Abstract
The number of studies that aims to apply host‐ or microbe‐derived biochemical biomarkers to periodontal disease diagnosis has increased significantly during the last three decades. The biochemical markers can reflect the presence, severity, and activity of periodontal diseases; however, heterogeneities in applied laboratory methods, data presentation, statistical analysis, and data interpretation prevent the translation of candidate host‐ or microbe‐derived biochemical biomarkers to clinical assay validation. Here, we propose a roadmap for making the research outcomes comparable and re‐analysable with the ultimate goal of translating research to clinical practice. This roadmap presents reporting recommendations for host‐ or microbe‐derived biochemical biomarker studies in periodontology. We aim to make essential elements of the research work (including diagnostic criteria, clinical endpoint definitions, participant recruitment criteria, sample collection and storage techniques, biochemical and microbiological detection methods, and applied statistical analysis) visible and comparable.
Keywords: gingivitis, gingival crevicular fluid, periodontal diseases, periodontitis, saliva
Clinical relevance.
Scientific rationale for study: Defining novel host‐ or microbe‐derived biochemical biomarkers for diagnosis and treatment planning is a rising trend in periodontal medicine. However, the heterogeneity in study designs, methods, data presentation, and lack of validation prevent their utilization in clinical practice.
Principal findings: We propose a roadmap for making the research outcomes comparable and re‐analysable with the ultimate goal of translating research to clinical practice.
Practical implications: Implementing the proposed seven‐point biochemical biomarker‐research roadmap into periodontal research will guide the publication of the results, interpretation of the outcomes with wide audiences, and utilization of biomarkers in clinical practice.
1. INTRODUCTION
The primary goal of periodontal treatment is to reduce morbidity and repair function of the dentition and enhance the quality of life by objectively diagnosing an individual's periodontal health condition and well‐being. An accurate periodontal diagnosis requires objective and comprehensive documentation of clinical signs and symptoms, usually obtained by direct clinical measurements. Oral clinical measurements aim to define the function and survival of the dentition and have been used since the dawn of modern dentistry. While clinical measurements are considered the gold standard in dental practice, their abilities to diagnose the periodontal disease at its early phases, prognosticate disease progression, or estimate treatment outcomes have shortcomings. For example, bleeding on probing (BOP), a widely used marker of gingival inflammation, may occur in the absence of the disease (Lang et al., 1991), and its sensitivity, specificity, and frequency are dependent on multiple factors (Karayiannis et al., 1992; Lang & Tonetti, 1996). Baseline pocket depth (PD) levels may have prognostic value in tooth loss (Petsos et al., 2021) or further attachment loss (Claffey et al., 1990); however, there is no information on the use of PD scores in the prediction of attachment gain or regeneration. In addition, the sensitivity of clinical measurements is low. Moreover, the change of symptoms over time may cause diagnostic uncertainties requiring a long‐term and well‐calibrated follow‐up. These limitations of clinical indices can be improved with biomarkers.
Biomarkers are objective and quantifiable determinants of normal biological or pathogenic processes and can be measured accurately and reproducibly (Biomarkers Definitions Working Group, 2001). Biomarkers can be roughly divided into three groups: biomarkers of exposure, biomarkers of effect, and biomarkers of susceptibility. A biomarker of exposure in infectious diseases is usually a pathogen (its presence or increased abundance) or its virulence factor (e.g., lipopolysaccharide, protease). A biomarker of effect is a part of the host response (e.g., proinflammatory cytokines, enzymes, degradation end products) resulting from the exposure to an exogenous agent. Finally, a biomarker of susceptibility defines individual factors that determine the severity of host response to exogenous agents (e.g., gene polymorphisms, gene copy‐number defects) (Chen et al., 2011). Diagnostic biomarkers guide physicians in discriminating the group of diseased individuals from the healthy ones, whereas monitoring and prognostic biomarkers improve their treatment planning towards personalized medicine and provide essential data for risk of disease recurrence during the maintenance phase of treatment (McLeod et al., 2019).
