Abstract
Ni2+-binding staphylococci were generated through surface display of combinatorially engineered variants of a fungal cellulose-binding domain (CBD) from Trichoderma reesei cellulase Cel7A. Novel CBD variants were generated by combinatorial protein engineering through the randomization of 11 amino acid positions, and eight potentially Ni2+-binding CBDs were selected by phage display technology. These new variants were subsequently genetically introduced into chimeric surface proteins for surface display on Staphylococcus carnosus cells. The expressed chimeric proteins were shown to be properly targeted to the cell wall of S. carnosus cells, since full-length proteins could be extracted and affinity purified. Surface accessibility for the chimeric proteins was demonstrated, and furthermore, the engineered CBDs, now devoid of cellulose-binding capacity, were shown to be functional with regard to metal binding, since the recombinant staphylococci had gained Ni2+-binding capacity. Potential environmental applications for such tailor-made metal-binding bacteria as bioadsorbents in biofilters or biosensors are discussed.
Bacterial surface display of heterologous proteins has in recent years become an increasingly active research area with a wide range of applications in immunology, vaccinology, and biotechnology (11, 49). An interesting application area is the display of metal-binding proteins on bacteria to create whole-cell tools for improved sequestration of toxic metals in wastewater (3), a field in which naturally occurring bacteria have been evaluated previously (10, 31). The heterologous expression of peptides and proteins with inherent metal-binding capacity have been employed to create bacteria with improved metalloadsorption properties (39), and surface display has been used as an attractive strategy in this context (22, 38, 47, 51).
Short metal-binding peptides, such as hexahistidyl peptides, have been introduced into bacterial surface proteins in order to create bacteria with improved metal-binding capacity (21, 46). Using this strategy, both Escherichia coli (46) and Staphylococcus carnosus (43) strains with increased ability to bind Ni2+ and Cd2+ ions have been generated. The introduction of combinatorial protein engineering has also made it possible to select peptides, from large peptide libraries with increased selectivity for certain metals (4, 30, 37, 44), and such peptides might become interesting for surface display applications (44).
Gram-positive surface display systems have been suggested to exhibit some advantages compared to gram-negative bacteria (28, 49): (i) the translocation involves only a single membrane, and (ii) gram-positive bacteria have been shown to be more rigid and thus less sensitive to shear forces (19, 36) due to the thick cell wall surrounding the cells, and thus potentially more suitable for field applications such as bioadsorption. For metal adsorption applications, gram-positive bacteria have the additional advantage of having inherent metal-binding capacity due to the thick peptidoglycan layer (31).
A gram-positive bacterium that has been investigated extensively for various surface display applications is the nonpathogenic S. carnosus (26, 42) which is used traditionally in starter cultures in meat fermentation applications (18). Recombinant S. carnosus strains with various proteins expressed on the surface have been successfully evaluated as live bacterial vaccine delivery vehicles (5, 6, 50), for potential diagnostic applications through the display of single-chain Fv antibody fragments (16) and engineered S. aureus protein A domains, called affibodies (15).
Recently, a fungal cellulose-binding domain (CBD) derived from the cellobiohydrolase Cel7A of Trichoderma reesei was subjected to a combinatorial protein engineering approach (23). A combinatorial library comprising 46 million variants of the 36-amino-acid CBD domain was constructed through the randomization of 11 amino acid positions, including the residues involved in cellulose binding. Using phage display technology, CBD variants that showed specific binding of and ability to inhibit the target enzyme porcine α-amylase (PPA) could be selected (23). Furthermore, a related CBD, derived from T. reesei cellulase Cel6A, was recently expressed in its nonengineered form on the surface of S. carnosus (24). Cellulose binding was demonstrated in different whole-cell assays in which the recombinant staphylococci were found to bind efficiently to cotton fibers.
The proven capacity of CBD to be engineered (23) and displayed on the surface of bacteria (24) inspired us to investigate the possibility of using the CBD scaffold for metal capture as well. In this study we have evaluated a strategy to generate bacteria with an increased affinity for nickel ions by combining phage display-based combinatorial protein engineering with subsequent surface expression on S. carnosus cells. Potential nickel ion-binding CBD variants were selected from the constructed library by biopanning against Ni2+-magnetic agarose beads. Eight such engineered CBD variants were selected and investigated for correct staphylococcal cell wall targeting, surface accessibility, and proteolytic stability. In addition, the Ni2+-binding ability of the generated recombinant S. carnosus cells with the surface-displayed engineered CBD variants was investigated in a whole-cell assay.
MATERIALS AND METHODS
Preparation and transformation of staphylococcal protoplasts.
The preparation and transformation of protoplasts from S. carnosus were performed as described by Götz and coworkers (13, 14).
Preparation of phage stocks.
E. coli cells, approximately 109 cells of bacterial strain RRIΔM15 (40), containing the constructed CBD library (23), were added to 100 ml of TSB medium supplemented with 100 mg of ampicillin per liter and 2% glucose, incubated at 37°C to an A600 nm of ≈0.5, and then infected with helper phage M13K07 (5 × 1010 particles) (New England Biolabs) at 37°C for 30 min. Superinfected cells were harvested and used to inoculate TSB medium supplemented with ampicillin (100 mg/liter), kanamycin (25 mg/liter), and isopropyl-β-d-thiogalactoside (IPTG) (100 μM). The culture was grown overnight at 30°C with shaking. Cells were pelleted by centrifugation, and phage particles were concentrated from the supernatant by polyethylene glycol-NaCl precipitation. The phages were suspended in phosphate-buffered saline (PBS; 50 mM phosphate, 100 mM NaCl, pH 7.2) and filtered through an 0.45-μm filter (Sartorius, Göttingen, Germany). The phage stock was titrated by transfection into exponentially growing E. coli RR1ΔM15.
