Summary
Abiotic and biotic environments influence a myriad of plant‐related processes, including growth, development, and the establishment and maintenance of interaction(s) with microbes. In the case of the latter, elevated temperature has been shown to be a key factor that underpins host resistance and pathogen virulence.
In this study, we elucidate a role for Arabidopsis NON‐RACE‐SPECIFIC DISEASE RESISTANCE1 (NDR1) by exploiting effector‐triggered immunity to define the regulation of plant host immunity in response to both pathogen infection and elevated temperature.
We generated time‐series RNA sequencing data of WT Col‐0, an NDR1 overexpression line, and ndr1 and ics1‐2 mutant plants under elevated temperature. Not surprisingly, the NDR1‐overexpression line showed genotype‐specific gene expression changes related to defense response and immune system function.
The results described herein support a role for NDR1 in maintaining cell signaling during simultaneous exposure to elevated temperature and avirulent pathogen stressors.
Keywords: Arabidopsis, effector‐triggered immunity (ETI), heat stress, NON‐RACE‐SPECIFIC DISEASE RESISTANCE‐1 (NDR1), Pst‐AvrRpt2, salicylic acid
Introduction
Plant response to abiotic and biotic stress requires the coordinated activity of numerous cellular processes, the vast majority of which share overlapping functions in basic physiological programs, including growth, development, and reproduction (Nejat & Mantri, 2017; Saijo & Loo, 2020). In recent years, the impact of elevated temperature on plant growth and defense has received increasing attention, due in part to ongoing changes in global climate and environmental stress (Havko et al., 2020; Zhao et al., 2020; Zhang et al., 2022). However, the precise mechanisms that govern immunity at elevated temperature remain undefined.
Plant growth, development, and immune signaling processes are each influenced by fluctuations in temperature and environment (Zhu et al., 2003, 2010; Cheng et al., 2013; Bahuguna & Jagadish, 2015), the outcome of which is a reduction in vegetative plant growth (Quint et al., 2016), impacts on flower development and fertility (Balasubramanian et al., 2006; Koini et al., 2009; McClung & Davis, 2010), and the inhibition of plant defense signaling in response to a range of biotic threats (Wang & Hua, 2009; Wang et al., 2009). Not surprisingly, plants have evolved mechanisms to cope with simultaneous exposure to biotic and abiotic stress, and in this they utilize overlapping mechanisms not only to respond to stress but also to anticipate environmental changes for the purpose of regulating the timing and amplitude of seemingly opposing signaling processes (Quint et al., 2016; Gimenez et al., 2018; Saijo & Loo, 2020; Iqbal et al., 2021).
As a point of convergence with biotic stress signaling, changes in the abiotic environment have been shown to profoundly impact the plant immune system, including the activation, duration, and attenuation of signaling (Venkatesh & Kang, 2019). Indeed, recent studies have demonstrated that the function of at least two key nodes of the plant immune system – namely, pathogen‐associated molecular‐pattern‐triggered immunity (PTI) and effector‐triggered immunity (ETI) – are intimately associated with processes required for response to abiotic stress (Tsuda et al., 2009). ETI, which is manifested following the recognition of pathogen race‐specific avirulence (Avr) proteins (aka, effectors), is regulated by host‐plant‐derived resistance (R) genes (Jones & Dangl, 2006; P. Li et al., 2020; Z. Li et al., 2020). As a highly conserved family of proteins found in all plants, nucleotide‐binding leucine‐rich repeat (NB‐LRR) protein molecules mediate the specific recognition of pathogens via the indirect and/or direct recognition of both conserved and race‐specific virulence factors (Elmore et al., 2011).
