Abstract
There has been considerable recent interest in the life cycle of Severe Acute Respiratory Syndrome Coronavirus‐2 (SARS‐CoV‐2), the causative agent of the Covid‐19 pandemic. Practically every step in CoV replication—from cell attachment and uptake via genome replication and expression to virion assembly has been considered as a specific event that potentially could be targeted by existing or novel drugs. Interference with cellular egress of progeny viruses could also be adopted as a possible therapeutic strategy; however, the situation is complicated by the fact that there is no broad consensus on how CoVs find their way out of their host cells. The viral nucleocapsid, consisting of the genomic RNA complexed with nucleocapsid proteins obtains a membrane envelope during virus budding into the lumen of the intermediate compartment (IC) at the endoplasmic reticulum (ER)–Golgi interface. From here, several alternative routes for CoV extracellular release have been proposed. Strikingly, recent studies have shown that CoV infection leads to the disassembly of the Golgi ribbon and the mobilization of host cell compartments and protein machineries that are known to promote Golgi‐independent trafficking to the cell surface. Here, we discuss the life cycle of CoVs with a special focus on different possible pathways for virus egress.
Keywords: rab1, rab11, the Golgi apparatus, the intermediate compartment (IC), the recycling endosome (RE)
A number of egress routes have been suggested for corona virions from the pre‐Golgi intermediate compartment (IC), where the virus capsids obtain their membrane envelope, in direction of the cell surface. We here discuss coronavirus egress routes in relation to the observed changes that occur to host cell intracellular compartments during coronavirus infection.
1. INTRODUCTION
Effective egress of mature virus particles from host cells is one of the important determinants of virus infectivity. While some viruses are released after mediating lysis of the infected cells, others acquire a membrane envelope by budding at the host cell plasma membrane (PM). Yet other viruses, like coronaviruses (CoVs), obtain their envelope as they bud into a membrane compartment inside the cell, and must therefore be transported from their site of assembly to the cell surface to be able to reach the extracellular milieu via exocytosis (Hernandez‐Gonzalez et al., 2021; Sturman & Holmes, 1983). The progeny CoVs assemble by budding into the lumen of the intermediate compartment (IC) (Klumperman et al., 1994; Stertz et al., 2007; Tooze et al., 1984), functionally situated between the endoplasmic reticulum (ER) and the Golgi apparatus in the early secretory pathway (Saraste & Kuismanen, 1984; Saraste & Marie, 2018). However, the IC also turns out to communicate directly with endocytic compartments (Saraste & Prydz, 2019), opening for unconventional modes of egress that bypass the Golgi stacks participating in the conventional secretory process (Ghosh et al., 2020; Saraste & Prydz, 2021). Assembly at the IC membranes is a common property of CoVs belonging to different genera (α‐, β‐, and γ‐CoVs), including the Severe Acute Respiratory Syndrome (SARS)‐CoV causing the serious outbreak in 2002 (SARS‐CoV) and the recent pandemic (SARS‐CoV‐2) in humans (Bracquemond & Muriaux, 2021; Stertz et al., 2007). In the assembly process, the virus nucleocapsid—consisting of the positive‐stranded genomic RNA complexed with nucleocapsid (N) proteins—is enclosed in a lipid bilayer derived from a subdomain of the IC membrane, that incorporates the viral membrane glycoproteins designated as S (spike), M (membrane), and E (envelope) into the forming virions (Figure 1) (Scherer et al., 2022).
FIGURE 1.
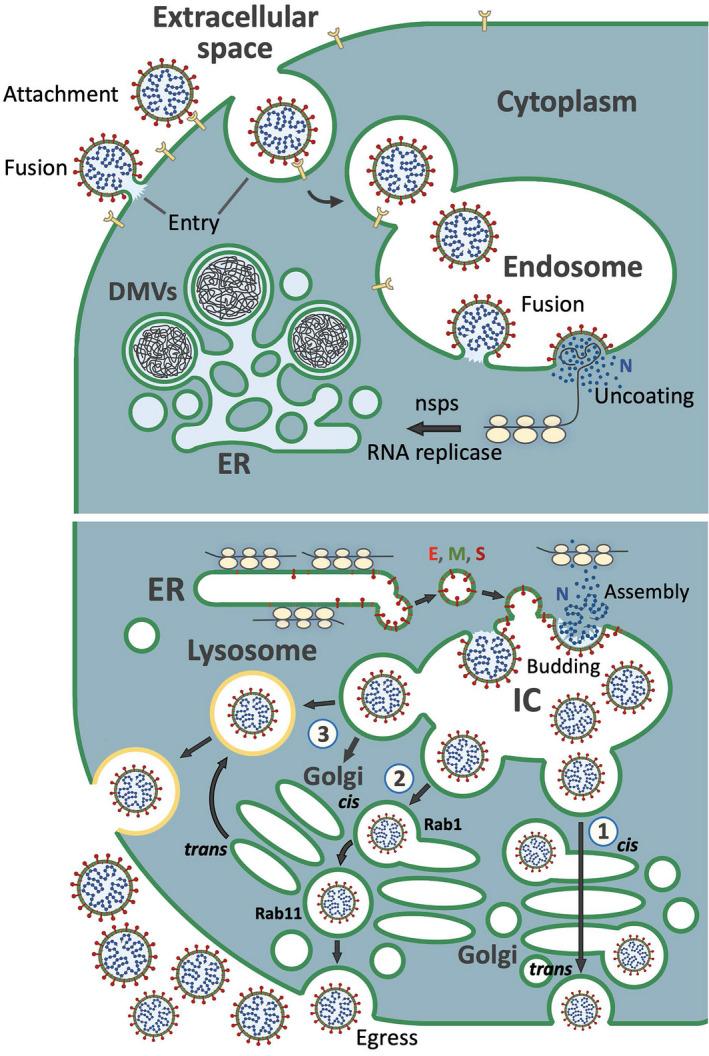
A cartoon illustrating early and late stages of the CoV life cycle. Upper panel: Following attachment to specific receptor(s)—such as ACE2—CoV enters cells either by fusing directly with the cell surface, or following its uptake into endosomes, where the viral envelope fuses with the endosomal membrane. In both cases, the viral nucleocapsid enters the cytoplasm and undergoes uncoating, resulting in the release of the viral RNA genome. The positive‐sense RNA associates with host cell ribosomes directing the synthesis of non‐structural proteins (nsps), which provide subunits of the viral RNA replicase or act in the biogenesis of an ER‐derived convoluted membrane compartment, which includes double‐membrane vesicles (DMVs)—the sites for viral RNA replication and transcription. Lower panel: Sub‐genomic mRNAs produced in the DMVs function in the synthesis of the viral structural proteins—the cytoplasmic nucleocapsid (N) protein and three membrane proteins (E, M, and S)—in free‐ or ER membrane‐bound ribosomes, respectively. Vesicle‐mediated transport and accumulation of the membrane proteins at the IC membranes sets, the stage for virus assembly by budding into the IC lumen. Three alternative pathways for CoV delivery from the IC to the extracellular space are depicted: Route 1) the progeny viruses highjack the constitutive secretory pathway as they segregate into the dilated rims of Golgi cisternae and pass across the Golgi stacks (cis‐to‐trans) based on cisternal progression. At trans‐Golgi, the viruses are sorted into post‐Golgi carriers which move to the PM and undergo exocytosis. Route 2) this pathway bypassing the Golgi stacks is based on a direct connection between the IC elements and REs, defined by Rab1 and Rab11, respectively. Prior to Golgi fragmentation, these compartments reside at the non‐compact zones of the Golgi ribbon, connecting the different Golgi stacks. In this case, the endocytic recycling system provides the carriers for the final delivery of the virus for exocytosis. Route 3) the progeny viruses are released from cells via lysosomal exocytosis. They may reach the lysosomes via trans‐Golgi; for example, following route 1, or employ a direct IC‐to‐lysosome pathway, which remains to be identified. for simplicity, only one CoV particle in the lumen of the carriers is shown, although many of them contain numerous viruses
For a long time, viral envelope glycoproteins have provided important tools for studies of intracellular transport of membrane proteins between the ER and the PM (Bergmann et al., 1981; Saraste & Kuismanen, 1984). Figuring out the transport routes for virus particles from their intracellular site of assembly to the site of release via exocytosis can be more challenging than studying conventional glycoprotein transport to the cell surface. Namely, the large size of intracellularly budding virus particles—for example, CoVs range from 80 to 120 nm in diameter—means that they have to be packaged into specialized transport carriers, which currently remain poorly characterized. Another reason is that virus infection typically leads to alterations in the structure and function of endomembrane compartments, resulting in the redistribution of traditional organelle markers. Indeed, viruses seem to have developed the ability to inhibit or reorganize secretory transport routes in a manner that is in their best interest.