Periodontal diseases present unique advantages and challenges in searching for host‐ or microbe‐derived biochemical biomarkers. Periodontitis is a chronic inflammatory disease of the tooth‐supporting tissues with an infectious origin and degenerative pathogenesis. Degradation of periodontal tissues is mainly the outcome of a biological process regulated by uncontrolled cytokine/chemokine expression and enzyme activation (Kurgan & Kantarci, 2018). Diagnosis of periodontal diseases requires the use of clinical measurements (BOP, PD, and clinical attachment loss) and radiographic determinations (alveolar bone loss). These clinical indices have been used widely in dental clinics for decades. Indeed, they are now presented as part of the American Academy of Periodontology and the European Federation of Periodontology guidelines in classifying periodontal health and disease (Caton et al., 2018). However, the clinical and radiographic indices indicate a disease only after it occurs and have limited prognostic capacity.
Diagnosing periodontal health and determining the disease stages with non‐invasive and accurate methods has been a long‐term goal in periodontal research and clinical practice. Technological advancements during past decades have expanded the search for biochemical biomarkers of periodontal diseases. Various cytokines or enzymes (gingipains of Porphyromonas gingivalis, interleukin‐1β, and matrix metalloproteinase‐8) in saliva and the gingival crevicular fluid (GCF) are now available as candidate biomarkers of periodontal disease initiation and progression (U. K. Gürsoy et al., 2022). As a circulating fluid, the non‐invasively collected saliva contains biomarkers that are useful to discriminate the periodontitis cases from periodontally healthy controls in a population, diagnose the initiation, healing, and recurrence of periodontitis at an individual level (personalized medicine), classify the extension of periodontal disease, identify oral infection in a non‐dental setting, and provide information about the overall health of the patient (Arias‐Bujanda et al., 2019). As an exudate, GCF reflects changes in the periodontal status at the site level, which can be used to predict post‐treatment healing response and monitor periodontal regeneration (Arias‐Bujanda et al., 2020).
The recent advancements have also significantly decreased costs in –omics and other high‐throughput technologies, which produce open‐ended data, and boosted the number of host‐ or microbe‐derived biochemical biomarker research publications. Thus, the list of potential biochemical biomarkers for assessing the disease processes is growing. On the other hand, systematic reviews and meta‐analyses of diagnostic markers of periodontal diseases have repeatedly demonstrated limited evidence of the validity of current analytes as reliable host‐ or microbe‐derived biochemical biomarkers (Arias‐Bujanda et al., 2019, 2020; Ghassib et al., 2019), suggesting several issues in design, execution, interpretation, validation, and translation of candidate biochemical markers.
2. BIOCHEMICAL BIOMARKERS OF PERIODONTAL DISEASES: FROM DISCOVERY TO CHAIRSIDE
Bringing a host‐ or microbe‐derived biochemical biomarker from bench to chairside requires four main steps: (1) descriptive (including hypothesis‐free –omics and high‐throughput methods for discovery) and mechanistic studies defining biochemical biomarker candidates and their roles in disease; (2) validating the power of the biochemical biomarker candidate in independent populations; (3) assay development for fast and reliable detection of the target of interest; and (4) clinical validation of the assay (Pavloum et al., 2013). The lack of confirmative or validating evidence in periodontal biomarker research has many reasons, including heterogeneities in diagnosis criteria, clinical endpoint definitions, participant recruitment criteria, sample collection, storage techniques, biochemical and microbiological detection methods, and applied statistical analyses. The heterogeneities in applied methods prevent the inclusion of published reports into the meta‐analyses. Ignorance of the importance of high‐quality reporting by dental researchers is also an important handicap. The lack of comparable methodologies, lack of transformation of population‐based data to the single individual level, and increased poor reporting delay the development of chairside assays and their use in clinical and non‐clinical settings.
3. GUIDELINE FOR BIOCHEMICAL BIOMARKER STUDIES IN PERIODONTAL MEDICINE
Here we propose a seven‐point roadmap to serve as the foundation for making the research outcomes comparable and re‐analysable with the ultimate goal of translating research to clinical practice. These seven criteria are shown as a checklist in Table 1. This checklist will function as a roadmap (Figure 1) to guide and encourage researchers to implement the essentials of host‐ or microbe‐derived biochemical biomarker research into the study design and serve as a guide to report the crucial features of their research work. A similar trend exists in other medical fields where reporting recommendations for biochemical biomarker studies are being published (Sauerbrei et al., 2018).