Phage selection of Ni2+-binding CBD variants.
Commercially available Ni-nitrilotriacetic acid (NTA) magnetic agarose beads (Qiagen, Hilden, Germany) were used for biopanning against Ni2+ ions. Beads were washed three times with PBS. Precipitated phage stock was preblocked by adding 2% gelatin (Difco, Detroit, Mich.) and incubating for 2 h in end-over-end rotation. Then 120 μl of the preblocked phage stock was suspended with 200 μl of a 5% suspension of Ni-NTA magnetic agarose beads. The suspension was incubated at room temperature for 2 h (end-over-end), and the beads were washed with PBS with 0.1% Tween 20 (PBST) and 20 mM imidazole: once in the first panning cycle, three times in the second cycle, six times in the third cycle, and eight times in the fourth and fifth cycles. Phage particles were eluted with 500 μl of 0.1 M glycine-HCl buffer (pH 2.2) by incubating for 10 min at room temperature. After elution, the beads were separated from the solution by magnetic sedimentation, and the supernatant containing eluted phage particles was neutralized by adding 50 μl of 1 M Tris-HCl (pH 8.5). Eluted phages were used to reinfect exponentially growing E. coli RR1ΔM15 (10 ml of cells, A600 ≈ 1), and a new phage stock was prepared as previously described. Five panning cycles were carried out. Randomly picked clones were sequenced by cycle sequencing with Big Dye terminators on the ABI377 (Perkin Elmer) system, and eight different CBD variants, CBD1 to CBD8, were selected for expression on bacterial surfaces.
DNA constructions.
Gene fragments encoding the eight novel CBD variants were PCR amplified using AmpliTaq polymerase (Perkin Elmer) and primers WeHe19 (5′-GGGGGTCGACGGATCCGGGTGCTAACCCAACCCAGTCTCACTACGGCCAG-3′) and WeHe20 (5′-GGGGGTCGACGGGTTGGCGCCGGGCAGGCACTGAG-3′) and pKN1 phagemids (32) carrying inserts corresponding to the selected CBD variants as templates. The generated gene fragments were ligated into the pGEMT vector system (Promega, Madison, Wis.). The CBD gene fragments were restricted from the pGEMT vector with endonucleases BamHI and SalI and ligated to pSPPmABPXM (42) previously restricted with the same enzymes. To determine the sequences of the selected CBD variants, the inserts were subjected to cycle sequencing as described above. The eight verified expression vectors pSPPCBD1ABPXM to pSPPCBD8ABPXM, designed for surface expression on S. carnosus, encode the surface-anchored fusion proteins PP-CBD1-ABP-XM′ to PP-CBD8-ABP-XM′, respectively. The expression vectors were used to transform S. carnosus TM300 (12) protoplasts to generate the eight different recombinant S. carnosus strains, which for simplicity were denoted Sc:CBD1 to Sc:CBD8.
Extraction and affinity purification of chimeric surface proteins.
The resulting recombinant staphylococci Sc:ABP (representing S. carnosus cells transformed with the parental vector pSPPmABPXM) and Sc:CBD1 to Sc:CBD8 were grown overnight at 37°C in 10 ml of tryptic soy broth (TSB) (Difco) (30 g/liter), supplemented with yeast extract (Difco) (5 g/liter) and chloramphenicol (Boehringer, Mannheim, Germany) (10 mg/liter). An aliquot of 0.5 ml of the overnight cultures was resuspended in 100 ml of the growth medium described above and grown to A578 of ≈ 0.8. Cells were harvested by centrifugation and washed twice with PBS (pH 7.5) before being resuspended in 5 ml of a modified SMMP medium (14) composed of 7.5 parts SMM (1 M sucrose, 0.04 M maleic acid, and 0.04 M MgCl2) and 2.5 parts of 7% Penassay antibiotic broth (Difco). The cells were then incubated with 50 U of lysostaphin (Sigma, St. Louis, Mo.) at 37°C for 2 h. The resulting protoplasts were pelleted by gentle centrifugation (6,000 × g, room temperature, 20 min), and the solubilized surface proteins PP-ABP-XM′ and PP-CBD1-ABP-XM′ to PP-CBD8-ABP-XM′ could be recovered from the supernatant by taking advantage of the albumin-binding protein ABP (42) from streptococcal protein G (34) via affinity chromatography on human serum albumin (HSA)-Sepharose columns (34). The solubilized chimeric surface proteins were diluted five times by adding water and 20× TST (0.5 M Trizma base-HCl [pH 8.0], 4 M NaCl, 20 mM EDTA, 1% Tween 20) to a final concentration of 1× TST, loaded onto HSA-Sepharose columns, and affinity purified as described by Ståhl and coworkers (48). Relevant purified fractions were pooled, lyophilized, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10 to 20% polyacrylamide) under reducing conditions. The gel was stained with Coomassie brilliant blue.
Enzymatic assay for detection of cell surface-exposed chimeric proteins.