In this Micro Review, we focus on the structural and functional changes occurring in the host cells during CoV infection and discuss how these changes may influence the mode of egress of newly assembled virus particles. Since the different stages of the virus life cycle are closely interconnected, we also briefly address earlier steps of virus infection.
2. VIRUS ENTRY AND EGRESS: GLYCAN BINDING AT THE RIGHT PLACE?
To be able to enter their host cells and release their genome to the cytoplasm, CoVs must first bind to transmembrane protein receptors at the cell surface. Subsequently, the viral membrane can either fuse directly with the host cell PM, or the virus is endocytosed and releases its genome from an endosomal compartment (Figure 1) (Fung & Liu, 2019; Jackson et al., 2022). The attachment and entry steps of the virus particles are mediated by the trimeric S glycoprotein. Direct fusion with the PM requires that the spike protein has been primed by the proprotein convertase furin during virus egress from a producer cell, and that the new host cell expresses the TMPRSS2 protease at its surface that introduces a second proteolytic cleavage. Alternatively, the two successive cleavages of the S protein may be created by proteases (cathepsins) after the virus has entered the lumen of the endosome (Hoffmann et al., 2020; Millet & Whittaker, 2015; Zhang & Zhang, 2021). For SARS‐CoV and SARS‐CoV‐2, angiotensin‐converting enzyme 2 (ACE2) has been shown to act as an obligate receptor for host cell entry, while for the Middle East Respiratory Syndrome (MERS)‐CoV the reported receptor is dipeptidyl peptidase IV (DDP4). In addition, several CoVs bind to the glycosaminoglycan (GAG) chains of heparan sulfate (HS) proteoglycans (de Haan et al., 2005; Milewska et al., 2014). SARS‐CoV‐2 depends both on ACE2 and HS for efficient S protein interaction with the host cell (Clausen et al., 2021), while HCoV‐NL63—another human CoV—seems to interact with HS via the M protein (Naskalska et al., 2019). It is important to note that the pattern of HS sulfation, which determines the biological specificity of the GAG chains, changes with age, which could significantly affect the susceptibility of different age groups to CoV infection (Feyzi et al., 1998; Kreuger et al., 1999). The diversity of cell surface glycan structures in a population is beneficial in an evolutionary perspective to ensure that certain individuals survive severe threats from disease‐causing microorganisms (Varki, 2011). The extent of variation in HS structure among individuals is not known in detail, and will also vary in different tissues. Studies of the receptor‐binding domain (RBD) of the S protein using glycan arrays and ACE2‐positive HEK cells demonstrated its additional affinity for sialic acid, preferentially in the context of mono‐sialylated gangliosides. The affinity was similar to that observed for binding to HS, and reduced levels of cell surface sialic acid were inhibitory to virus attachment and cell entry (Nguyen et al., 2022).
Many viruses depend on glycans as receptors or co‐receptors for efficient binding to and entry into their host cells (Aquino & Park, 2016; Russell et al., 2006). Since the same glycans are synthesized and modified in the secretory pathway, progeny viruses undergoing egress must either avoid binding to the glycan receptors, or be able to detach from the bound glycan at the cell surface by an appropriate enzymatic activity. Influenza viruses bind to variants of sialic acid both during entry into and egress from their host cells, and utilize the activity of viral neuraminidase to promote the release of newly synthesized virions from cell surface glycans (McAuley et al., 2019). Enzymatic release of SARS‐CoV‐2 from host cell HS GAGs has not been demonstrated, but has been described for instance in the case of Herpes simplex virus 1, which is released from the cell surface by heparanase degradation of HS GAGs attached to syndecan‐1 (de Pasquale et al., 2021; Hadigal et al., 2020). As discussed below, CoV infection causes disassembly of the Golgi apparatus (Cortese et al., 2020; Hackstadt et al., 2021; Lavi et al., 1996; Ruch & Machamer, 2012; Ulasli et al., 2010), where HS synthesis normally takes place (Prydz & Dalen, 2000). How CoV‐mediated Golgi disassembly influences the biosynthesis and transport of HS is not known in detail, but Golgi fragmentation is caused by the depletion of two Golgi‐associated peripheral membrane proteins, GRASP55 and GRASP65, leads to a reduction in HS synthesis (Ahat et al., 2022). In the early phase of infection, HS chains are most likely still normally synthesized, but the SARS‐CoV‐2 virions may prefer egress route(s) where the S protein avoids encountering and binding to the newly synthesized glycosaminoglycans.
3. VIRUS RNA REPLICATION AND PROTEIN SYNTHESIS
Virus‐infected cells rearrange their endomembranes to establish viral factories, where the viral genome is replicated and transcribed (Blanchard & Roingeard, 2015; Hernandez‐Gonzalez et al., 2021; Miller & Krijnse‐Locker, 2008; Sachse et al., 2019; Snijder et al., 2020; Wong et al., 2021). CoV infection leads to the formation of double‐membrane vesicles (DMVs) that are continuous with ER‐derived convoluted membranes (Figure 1) (Cortese et al., 2020; Eymieux, Rouillé, et al., 2021; Fehr & Perlman, 2015; Klein et al., 2020; Knoops et al., 2008; Mendonca et al., 2021; Stertz et al., 2007; Wong et al., 2021) and may develop into structures called vesicle packages. These membrane‐enclosed environments are thought to protect the viral RNA from recognition by host cell innate immunity mechanisms, thus providing safe havens for viral RNA replication (Malone et al., 2022; Sachse et al., 2019). Interestingly, there appears to be a close connection between the DMVs and the IC subdomains where CoV assembly takes place (Cortese et al., 2020; Mendonca et al., 2021; Scherer et al., 2022); however, how the genomic RNAs actually reach the sites of assembly remains poorly understood.
