemo‐refractory CRC and NSCLC patients. The major limitation of these findings is the exploratory nature of the analysis with the limited number of patients. The results of this pilot study need to be extended and validated in further clinical studies. In this respect, we are currently starting a large multicenter randomized phase II trial (CAVE‐mCRC 2, EudraCT Number: 2021‐004593‐36) to investigate rechallenge with cetuximab plus avelumab vs cetuximab in chemo‐refractory RAS WT mCRC patients with basal plasma ctDNA RAS/BRAF WT tumor. Our study could allow the prospective evaluation of the predictive role of gut microbiota for cetuximab plus avelumab treatment.
CONFLICT OF INTEREST
Carminia Maria Della Corte has served as an advisor for MSD. Morena Fasano has a family relationship with Merck. Evaristo Maiello has served as advisor and speaker for Astra Zeneca, Eli Lilly, Servier, Sanofi Genzyme, Roche, Merck, Eisai and Pfizer. Floriana Morgillo has served as an advisor for MSD, Lilly; Institutional Research Grants: AstraZeneca. Teresa Troiani has served as an advisor and speaker for Roche, Merck‐Serono, Sanofi, Servier, Novartis and Bayer. Erika Martinelli has served as advisor and speaker for Astra Zeneca, Amgen, Bayer, Merck‐Serono, Roche, Sanofi, Servier and Pierre Fabre. Massimo Di Maio has served as an advisor for AstraZeneca, Merck Sharp & Dohme, Pfizer, Novartis, Roche, Takeda Pharmaceuticals, Janssen Pharmaceuticals, Mediolanum Farmaceutici, Eisai, Amgen, Merck Serono and received institutional Research Grant from Tesaro ‐ GlaxoSmithKline. Fortunato Ciardiello has served as advisor and speaker for Roche, Amgen, Merck‐Serono, Pfizer, Sanofi, Bayer, Servier, BMS, Cellgene and Lilly. Received institutional Research Grants from Bayer, Roche, Merck‐Serono, Amgen, AstraZeneca and Takeda. The other authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Giulia Martini, Davide Ciardiello, Erika Martinelli and Fortunato Ciardiello: Conceptualization, data curation, methodology, writing‐original draft. Giulia Martini, Davide Ciardiello, Marcello Dallio, Vincenzo Famiglietti, Lucia Esposito, Massimo Di Maio, Erika Martinelli, Fortunato Ciardiello: Data curation, statistical analysis, interpretation of data. Carminia Maria Della Corte, Stefania Napolitano, Morena Fasano, Floriana Morgillo, Evaristo Maiello, Concetta Tuccillo, Alessandro Federico, Antonietta Gerarda Gravina, Marco Romano, Teresa Troiani, Carmelina Loguercio, Massimo Di Maio, Fortunato Ciardiello: Supervision, Writing‐Original Draft Revision. The work reported in the study has been performed by the authors, unless clearly specified in the text.
ETHICS STATEMENT
The CAVE‐mCRC and CAVE‐LUNG trials were approved by the Ethic Committee of Università degli Studi della Campania Luigi Vanvitelli, Azienda Ospedaliera Universitaria‐AORN “Ospedali dei Colli”, Napoli, Italy. CAVE‐mCRC trial is registered with Eudract.ema.europa.eu, EudraCT number: 2017‐004392‐32 and ClinicalTrial.gov identifier: NCT04561336. CAVE‐LUNG trial is registered with Eudract.ema.europa.eu, EudraCT number 2017‐004195‐58. All patients provided written informed consent before entering the trial. The study was conducted in accordance with the principles of the Declaration of Helsinki and the International Conference on Harmonization and Good Clinical Practice guidelines.
Supporting information
Data S1 Supporting Information
ACKNOWLEDGMENTS
Open Access Funding provided by Universita degli Studi della Campania Luigi Vanvitelli within the CRUI‐CARE Agreement.
Martini G, Ciardiello D, Dallio M, et al. Gut microbiota correlates with antitumor activity in patients with mCRC and NSCLC treated with cetuximab plus avelumab. Int J Cancer. 2022;151(3):473‐480. doi: 10.1002/ijc.34033
Giulia Martini and Davide Ciardiello contributed equally to this study.