Overnight cultures of recombinant and wild-type staphylococci were diluted 1:200 in growth medium (containing chloramphenicol when appropriate) and grown at 37°C to an A578 of ≈1. The cells were harvested and washed twice with PBS supplemented with 0.05% Tween 20 (PBST) (pH 7.5) before being resuspended in PBST to an A578 of 1. One-milliliter aliquots from these suspensions were incubated with biotinylated HSA (biotinylated with d-biotinoyl-Ε-aminocaproic acid N-hydroxysuccinimide ester [Boehringer, Mannheim, Germany] according to the supplier's recommendations) at a final concentration of 60 nM for 30 min at room temperature. Cells were washed three times in PBST prior to resuspension in 1 ml of PBST containing 0.5 U of streptavidin-alkaline phosphatase conjugate (Boehringer) and then incubated for another 30 min at room temperature. After three additional washes, the cells were diluted 1:5 in substrate buffer, and five aliquots of 100 μl of each cell type were loaded into a microtiter plate before addition of 100 μl of the substrate solution p-nitrophenyl phosphate (Sigma). The change in A405 was measured for 10 min in an enzyme-linked immunosorbent assay (ELISA) plate reader (Sunrise: Tecan, Grödingen, Austria).
Functional analysis of surface-displayed polyhistidyl peptides.
The Ni2+-binding assay was performed essentially in accordance to the colorimetric assay described above. However, instead of incubating the cells with biotinylated-HSA, Qiaexpress Ni-NTA-alkaline phosphatase conjugate (Qiagen, Hilden, Germany) was used. One-milliliter aliquots of cell suspension (A578 = 1) in PBST were incubated for 60 min at room temperature with Ni-NTA-alkaline phosphatase conjugate at a dilution of 1:500. The cells were then washed four times (the three first in PBST and the last in substrate buffer) before being resuspended in 1 ml of substrate buffer. Five aliquots of 100 μl of each cell type were loaded into a microtiter plate, after which 100 μl of the substrate solution (p-nitrophenyl phosphate) was added. The change in A405 was measured for 60 min in an ELISA reader.
RESULTS
Expression vectors for surface display of CBD variants on S. carnosus
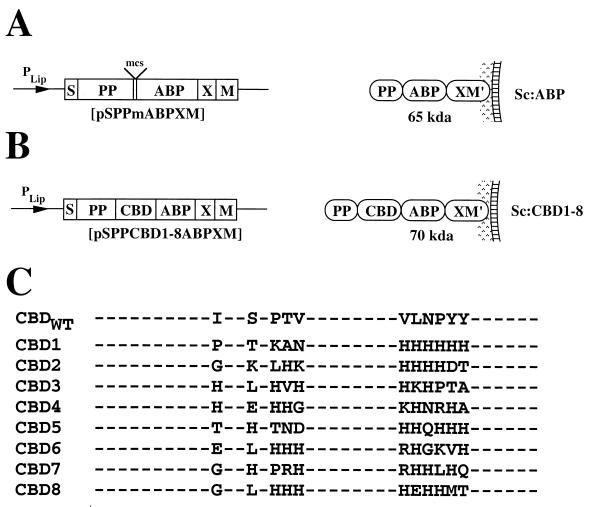
Eight randomly picked clones were sequenced after five consecutive rounds of biopanning. The results from the DNA sequencing of the selected CBD variants are shown in Fig. 1. Sequence analysis of selected CBD variants (Fig. 1C) revealed a striking preference for histidine residues at the randomized positions, with Sc:CBD1 containing six consecutive histidine residues and Sc:CBD6 also being rich in histidine residues. Other frequently occurring amino acids were, in descending order, Arg, Thr, Lys, Asn, Gly, Pro, Asp, and Glu. Eight novel E. coli-Staphylococcus shuttle vectors were constructed for display of chimeric surface proteins containing the phage-selected engineered CBD variants on S. carnosus. The gene fragments encoding the eight different CBD variants, here denoted CBD1 to CBD8, were introduced by PCR-based subcloning into the general surface expression vector pSPPmABPXM (42), designed for surface display on S. carnosus. The parental vector (Fig. 1A) and the eight novel expression vectors (Fig. 1B) are schematically depicted together with the encoded gene products PP-ABP-XM′ and PP-CBD1-ABP-XM′ through PP-CBD8-ABP-XM′.
FIG. 1.
Schematic representation of expression cassettes, encoded gene products, and identified amino acid residues at randomized positions in the engineered CBD variants. (A and B) Expression cassettes of the different expression vectors designed for surface display on S. carnosus, shown with their encoded gene products illustrated as anchored to the staphylococcal cell wall. The names of the constructed vectors are given below the expression cassettes, and the molecular masses of the encoded proteins are indicated together with the abbreviated names of the recombinant staphylococci. (C) Amino acid sequences of the wild-type and engineered CBD at the randomized positions.
The surface display system of S. carnosus takes advantage of the promoter, signal sequence, and propeptide region (PP) from a Staphylococcus hyicus lipase gene construct (25). For efficient anchoring to the cell wall, the vector also contains the cell wall-anchoring region of Staphylococcus aureus protein A (SpA). This consists of X, a charged repetitive region postulated to interact with the peptidoglycan cell wall (17), and M, a region common to many gram-positive cell surface-bound proteins that is required for cell surface anchoring (29, 45). M′ represents the processed and covalently anchored form of the M sequence of S. aureus protein A (29, 45). Also present in the vector is a multifunctional albumin-binding protein (ABP) derived from streptococcal protein G (34, 42). The ABP domain is expressed as the part of the chimeric surface protein closest to the cell wall-anchoring regions. This ABP region has proven to be useful as a reporter molecule in a colorimetric assay to analyze the surface accessibility of the expressed chimeric surface proteins (42), as an affinity tag for recovery of recombinant surface proteins (42), and also as a spacer protein to increase the surface accessibility of displayed proteins (50). Note that the PP from S. hyicus is not processed in S. carnosus (12), while it is cleaved off in its homologous host S. hyicus (1, 2). The PP has been shown to be essential for efficient translocation of heterologous gene fusions through the cell wall in S. carnosus when the S. hyicus promoter system is used (8, 41). The recombinant Staphylococcus strains will for simplicity be called Sc:ABP and Sc:CBD1 to Sc:CBD8, respectively.