Interestingly, the biogenesis of the DMVs engages the machineries operating in autophagy (Blanchard & Roingeard, 2015; Twu et al., 2021), with possible additional contribution from peroxisomes (Cortese et al., 2020). The autophagic pathway has also been implicated as an egress route for SARS‐CoV‐2 virus particles in light of ubiquitination of the M protein (Yuan et al., 2022). Following replication and transcription of viral RNAs, the genomic and sub‐genomic mRNAs leave the DMVs, most likely through special pores (Wolff et al., 2020). After entering the cytoplasm they are ready to be translated in ER‐associated ribosomes to yield the viral envelope proteins E, M, and S, which contain N‐terminal signal sequences for ER translocation. By contrast, the nucleocapsid protein N is synthesized on free ribosomes (Figure 1). The non‐structural proteins (nsps) and accessory proteins (ORFs) of CoVs are not included in the virus particles, but by interacting with specific host proteins (Gordon et al., 2020; Stukalov et al., 2021) can these proteins participate not only in viral RNA replication (Figure 1), but also in the virus‐induced organelle rearrangements, such as Golgi disassembly (see below). Consequently, these proteins may also be linked to the mechanisms of virus egress.
4. VIRUS ASSEMBLY
CoV assembly is initiated by coating of the RNA genome by N proteins, leading to the formation of phase‐separated condensates in association with the M protein—the major viral membrane protein present in the IC membranes—and virus budding (Lu et al., 2021). Co‐expression of the structural proteins of CoVs has demonstrated that the formation of virus‐like particles (VLPs) also requires the E protein (Fischer et al., 1998; Vennema et al., 1996; Xu et al., 2020). The S protein is not required for VLP formation, but is essential for virus infectivity; that is, all three proteins must co‐localize in the IC to ensure the formation of fully functional CoV particles. The three proteins—the E protein (Cohen et al., 2011; Corse & Machamer, 2000; Li et al., 2014), the M protein (Klumperman et al., 1994; Krijnse‐Locker et al., 1992; Machamer & Rose, 1987; Swift & Machamer, 1991) and the S protein (Lontok et al., 2004; McBride et al., 2007)—are all transmembrane proteins that following their insertion into the ER membrane are transported to the IC (Figure 1). Efficient incorporation of these proteins into the virus envelope requires that they harbor signals for retention or retrieval to the perinuclear Golgi region of the host cell, which has been demonstrated by studies of individually expressed proteins. Generally, receptor‐mediated retrieval of endogenous ER proteins via C‐terminal KDEL signals functions throughout the Golgi apparatus (Miesenböck & Rothman, 1995), and likewise, membrane proteins with a terminal double lysine motif (KKXX) in their cytoplasmic tails are retrieved retrogradely from the PM and distal regions of the Golgi apparatus to the IC and the ER (Itin et al., 1995; Jackson et al., 1990; Nilsson et al., 1989; Townsley & Pelham, 1994).
During the early stages of CoV infection, before the Golgi apparatus is severely affected, viral proteins can be returned to the IC from more distal compartments by well‐known mechanisms (Bracquemond & Muriaux, 2021). Both retention and retrieval signals operate to maintain their concentration in the perinuclear Golgi region (reviewed by Ujike & Taguchi, 2015). For instance, the M proteins of certain CoVs localize to secretory compartments that lie beyond the sites of CoV assembly at the IC (Klumperman et al., 1994; Perrier et al., 2019). The S protein forms trimers in the ER, which are incorporated into virions at the IC through their interactions with the highly abundant M protein (Godeke et al., 2000). When expressed in BHK cells, the S protein displays a more widespread distribution in the secretory compartments, and is also detected at the PM (Nal et al., 2005; Vennema et al., 1990). The S protein also contains a di‐basic signal in its cytoplasmic tail that mediates its COPI‐mediated retrieval to the IC (McBride et al., 2007), where the protein can be retained through its interaction with the M protein (Opstelten et al., 1995). Furthermore, the E protein contains intrinsic information that retains it in the perinuclear IC/Golgi region (Corse & Machamer, 2000, 2002), where the E and M proteins interact via their cytoplasmic tails (Corse & Machamer, 2003).
The E and the S proteins of CoVs are both S‐acylated/palmitoylated at cysteines in their cytoplasmic domains (Lopez et al., 2008; McBride & Machamer, 2010). Inhibition of acylation of the S protein reduced its interaction with the M protein (Thorp et al., 2006) and inhibited fusion between viral and cellular membranes (Li et al., 2022; Petit et al., 2007), suggesting that its association with particular lipid domains is important at different stages of the virus life cycle. Interestingly, it has been recently reported that the cytoplasmic domain of the S protein is acylated at a total of 10 cysteines. The hyper‐acylation process starts in the ER with palmitate addition to cysteines close to the transmembrane domain, and continues at additional cysteines after ER exit, with each S protein trimer arriving at the IC being decorated by up to 30 acyl chains (Mesquita et al., 2021). Based on its extensive acylation the S protein triggers the formation of cholesterol‐rich membrane nanodomains in the IC membranes, thereby facilitating virus budding (Mesquita et al., 2021) possibly by promoting membrane curvature (Ernst et al., 2019). The formation of “lipid rafts” at the IC during SARS‐CoV‐2 infection may also play a role in the formation of specialized transport carriers mediating the egress of progeny viruses.
5. ENDOMEMBRANE ALTERATIONS SUPPORT CoV REPLICATION, ASSEMBLY, AND EGRESS
As mentioned above, the first observable change occurring intracellularly in CoV‐infected cells is the formation of DMVs—the sites of RNA replication—which appear already at 3 hours of post‐infection (Cortese et al., 2020; Eymieux, Rouillé, et al., 2021; Mendonca et al., 2021; Stertz et al., 2007). Other early membrane rearrangements in CoV‐infected cells include alterations in the appearance of mitochondria and the recruitment of peroxisomes to the vicinity of the DMVs (Cortese et al., 2020).
It has been recognized for some time that virus infection impacts autophagy, a key process that regulates cellular homeostasis by directing dysfunctional organelles and proteins toward degradation, thereby providing building blocks for biosynthesis during starvation. The initiation of autophagy involves the formation of a double‐membrane structure called the phagophore (Seglen et al., 1990), which grows to form the autophagosome, enclosing in a selective or non‐selective manner cytoplasmic material for delivery to lysosomes for degradation. Autophagy can be activated in response to virus infection, to shield the invading virus, and to deliver it to pre‐lysosomes or lysosomes for proteolytic degradation and presentation of peptide fragments to the adaptive immune system (Liang et al., 2021). However, many viruses encode proteins that inhibit autophagy, redirecting membrane sources normally used for this process to alternative purposes for their benefit (Blanchard & Roingeard, 2015; Roth et al., 2020). The IC/cis‐Golgi membranes that are known to provide such a membrane source (Ge et al., 2015) were recently shown to co‐operate with the endosomal system in the formation of a precursor membrane structure designated as the pro‐phagophore. Moreover, the formation of this hybrid compartment that initiates autophagy was reported to be inhibited by the nsp6 of SARS‐CoV‐2 (Kumar et al., 2021).