TABLE 1.
Checklist for periodontal biochemical biomarker research
| Check points | Description | Page number/lines |
|---|---|---|
| Biology | Biological specificity of tested analyte and sample material | |
| Clinical definitions | Definitions of primary outcome variables (disease and health) | |
| Recruitment | Recruitment cohort | |
| Sampling | Sample storage and handling | |
| Freeze–thaw cycles | ||
| Analyse |
Description of applied method (incl. Catalog number company) Use of internal controls and number of replicates LOD, percentage of samples (for each study group separately) under LOD values and substitution methods |
|
| Statistics | Cut‐off, sensitivity, and specificity values | |
| Interpretation | Implementation of the confounders into statistical analyses (univariable and multivariable analyses) |
Abbreviations: LOD, limit of detection; Catalog number.
FIGURE 1.
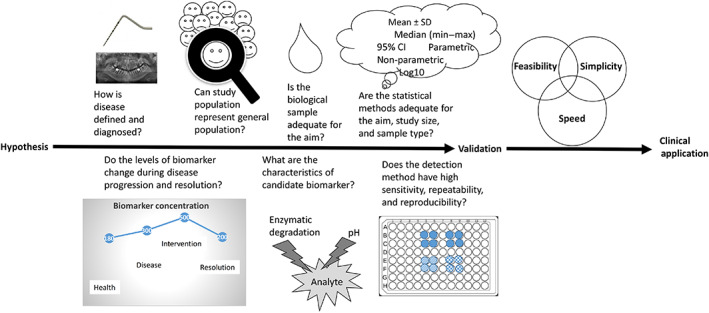
A road map from hypothesis to biomarker validation, assay development, and clinical application with critical questions [Colour figure can be viewed at wileyonlinelibrary.com]
The seven‐point roadmap is explained below. Genetic susceptibility markers, which aim to predict the risk of disease onset, are not included in this work, as evidence on the application of such surrogates are scarce. Indeed, the dynamics of the genetic and biochemical biomarkers vary in body fluids, and their translational value in prediction, diagnosis, and prognosis differs. Nevertheless, predicting the occurrence of periodontitis by detecting the genetic variations has been in the interest of various research groups for the last decades. Indeed, genome‐wide association studies have produced some evidence that periodontitis is associated with susceptibility alleles (Schaefer, 2018). However, these studies have been of limited predictive value. The main reason behind this outcome is the interplay between the internal and external risk factors where genetic background acts as one of the determinants of periodontal disease initiation and development not being due to an individual risk allele alone (Nibali et al., 2017). For the same reason, commercialized genetic tests have failed to bring generalized and additional benefit to the early diagnosis of periodontitis or the prognosis of treatment outcomes.
Likewise, the definition of microbiological biomarkers is limited to microbe‐derived biochemical markers found in periodontal and oral fluids and does not include species‐level analyses of microbial communities of the oral microbiome.
3.1. Definition of health and disease
A universally accepted definition of disease and health need to be implemented. Associating the host‐ or microbe‐derived biochemical biomarker levels solely with clinical indices (i.e., PD, BOP%) without a clear diagnosis or applying self‐defined disease definitions (PD > 4 mm at more than one site) has been commonly done in biomarker research. However, these approaches have limited the comparability of outcomes with other studies. In line with the most recent classification of periodontal diseases (Papapanou et al., 2018), the definitions for disease versus health need to be universally adopted. Considering that the current classification may evolve with time and that population‐based studies may require different disease definition criteria than observational analytical studies, presenting clinical parameters (PD, clinical attachment level, and BOP%) of the study groups based on the standardized case definitions will be beneficial (Holtfreter et al., 2015).
3.2. Variation or change of the candidate biochemical biomarker between health and disease
Host‐ or microbe‐derived biochemical biomarkers are used to detect pathogenic processes and follow the response to a therapeutic or medical intervention. The change in concentrations of a candidate biochemical biomarker during the development of the disease should return to the levels associated with normal (healthy) values after periodontal treatment. Therefore, implementing an intervention to the study design is critical in demonstrating the post‐treatment response of host‐ or microbe‐derived biochemical biomarker levels. Universally accepted endpoints of periodontal therapy (shallow periodontal pockets [≤4 mm] that do not bleed on probing in patients with full‐mouth bleeding scores <30%) are crucial in creating comparable data (Loos & Needleman, 2020).
