Abstract
The membrane integrity of a cell is a well-accepted criterion for characterizing viable (active or inactive) cells and distinguishing them from damaged and membrane-compromised cells. This information is of major importance in studies of the function of microbial assemblages in natural environments, in order to assign bulk activities measured by various methods to the very active cells that are effectively responsible for the observations. To achieve this task for bacteria in freshwater and marine waters, we propose a nucleic acid double-staining assay based on analytical flow cytometry, which allows us to distinguish viable from damaged and membrane-compromised bacteria and to sort out noise and detritus. This method is derived from the work of S. Barbesti et al. (Cytometry 40:214–218, 2000) which was conducted on cultured bacteria. The principle of this approach is to use simultaneously a permeant (SYBR Green; Molecular Probes) and an impermeant (propidium iodide) probe and to take advantage of the energy transfer which occurs between them when both probes are staining nucleic acids. A full quenching of the permeant probe fluorescence by the impermeant probe will point to cells with a compromised membrane, a partial quenching will indicate cells with a slightly damaged membrane, and a lack of quenching will characterize intact membrane cells identified as viable. In the present study, this approach has been adapted to bacteria in freshwater and marine waters of the Mediterranean region. It is fast and easy to use and shows that a large fraction of bacteria with low DNA content can be composed of viable cells. Admittedly, limitations stem from the unknown behavior of unidentified species present in natural environments which may depart from the established permeability properties with respect to the fluorescing dyes.
Bacteria are a key component in aquatic and terrestrial ecosystems due to their wide biodiversity, their capacity to survive extreme environments, and their large variety of metabolic activities. In aquatic environments, with elevated abundances in the range of 105 to 106 cells cm−3 (39), bacteria play a major role in recycling organic matter and subsequently in sustaining the nutrient turnover. The understanding of bacterial activity with respect to the fate of organic matter in marine water and freshwater is ecologically important, and many studies are dedicated to this objective (7, 9, 10, 37, 38, 40, 51). Bacteria are able to metabolize part of the dissolved organic matter (DOM) present in micro- or nanomolar concentrations in marine environments (5, 25) and to convert it into particulate organic matter through bacterial biomass production, making it available to higher trophic levels (38). The rate at which the biological oxidation of DOM by bacteria occurs is dependent on DOM biodegradability (2, 18). Conversely, viral lysis of bacteria releases cellular constituents into the environment and supplies a new source of carbon and nitrogen for primary producers and consumers (1, 40).
The presence of bacteria in aquatic environments is also of special concern with respect to human health problems and water quality. Indeed, several bacterial strains in freshwater and marine water have been demonstrated to be pathogenic. For instance, Vibrio vulnificus in estuarine and marine waters is responsible for fatal infections following ingestion (typically of wild oysters) or contamination of a wound (28, 36, 52). Other species, such as Escherichia coli and Salmonella enterica serovar Enteritidis have been shown to be indicators of human pollution through fecal contamination (14). In response to their environmental conditions, bacteria may be present in a viable but nonculturable state by classical microbiological methods (6, 26, 32). When bacteria are monitored, it is very important to determine the viability of cells in addition to their abundance, since this is a critical issue in assessing water quality and preventing sanitation problems (44). New approaches have been developed to assess bacterial viability (29, 33, 39). Most are based on the ability to detect metabolic or functional activity by using different fluorochromes: (i) detection of cell divisions by counts of CFU on plates (34), (ii) assessment of membrane potential with dyes of the carbocyanine and oxonol families (31), (iii) dye (propidium iodide [PI] or ethidium bromide) exclusion due to membrane integrity (19, 43), (iv) intracellular reduction of 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) and 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium chloride (INT) by dehydrogenases (8, 11, 41, 46, 47, 49, 50), (v) reduction of fluorescein diacetate (FDA) by esterases (17, 22), (vi) radiolabeled substrate uptake analyzed by microautoradiography (24, 47), (vii) the presence of nucleoids (55), and (viii) nucleic acid content heterogeneity (12). Molecular analysis of components such as ribosomes can also reveal the state of growth (13). The 16S rRNA probe count (24, 42, 48, 53) is one example of molecular approaches.
In investigations of bacteria in natural environments, it is necessary to distinguish them from other particles and to characterize them by their properties, such as morphology, physiology, and taxonomy. All of these aspects imply analyses at the individual cell level, but methods involving cell enumeration by optical microscopy are both time-consuming and subjective (39). The development of analytical flow cytometry (45), together with advances in the field of fluorescent probes (21, 39), have enabled the application of this rapid and semiautomatic technique to the study of bacteria in natural aquatic environments. Barbesti et al. (3) took advantage of flow cytometry to devise a DNA double-staining protocol based on energy transfer between specific probes to distinguish viable, dead, and compromised cells in bacterial cultures. In the present study, we report the use of this nucleic acid double-staining (NADS) protocol with natural freshwater and marine water to count bacteria and to rapidly determine clusters according to their cell membrane permeability by using the same assay.
MATERIALS AND METHODS
Sampling sites.
Freshwater samples were collected from three rivers in northern Italy: the Adda, the Lambro, and the Seveso. A monitoring experiment was conducted along the Adda River, and samples were collected ca. 1 km upstream from a sewage outflow, at the site of sewage outflow, and at ca. 1 and 6 km downstream from the sewage outflow. Sampling was carried out once a week for 1 month.
Seawater samples were collected in surface waters from two different environments: at site 1 inside Pointe-Rouge, a pleasure-boat harbor in Marseilles, France, and site 2 facing Maire Island, at the southern edge of the city and not under its direct influence. Samples were prefiltered on a 100-μm-mesh-size net to avoid clogging of the instrument.
Instruments.