Funding InformationTwo research grants, that partially covered the costs of the study, were provided by Merck and by Regione Campania (I‐Cure Research Project, Grant number: Cup 21C17000030007)
DATA AVAILABILITY STATEMENT
The data that support the findings of our study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non‐small‐cell lung cancer. N Engl J Med. 2015;372:2018‐2028. [DOI] [PubMed] [Google Scholar]
- 2. Robert C, Schachter J, Long GV, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med. 2015;372:2521‐2532. [DOI] [PubMed] [Google Scholar]
- 3. Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320‐330. [DOI] [PubMed] [Google Scholar]
- 4. Le DT, Uram JN, Wang H, et al. PD‐1 blockade in tumors with mismatch‐repair deficiency. N Engl J Med. 2015;372:2509‐2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khan S, Zaidi S, Alouffi AS, Hassan I, Imran A, Khan RA. Computational proteome‐wide study for the prediction of Escherichia coli protein targeting in host cell organelles and their implication in development of colon cancer. ACS Omega. 2020;5:7254‐7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang X, Siddique A, Khan AA, et al. Chlamydia trachomatis infection: their potential implication in the etiology of cervical cancer. J Cancer. 2021;12:4891‐4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khan S, Zakariah M, Rolfo C, Robrecht L, Palaniappan S. Prediction of mycoplasma hominis proteins targeting in mitochondria and cytoplasm of host cells and their implication in prostate cancer etiology. Oncotarget. 2017;8:30830‐30843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zakariah M, Khan S, Chaudhary AA, Rolfo C, Ben IMM, Alotaibi YA. To decipher the mycoplasma hominis proteins targeting into the endoplasmic reticulum and their implications in prostate cancer etiology using next‐generation sequencing data. Molecules. 2018;23:994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alshamsan A, Khan S, Imran A, Aljuffali IA, Alsaleh K. Prediction of chlamydia pneumoniae protein localization in host mitochondria and cytoplasm and possible involvements in lung cancer etiology: a computational approach. Saudi Pharm J. 2017;25:1151‐1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y, Imran A, Shami A, Chaudhary AA, Khan S. Decipher the helicobacter pylori protein targeting in the nucleus of host cell and their implications in gallbladder cancer: an in silico approach. J Cancer. 2021;12:7214‐7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martinelli E, Martini G, Famiglietti V, et al. Cetuximab rechallenge plus avelumab in pretreated patients with RAS wild‐type metastatic colorectal cancer: the phase 2 single‐arm clinical CAVE trial. JAMA Oncol. 2021;7:1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferris RL, Lenz HJ, Trotta AM, et al. Rationale for combination of therapeutic antibodies targeting tumor cells and immune checkpoint receptors: harnessing innate and adaptive immunity through IgG1 isotype immune effector stimulation. Cancer Treat Rev. 2018;63:48‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bourhis J, Stein A, Paul de Boer J, et al. Avelumab and cetuximab as a therapeutic combination: an overview of scientific rationale and current clinical trials in cancer. Cancer Treat Rev. 2021;97:102172. [DOI] [PubMed] [Google Scholar]
- 14. Fasano M, Della Corte CM, Di Liello R, et al. Induction of natural killer antibody‐dependent cell cytotoxicity and of clinical activity of cetuximab plus avelumab in non‐small cell lung cancer. ESMO Open. 2020;5:e000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ciardiello D, Vitiello PP, Cardone C, et al. Immunotherapy of colorectal cancer: challenges for therapeutic efficacy. Cancer Treat Rev. 2019;76:22‐32. [DOI] [PubMed] [Google Scholar]
- 16. Skelly AN, Sato Y, Kearney S, Honda K. Mining the microbiota for microbial and metabolite‐based immunotherapies. Nat Rev Immunol. 2019;19:305‐323. [DOI] [PubMed] [Google Scholar]
- 17. Siddiqui MT, Cresci GA. The immunomodulatory functions of butyrate. J Inflamm Res. 2021;14:6025‐6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next‐generation sequencing‐based diversity studies. Nucleic Acids Res. 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Claesson MJ, O'Sullivan O, Wang Q, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One. 2009;4:e6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vitiello PP, De Falco V, Giunta EF, et al. Clinical practice use of liquid biopsy to identify RAS/BRAF mutations in patients with metastatic colorectal cancer (mCRC): a single institution experience. Cancers (Basel). 2019;11:1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16:35‐56. [DOI] [PubMed] [Google Scholar]
- 22. Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kroemer G, Zitvogel L. Cancer immunotherapy in 2017: the breakthrough of the microbiota. Nat Rev Immunol. 2018;18:87‐88. [DOI] [PubMed] [Google Scholar]
- 25. Paulos CM, Wrzesinski C, Kaiser A, et al. Microbial translocation augments the function of adoptively transferred self/tumor‐specific CD8+ T cells via TLR4 signaling. J Clin Invest. 2007;117:2197‐2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14:356. [DOI] [PubMed] [Google Scholar]
- 27. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science (80‐). 2018;359:91‐97. [DOI] [PubMed] [Google Scholar]
- 28. Sivan A, Corrales L, Hubert N, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti‐PD‐L1 efficacy. Science. 2015;350:1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lonardi S, Prete AA, Morano F, et al. Randomized phase II trial of avelumab alone or in combination with cetuximab for patients with previously treated, locally advanced, or metastatic squamous cell anal carcinoma: the CARACAS study. J Immunother Cancer. 2021;9:e002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okumura S, Konishi Y, Narukawa M, et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Commun. 2021;12:5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Derosa L, Hellmann MD, Spaziano M, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non‐small‐cell lung cancer. Ann Oncol. 2018;29:1437‐1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hakozaki T, Richard C, Elkrief A, et al. The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non‐small cell lung cancer. Cancer Immunol Res. 2020;8:1243‐1250. [DOI] [PubMed] [Google Scholar]
- 33. Kespohl M, Vachharajani N, Luu M, et al. The microbial metabolite butyrate induces expression of Th1‐associated factors in cD4+ T cells. Front Immunol. 2017;8:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang SL, Mao YQ, Zhang ZY, et al. Pectin supplement significantly enhanced the anti‐PD‐1 efficacy in tumor‐bearing mice humanized with gut microbiota from patients with colorectal cancer. Theranostics. 2021;11:4155‐4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Supporting Information
Data Availability Statement
The data that support the findings of our study are available from the corresponding author upon reasonable request.