3.3. Biological characteristics of a candidate host‐ or microbe‐derived biochemical biomarker
The severity, prevalence, and treatment response of periodontal diseases are affected by local (e.g., smoking) and systemic (e.g., systemic diseases and conditions) factors. The same factors also affect the concentrations of biochemical biomarker candidates (e.g., interleukin‐1β, lactate dehydrogenase, matrix metalloproteinase‐8) either directly or indirectly (U. K. Gürsoy et al., 2009; M. Gürsoy et al., 2010). The extracellular release of the analyte, its enzymatic degradation by the host or bacterial proteases, and its half‐life in the sample medium will significantly affect the study outcomes. Therefore, the analyte's cellular source, its role in the pathogenesis of periodontal diseases, its relation to periodontal risk factors and exposure profiles, and its stability in samples need to be carefully determined during the study planning and data interpretation.
3.4. Participant recruitment
Characteristics of the study participants will determine the outcomes. For example, the recruitment of study groups from different sources, which may actually reflect a more homogenous cohort not necessarily representative of the general population (e.g., periodontitis patients from dental clinics and periodontally healthy controls from dental students), will produce significant variations in group characteristics (Sedgwick, 2015). Descriptive characteristics of the study population (e.g., socioeconomic status, gender distribution, age, smoking, diabetes, body mass index) need to be presented in a baseline table to bring confounding and nuisance variables to the front. Recruitment bias, which may directly (e.g., age) or indirectly (e.g., educational level) affect the study outcomes, needs to be addressed in the interpretation and discussion. A rationale for sample size needs to be defined. The flow of patients and the reasons for drop‐outs should be given in a flow chart for clarity. Indeed, authors need to follow validated checklists (i.e., STROBE) or guidelines (Holtfreter et al., 2015).
3.5. Sample handling and sample‐specific characteristics
Sample type (e.g., resting vs. flow GCF), collection method (e.g., stimulated vs. unstimulated saliva), and storage conditions (e.g., −20 vs. –70°C) are usually presented in all biochemical biomarker studies. However, other important factors such as repeated freeze–thaw cycles or biological functions of applied enzyme inhibitor cocktails, which substantially affect sample protein concentrations and activations, are rarely discussed. Biological fluids have different matrices (Chiu et al., 2010). Sample dilutions may lead to unpredicted variations in antigen–antibody interactions, which may be analyte‐ and matrix‐specific and thus induce non‐linear changes in protein concentrations (Browne et al., 2013). If sample dilution was applied, dilution factor and dilution buffer need to be clearly given, and their possible effects must be discussed. Researchers are highly advised to follow the guidelines and recommendations for clinical biomarker preservation and assessment (Vaught & Henderson, 2011; Dakappagari et al., 2017). Finally, the selected sample material must fit with the intended purpose of the study.
3.6. Detection methodology
Advanced laboratory detection methods for host‐ or microbe‐derived biochemical biomarkers have been developed over the last decades. However, comparative analyses of methods are limited. For example, antibody‐ or bead‐based assays that are widely used rely on standard curves with different mathematical formulae and antibody‐specific performances. In addition, antibody‐based platforms present strong batch effects. Proteomics also has a strong batch effect, which introduces noise that reduces the statistical power and decreases the validity of the conclusions (Čuklina et al., 2021). Therefore, a direct comparison of the outcomes is usually inaccurate. Furthermore, depending on the assay of choice, the concentrations of target analytes may be left under the limit of detection (LOD), especially in periodontally healthy groups. Thus, LOD levels, the percentage of samples left under LOD levels, and the missing data substitution methods need to be presented (Whitcomb & Schisterman, 2008).