Different flow cytometers were used in this study as follows. (i) We used a Bryte-HS (Bio-Rad, Hercules, Calif.) flow cytometer equipped with a 75-W Xenon lamp, and a 470- to 490-nm excitation filter was used for the analysis of freshwater samples. The emitted fluorescence was split into two different channels: FL1 (green) at 515 to 565 nm and FL2 (orange-red) at 590 to 650 nm. All of the data related to freshwater samples were analyzed by using Bio-Rad WinBryte software. (ii) Wed also used a Cytoron Absolute (Ortho Diagnostic Systems) flow cytometer, equipped with an air-cooled argon laser (excitation wavelength at 488 nm) to analyze seawater samples. Each cell was characterized by five optical parameters: two diffraction parameters, namely, forward-angle scatter (related to the particle size) and right-angle scatter (related to the cell structure), and three fluorescence parameters measuring emissions in the red (>620-nm), orange (565- to 592-nm), and green (515- to 530-nm) wavelength ranges. Data were collected and stored in list mode with Immunocount software (Ortho Diagnostic Systems). This software provides directly the cell concentration (as cells millimeter−3) of the resolved populations. (iii) Finally, we used an Elite EPS (Beckman-Coulter) equipped with a 488-nm argon laser for cell sorting in Gif-sur-Yvette, France. The emitted fluorescence was split into three different channels: PMT3 (green) at 520 to 530 nm, PMT4 (orange) at 615 to 640 nm, and PMT5 (red) at 645 to 795 nm. Sorting was done with a sample rate of ca. 1,000 events s−1 into Eppendorf tubes. Visual checks for the identification of bacteria from detritus, as well as sorting and staining, were made on a Polyvar (Reichert, Vienna, Austria) microscope with epifluorescence and interference contrast.
Fluorescent probes.
SYBR Green I or II (Molecular Probes, Eugene, Oreg.) and PI (Sigma Chemical Co.) were used for the double staining of nucleic acids. SYBR Green and PI fluoresce in the green (maximum at 521 nm) and the red (maximum at 617 nm) wavelength ranges, respectively, when excited with a 488-nm argon laser. They both stain RNA and DNA (15). 5 (and 6)-carboxyfluorescein diacetate (CFDA; Molecular Probes) was employed to detect esterase activity in living cells (20, 22). A stock solution of 5 mg cm−3 was made in dimethyl sulfoxide (Sigma) and stored at 4°C. This stock solution was thawed and diluted 100-fold in distilled water to prepare daily working solutions kept for a maximum of 3 h to avoid CFDA degradation. Samples (3 cm3) were supplemented with 100-mm3 CFDA working solution and were analyzed after 5 min of incubation in the dark at room temperature.
Flow cytometric analysis.
For each freshwater sample, three dilutions were prepared and stained with 1:10,000 (vol/vol) SYBR Green I (neither the molecular weight nor the chemical formula are provided by the manufacturer) and a 10-μg cm−3 PI commercial solution. The final fluorochrome concentrations were as defined by Barbesti et al. (3) for bacterial cultures. Seawater samples were stained without dilution with 1:1,000 (vol/vol) SYBR Green II (neither the molecular weight nor the chemical formula are provided by the manufacturer) and a 10-μg cm−3 PI commercial solution. For both types of samples, flow cytometric analyses were processed after 30 min of incubation in the dark.
CFU.
The culturability of bacteria from freshwater was assessed by mixing 0.1 cm3 of the sample with ca. 25-cm3 of plate count agar medium [PCA; peptone from 5 g of casein dm−3, 2.5 g of yeast extract, 1 g of d-(+)-glucose dm−3, 14 g of agar dm−3 (pH 7.0 at 25°C)] at 45°C. After medium solidification, plates were incubated for 72 h at 22°C. The total bacterial charge was estimated as the CFU per cubic centimeter of sample.
Ozone and heat treatments.
One liter of freshwater was treated with ozone (ca. 30 mg dm−3) at a laboratory scale for 90 min in order to kill the sample cells. After ozone treatment, the bacterial viability and culturability was checked by analyzing 1 cm3 of ozonized sample by flow cytometry and plating 0.1 cm3 on PCA, respectively.
The flow cytometric characterization of membrane-compromised bacteria was also done by exposing cells of seawater samples to heat treatment (40 min in a 60 to 70°C water bath).
RESULTS
NADS protocol.
The live-dead protocol developed by Barbesti et al. (3) for cultures of E. coli is based on the simultaneous staining of bacteria nucleic acids by a permeant (SYBR Green I) and an impermeant (PI) fluorescent probe and the interpretation of the green versus the red fluorescence cytograms. Green, green plus orange-red, and orange-red E. coli cells were identified as live, damaged, and dead cells, respectively (3). In order to extend this approach to natural samples from aquatic environments, several experiments were conducted with bacteria from freshwater and seawater.