Another link between autophagy and SARS‐CoV‐2 infection is provided by phosphatidylinositol‐3‐kinase (PI3K), which besides being involved in autophagosome biogenesis, is required for the formation of DMVs (Twu et al., 2021; Williams et al., 2021). In addition, the ORF3a protein of SARS‐CoV‐2 has been shown to block autophagy by inhibiting the machinery—including the tethering complex (HOPS) and the SNARE (syntaxin 17)—that mediates the fusion between autophagosomes and lysosomes (Miao et al., 2021). Besides escaping engulfment by the autophagic pathway, a potential benefit for the virus could be ensuring that the IC membranes are preferentially used for virus assembly, instead of being depleted by autophagosome formation. The egress of β‐CoVs was shown to be enhanced by ORF3a, which also contributes to Golgi fragmentation (see below), while the progeny CoVs were suggested to follow an exit route that passes via late endosomes and/or lysosomes (Figure 1; Chen et al., 2021).
CoV infection, like infection of cells by a variety of other viruses, has been shown to induce fragmentation of the Golgi apparatus (Cortese et al., 2020; Glingston et al., 2019; Hackstadt et al., 2021; Lavi et al., 1996; Ruch & Machamer, 2012; Ulasli et al., 2010). This striking organelle alteration can be observed already at 6 hours of post‐infection when the first progeny viruses are released into the medium of cultured cells (Cortese et al., 2020; Hackstadt et al., 2021; Lavi et al., 1996; Ulasli et al., 2010). Expression of individual CoV proteins has also been shown to induce Golgi fragmentation, with both the E protein and ORF3a having this ability. Notably, both proteins have been reported to function as ion channels (Freundt et al., 2010; Hackstadt et al., 2021; Ruch & Machamer, 2011), indicating that the induction of Golgi fragmentation is an intrinsic property of viral proteins. However, it may involve additional host factors (Ruch & Machamer, 2011; Westerbeck & Machamer, 2019).
Strikingly, instead of interfering with virus release, the dramatic Golgi alterations seem to facilitate this process (Ruch & Machamer, 2012). Indeed, Golgi‐independent egress via lysosomes (Figure 1), as shown by its resistance to Brefeldin A (BFA) treatment, has been suggested to dominate during β‐CoV infection (Ghosh et al., 2020. This mode of egress appears to be stimulated by the ORF3a protein of SARS‐CoV‐2 which redirects the lysosomal membrane proteins LAMP1 and LAMP2 to the PM (Chen et al., 2021), in analogy to observations made with reovirus‐infected cells (Fernandez de Castro et al., 2020). SARS‐CoV ORF3a does not seem to promote the same mechanism, but is still stimulatory to virus egress, possibly by forming ion channels in the cell membrane (Lu et al., 2006). What has been designated as lysosomal egress is reported to involve a pathway traversing late endosomes and/or lysosomes (Chen et al., 2021; Ducatelle & Hoorens, 1984; Freundt et al., 2010; Ghosh et al., 2020); however, the detailed intracellular route remains to be characterized. The same holds also for virus exit at the cell surface, where two modes of release to the cell exterior have been reported, one via membrane tunnels providing openings for large virus‐containing carriers (Mendonca et al., 2021) and another involving secretory vesicles that generally contain a single virus particle (Eymieux, Uzbekov, et al., 2021).
We have previously drawn the attention to the direct connection between the IC and the endocytic recycling system—consisting of the widely distributed recycling endosomes (REs) and the pericentrosomal endocytic recycling compartment (ERC)—in providing a pathway for the delivery of CoVs from the IC to the extracellular space (Figure 1; Saraste & Prydz, 2021). These compartments—the IC and REs, defined by the GTPases Rab1 and Rab11, respectively—also appear to co‐exist in the non‐compact zones of the Golgi ribbon, adjacent to the dilated rims of the stacked Golgi cisternae, with which they share structural similarity (Figure 1; Saraste & Prydz, 2019). Interestingly, besides its role in ER‐Golgi trafficking, the IC has been implicated in Golgi‐independent transport routes, that can be defined, for instance, by their resistance to BFA (Marie et al., 2009; Prydz et al., 2008, 2013; Sannerud et al., 2006; Tveit et al., 2009; Zhang et al., 2020). A secretory Golgi‐bypass route via RE has also been demonstrated in neuronal dendrites (Kennedy & Hanus, 2019).
Interestingly, an increasing number of viruses turn out to exploit the endocytic recycling apparatus defined by Rab11 for their assembly and/or release, regardless of whether they bud intracellularly or at the PM (Bruce et al., 2012; Coller et al., 2012; Lučin et al., 2018; Pereira et al., 2014; Rowe et al., 2008; Vale‐Costa & Amorim, 2016). For example, infection of cells with influenza virus results in the accumulation of Rab11‐positive REs at the pericentrosomal ERC, creating cholesterol‐rich membrane domains for the binding of viral genome‐containing ribo‐nucleoproteins (vRNPs) (Kawaguchi et al., 2015). Subsequently, the vRNPs are delivered in a Rab11‐ and microtubule‐dependent fashion to the PM where virus assembly by budding is completed (Bruce et al., 2012). The functional association of IC membranes with the REs/ERC (Marie et al., 2009; Saraste & Marie, 2018) raises the possibility that this pericentrosomal membrane system at the crossroads of the endo‐ and exocytic transport routes also plays a role in the release of CoVs (Saraste & Prydz, 2021).
6. SUMMARY AND OUTLOOK
Taking into consideration the recently expanding literature, we summarize here the available information on the pathways and mechanisms that the intracellularly budding CoVs employ as they are exported from their host cells (Figure 1). As is often the case with recently emerged “hot topics”, the situation is quite puzzling and further research is required to obtain a better picture of how these viruses take advantage of the more or less conventional cellular machineries to promote their release. Nevertheless, it is clear that the long‐prevailing concept on the role of the classical secretory pathway in CoV egress has been seriously questioned. Indeed, the lack of inhibition of β‐CoV release by BFA set the stage for the search and characterization of an alternative Golgi‐independent exit route via the endo‐lysosomal system (Ghosh et al., 2020). One potential problem with this pathway is that the progeny viruses would undergo proteolytic degradation as they pass through lysosomes and become non‐infectious. This could probably be partially avoided if CoV infection leads to the reported neutralization of the acidic lumen of endosomes and lysosomes over time (Ghosh et al., 2020). However, recent cryo‐EM studies indicated that the spikes of SARS‐CoV‐2 particles residing in lysosome‐like organelles have undergone proteolysis, although it remained unclear whether they represent newly made or re‐internalized virus particles (Mendonca et al., 2021). Another problem with this pathway is to understand how viruses are delivered from the IC to lysosomes, since they evidently cannot follow the BFA‐sensitive pathway via the Golgi stacks and the trans‐Golgi/TGN (Figure 1). Indeed, the effects of BFA and other transport inhibitors on the release of the various CoVs from different host cells, including epithelial cells, deserve further analysis.