3.7. Statistics and data interpretation
Applied statistical methods must be specified for each variable clearly. Rather than simply reporting an observed difference by stating “a statistical significance was observed (p < .05)”, actual p‐values for each comparison need to be presented. Statistical difference between disease and control groups in the concentration of biomarker candidates is not enough to define the tested marker as a biomarker. A cut‐off must be defined to produce comparable thresholds for disease and health. Indeed, by presenting the sensitivity and specificity values of the given cut‐off value, the effect of covariates (age, systemic diseases, and smoking) on the diagnostic power of biomarkers will become visible (Subtil & Rabilloud, 2014). A common practice is to define subgroups during the statistical analysis. At the same time, such subgroups have usually been not a part of the original study design and not considered during the power analysis. As a result, poorly defined and performed subgroup analyses may lead to false positives or false negatives due to inadequate power (Burke et al., 2015).
4. SUMMARY
The seven criteria and the checklist we presented are intended for host‐ or microbe‐derived biochemical biomarkers of diagnosis and prognosis. Here, our aim was not to evaluate the advantages and the disadvantages of site‐specific or patient‐specific biomarkers over each other. Site‐ and patient‐specific biomarkers and their contribution to precision medicine have been discussed elsewhere (Steigmann et al., 2020). Periodontology has been a pioneer in biomarker research in dental medicine. As a result, there has been an overflow in the number of published articles on host‐ or microbe‐derived biochemical biomarker research, which has presented a wealth of information on the biological mechanisms underlying health and disease in periodontal tissue responses. It is time to translate the associations into clinical use. We proposed implementing a seven‐point biochemical biomarker‐related roadmap into periodontal biomarker research to guide publication, interpretation, and utilization in clinical practice. Utilizing the proposed roadmap into periodontal biomarker research will improve data quality, validation, reproducibility, and, most importantly, help researchers utilize their research outcomes in clinical practice.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Ulvi Kahraman Gürsoy and Alpdogan Kantarci contributed to electronic and hand search, performed the data extraction and interpretation, and performed drafting and critically revising the manuscript.
Gürsoy, U. K. , & Kantarci, A. (2022). Molecular biomarker research in periodontology: A roadmap for translation of science to clinical assay validation. Journal of Clinical Periodontology, 49(6), 556–561. 10.1111/jcpe.13617
Contributor Information
Ulvi Kahraman Gürsoy, Email: ulvi.gursoy@utu.fi.
Alpdogan Kantarci, Email: AKantarci@forsyth.org.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article, as no datasets were generated or analysed during the current study.
REFERENCES
- Arias‐Bujanda, N. , Regueira‐Iglesias, A. , Balsa‐Castro, C. , Nibali, L. , Donos, N. , & Tomás, I. (2019). Accuracy of single molecular biomarkers in gingival crevicular fluid for the diagnosis of periodontitis: A systematic review and meta‐analysis. Journal of Clinical Periodontology, 46, 1166–1182. [DOI] [PubMed] [Google Scholar]
- Arias‐Bujanda, N. , Regueira‐Iglesias, A. , Balsa‐Castro, C. , Nibali, L. , Donos, N. , & Tomás, I. (2020). Accuracy of single molecular biomarkers in saliva for the diagnosis of periodontitis: A systematic review and meta‐analysis. Journal of Clinical Periodontology, 47, 2–18. [DOI] [PubMed] [Google Scholar]
- Biomarkers Definitions Working Group . (2001). Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clinical Pharmacology & Therapeutics, 69, 89–95. [DOI] [PubMed] [Google Scholar]