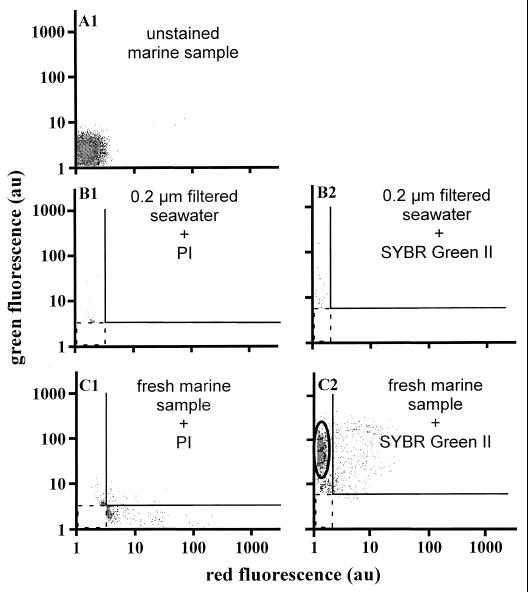
Figure 1 shows the green versus the red fluorescence cytograms from marine samples under different conditions. Cytogram A1 is representative of marine samples in the absence of staining. The weak signals close to the origin are due to the instrument's electronic noise and the background signals of nonfluorescent heterotrophic bacteria. Stained cells will therefore yield fluorescence signals above this background. Cytograms B1 and B2 were obtained with samples of 0.2-μm-pore-size-filtered seawater supplemented with PI (cytogram B1) or SYBR Green II (cytogram B2). With these cell-free samples, signals are produced by the instrument noise and the free dyes in solution which may generate a large number of events and quickly saturate the data acquisition. Consequently, a fluorescence threshold (dotted square) was systematically introduced. Cytograms C1 and C2 correspond to fresh marine samples stained with PI and SYBR Green II, respectively. Few cells appeared to be stained by PI (red fluorescence signals) in cytogram C1, and two populations were distinguished upon staining by SYBR Green II (green fluorescence signals) in cytogram C2.
FIG. 1.
Green versus red fluorescence cytograms corresponding to different test experiments. Panels: A1, unstained seawater sample; B1 and B2: 0.2-μm-filtered seawater sample supplemented with PI and SYBR Green II, respectively; C1 and C2, seawater samples supplemented with PI and SYBR Green, respectively. The final probe concentrations were 10 μg of PI cm−3 and 1:1,000 (vol/vol) SYBR Green II.
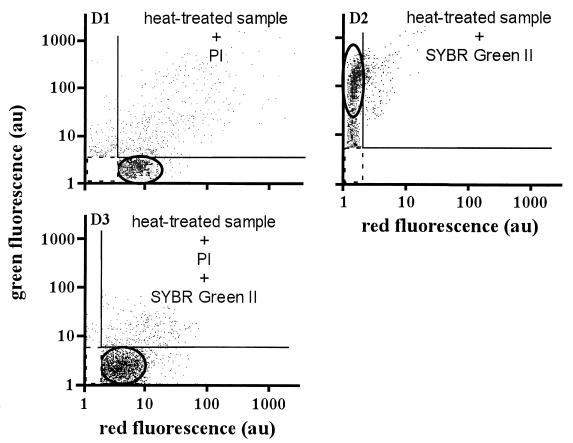
In Fig. 2, cytograms D1 and D2 concern a seawater sample subjected to the heat treatment described in Materials and Methods to kill all cells, including bacteria. Membrane-compromised bacteria are identified by their PI staining in cytogram D1, whereas staining with SYBR Green II (cytogram D2) resulted in a signature similar to that of the fresh sample in cytogram C2. When both stains were simultaneously incubated in the heat-treated sample, cells essentially exhibited red fluorescence as expected (cytogram D3).
FIG. 2.
Single and dual staining of heat-treated seawater samples. Panels: D1 and D2, heat-treated seawater samples supplemented with PI and SYBR Green II, respectively; D3, heat-treated (40 min at 60 to 70°C) seawater sample simultaneously stained with PI and SYBR Green. Killed bacteria essentially appeared as red fluorescent cells. The final probe concentrations were 10 μg of PI cm−3 and 1:1,000 (vol/vol) SYBR Green II.
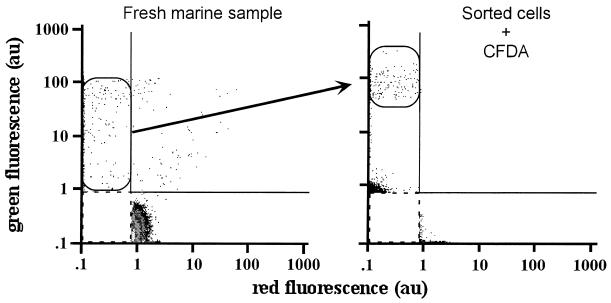
In an experiment in which a seawater sample was double stained with SYBR Green II and PI, cells fluorescing preferentially in the green (gated cells in the left cytogram of Fig. 3) were sorted as described in Materials and Methods and stained with CFDA. This additional staining resulted in a significant increase in the green fluorescence of all sorted cells. In contrast, heat-treated cells, which only yielded red fluorescence after double staining by SYBR Green II and PI, did not emit green fluorescence after being stained with CFDA (not shown).
FIG. 3.
Cell sorting and esterase activity assay. Green fluorescent bacteria resolved by the NADS protocol (gated cells on the fresh-marine-sample cytogram) were sorted and further stained with CFDA as described in Materials and Methods. The cells exhibited an increase in green fluorescence due to esterase activity and carboxyfluorescein retention (gated cells on the sorted cells-plus-CFDA cytogram).