Recovery and characterization of chimeric surface proteins.
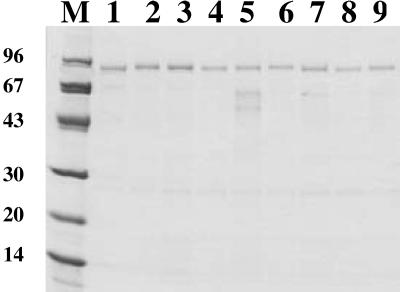
In order to investigate whether the expressed chimeric surface proteins (Fig. 1A and 1B) were successfully produced and targeted to the cell surface, the recombinant staphylococci Sc:ABP and Sc:CBD1 through Sc:CBD8 were cultivated to equal cell densities, harvested, and subjected to lysostaphin treatment to release cell wall-bound proteins without disrupting the bacterial membrane. After centrifugation, the supernatant containing the released proteins was loaded onto HSA columns for ABP-mediated affinity purification (34). The eluted proteins were then subjected to SDS-PAGE analysis followed by Coomassie staining of the gel (Fig. 2). Full-length proteins with little or no proteolytic degradation could be recovered from the cell wall fractions of the recombinant staphylococci. In accordance with earlier observations for proteins containing the lipase propeptide, the recovered cell wall proteins all migrated as somewhat larger than their calculated theoretical values (12, 42). The wild-type strain does not express any serum albumin-binding surface protein (42). These results demonstrate that the hybrid receptors were properly expressed and localized to the cell wall of the recombinant S. carnosus cells. Furthermore, the extracted hybrid surface proteins display serum albumin-binding capacity, since full-length fusion proteins could be recovered by HSA affinity chromatography. However, surface accessibility and functionality of the hybrid receptors on intact recombinant staphylococci remained to be demonstrated.
FIG. 2.
Characterization of gene products by SDS-PAGE (10 to 20% polyacrylamide) analysis under reducing conditions. The chimeric surface proteins were extracted from the cell wall of the recombinant staphylococci and subjected to ABP-mediated HSA affinity chromatography. Sc:ABP (lane 1), Sc:CBD1 (lane 2), Sc:CBD2 (lane 3), Sc:CBD3 (lane 4), Sc:CBD4 (lane 5), Sc:CBD5 (lane 6), Sc:CBD6 (lane 7), Sc:CBD7 (lane 8), Sc:CBD8 (lane 9); lane M, marker proteins, with molecular masses shown in kilodaltons.
Surface accessibility of chimeric surface proteins.
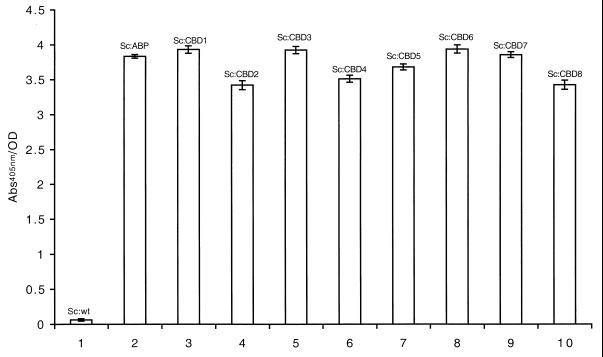
The surface accessibility of the displayed chimeric proteins on whole-cell staphylococci was analyzed by a previously described colorimetric assay (42), again by taking advantage of the ABP region present in the chimeric surface proteins as a reporter peptide. Recombinant and wild-type staphylococci were grown to early logarithmic phase, harvested, and then incubated with biotinylated HSA, followed by incubation with a streptavidin-alkaline phosphatase conjugate. The presence of surface-displayed ABP-containing surface proteins was detected using a chromogenic substrate. All recombinant staphylococci showed a significant positive response of similar magnitude (Fig. 3, bars 2 to 9), while wild-type S. carnosus (Fig. 3, bar 1), as expected, was negative in this assay. The chimeric surface proteins were thereby shown to be targeted and anchored, in an accessible form, to the outer surface of the recombinant staphylococci.
FIG. 3.
Histogram representation of results from the colorimetric assay for detection of surface-displayed receptors containing ABP. Wild-type (wt) and recombinant S. carnosus cells were incubated with biotinylated HSA (for binding to surface-exposed ABP-containing surface proteins). After addition of streptavidin-alkaline phosphatase and a chromogenic substrate, the color shift is monitored at 405 nm. Sc:wt (bar 1), Sc:ABP (bar 2), Sc:CBD1 (bar 3), Sc:CBD2 (bar 4), Sc:CBD3 (bar 5), Sc:CBD4 (bar 6), Sc:CBD5 (bar 7), Sc:CBD6 (bar 8), Sc:CBD7 (bar 9), Sc:CBD8 (bar 10). Error bars show standard deviation.