In line with data obtained with other viruses, the role of secretory autophagy in CoV egress has also been considered (Yuan et al., 2022). However, the detailed mechanisms of this process, which until now have been predominantly described in the case of cytosolic proteins, remain poorly understood. Indeed, one may ask how it can be responsible for the efficient secretion of large‐sized intralumenal virus particles. Finally, the collective results showing that a number of viruses highjack the endocytic recycling apparatus and the master GTPase Rab11 during the late stages of their replication makes the direct IC‐RE pathway a particularly attractive candidate to consider in the case of CoVs (Figure 1). Moreover, this pathway can readily explain the puzzling findings regarding efficient CoV release after the Golgi apparatus has been subjected to extensive reorganization due to virus infection (Saraste & Prydz, 2021). In fact, it may even be beneficial for the virus to be able to down‐regulate alternative paths or remove obstacles as it embarks on its “unconventional journey” from the IC via REs to the extracellular space.
Prydz, K. & Saraste, J. (2022). The life cycle and enigmatic egress of coronaviruses. Molecular Microbiology, 117, 1308–1316. 10.1111/mmi.14907
Contributor Information
Kristian Prydz, Email: kristian.prydz@ibv.uio.no.
Jaakko Saraste, Email: jaakko.saraste@uib.no.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
REFERENCES
- Ahat, E. , Song, Y. , Xia, K. , Reid, W. , Li, J. , Bui, S. et al. (2022) GRASP depletion‐mediated Golgi fragmentation impairs glycosaminoglycan synthesis, sulfation, and secretion. Cellular and Molecular Life Sciences, 79, 199. 10.1007/s00018-022-04223-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino, R.S. & Park, P.W. (2016) Glycosaminoglycans and infection. Frontiers in Bioscience, 21, 1260–1277. 10.1002/hep.29265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann, J.E. , Tokuyasu, K.T. & Singer, S.J. (1981) Passage of an integral membrane protein, the vesicular stomatitis virus glycoprotein, through the Golgi apparatus en route to the plasma membrane. Proceedings of the National Academy of Sciences of the United States of America, 7, 1746–1750. 10.1073/pnas.78.3.1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard, E. & Roingeard, P. (2015) Virus‐induced double‐membrane vesicles. Cellular Microbiology, 17, 45–50. 10.1111/cmi.12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracquemond, D. & Muriaux, D. (2021) Beta‐coronavirus assembly: clues and perspectives for elucidating SARS‐CoV‐2 particle formation and egress. mBio, 12, e02371‐21. 10.1128/mBio.02371-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, E.A. , Stuart, A. , McCaffrey, M.W. & Digard, P. (2012) Role of the Rab11 pathway in negative‐strand virus assembly. Biochemical Society Transactions, 44, 1409–1425. 10.1042/BST20120166 [DOI] [PubMed] [Google Scholar]
- Chen, D. , Zhen, Q. , Sun, L. , Ji, M. , Li, Y. , Deng, H. et al. (2021) ORF3a of SARS‐CoV‐2 promotes lysosomal exocytosis‐mediated viral egress. Developmental Cell, 56, 1–14. 10.1016/j.devcel.2021.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, T.M. , Sandoval, D.R. , Spliid, C.B. , Pihl, J. , Perrett, H.R. , Painter, C.D. et al. (2021) SARS‐CoV‐2 infection depends on cellular heparan sulfate and ACE2. Cell, 183, 1043–1057. 10.1016/j.cell.2020.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J.R. , Lin, L.D. & Machamer, C.E. (2011) Identification of a Golgi‐complex‐targeting signsal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. Journal of Virology, 85, 5794–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, K.E. , Heaton, N.S. , Berger, K.L. , Cooper, J.D. , Saunders, J.L. & Randall, G. (2012) Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathogens, 8, e1002466. 10.1371/journal.ppat.1002466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse, E. & Machamer, C.E. (2000) Infectous bronchitis virus E protein is targeted to the Golgi complex and directs release of virus‐like particles. Journal of Virology, 74, 4319–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse, E. & Machamer, C.E. (2002) The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. Journal of Virology, 76, 1273–1284. 10.1128/jvi.76.3.1273-1284.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse, E. & Machamer, C.E. (2003) The cytoplasmic tails of bronchitis virus E and M proteins mediate their interaction. Virology, 312, 25–35. 10.1016/s0042-6822(03)00175-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese, M. , Lee, J.Y. , Cerikan, B. , Neufeldt, C.J. , Oorschot, V.M.J. , Köhrer, S. et al. (2020) Integrative imaging reveals SARS‐CoV‐2‐induced reshaping of subcellular morphologies. Cell Host & Microbe, 28, 853–866. 10.1016/j.chom.2020.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haan, C.A. , Li, Z. , te Lintelo, E. , Bosch, B.J. , Haijema, B.J. & Rottier, P.J. (2005) Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. Journal of Virology, 79, 14451–14456. 10.1128/JVI.79.22.14451-14456.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pasquale, V. , Quiccione, M.S. , Tafuri, S. , Avallone, L. & Pavone, L.M. (2021) Heparan sulfate proteoglycans in viral infection and treatment: a special focus on SARS‐CoV‐2. International Journal of Molecular Sciences, 22, 6574. 10.3390/ijms22126574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatelle, R. & Hoorens, J. (1984) Significance of lysosomes in the morphogenesis of coronaviruses. Archives of Virology, 79, 1–12. 10.1007/BF01314299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, A.M. , Toomre, D. & Bogan, J.S. (2019) Acylation—a new means to control traffic through the Golgi. Frontiers in Cell and Development Biology, 7, 109. 10.3389/fcell.2019.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymieux, S. , Rouillé, Y. , Terrier, O. , Seron, K. , Blanchard, E. , Rosa‐Calatrava, M. et al. (2021) Ultrastructural modifications induced by SARS‐CoV‐2 in Vero cells: a kinetic analysis of viral factory formation, viral particle morphogenesis and virion release. Cellular and Molecular Life Sciences, 78, 3565–3576. 10.1007/s00018-020-03745-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymieux, S. , Uzbekov, R. , Rouillé, Y. , Blanchard, E. , Hourioux, C. , Dubuisson, J. et al. (2021) Secretory vesicles are the principal means of SARS‐CoV‐2 egress. Cell, 10, 2047. 10.3390/cells10082047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A.R. & Perlman, S. (2015) Coronaviruses: an overview of their replication and pathogenesis. Methods in Molecular Biology, 1282, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de Castro, I. , Tenorio, R. , Ortega‐Gonzales, P. , Knowlton, J.J. , Zamora, P.F. , Lee, C.H. et al. (2020) A modified lysosomal organelle mediates nonlytic egress of reovirus. The Journal of Cell Biology, 219, e201910131. 10.1083/jcb.201910131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyzi, E. , Saldeen, T. , Larsson, E. , Lindahl, U. & Salmivirta, M. (1998) Age‐dependent modulation of heparan sulfate structure and function. The Journal of Biological Chemistry, 273, 13395–13398. [DOI] [PubMed] [Google Scholar]