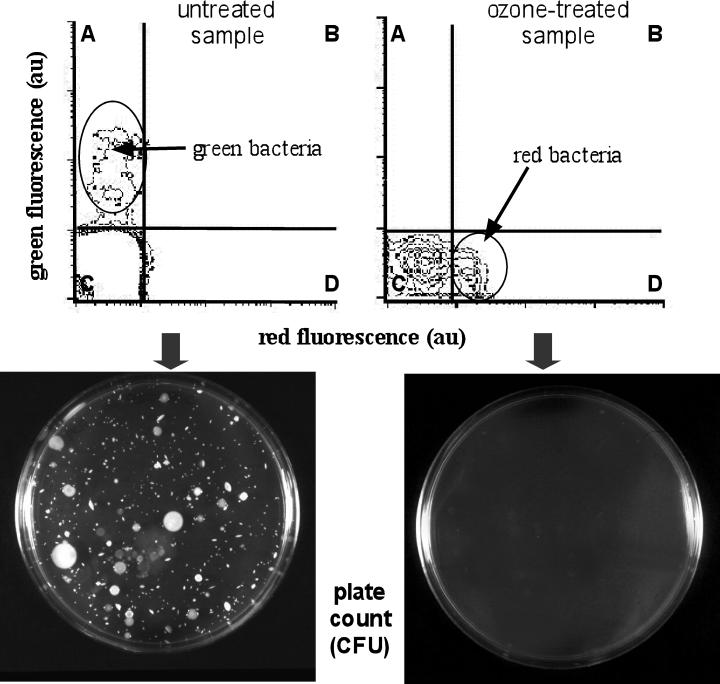
Ozone treatment of freshwater samples led to complementary results. In Fig. 4, a freshwater sample collected from Seveso River and double stained with SYBR Green I and PI essentially gave cells with green fluorescence (untreated-sample cytogram), whereas all cells fluoresced red after ozone treatment (ozone-treated-sample cytogram). After 0.1 cm3 of the fresh sample was plated on PCA, the formation of numerous colonies was observed (Fig. 4, left plate). In contrast, no colonies were observed after being plated on PCA with 0.1 cm3 of the fraction submitted to ozone treatment (Fig. 4, right plate).
FIG. 4.
Application of the NADS protocol to a Seveso River (Italy) sample before and after ozone treatment. The untreated-sample cytogram corresponds to a freshwater subsample mainly containing green bacteria. The ozone-treated-sample cytogram was obtained after ozone treatment to kill cells, and it shows essentially red bacteria. Quadrants are identified as A to D. The plate count obtained with green bacteria from the untreated sample shows abundant colonies, whereas the plate count made with red cells from the ozone-treated sample yielded no colonies.
Freshwater. (i) Protocol optimization.
Bacterial abundance in freshwaters varies significantly, depending upon the level of eutrophication and various discharges produced by human activity. We have therefore tested the adequacy of NADS, devised for bacterial cultures (1:10,000 [vol/vol] SYBR Green I final dilution and a 10-μg cm−3 final concentration of PI ) (3), when applied to freshwater samples. Adda River samples were stained as collected and after a 10-fold dilution. For the diluted subsample, the background fluorescence of SYBR Green I in solution was too high to be eliminated in the orange-red channel by electronic compensation (not shown). In addition, an increase in background fluorescence, affecting signal resolution, was observed for high PI concentration. To avoid these problems, it was necessary to adapt the concentrations of the fluorescent probes to that of bacteria. A final concentration of 1 μg of PI cm−3 and a final dilution of 1:100,000 for SYBR Green I were found to be optimal for freshwater samples with less than 0.5 × 106 bacteria cm−3. For more concentrated freshwater samples (up to 20 × 106 bacteria cm−3), the fluorochrome concentrations (1:10,000 [vol/vol] SYBR Green I final dilution and 10-μg cm−3 final concentration of PI) determined for bacterial cultures remain the best. The staining efficiency was also checked with respect to incubation time for 60 min. Staining was stable for each class after 30 min of incubation.
(ii) Adda River monitoring.
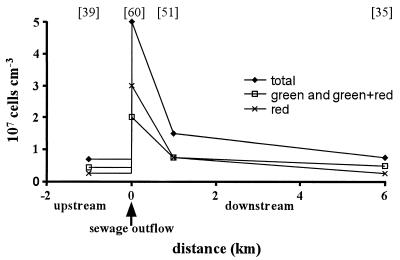
The Adda River (Italy) was sampled as described in Materials and Methods. The different bacterial states were resolved using the NADS protocol, and a typical distribution is displayed in Fig. 5. At 1 km upstream from the sewage outflow, the green and green-plus-red bacterial fractions corresponded to 61% of the whole bacterial population, whereas the red fluorescent bacteria amounted to 39% (Fig. 5). The strong bacterial input at the site of the sewage outflow (5 × 107 cells cm−3) was reduced by ca. 80% 1 km downstream and by ca. 90% 5 km farther downstream. The sewage outflow also corresponded to an inversion of the ratio of green and green-plus-red cells to red cells, leading to 60% red fluorescent cells. This effect was reduced by half 1 km downstream and fully disappeared 6 km downstream, where the percentage of red fluorescent cells decreased to 35% as upstream.
FIG. 5.
Bacteria distribution in the Adda River (Italy) at four different stations: 1 km upstream from a sewage outflow, at the sewage outflow site, and at 1 and 6 km downstream from the sewage outflow point. Bacteria were resolved according to their fluorescence (green and green-plus-red bacteria and red fluorescent bacteria) generated by the NADS protocol. The values in brackets are the percentages of red fluorescent bacteria relative to the total count.
Marine water. (i) Protocol optimization.
In the SYBR Green dye family, SYBR Green II expresses a higher selectivity for RNA, with a quantum yield of 0.54, while keeping a strong affinity for double-stranded DNA with a quantum yield of 0.36, about half that of SYBR Green I (15). SYBR Green II was therefore tested on marine samples to better use the energy transfer phenomenon when both dyes (SYBR Green II and PI) bind to nucleic acids. The optimum concentration of SYBR Green II to detect bacteria in seawater was found to be a 1:1,000 (vol/vol) SYBR Green II final dilution after we tested the concentration range from 0.025:1,000 to 2:1,000 (vol/vol). This threshold for a maximum detection of bacteria is effective after only 10 min of incubation. The concentration of PI was set at 10 μg cm−3. The appropriate incubation time for the NADS procedure when SYBR Green II and PI were used was also found to be 30 min by monitoring the time course of the different bacterial states resolved by NADS during 1 h of incubation.