- Browne, R. W. , Kantarci, A. , LaMonte, M. J. , Andrews, C. A. , Hovey, K. M. , Falkner, K. L. , Cekici, A. , Stephens, D. , Genco, R. J. , Scannapieco, F. A. , Van Dyke, T. E. , & Wactawski‐Wende, J. (2013). Performance of multiplex cytokine assays in serum and saliva among community‐dwelling postmenopausal women. PLoS One, 8, e59498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J. F. , Sussman, J. B. , Kent, D. M. , & Hayward, R. A. (2015). Three simple rules to ensure reasonably credible subgroup analyses. BMJ, 4, h5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton, J. G. , Armitage, G. , Berglundh, T. , Chapple, I. L. C. , Jepsen, S. , Kornman, K. S. , Mealey, B. L. , Papapanou, P. N. , Sanz, M. , & Tonetti, M. S. (2018). A new classification scheme for periodontal and peri‐implant diseases and conditions ‐ Introduction and key changes from the 1999 classification. Journal of Clinical Periodontology, 45, S1–S8. [DOI] [PubMed] [Google Scholar]
- Chen, X. H. , Huang, S. , & Kerr, D. (2011). Biomarkers in clinical medicine. IARC Scientific Publications, 163, 303–322. [PubMed] [Google Scholar]
- Chiu, M. L. , Lawi, W. , Snyder, S. T. , Wong, P. K. , Liao, J. C. , & Gau, V. (2010). Matrix effects—A challenge toward automation of molecular analysis. JALA: Journal of the Association for Laboratory Automation, 15, 233–242. [Google Scholar]
- Claffey, N. , Nylund, K. , Kiger, R. , Garrett, S. , & Egelberg, J. (1990). Diagnostic predictability of scores of plaque, bleeding, suppuration and probing depth for probing attachment loss. 3 1/2 years of observation following initial periodontal therapy. Journal of Clinical Periodontology, 17, 108–114. [DOI] [PubMed] [Google Scholar]
- Čuklina, J. , Lee, C. H. , Williams, E. G. , Sajic, T. , Collins, B. C. , Rodríguez Martínez, M. , Sharma, V. S. , Wendt, F. , Goetze, S. , Keele, G. R. , Wollscheid, B. , Aebersold, R. , & Pedrioli, P. G. A. (2021). Diagnostics and correction of batch effects in large‐scale proteomic studies: A tutorial. Molecular Systems Biology, 17, e10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakappagari, N. , Zhang, H. , Stephen, L. , Amaravadi, L. , & Khan, M. U. (2017). Recommendations for clinical biomarker specimen preservation and stability assessments. Bioanalysis, 9, 643–653. [DOI] [PubMed] [Google Scholar]
- Ghassib, I. , Chen, Z. , Zhu, J. , & Wang, H. L. (2019). Use of IL‐1 beta, IL‐6, TNF‐alpha, and MMP‐8 biomarkers to distinguish peri‐implant diseases: A systematic review and meta‐analysis. Clinical Implant Dentistry and Related Research, 21, 190–207. [DOI] [PubMed] [Google Scholar]
- Gürsoy, M. , Könönen, E. , Tervahartiala, T. , Gürsoy, U. K. , Pajukanta, R. , & Sorsa, T. (2010). Longitudinal study of salivary proteinases during pregnancy and postpartum. Journal of Periodontal Research, 45, 496–503. [DOI] [PubMed] [Google Scholar]
- Gürsoy, U. K. , Gursoy, M. , & Kononen, E. (2022). Biomarkers and periodontal regenerative approaches. Dental Clinics of North America, 66, 157–167. [DOI] [PubMed] [Google Scholar]
- Gürsoy, U. K. , Könönen, E. , Uitto, V. J. , Pussinen, P. J. , Hyvärinen, K. , Suominen‐Taipale, L. , & Knuuttila, M. (2009). Salivary interleukin‐1beta concentration and the presence of multiple pathogens in periodontitis. Journal of Clinical Periodontology, 36, 922–927. [DOI] [PubMed] [Google Scholar]
- Holtfreter, B. , Albandar, J. M. , Dietrich, T. , Dye, B. A. , Eaton, K. A. , Eke, P. I. , Papapanou, P. N. , Kocher, T. , & Joint EU/USA Periodontal Epidemiology Working Group . (2015). Standards for reporting chronic periodontitis prevalence and severity in epidemiologic studies: Proposed standards from the Joint EU/USA Periodontal Epidemiology Working Group. Journal of Clinical Periodontology, 42, 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiannis, A. , Lang, N. P. , Joss, A. , & Nyman, S. (1992). Bleeding on probing as it relates to probing pressure and gingival health in patients with a reduced but healthy periodontium. A clinical study. Journal of Clinical Periodontology, 19, 471–475. [DOI] [PubMed] [Google Scholar]