- Fischer, F. , Stegen, C.F. , Masters, P.S. & Samsonoff, W.A. (1998) Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. Journal of Virology, 72, 7885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt, E.C. , Yu, L. , Goldsmith, C.S. , Welsh, S. , Cheng, A. , Yount, B. et al. (2010) The open reading frame 3a protein of severe acute respiratory syndrome‐associated coronavirus promotes membrane rearrangement and cell death. Journal of Virology, 84, 1097–1109. 10.1128/JVI.01662-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, T.S. & Liu, D.X. (2019) Human coronaviruses: host‐pathogen interaction. Annual Review of Microbiology, 73, 529–557. 10.1146/annurev-micro-020518-115759 [DOI] [PubMed] [Google Scholar]
- Ge, L. , Wilz, L. & Schekman, R. (2015) Biogenesis of autophagosomal precursors for LC3 lipidation from the ER‐Golgi intermediate compartment. Autophagy, 11, 2372–2374. 10.1080/15548627.2015.1105422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S. , Dellibovi‐Ragheb, T.A. , Kerviel, A. , Pak, E. , Qiu, Q. , Fisher, M. et al. (2020) β‐Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell, 183, 1520–1523. 10.1016/j.cell.2020.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glingston, R.S. , Deb, R. , Kumar, S. & Nagotu, S. (2019) Organelle dynamics and viral infection: at cross roads. Microbes and Infection, 21, 20–32. 10.1016/j.micinf.2018.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeke, G.J. , de Haan, C.A.M. , Rossen, J.W.A. , Vennema, H. & Rottier, P.J.M. (2000) Assembly of spikes into coronavirus particles is mediated by the carboxy‐terminal domain of the spike protein. Journal of Virology, 74, 1566–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D.E. , Jang, G.M. , Bouhaddou, M. , Xu, J. , Obernier, K. , White, K.M. et al. (2020) A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature, 583, 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt, T. , Chiramel, A.I. , Hoyt, F.H. , Williamson, B.N. , Dooley, C.A. , Beare, P.A. et al. (2021) Disruption of the Golgi apparatus and contribution of the endoplasmic reticulum to the SARS‐CoV‐2 replication complex. Viruses, 13, 1798. 10.3390/v13091798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadigal, S. , Koganti, R. , Ydavalli, T. , Agelidis, A. , Suryawanshi, R. & Shukla, D. (2020) Heparanase‐regulated syndecan‐1 shedding facilitates herpes simplex virus 1 egress. Journal of Virology, 94, e01672‐19. 10.1128/JVI.01672-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Gonzalez, M. , Larocque, G. & Way, M. (2021) Virus use and subversion of membrane organization and trafficking. Journal of Cell Science, 134, jcs252676. 10.1242/jcs.252676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Krüger, N. , Herrler, T. , Erichsen, S. et al. (2020) SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181, 271–280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itin, C. , Kappeler, F. , Linstedt, A.D. & Hauri, H.P. (1995) A novel endocytosis signal related to the KKXX ER‐retrieval signal. The EMBO Journal, 14, 2250–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C.B. , Farzan, M. , Chen, B. & Choe, H. (2022) Mechanisms of SARS‐CoV‐2 entry into cells. Nature Reviews Molecular Cell Biology, 23, 3–20. 10.1038/s41580-021-00418-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M.R. , Nilsson, T. & Peterson, P.A. (1990) Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. The EMBO Journal, 9, 3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi, A. , Hirohama, M. , Osari, S. & Nagata, K. (2015) Influenza virus induces cholesterol‐enriched endocytic recycling compartments for budozone formation via cell cycle‐independent centrosome maturation. PLoS Pathogens, 11, e1005284. 10.1371/journal.ppat.1005284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, M.J. & Hanus, C. (2019) Architecture and dynamics of the neuronal secretory network. Annual Review of Cell and Developmental Biology, 35, 543–566. 10.1146/annurev-cellbio-100818-125418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, S. , Cortese, M. , Winter, S.L. , Wachsmuth‐Melm, M. , Neufeldt, C.J. , Cerikan, B. et al. (2020) SARS‐CoV‐2 structure and replication characterized by in situ cryo‐electron tomography. Nature Communications, 11, 5885. 10.1038/s41467-020-19619-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman, J. , Krijnse‐Locker, J. , Meijer, A. , Horzinek, M.C. , Geuze, H.J. & Rottier, P.J.M. (1994) Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. Journal of Virology, 68, 6523–6534. 10.1128/JVI.68.10.6523-6534.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops, K. , Kikkert, M. , van den Worm, S.H. , Zevenhoven‐Dobbe, J.C. , van der Meer, Y. , Koster, A.J. et al. (2008) SARS‐coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biology, 6, 1957–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuger, J. , Prydz, K. , Pettersson, R.F. , Lindahl, U. & Salmivirta, M. (1999) Characterization of fibroblast growth factor 1 binding heparan sulfate domain. Glycobiology, 9, 723–729. [DOI] [PubMed] [Google Scholar]
- Krijnse‐Locker, J. , Griffiths, G. , Horzinek, M.C. & Rottier, P. (1992) O‐glycosylation of the coronavirus M protein. The Journal of Biological Chemistry, 267, 14094–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Javed, R. , Mudd, M. , Pallikkuth, S. , Lidke, K.A. , Jain, A. et al. (2021) Mammalian hybrid pre‐autophagosomal structure HyPAS generates autophagosomes. Cell, 184, 1–20. 10.1016/j.cell.2021.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi, E. , Wang, Q. , Weiss, S.R. & Gonatas, N.K. (1996) Syncytia formation induced by coronavirus infection is associated with fragmentation and rearrangement of the Golgi apparatus. Virology, 221, 325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Liu, Y. , Lu, Y. , Gao, S. & Zhang, L. (2022) Palmitoylation of SARS‐CoV‐2 S protein is critical for S‐mediated syncytia formation and virus entry. Journal of Medical Virology, 94, 342–348. 10.1002/jmv.27339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Surya, W. , Claudine, S. & Torres, J. (2014) Structure of a conserved Golgi complex‐targeting signal in coronavirus envelope proteins. The Journal of Biological Chemistry, 289, 12535–12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, S. , Wu, Y.S. , Li, D.Y. , Tang, J.X. & Lu, H.F. (2021) Autophagy in viral infection and pathogenesis. Frontiers in Cell and Development Biology, 9, 766142. 10.3389/fcell.2021.766142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lontok, E. , Corse, E. & Machamer, C.E. (2004) Intracellular targeting signals contribute to localization of coronavirus spike proteins near the assembly site. Journal of Virology, 78, 5913–5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, L.A. , Riffle, A.J. , Pike, S.L. , Gardner, D. & Hogue, B. (2008) Importance of conserved cysteine residues in coronavirus envelope protein. Journal of Virology, 82, 3000–3010. 10.1128/JVI.01914-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. , Ye, Q. , Singh, D. , Cao, Y. , Diedrich, J.K. , Yates, J.R., 3rd et al. (2021) The SARS‐CoV‐2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane‐associated M protein. Nature Communications, 12, 502. 10.1038/241467-020-20768-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W. , Zheng, B.J. , Xu, K. , Schwarz, W. , Du, L. , Wong, C.K.L. et al. (2006) Severe acute respiratory syndrome‐associated coronavirus 3a protein forms an ion channel and modulates virus release. Proceedings of the National Academy of Sciences USA, 103, 12540–12545. 