The flow cytometric analysis of bacteria in seawater by the NADS protocol yielded typical cytograms characterizing cells by their fluorescence in panels A, B, and D (Fig. 4) as green, green-plus-red, and red, respectively, as for freshwater bacteria.
(ii) Northwestern Mediterranean coast.
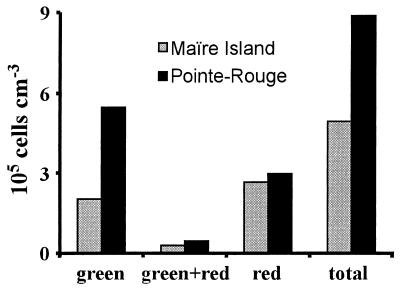
The comparison of the bacterial abundance at both marine sampling sites and in the different fluorescent states is reported in Fig. 6. The overall abundance was higher by 80% at the Pointe-Rouge site (site 1; 8.87 × 105 cells cm−3) than at the site facing Maire Island (site 2; 4.94 × 105 cells cm−3). The amount of red fluorescent bacteria was similar at both sites (ca. 3 × 105 cells cm−3), which led to a lower percentage of red fluorescent bacteria at site 1 than at site 2 (33 versus 53%). Consistently, there were more green bacteria at site 1 than at site 2, both in absolute and in relative terms (62 and 41%, respectively). The levels of green-plus-red bacteria were similar at both sites (5 and 6%, respectively).
FIG. 6.
Staining classes for bacteria collected from surface water inside Pointe-Rouge harbor (Marseilles, France) and a few kilometers away from the town, at the site facing Maire Island.
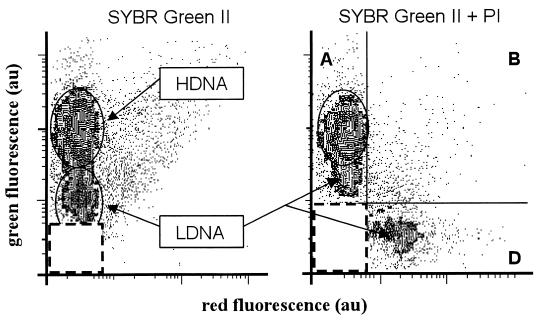
The relationship between the DNA content and the bacterial response to the NADS assay is illustrated in Fig. 7 with different staining protocols applied to a seawater sample collected at site 2. As previously reported (12), two different populations are distinguished by their DNA content (Fig. 7) when bacteria were stained with SYBR Green II alone: a population with a low DNA content (Fig. 7, LDNA) characterized by a low green fluorescence and a population with a high DNA content (HDNA) characterized by a higher green fluorescence (Fig. 7). The NADS protocol applied to a subsample resolved two populations within the green fluorescent cells (Fig. 7). The bacterial abundances yielded by the two staining protocols are summarized in Table 1. Both protocols gave the same amount of total bacteria (5.4 × 105 to 5.5 × 105 cells cm−3). In addition, all HDNA bacteria characterized upon staining with SYBR Green II alone remained as green bacteria after the NADS assay, whereas LDNA bacteria were resolved as green (∼68%), green-plus-red (2%), and red cells (∼30%).
FIG. 7.
Comparison of single (SYBR Green II; 1:1,000 [vol/vol] final dilution) and double (NADS protocol) staining of bacteria in a seawater sample collected off Marseilles at the site facing Maire Island. The green versus the red fluorescence cytograms corresponding to SYBR Green II staining reveal the presence of bacteria with high (H) and low (L) DNA contents. The cytogram associated with the NADS protocol shows a significant amount of LDNA bacteria appearing as red-negative cells in quadrant A. At the same time, cells that were negative with SYBR Green II were apparent with PI staining in quadrant D. Refer to Table 1 for the counts.
TABLE 1.
Comparison of single (SYBR Green II) and double (NADS protocol) staining of bacteria in a seawater sample collected off Marseilles at the site facing Maire Island
| Group | SYBR Green II method
|
NADS protocol
|
|||||
|---|---|---|---|---|---|---|---|
| Count (105 cells cm−3) | % | Count (105 cells cm−3) | % | Cell typea | Count (105 cells cm−3) | % | |
| Total bacteria | 5.54 | 5.43 | |||||
| HDNA bacteria | 2.60 | 46.9 | 2.63 | 48.5 | Green | 2.63 | 100.0 |
| Green plus red | 0 | ||||||
| Red | 0 | ||||||
| LDNA bacteria | 2.94 | 53.1 | 2.80 | 51.5 | Green | 1.91 | 68.2 |
| Green plus red | 0.06 | 2.0 | |||||
| Red | 0.83 | 29.8 | |||||
As determined by fluorescence.
DISCUSSION
This NADS flow cytometric protocol is based on the simultaneous use of permeant (SYBR Green dyes) (27) and impermeant (PI) fluorescent probes (22, 30, 43, 53). Both dyes can be readily excited with the blue light from a laser or arc lamp of relatively simple and portable flow cytometers, which makes the assay suitable for field control. The efficiency of the combined staining is magnified by the energy transfer from SYBR Green to PI when both are bound to the nucleic acids as described by Barbesti et al. (3). Consequently, membrane-compromised bacterial cells are expected to fluoresce in the red-wavelength range, whereas the fluorescence emitted by membrane intact cells will be restricted to the green wavelengths. Cells with a partially damaged membrane will enable various amounts of PI to bind some nucleic acids that will result in a corresponding increase of the red fluorescence and a decrease of the green fluorescence depending on the extent of the energy transfer from SYBR Green to PI. SYBR Green I performed adequately with freshwater, but SYBR Green II has been shown to perform better in seawater (27).