Whole-cell Ni2+ binding.
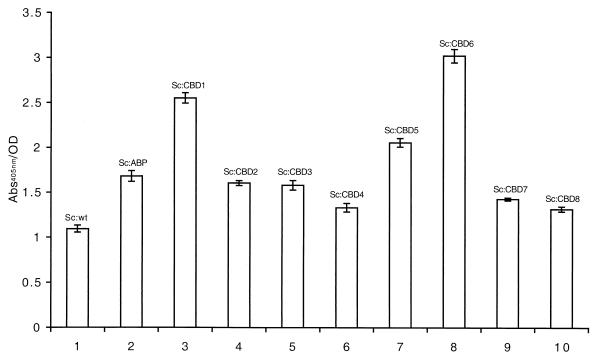
In order to investigate the metal-binding capacity of recombinant S. carnosus strains expressing the selected CBD variants, a whole-cell assay was developed. Recombinant and wild-type staphylococcal cells were grown to equal cell densities and incubated with a nickel-chelating alkaline phosphatase conjugate. After subsequent washing steps, a chromogenic substrate was added, and the color response was monitored (Fig. 4). It was demonstrated that recombinant S. carnosus cells carrying surface-displayed CBD variants (Fig. 4, bars 3 to 10) all showed a higher Ni2+-binding capacity than did wild-type S. carnosus (Fig. 4, bar 1). While it was expected that wild-type S. carnosus cells would show significant background binding to Ni2+ (Fig. 4, bar 1), most probably related to the inherent metal-binding capacity of the thick peptidoglycan layer (31), it is not evident why the staphylococci transformed with the parental vector (Fig. 4, bar 2) have gained improved metal-binding capacity. Similar observations have, however, been demonstrated previously (43).
FIG. 4.
Histogram representation of results from the whole-cell Ni2+-binding assay. Wild-type (wt) and recombinant staphylococci were incubated with a nickel-chelated alkaline phosphatase conjugate. After addition of substrate, the color shift was monitored at 405 nm. Sc:wt (bar 1), Sc:ABP (bar 2), Sc:CBD1 (bar 3), Sc:CBD2 (bar 4), Sc:CBD3 (bar 5), Sc:CBD4 (bar 6), Sc:CBD5 (bar 7), Sc:CBD6 (bar 8), Sc:CBD7 (bar 9), Sc:CBD8 (bar 10). Error bars show standard deviation.
Interestingly, two of the eight recombinant strains, Sc:CBD1 (Fig. 4, bar 3) and Sc:CBD6 (Fig. 4, bar 8), exhibited a substantial increase in Ni2+-binding capacity compared to Sc:ABP, while the remaining strains displayed intermediate capacities or no increase in Ni2+ binding. Taken together, these results demonstrate that 25% of the new recombinant staphylococcal strains generated through a combinatorial engineering approach and expressing the selected CBD variants CBD1 and CBD6 had significantly improved Ni2+-binding capacity compared to the parental strain.
DISCUSSION
We have in this study described how Ni2+-binding CBD selected by phage display technology from a combinatorial CBD protein library have been expressed as part of a chimeric surface protein on the gram-positive bacterium S. carnosus. The surface localization of the expressed CBDs were analyzed by lysostaphin treatment to release cell wall-bound proteins, and the extracted proteins were recovered by ABP-mediated HSA affinity chromatography. The cell wall localization of recombinant proteins was verified by SDS-PAGE analysis. The purified proteins were shown to be proteolytically stable, as they were produced in a full-length form with little or no degradation products present, indicating that the native cysteine bridges have been formed correctly. Furthermore, the displayed CBDs were shown to be accessible at the cell surface by a colorimetric assay based on the ABP domain. This demonstrates that the engineered CBD variants had also retained their capacity to be secreted and anchored at the bacterial cell surface.
The functionality of the recombinant S. carnosus strains, in terms of Ni2+-binding capacity, was evaluted using a whole-cell enzymatic assay involving a nickel-chelating alkaline phosphatase conjugate. The results clearly indicated that all recombinant strains had gained metal-binding capacity compared to the wild-type staphylococcal strain. Furthermore, two of the strains, Sc:CBD1 and Sc:CBD6, also showed a significant increase in Ni2+ binding compared to the cells expressing only the ABP, while the remaining six recombinant strains showed an intermediate or no increase in metal-binding properties compared to the ABP variant. Sequence analysis of all selected CBD variants revealed a marked preference for histidine residues at the randomized positions, corresponding to 41% of the total amino acid content at substituted codons. There does not seem to be a marked preference for certain positions for the histidine residues. In fact, histidine residues have been found in all of the randomized positions, taking into account all eight CBD variants investigated. This would indicate that the principle of phage-mediated biopanning against the Ni2+-NTA-agarose beads was indeed functioning, but that only two of the eight selected CBD clones could improve Ni2+ binding in the whole-cell format. A potential strategy to improve the Ni2+-binding capacity of the bacteria might be to use multimeric copies of the selected CBD variants in the chimeric surface proteins. This would increase the stoichiometric potential for metal binding but could also be advantageous from a steric perspective.
The obtained results would furthermore suggest that it might be possible to select CBDs with selective binding for specific metal ions if solid supports (such as agarose beads) with different chelated metal ions were available. Furthermore, if fluorescent reagents with chelated metal ions were available, the biopanning might be more conveniently performed using fluorescence-activated cell sorting of staphylococcal cells carrying a surface-displayed CBD library, as has been demonstrated in antibody maturation and enzyme evolution strategies (7, 35). Such a strategy would obviously have the advantage of eliminating the initial phage display selection procedure.