10.1073/pnas.0605402103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lučin, P. , Kareluša, L. , Blagojević Zagorac, G. , Mahmutefendić Lučin, H. , Pavišić, V. , Jug Vučko, N. et al. (2018) Cytomegaloviruses exploit recycling Rab proteins in the sequential establishment of the assembly compartment. Frontiers in Cell and Development Biology, 6, 65. 10.3389/fcell.2018.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer, C.E. & Rose, J.K. (1987) A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. The Journal of Cell Biology, 105, 1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, B. , Urakova, N. , Snijder, E.J. & Campbell, E.A. (2022) Structures and functions of coronavirus replication‐transcription complexes and their relevance for SARS‐CoV‐2 drug design. Nature Reviews Molecular Cell Biology, 23, 21–39. 10.1038/s41580-021-00432-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie, M. , Dale, H.A. , Sannerud, R. & Saraste, J. (2009) The function of the intermediate compartment in pre‐Golgi trafficking involves its stable connection with the centrosome. Molecular Biology of the Cell, 20, 4458–4470. 10.1091/mbc.e08-12-1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley, J.L. , Gilbertson, B.P. , Trifkovic, S. , Brown, L.E. & McKimm‐Breschkin, J.L. (2019) Influenza virus neuraminidase structure and functions. Frontiers in Microbiology, 10, 39. 10.3389/fmicb.2019.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, C.E. , Li, J. & Machamer, C.E. (2007) The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. Journal of Virology, 81, 2418–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, C.E. & Machamer, C.E. (2010) Palmitoylation of SARS‐CoV S protein is necessary for partitioning into detergent‐resistant membranes and cell‐cell fusion but not interaction with M protein. Virology, 405, 139–148. 10.1016/j.virol.2010.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonca, L. , Howe, A. , Gilchrist, J.B. , Sheng, Y. , Sun, D. , Knight, M.L. et al. (2021) Correlative multi‐scale cryo‐imaging unveils SARS‐CoV‐2 assembly and egress. Nature Communications, 12(1), 4629. 10.1038/s41467-021-24887-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita, F.S. , Abrami, L. , Sergeeva, O. , Turelli, P. , Qing, E. , Kunz, B. et al. (2021) S‐acylation controls SARS‐CoV‐2 membrane lipid organization and enhances infectivity. Developmental Cell, 56, 2790–2807. 10.1016/j.devcel.2021.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, G. , Zhao, H. , Bi, Y. , Wang, P. & Zhang, H. (2021) ORF3a of the COVID‐19 virus SARS‐CoV‐2 blocks HOPS complex‐mediated assembly of the SNARE complex required for autolysosome formation. Developmental Cell, 56, 427–442. 10.1016/j.devcel.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenböck, G. & Rothman, J.E. (1995) The capacity to retrieve escaped ER proteins extends to the trans‐most cisternae of the Golgi stack. The Journal of Cell Biology, 129, 309–319. 10.1083/jcb.129.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska, A. , Zarebski, M. , Nowak, P. , Stozek, K. , Potempa, J. & Pyrc, K. (2014) Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. Journal of Virology, 88, 13221–13230. 10.1128/JVI.02078-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S. & Krijnse‐Locker, J. (2008) Modification of intracellular membrane structures for virus replication. Nature Reviews Microbiology, 6, 363–374. 10.1038/nrmicro1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet, J.K. & Whittaker, G.R. (2015) Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Research, 202, 120–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal, B. , Chan, C. , Kien, F. , Siu, L. , Tse, J. , Chu, K. et al. (2005) Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. The Journal of General Virology, 86, 1423–1434. 10.1099/vir.0.80671-0 [DOI] [PubMed] [Google Scholar]
- Naskalska, A. , Dabrowska, A. , Szczepanski, A. , Milewska, A. , Jasik, K.P. & Pyrc, K. (2019) Membrane protein of human coronavirus NL63 is responsible for interaction with the adhesion receptor. Journal of Virology, 93, e00355–e00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L. , McCord, K.A. , Bui, D.T. , Bouwman, K.M. , Kitova, E.N. , Elaish, M. et al. (2022) Sialic acid‐containing glycolipids mediate binding and viral entry of SARS‐CoV‐2. Nature Chemical Biology, 18, 81–90. 10.1038/s41589-021-00924-1 [DOI] [PubMed] [Google Scholar]
- Nilsson, T. , Jackson, M. & Peterson, P.A. (1989) Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell, 58, 707–718. 10.1016/0092-8674(89)90105-0 [DOI] [PubMed] [Google Scholar]
- Opstelten, D.J.E. , Raamsman, M.J.B. , Wolfs, K. , Horzinek, M.C. & Rottier, P.J.M. (1995) Envelope glycoprotein interactions in coronavirus assembly. The Journal of Cell Biology, 131, 339–349. 10.1083/jcb.131.2.339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, L.E. , Clark, J. , Grznarova, P. , Wen, X. , LaCasse, R. , Ruml, T. et al. (2014) Direct evidence for intracellular anterograde co‐transport of M‐PMV gag and env on microtubules. Virology, 449, 109–119. 10.1016/j.virol.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier, A. , Bonnin, A. , Desmarets, L. , Danneels, A. , Goffard, A. , Rouillé, Y. et al. (2019) The C‐terminal domain of the MERS coronavirus M protein contains a trans‐Golgi network localization signal. The Journal of Biological Chemistry, 294, 14406–14421. 10.1074/jbc.RA119.008964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, C.M. , Chouljenko, V.N. , Iyer, A. , Colgrove, R. , Farzan, M. , Knipe, D.M. et al. (2007) Palmitoylation of the cysteine‐rich endodomain of the SARS‐coronavirus spike glycoprotein is important for spike‐mediated cell fusion. Virology, 360, 264–274. 10.1016/j.virol.2006.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prydz, K. & Dalen, K.T. (2000) Synthesis and sorting of proteoglycans. Journal of Cell Science, 113, 193–205. [DOI] [PubMed] [Google Scholar]
- Prydz, K. , Dick, G. & Tveit, H. (2008) How many ways through the Golgi maze? Traffic, 9, 299–304. [DOI] [PubMed] [Google Scholar]
- Prydz, K. , Tveit, H. , Vedeler, A. & Saraste, J. (2013) Arrivals and departures at the plasma membrane: direct and indirect transport routes. Cell and Tissue Research, 352, 5–20. [DOI] [PubMed] [Google Scholar]
- Roth, A.N. , Aravamudhan, P. , Fernandez de Castro, I. , Tenorio, R. , Risco, C. & Dermody, T.S. (2020) Ins and outs of reovirus: vesicular trafficking in viral entry and egress. Trends in Microbiology, 29, 363–375. 10.1016/j.tim.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, R.K. , Suszko, J.W. & Pekosz, A. (2008) Roles for the recycling endosome, Rab8, and Rab11 in hantavirus release from epithelial cells. Virology, 382, 239–249. 10.1016/j.virol.2008.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch, T.R. & Machamer, C.E. (2011) The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. Journal of Virology, 85, 675–685. 10.1128/JVI.01570-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch, T.R. & Machamer, C.E. (2012) The coronavirus E protein: assembly and beyond. Viruses, 4, 363–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, R.J. , Stevens, D.J. , Haire, L.F. , Gamblin, S.J. & Skehel, J.J. (2006) Avian and human receptor binding by hemagglutinins of influenza a viruses. Glycoconjugate Journal, 23, 85–92. 10.1007/s10719-006-5440-1 [DOI] [PubMed] [Google Scholar]