Analogous fluorescence quenching could be achieved by using Hoechst stain and PI to test the membrane integrity (35) in a double-staining protocol. However, Hoechst fluorochromes require UV excitation, which generally necessitates the use of expensive flow cytometers.
Evidence for esterase activity and fluorochrome retention in all green fluorescent cells sorted after the NADS assay (Fig. 3), as well as the plate count experiment (Fig. 4), lend independent support to the identification of the green cells as viable. In addition, the heat (Fig. 2) and ozone (Fig. 3) treatments used to artificially kill cells in the analyzed samples also provided independent support for the identification of red cells as membrane-compromised cells, qualifying our NADS protocol as a membrane integrity assay for bacteria in freshwater and marine water. However, if dead (heat- or ozone-killed) cells undoubtedly appear as red cells, the full identification of red cells in natural samples as dead cells remains to be further substantiated. Indeed, some unknown species might be able to accumulate PI without the prerequisite of having a compromised membrane. The NADS assay descibed here requires an incubation time of 30 min to achieve a full staining, both for freshwater and seawater samples, which is consistent with other published staining protocols (27), wherein reported incubations last 15 and 30 min for fixed and unfixed bacteria, respectively. In the present protocol, only bacteria containing nucleic acids are stained either by both PI and SYBR Green I or II in the case of membrane-compromised cells or by SYBR Green I or II alone when the insertion of PI is prevented by the membrane integrity. Under these conditions, bacterium-like detritus without nucleic acids cannot be confused with real bacteria (55). The assay can be processed in <1 h, and it yields both quantitative (i.e., cell count) and qualitative (i.e., for viable, membrane-damaged, and membrane-compromised cells) information for each analyzed sample. The assessment of viability by epifluorescence microscopy after acridine orange or DAPI (4′,6′-diamidino-2-phenylindole) staining on membrane filters (16, 54)—a more time-consuming, less accurate, and less objective approach than flow cytometric assay—is incompatible with routine sampling and analyses.
The NADS protocol offers higher resolution than the HDNA-LDNA classification resulting from staining DNA with a single fluorescent probe (12), in which cells with a high DNA content were identified as the fraction containing active bacteria and those with a low DNA content were representative of inactive and dead bacteria. Damaged cells were not resolved in this approach, whereas the NADS protocol clearly separates inactive cells (i.e., membrane compromised, in red) from damaged cells (in red-plus-green).
The above-described results related to the NADS protocol support the hypothesis that HDNA bacteria are viable cells. However, the assimilation of LDNA bacteria with dead cells should be revised, since in the above example two-thirds of the LDNA bacteria were found to be viable and only one-third compromised membrane cells were found to be viable (Table 1). The remaining 2% were membrane-damaged cells, a transient step between viable and membrane-compromised states. We recall that the NADS protocol is based only on the cell membrane integrity, which is a widely accepted criterion for viability (23, 35). Indeed, in spite of the long debate about live and dead bacteria (4), the loss of membrane integrity results in the collapse of the cell energetics and active transports, that is to say, the death of the cell (35). To the extent that the red fluorescing (PI permeable) cells have irreversibly damaged membranes, this parameter can provide a rough approximation of nonviable cells. On the other hand, a cell with preserved membrane integrity (PI impermeable and therefore only stained by green fluorescent SYBR Green) can be considered as viable, which covers active (enzyme activities, metabolism, compact DNA, and high content in rRNA) or inactive states (e.g., the lack of enzyme activity in nutrient-depleted environments) (35). In addition, active cells include culturable and nonculturable bacteria. The reported NADS assay is not, strictly speaking, a live-dead assay but rather a proxy, whose efficiency depends on cell types and physiologic states occurring in natural samples composed of very diverse populations in physiologic and cytostructural terms. Indeed, present limitations to the NADS assay stem from the large biodiversity of bacteria in natural environments, where most of the species are still unknown. Therefore, it is possible that some as-yet-unknown species may not be consistent with observations made thus far regarding the bacterial membrane permeability with respect to the fluorescent probes used in the NADS assay (PI and SYBR Green). This must be kept in mind when one is dealing with natural samples. The NADS assay, which only addresses membrane integrity, provides the resolution of a bacterial population into viable (active, inactive, culturable, and nonculturable), membrane-damaged, and membrane-compromised cells, but complementary approaches are needed to independently demonstrate the presence of specific activity or metabolism, as well as the capacity for a cell to undergo division. Because of its simplicity, NADS should become an efficient tool in field sample analysis and a valuable source of information concerning the functioning of the microbial compartment. Indeed, until now, bacterial activity and production have referred to total bacteria counts, wherein dead cells may represent a large fraction (12; this study), even though these cells contribute neither to remineralization nor to bacterial biomass production. The application of the flow cytometric NADS protocol in field studies will bring quantitative and qualitative improvements to the basic information characterizing the role of bacteria in the environment. This should in turn clarify our understanding of the part played by bacteria in biogeochemical cycles and their modeling.
ACKNOWLEDGMENTS
This study was supported by the Integrated Action Programme “GALILEE” between France and Italy. G.G. is the recipient of a doctoral fellowship from the Council of the Provence-Alpes-Côte-d'Azur Region.