- Kurgan, S. , & Kantarci, A. (2018). Molecular basis for immunohistochemical and inflammatory changes during progression of gingivitis to periodontitis. Periodontology 2000, 76, 51–67. [DOI] [PubMed] [Google Scholar]
- Lang, N. P. , Nyman, S. , Senn, C. , & Joss, A. (1991). Bleeding on probing as it relates to probing pressure and gingival health. Journal of Clinical Periodontology, 18, 257–261. [DOI] [PubMed] [Google Scholar]
- Lang, N. P. , & Tonetti, M. S. (1996). Periodontal diagnosis in treated periodontitis. Why, when and how to use clinical parameters. Journal of Clinical Periodontology, 23, 240–250. [DOI] [PubMed] [Google Scholar]
- Loos, B. G. , & Needleman, I. (2020). Endpoints of active periodontal therapy. Journal of Clinical Periodontology, 47, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod, C. , Norman, R. , Litton, E. , Saville, B. R. , Webb, S. , & Snelling, T. L. (2019). Choosing primary endpoints for clinical trials of health care interventions. Contemporary Clinical Trials Communications, 12, 100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibali, L. , Di Iorio, A. , Tu, Y. K. , & Vieira, A. R. (2017). Host genetics role in the pathogenesis of periodontal disease and caries. Journal of Clinical Periodontology, 44(Suppl. 18), S52–S78. [DOI] [PubMed] [Google Scholar]
- Papapanou, P. N. , Sanz, M. , Buduneli, N. , Dietrich, T. , Feres, M. , Fine, D. H. , Flemmig, T. F. , Garcia, R. , Giannobile, W. V. , Graziani, F. , Greenwell, H. , Herrera, D. , Kao, R. T. , Kebschull, M. , Kinane, D. F. , Kirkwood, K. L. , Kocher, T. , Kornman, K. S. , Kumar, P. S. , … Tonetti, M. S. (2018). Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri‐Implant Diseases and Conditions. Journal of Clinical Periodontology, 45, 162–S170. [DOI] [PubMed] [Google Scholar]
- Pavloum, M. P. , Diamandis, E. P. , & Blasutig, I. M. (2013). The long journey of cancer biomarkers from the bench to the clinic. Clinical Chemistry, 59, 147–157. [DOI] [PubMed] [Google Scholar]
- Petsos, H. , Ramich, T. , Nickles, K. , Dannewitz, B. , Pfeifer, L. , Zuhr, O. , & Eickholz, P. (2021). Tooth loss in periodontally compromised patients: Retrospective long‐term results 10 years after active periodontal therapy ‐ tooth‐related outcomes. Journal of Periodontology, 92, 1761–1775. [DOI] [PubMed] [Google Scholar]
- Sauerbrei, W. , Taube, S. E. , McShane, L. M. , Cavenagh, M. M. , & Altman, D. G. (2018). Reporting recommendations for tumor marker prognostic studies (REMARK): An abridged explanation and elaboration. Journal of the National Cancer Institute, 110, 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, A. S. (2018). Genetics of periodontitis: Discovery, biology, and clinical impact. Periodontology 2000, 78, 162–173. [DOI] [PubMed] [Google Scholar]
- Sedgwick, P. (2015). Bias in observational study designs: Case‐control studies. BMJ, 30, h560. [DOI] [PubMed] [Google Scholar]
- Steigmann, L. , Maekawa, S. , Sima, C. , Travan, S. , Wang, C. W. , & Giannobile, W. V. (2020). Biosensor and lab‐on‐a‐chip biomarker‐identifying technologies for oral and periodontal diseases. Frontiers in Pharmacology, 9, 588480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil, F. , & Rabilloud, M. (2014). Estimating the optimal threshold for a diagnostic biomarker in case of complex biomarker distributions. BMC Medical Informatics and Decision Making, 14, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaught, J. B. , & Henderson, M. K. (2011). Biological sample collection, processing, storage and information management. IARC Scientific Publications, 1, 23–42. [PubMed] [Google Scholar]
- Whitcomb, B. W. , & Schisterman, E. F. (2008). Assays with lower detection limits: Implications for epidemiological investigations. Paediatric and Perinatal Epidemiology, 22, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article, as no datasets were generated or analysed during the current study.