- Sachse, M. , Fernandez de Castro, I. , Tenorio, R. & Risco, C. (2019) The viral replication organelles within cells studied by electron microscopy. Advances in Virus Research, 105, 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud, R. , Marie, M. , Nizak, C. , Dale, H.A. , Pernet‐Gallay, K. , Perez, F. et al. (2006) Rab1 defines a novel pathway connecting the pre‐Golgi intermediate compartment with the cell periphery. Molecular Biology of the Cell, 20, 4458–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, J. & Kuismanen, E. (1984) Pre‐ and post‐Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell, 38, 535–549. [DOI] [PubMed] [Google Scholar]
- Saraste, J. & Marie, M. (2018) Intermediate compartment (IC): from pre‐Golgi vacuoles to a semi‐autonomous membrane system. Histochemistry and Cell Biology, 150, 407–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, J. & Prydz, K. (2019) A new look at the functional organization of the Golgi ribbon. Frontiers in Cell and Development Biology, 7, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, J. & Prydz, K. (2021) Assembly and exit of coronaviruses: hijacking an unconventional secretory pathway from the pre‐Golgi intermediate compartment via the Golgi ribbon to the extracellular space. Cell, 10, 503. 10.3390/cells10030503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, K.M. , Mascheroni, L. , Carnell, G.W. , Wunderlich, L.C.S. , Makarchuk, S. , Brockhoff, M. et al. (2022) SARS‐CoV‐2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome‐mediated egress. Science Advances, 8, eabl4895. 10.1126/sciadv.abl4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen, P.O. , Gordon, P.B. & Holen, I. (1990) Non‐selective autophagy. Seminars in Cell Biology, 1, 441–448. [PubMed] [Google Scholar]
- Snijder, E.J. , Limpens, R.W.A.L. , de Wilde, A.H. , de Jong, A.W.H. , Zevenhoven‐Dobbe, J.C. , Maier, H.J. et al. (2020) A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biology, 18, e3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz, S. , Reichelt, M. , Spiegel, M. , Kuri, T. , Martinez‐Sobrido, L. , Garcia‐Sastre, A. et al. (2007) The intracellular sites of early replication and budding of SARS‐coronavirus. Virology, 361, 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukalov, A. , Girault, V. , Grass, V. , Karayel, O. , Bergant, V. , Urban, C. et al. (2021) Multilevel proteomics reveals host perturbations by SARS‐CoV‐2 and SARS‐CoV. Nature, 594, 246–252. 10.1038/s41586-021-03493-4 [DOI] [PubMed] [Google Scholar]
- Sturman, L.S. & Holmes, K.V. (1983) The molecular biology of coronaviruses. Advances in Virus Research, 28, 35–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift, A.M. & Machamer, C.E. (1991) A Golgi retention signal in a membrane‐spanning domain of coronavirus E1 protein. The Journal of Cell Biology, 115, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp, E.B. , Boscarino, J.A. , Logan, H.L. , Goletz, J.T. & Gallagher, T.M. (2006) Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. Journal of Virology, 80, 1280–1289. 10.1128/JVI.80.3.1280-1289.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze, S.A. , Tooze, J. & Warren, G. (1984) Replication of coronavirus MHV‐A59 in sac cells: determination of the first site of budding of progeny virions. European Journal of Cell Biology, 33, 281–293. [PubMed] [Google Scholar]
- Townsley, F.M. & Pelham, H.R. (1994) The KKXX signal mediates retrieval of membrane proteins from the Golgi to the ER in yeast. European Journal of Cell Biology, 64, 211–216. [PubMed] [Google Scholar]
- Tveit, H. , Akslen, L.K.A. , Fagereng, G.L. , Tranulis, M.A. & Prydz, K. (2009) A secretory Golgi bypass route to the apical surface domain of epithelial MDCK cells. Traffic, 10, 1685–1695. [DOI] [PubMed] [Google Scholar]
- Twu, W.I. , Lee, J.Y. , Kim, H. , Prasad, V. , Cerikan, B. , Haselmann, U. et al. (2021) Contribution of autophagy machinery factors to HCV and SARS‐CoV‐2 replication organelle formation. Cell Reports, 37, 110049. 10.1016/j.celrep.2021.110049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike, M. & Taguchi, F. (2015) Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses, 7, 1700–1725. 10.3390/v7041700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulasli, M. , Verheije, M.H. , de Haan, C.A. & Reggiori, F. (2010) Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cellular Microbiology, 12, 844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale‐Costa, S. & Amorim, J. (2016) Recycling endosomes and viral infection. Viruses, 8, 64. 10.3390/v8030064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki, A. (2011) Evolutionary forces shaping the Golgi glycosylation machinery: why cell surface glycans are universal to living cells. Cold Spring Harbor Perspectives in Biology, 3, a005462. 10.1101/cshperspect.a005462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema, H. , Godeke, G.J. , Rossen, J.W.A. , Voorhout, W.F. , Horzinek, M.C. , Opstelten, D.J.E. et al. (1996) Nucleocapsid‐independent assembly of coronavirus‐like particles by coexpression of viral envelope proteins. The EMBO Journal, 15, 2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema, H. , Heijnen, L. , Zijderveld, A. , Horzinek, M.C. & Spaan, W.J. (1990) Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. Journal of Virology, 64, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck, J.W. & Machamer, C.E. (2019) The infectious bronchitis coronavirus envelope protein alters Golgi pH to protect the spike protein and promote the release of infectious virus. Journal of Virology, 93, e00015‐19. 10.1128/JVI.00015-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C.G. , Jureka, A.S. , Silvas, J.A. , Nicolini, A.M. , Chvatal, S.A. , Carlson‐Stevermer, J. et al. (2021) Inhibitors of VPS34 and fatty‐acid metabolism suppress SARS‐CoV‐2 replication. Cell Reports, 36, 109479. 10.1016/j.celrep.2021.109479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, G. , Limpens, R.W.A.L. , Zevenhoven‐Dobbe, J.C. , Laugks, U. , Zheng, S. , de Jong, A.W.M. et al. (2020) A molecular pore spans the double membrane of the corona replication organelle. Science, 369, 1395–1398. 10.1126/science.abd3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, L.H. , Edgar, J.R. , Martello, A. , Ferguson, B.J. & Eden, E.R. (2021) Exploiting connections for viral replication. Frontiers in Cell and Development Biology, 9, 640456. 10.3389/fcell.2021.640456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R. , Shi, M. , Li, L. , Song, P. & Li, N. (2020) Construction of SARS‐CoV‐2 virus‐like particles by mammalian expression system. Frontiers in Bioengineering and Biotechnology, 8, 862. 10.3389/fbioe.2020.00862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Z. , Hu, B. , Xiao, H. , Tan, X. , Li, Y. , Tang, K. et al. (2022) The E3 ubiquitin ligase RNF5 facilitates SARS‐CoV‐2 membrane protein‐mediated virion release. mBio, 1, e0316821. 10.1128/mbio.03168-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Liu, L. , Lin, X. , Wang, Y. , Li, Y. , Guo, Q. et al. (2020) A translocation pathway for vesicle‐mediated unconventional protein secretion. Cell, 181, 637–652. [DOI] [PubMed] [Google Scholar]
- Zhang, H. & Zhang, H. (2021) Entry, egress and vertical transmission of SARS‐CoV‐2. Journal of Molecular Cell Biology, 13, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