REFERENCES
- 1.Alonso M C, Rodriguez V, Rodriguez J, Borrego J J. Role of ciliates, flagellates, and bacteriophages on the mortality of marine bacteria and on dissolved-DNA concentration in laboratory experimental systems. J Exp Mar Biol Ecol. 2000;244:239–252. [Google Scholar]
- 2.Arnosti C, Repeta D J, Blough N V. Rapid bacterial degradation of polysaccharides in anoxic marine systems. Geochim Cosmochim Acta. 1994;58:2639–2652. [Google Scholar]
- 3.Barbesti S, Citterio S, Labra M, Baroni M D, Neri M G, Sgorbati S. Two- and three-color fluorescence flow cytometric analysis of immunoidentified viable bacteria. Cytometry. 2000;40:214–218. [PubMed] [Google Scholar]
- 4.Barer M R, Harwood C R. Bacterial viability and culturability. Adv Microb Physiol. 1999;41:94–137. doi: 10.1016/s0065-2911(08)60166-6. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi M. Nouvelles approches d'étude des réseaux microbiens. Ann Limnol. 1998;34:465–473. [Google Scholar]
- 6.Button D K, Schut F, Quang P, Martin R, Robertson B R. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho B C, Azam F. Major role of bacteria in biogeochemical fluxes in the ocean's interior. Nature. 1988;332:441–443. [Google Scholar]
- 8.Choi J W, Sherr B F, Sherr E B. Dead or alive? A large fraction of ETS-inactive marine bacterioplankton cells, as assessed by reduction of CTC, can become ETS-active with incubation and substrate addition. Aquat Microb Ecol. 1999;18:105–115. [Google Scholar]
- 9.Del Giorgio P A, Cole J J, Cimbleris A. Respiration rates in bacteria exceed phytoplankton production in unproductive aquatic systems. Nature. 1997;385:148–151. [Google Scholar]
- 10.Del Giorgio P A, Peters R H. Balance between phytoplankton production and plankton respiration in lakes. Can J Fish Aquat Sci. 1993;50:282–289. [Google Scholar]
- 11.Dufour P, Torreton J P, Colon M. Advantages of distinguishing the active fraction in bacterioplankton assemblages: some examples. Hydrobiologia. 1990;207:295–301. [Google Scholar]
- 12.Gasol J M, Zweifel U L, Peters F, Fuhrman J A, Hagström A. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl Environ Microbiol. 1999;65:4475–4483. doi: 10.1128/aem.65.10.4475-4483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gausing K. Regulation of ribosome production in Escherichia coli: synthesis and stability of ribosomal RNA and of ribosomal protein messenger RNA at different growth rates. J Mol Biol. 1977;115:335–354. doi: 10.1016/0022-2836(77)90158-9. [DOI] [PubMed] [Google Scholar]
- 14.Grimes D J, Colwell R R. Viability and virulence of Escherichia coli suspended by membrane chamber in semitropical ocean water. FEMS Microbiol Lett. 1986;34:161–165. [Google Scholar]
- 15.Haugland R P. Handbook of fluorescent probes and research chemicals. Eugene, Oreg: Molecular Probes, Inc.; 1998. p. 680. [Google Scholar]
- 16.Hobbie J E, Daley R J, Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphreys M J, Allman R, Lloyd D. Determination of the viability of Trichomonas vaginalis using flow cytometry. Cytometry. 1994;15:343–348. doi: 10.1002/cyto.990150410. [DOI] [PubMed] [Google Scholar]
- 18.Jahnke R A, Craven D B. Quantifying the role of heterotrophic bacteria in the carbon cycle: a need for respiration rate measurements. Limnol Oceanogr. 1995;40:436–441. [Google Scholar]
- 19.Jernaes M W, Steen H B. Staining of Escherichia coli for flow cytometry: influx and efflux of ethidium bromide. Cytometry. 1994;17:302–309. doi: 10.1002/cyto.990170405. [DOI] [PubMed] [Google Scholar]
- 20.Jochem F J. Dark survival strategies in marine phytoplankton assessed by cytometric measurement of metabolic activity with fluorescein diacetate. Mar Biol. 1999;135:721–728. [Google Scholar]
- 21.Johnson I. Fluorescent probes for living cells. Histochem J. 1998;30:123–140. doi: 10.1023/a:1003287101868. [DOI] [PubMed] [Google Scholar]
- 22.Jones K H, Senft J A. An improved method to determine cell viability by simultaneous staining with fluorescein diacetate-propidium iodide. J Histochem Cytochem. 1985;33:77–79. doi: 10.1177/33.1.2578146. [DOI] [PubMed] [Google Scholar]
- 23.Joux F, Lebaron P. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microbes Infect. 2000;2:1523–1535. doi: 10.1016/s1286-4579(00)01307-1. [DOI] [PubMed] [Google Scholar]
- 24.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchman D L, Suzuki Y, Garside C, Ducklow H W. High turnover rates of dissolved organic carbon during a spring phytoplankton bloom. Nature. 1991;352:612–614. [Google Scholar]
- 26.Kogure K, Simidu U, Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979;25:415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- 27.Lebaron P, Parthuisot N, Catala P. Comparison of blue nucleic acid dyes for flow cytometric enumeration of bacteria in aquatic systems. Appl Environ Microbiol. 1998;64:1725–1730. doi: 10.1128/aem.64.5.1725-1730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linkous D A, Oliver J D. Pathogenesis of Vibrio vulnificus. FEMS Microbiol Lett. 1999;174:207–214. doi: 10.1111/j.1574-6968.1999.tb13570.x. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd D, Hayes A J. Vigor, vitality, and viability of microorganisms. FEMS Microbiol Lett. 1995;133:1–7. [Google Scholar]