For the generation of tailor-made bacteria with specific binding properties, it would perhaps be interesting to explore other protein scaffolds with the capacity to selectively bind metal for surface display and metal adsorption. De novo design of mercury-binding two- and three-helix bundle protein domains has recently been demonstrated (9), and similar examples have also been presented (27). Metal-binding capacity has been engineered into protein domains occurring naturally on gram-positive bacteria, such as the B1 domain of streptococcal protein G (20). This might suggest that the combinatorial protein libraries based on a three-helical bundle S. aureus protein A domain (32, 33) could be used for selection of metal-binding domains, and such domains should be particularly suited to create recombinant staphylococci with selective metal-binding ability.
Although S. carnosus is a food-grade bacterium (18) and nonpathogenic (50), and thus safe to use as a microbial bioadsorbent in the development of biosensors, it is not evident that S. carnosus is optimal for treatment of wastewater and at other environmental sites. Further experimentation is needed to investigate how well the bacteria survive in different situations. Nevertheless, this investigation, in which metal binding has been engineered into a staphylococcal surface, should be of importance since it presents a novel strategy for recruiting new types of bacteria into environmental research.
In conclusion, we have demonstrated the possibility of using combinatorial protein engineering in the phage display format to select novel Ni2+-binding CBD variants and to use these binders to create recombinant staphylococci with increased Ni2+-binding capacity. This is the first study in which metal binding has been engineered into a gram-positive bacterium through a combinatorial protein engineering approach.
ACKNOWLEDGMENTS
We are grateful to Patrik Samuelson for valuable discussions.
This work was financially supported in part by the Swedish National Board for Technical and Industrial Development (NUTEK) and in part by the program Cell Factory for Functional Genomics within the Swedish Foundation for Strategic Research.
REFERENCES
- 1.Ayora S, Götz F. Genetic and biochemical properties of an extracellular neutral metalloprotease from Staphylococcus hyicus subsp. hyicus. Mol Gen Genet. 1994;242:421–430. doi: 10.1007/BF00281792. [DOI] [PubMed] [Google Scholar]
- 2.Ayora S, Lindgren P E, Götz F. Biochemical properties of a novel metalloprotease from Staphylococcus hyicus subsp. hyicus involved in extracellular lipase processing. J Bacteriol. 1994;176:3218–3223. doi: 10.1128/jb.176.11.3218-3223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brower J B, Ryan R L, Pazirandeh M. Comparison of ion-exchange resins and biosorbents for the removal of heavy metals from plating factory wastewater. Environ Sci Technol. 1997;31:2910–2914. [Google Scholar]
- 4.Brown S. Metal-recognition by repeating polypeptides. Nat Biotechnol. 1997;15:269–272. doi: 10.1038/nbt0397-269. [DOI] [PubMed] [Google Scholar]
- 5.Cano F, Liljeqvist S, Nguyen T N, Samuelson P, Bonnefoy J Y, Ståhl S, Robert A. A surface-displayed cholera toxin B peptide improves antibody responses using food-grade staphylococci for mucosal subunit vaccine delivery. FEMS Immunol Med Microbiol. 1999;25:289–298. doi: 10.1111/j.1574-695X.1999.tb01354.x. [DOI] [PubMed] [Google Scholar]
- 6.Cano F, Plotnicky-Gilquin H, Nguyen T N, Liljeqvist S, Samuelson P, Bonnefoy J, Ståhl S, Robert A. Partial protection to respiratory syncytial virus (RSV) elicited in mice by intranasal immunization using live staphylococci with surface-displayed RSV-peptides. Vaccine. 2000;18:2743–2752. doi: 10.1016/s0264-410x(00)00063-3. [DOI] [PubMed] [Google Scholar]
- 7.Daugherty P S, Chen G, Olsen M J, Iverson B L, Georgiou G. Antibody affinity maturation using bacterial surface display. Protein Eng. 1998;11:825–832. doi: 10.1093/protein/11.9.825. [DOI] [PubMed] [Google Scholar]
- 8.Demleitner G, Götz F. Evidence for importance of the Staphylococcus hyicus lipase pro-peptide in lipase secretion, stability and activity. FEMS Microbiol Lett. 1994;121:189–197. doi: 10.1111/j.1574-6968.1994.tb07098.x. [DOI] [PubMed] [Google Scholar]
- 9.Dieckmann G R, McRorie D K, Tierney D L, Utschig L M, Singer C P, O'Halloran T V, Penner-Hahn J E, DeGrado W F, Pecoraro V L. De novo design of mercury-binding two- and three-helical bundles. J Am Chem Soc. 1997;119:6195–6196. [Google Scholar]
- 10.Gadd G M. Bioremedial potential of microbial mechanisms of metal mobilization and immobilization. Curr Opin Biotechnol. 2000;11:271–279. doi: 10.1016/s0958-1669(00)00095-1. [DOI] [PubMed] [Google Scholar]