- 30.Lopez-Amoros R, Castel S, Comas-Riu J, Vives-Rego J. Assessment of Escherichia coli and Salmonella viability and starvation by confocal laser microscopy and flow cytometry using rhodamine 123:DiBAC4(3), propidium iodide, and CTC. Cytometry. 1997;29:298–305. doi: 10.1002/(sici)1097-0320(19971201)29:4<298::aid-cyto6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 31.Mason D J, Lopez-Amoros R, Allman R, Stark J M, Lloyd D. The ability of membrane potential dyes and calcafluor white to distinguish between viable and nonviable bacteria. J Appl Bacteriol. 1995;78:309–315. doi: 10.1111/j.1365-2672.1995.tb05031.x. [DOI] [PubMed] [Google Scholar]
- 32.McDougald D, Rice S A, Weichart D, Kjelleberg S. Nonculturability: adaptation or debilitation. FEMS Microbiol Ecol. 1998;25:1–9. [Google Scholar]
- 33.McFeters G A, Yu F P, Pyle B H, Stewart P S. Physiological assessment of bacteria using fluorochromes. J Microbiol Methods. 1995;21:1–13. doi: 10.1016/0167-7012(94)00027-5. [DOI] [PubMed] [Google Scholar]
- 34.Miles A A, Misra S S. The estimation of the bactericidal power of the blood. J Hyg. 1938;38:732–737. doi: 10.1017/s002217240001158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nebe-Von G, Caron, Badley R A. Viability assessment of bacteria in mixed populations using flow cytometry. J Microsc. 1995;179:55–66. [Google Scholar]
- 36.Oliver J D, Bockian R. In vivo resuscitation, and virulence towards mice, of viable but nonculturable cells of Vibrio vulnificus. Appl Environ Microbiol. 1995;61:2620–2623. doi: 10.1128/aem.61.7.2620-2623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Packard T, Denis M, Rodier M, Gardfield P. Deep-ocean metabolic CO2 production calculations from ETS activity. Deep-Sea Res. 1988;35:371–382. [Google Scholar]
- 38.Pomeroy L R. Significance of microorganisms in carbon and energy flow in marine ecosystems. In: Klug M J, Reddy C A, editors. Current perspectives in microbial ecology. Washington, D.C.: American Society for Microbiology; 1984. pp. 405–411. [Google Scholar]
- 39.Porter J, Deere D, Pickup R, Edwards C. Fluorescent probes and flow cytometry: new insights into environmental bacteriology. Cytometry. 1996;23:91–96. doi: 10.1002/(SICI)1097-0320(19960201)23:2<91::AID-CYTO1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 40.Proctor L M, Fuhrman J A. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343:60–62. [Google Scholar]
- 41.Rodriguez G G, Phipps D, Ishiguro K, Ridgway H F. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl Environ Microbiol. 1992;58:1801–1808. doi: 10.1128/aem.58.6.1801-1808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruimy R, Breittmayer V, Boivin V, Christen R. Assessment of the state of activity of individual bacterial cells by hybridization with a ribosomal RNA targeted fluorescently labeled oligonucleotidic probe. FEMS Microbiol Ecol. 1994;15:207–214. [Google Scholar]
- 43.Sgorbati S, Barbesti S, Citterio S, Bestetti G, De Vecchi R. Characterization of number, DNA content, viability and cell size of bacteria from natural environments using DAPI PI dual staining and flow cytometry. Minerva Biotech. 1996;8:9–15. [Google Scholar]
- 44.Sgorbati S, Barbesti S, Neri M G, Davegna C, Citterio S. Rapid flow cytometric immunodetection of bacteria to monitor aquatic environments. Eur J Histochem. 1997;41:187–188. [PubMed] [Google Scholar]
- 45.Shapiro H M. Practical flow cytometry. New York, N.Y: Alan R. Liss, Inc.; 1988. p. 353. [Google Scholar]
- 46.Smith E M. Coherence of microbial respiration rate and cell-specific bacterial activity in a coastal planktonic community. Aquat Microb Ecol. 1998;16:27–35. [Google Scholar]
- 47.Tabor P S, Neihof R A. Improved method for determination of respiring individual microorganisms in natural waters. Appl Environ Microbiol. 1982;43:1249–1255. doi: 10.1128/aem.43.6.1249-1255.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiem S M, Krumme M L, Smith R L, Tiedje J M. Use of molecular techniques to evaluate the survival of a microorganism injected into an aquifer. Appl Environ Microbiol. 1994;60:1059–1067. doi: 10.1128/aem.60.4.1059-1067.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ullrich S, Karrasch B, Hoppe H G. Is the CTC dye technique an adequate approach for estimating active bacterial cells? Aquat Microb Ecol. 1999;17:207–209. [Google Scholar]
- 50.Ullrich S, Karrasch B, Hoppe H G, Jeskulke K, Mehrens M. Toxic effects on bacterial metabolism of the redox dye 5-cyano-2:3-ditolyl tetrazolium chloride. Appl Environ Microbiol. 1996;62:4587–4593. doi: 10.1128/aem.62.12.4587-4593.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Wambeke F. Influence of phytoplankton lysis or grazing on bacterial metabolism and trophic relationships. Microb Ecol. 1994;27:143–158. doi: 10.1007/BF00165814. [DOI] [PubMed] [Google Scholar]
- 52.Whitesides M D, Oliver J D. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. Appl Environ Microbiol. 1997;63:1002–1005. doi: 10.1128/aem.63.3.1002-1005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams S C, Hong Y, Danavall D C A, Howard-Jones M H, Gibson D, Frisher M E, Verity P G. Distinguishing between living and nonliving bacteria: evolution of the vital stain propidium iodide and its combined use with molecular probes in aquatic samples. J Microbiol Methods. 1998;32:225–236. [Google Scholar]
- 54.Zimmerman R, Meyer-Reil L. A new method for fluorescence staining of bacterial populations on membrane filters. Kiel Meeresforsch. 1974;30:24–27. [Google Scholar]
- 55.Zweifel U L, Hagstrom A. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts) Appl Environ Microbiol. 1995;61:2180–2185. doi: 10.1128/aem.61.6.2180-2185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]