- 11.Georgiou G, Stathopoulos C, Daugherty P S, Nayak A R, Iverson B L, Curtiss R., 3rd Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat Biotechnol. 1997;15:29–34. doi: 10.1038/nbt0197-29. [DOI] [PubMed] [Google Scholar]
- 12.Götz F. Staphylococcus carnosus: a new host organism for gene cloning and protein production. Soc Appl Bacteriol Symp Ser. 1990;19:49S–53S. doi: 10.1111/j.1365-2672.1990.tb01797.x. [DOI] [PubMed] [Google Scholar]
- 13.Götz F, Ahrne S, Lindberg M. Plasmid transfer and genetic recombination by protoplast fusion in staphylococci. J Bacteriol. 1981;145:74–81. doi: 10.1128/jb.145.1.74-81.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Götz F, Schumacher B. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol Lett. 1987;40:285–288. [Google Scholar]
- 15.Gunneriusson E, Samuelson P, Ringdahl J, Grönlund H, Nygren P-Å, Ståhl S. Staphylococcal surface-display of immunoglobulin A (IgA) and IgE-specific in vitro selected binding proteins (affibodies) based on Staphylococcus aureus protein A. Appl Environ Microbiol. 1999;65:4134–4140. doi: 10.1128/aem.65.9.4134-4140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunneriusson E, Samuelson P, Uhlén M, Nygren P-Å, Ståhl S. Surface display of a functional single-chain Fv antibody on staphylococci. J Bacteriol. 1996;178:1341–1346. doi: 10.1128/jb.178.5.1341-1346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guss B, Uhlén M, Nilsson B, Lindberg M, Sjöquist J, Sjödahl J. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur J Biochem. 1984;138:413–420. doi: 10.1111/j.1432-1033.1984.tb07931.x. [DOI] [PubMed] [Google Scholar]
- 18.Hammes W P, Bosch I, Wolf G. Contribution of Staphylococcus carnosus and Staphylococcus piscifermentans to the fermentation of protein foods. J Appl Bacteriol Symp Suppl. 1995;79:76S–83S. [Google Scholar]
- 19.Kelemen M V, Sharpe J E. Controlled cell disruption: a comparison of the forces required to disrupt different microorganisms. J Cell Sci. 1979;35:431–441. doi: 10.1242/jcs.35.1.431. [DOI] [PubMed] [Google Scholar]
- 20.Klemba M, Gardner K H, Marino S, Clarke N D, Regan L. Novel metal-binding proteins by design. Nat Struct Biol. 1995;2:368–373. doi: 10.1038/nsb0595-368. [DOI] [PubMed] [Google Scholar]
- 21.Kotrba P, Doleckova L, de Lorenzo V, Ruml T. Enhanced bioaccumulation of heavy metal ions by bacterial cells due to surface display of short metal binding peptides. Appl Environ Microbiol. 1999;65:1092–1098. doi: 10.1128/aem.65.3.1092-1098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotrba P, Pospisil P, de Lorenzo V, Ruml T. Enhanced metallosorption of Escherichia coli cells due to surface display of beta- and alpha-domains of mammalian metallothionein as a fusion to LamB protein. J Recept Signal Transduct Res. 1999;19:703–715. doi: 10.3109/10799899909036681. [DOI] [PubMed] [Google Scholar]
- 23.Lehtiö J, Teeri T T, Nygren P-Å. Alpha-amylase inhibitors selected from a combinatorial library of a cellulose binding domain scaffold. Proteins. 2000;41:316–322. doi: 10.1002/1097-0134(20001115)41:3<316::aid-prot40>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 24.Lehtiö J, Wernérus H, Samuelson P, Teeri T T, Ståhl S. Directed immobilization of recombinant staphylococci on cotton fibers by functional display of a fungal cellulose-binding domain. FEMS Microbiol Lett. 2001;195:197–204. doi: 10.1111/j.1574-6968.2001.tb10521.x. [DOI] [PubMed] [Google Scholar]
- 25.Liebl W, Götz F. Studies on lipase directed export of Escherichia coli beta-lactamase in Staphylococcus carnosus. Mol Gen Genet. 1986;204:166–173. doi: 10.1007/BF00330205. [DOI] [PubMed] [Google Scholar]
- 26.Liljeqvist S, Samuelson P, Hansson M, Nguyen T N, Binz H, Ståhl S. Surface display of the cholera toxin B subunit on Staphylococcus xylosus and Staphylococcus carnosus. Appl Environ Microbiol. 1997;63:2481–2488. doi: 10.1128/aem.63.7.2481-2488.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Valentine J S. Engineering metal-binding sites in proteins. Curr Opin Struct Biol. 1997;7:495–500. doi: 10.1016/s0959-440x(97)80112-1. [DOI] [PubMed] [Google Scholar]
- 28.Malik P, Terry T D, Bellintani F, Perham R N. Factors limiting display of foreign peptides on the major coat protein of filamentous bacteriophage capsids and a potential role for leader peptidase. FEBS Lett. 1998;436:263–266. doi: 10.1016/s0014-5793(98)01140-5. [DOI] [PubMed] [Google Scholar]
- 29.Mazmanian S K, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 30.Mejàre M, Ljung S, Bülow L. Selection of cadmium specific hexapeptides and their expression as OmpA fusion proteins in Escherichia coli. Protein Eng. 1998;11:489–494. doi: 10.1093/protein/11.6.489. [DOI] [PubMed] [Google Scholar]
- 31.Mullen M D, Wolf D C, Ferris F G, Beveridge T J, Flemming C A, Bailey G W. Bacterial sorption of heavy metals. Appl Environ Microbiol. 1989;55:3143–3149. doi: 10.1128/aem.55.12.3143-3149.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nord K, Gunneriusson E, Ringdahl J, Ståhl S, Uhlén M, Nygren P-Å. Binding proteins selected from combinatorial libraries of an alpha- helical bacterial receptor domain. Nat Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 33.Nord K, Nilsson J, Nilsson B, Uhlén M, Nygren P-Å. A combinatorial library of an alpha-helical bacterial receptor domain. Protein Eng. 1995;8:601–608. doi: 10.1093/protein/8.6.601. [DOI] [PubMed] [Google Scholar]
- 34.Nygren P-Å, Palmcrantz E, Eliasson M, Abrahmsén L, Uhlén M. Analysis and use of the serum albumin binding domains of streptococcal protein G. J Mol Recognit. 1988;1:69–74. doi: 10.1002/jmr.300010204. [DOI] [PubMed] [Google Scholar]
- 35.Olsen M J, Stephens D, Griffiths D, Daugherty P, Georgiou G, Iverson B L. Function-based isolation of novel enzymes from a large library. Nat Biotechnol. 2000;18:1071–1084. doi: 10.1038/80267. [DOI] [PubMed] [Google Scholar]
- 36.Pagan R, Manas P, Raso J, Condon S. Bacterial resistance to ultrasonic waves under pressure at nonlethal (manosonication) and lethal (manothermosonication) temperatures. Appl Environ Microbiol. 1999;65:297–300. doi: 10.1128/aem.65.1.297-300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patwardhan A V, Goud G N, Koepsel R R, Ataai M M. Selection of optimum affinity tags from a phage-displayed peptide library. Application to immobilized copper(II) affinity chromatography. J Chromatogr A. 1997;787:91–100. doi: 10.1016/s0021-9673(97)00580-3. [DOI] [PubMed] [Google Scholar]
- 38.Pazirandeh M, Chrisey L A, Mauro J M, Campbell J R, Gaber B P. Expression of the Neurospora crassa metallothionein gene in Escherichia coli and its effect on heavy-metal uptake. Appl Microbiol Biotechnol. 1995;43:1112–1117. doi: 10.1007/BF00166934. [DOI] [PubMed] [Google Scholar]
- 39.Romeyer F M, Jacobs F A, Brousseau R. Expression of a Neurospora crassa metallothionein and its variants in Escherichia coli. Appl Environ Microbiol. 1990;56:2748–2754. doi: 10.1128/aem.56.9.2748-2754.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rüther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982;10:5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samuelson P, Cano F, Robert A, Ståhl S. Engineering of a Staphylococcus carnosus surface display system by substitution or deletion of a Staphylococcus hyicus lipase propeptide. FEMS Microbiol Lett. 1999;179:131–139. doi: 10.1111/j.1574-6968.1999.tb08718.x. [DOI] [PubMed] [Google Scholar]
- 42.Samuelson P, Hansson M, Ahlborg N, Andréoni C, Götz F, Bächi T, Nguyen T N, Binz H, Uhlén M, Ståhl S. Cell surface display of recombinant proteins on Staphylococcus carnosus. J Bacteriol. 1995;177:1470–1476. doi: 10.1128/jb.177.6.1470-1476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samuelson P, Wernérus H, Svedberg M, Ståhl S. Staphylococcal surface display of metal-binding polyhistidyl peptides. Appl Environ Microbiol. 2000;66:1243–1248. doi: 10.1128/aem.66.3.1243-1248.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schembri M A, Kjaergaard K, Klemm P. Bioaccumulation of heavy metals by fimbrial designer adhesins. FEMS Microbiol Lett. 1999;170:363–371. doi: 10.1111/j.1574-6968.1999.tb13396.x. [DOI] [PubMed] [Google Scholar]
- 45.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 46.Sousa C, Cebolla A, de Lorenzo V. Enhanced metalloadsorption of bacterial cells displaying poly-His peptides. Nat Biotechnol. 1996;14:1017–1020. doi: 10.1038/nbt0896-1017. [DOI] [PubMed] [Google Scholar]
- 47.Sousa C, Kotrba P, Ruml T, Cebolla A, De Lorenzo V. Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J Bacteriol. 1998;180:2280–2284. doi: 10.1128/jb.180.9.2280-2284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ståhl S, Sjölander A, Nygren P-Å, Berzins K, Perlmann P, Uhlén M. A dual expression system for the generation, analysis and purification of antibodies to a repeated sequence of the Plasmodium falciparum antigen Pf155/RESA. J Immunol Methods. 1989;124:43–52. doi: 10.1016/0022-1759(89)90184-1. [DOI] [PubMed] [Google Scholar]
- 49.Ståhl S, Uhlén M. Bacterial surface display: trends and progress. Trends Biotechnol. 1997;15:185–192. doi: 10.1016/S0167-7799(97)01034-2. [DOI] [PubMed] [Google Scholar]
- 50.Ståhl S, Samuelson P, Hansson M, Andréoni C, Goetsch L, Libon C, Liljeqvist S, Gunneriusson E, Binz H, Nguyen N, Uhlén M. Development of nonpathogenic staphylococci as vaccine delivery vehicles. In: Pozzi G, Wells J, editors. Gram-positive bacteria: vaccine vehicles for mucosal immunization. Georgetown, Tex: Landes Bioscience; 1997. pp. 62–81. [Google Scholar]
- 51.Valls M, Atrian S, de Lorenzo V, Fernandez L A. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat Biotechnol. 2000;18:661–665. doi: 10.1038/76516. [DOI] [PubMed] [Google Scholar]