In addition to NB‐LRR proteins, numerous additional processes have been identified as critical components of the immune signaling network (De Vleesschauwer et al., 2014; Tsuda & Somssich, 2015; Li & Day, 2019; Maier et al., 2021). Interestingly, research has also demonstrated a role for these (i.e. phytohormones, transcription factors) in abiotic stress signaling (Berens et al., 2019; Saijo & Loo, 2020). Among these, NON‐RACE‐SPECIFIC DISEASE RESISTANCE‐1 (NDR1) was identified nearly three decades ago as a critical component of plant immune system function (Century et al., 1995), with key functions associated with ETI and salicylic acid (SA)‐dependent, signaling networks in Arabidopsis (Lu, 2009; Lu et al., 2013). As a broader role for NDR1 in plant processes, recent work has shown that NDR1 and NDR1‐like genes (i.e. HIN; Bao et al., 2016) play important roles in stress response signaling (Lu et al., 2021). Among the best characterized examples of NDR1‐dependent immune signaling cascades is RESISTANCE TO PSEUDOMONAS SYRINGAE‐2 (RPS2) (Kunkel et al., 1993), an NB‐LRR‐encoding gene required for the recognition and activation of resistance in response to the Gram‐negative bacterial phytopathogen Pseudomonas syringae expressing the type III effector (T3E) protein AvrRpt2. Cleavage of RPM1‐interacting protein 4 (RIN4) by AvrRpt2 is required for the activation of RPS2‐based ETI (Mackey et al., 2003). In the absence of RPS2, AvrRpt2 promotes pathogen virulence in host cells (Mudgett, 2005). As a function for the role of NDR1 in RPS2 signaling, previous work demonstrated that RPS2‐mediated resistance is NDR1 dependent (Axtell et al., 2003). Furthermore, enhanced level of disease resistance has been observed in the NDR1‐overexpression line (Coppinger et al., 2004), indicating cleavage of RIN4 by AvrRpt2 would occur more rapidly in this line, thereby leading to the release of negative regulation on the R‐proteins RPS2 (Axtell & Staskawicz, 2003, 2010) and RPM1 (Mackey et al., 2003) and the subsequent activation of ETI.
Herein, we describe a role for NDR1 in plant immunity under heat stress conditions. Using a combination of physiological and transcriptome‐based approaches, we observed that in contrast to the temperature‐sensitive SA defense pathway gene ISOCHRISMATE SYNTHASE 1 (ICS1) (Huot et al., 2017), the NDR1‐overexpression line stabilizes ETI‐specific RPS2 messenger RNA (mRNA) accumulation at elevated temperature. Our findings suggest pathogen resistance at elevated temperature is mediated through crosstalk between NDR1 and RPS2, a mechanism that requires robust signaling of SA‐dependent processes.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh. seeds ecotype Columbia‐0 (Col‐0) were used as wild‐type (WT) plants, together with mutant plants (ndr1, ndr1/35S::NDR1, and ics1‐2), all of which are in the Col‐0 background. Plants were grown in Arabidopsis soil mix comprised of equal parts of Sure‐Mix (Sure, Galesburg, MI, USA), Perlite (PVP Industries, Orwell, OH, USA), and Vermiculite (PVP Industries). Plants were grown for 3–4 wk at 21°C under a 12 h : 12 h, light : dark cycle with 60% relative humidity and a light intensity of 120 μmol m−2 s−1 prior to heat stress in a BigFoot Series growth chamber (BioChambers, Winnipeg, MB, Canada). For temperature assays, plants were separated into two chambers set to either 21°C (permissive) or 29°C (elevated). Plants were subjected to heat stress treatment for 48 h at 29°C prior to pathogen treatment. The area of infiltration was marked to ensure that the leaf tissue subsequently collected for the assays contained bacterial inoculum.
Bacterial strains and disease assays
Pseudomonas syringae pv tomato (Pst) DC3000 harboring the open‐reading frames of the T3E, AvrRpt2, AvrPphB, AvrRpm1, as well as the empty vector (EV; pVSP61; Kunkel et al., 1993) were grown on NYGA (5 g l−1 Bacto‐peptone, 3 g l−1 yeast extract, and 20 ml l−1 glycerol, with 15 g l−1 agar for solid medium) containing 25 μg ml−1 kanamycin (kan) and 100 μg ml−1 rifampicin (rif) for 2 d at 28°C. After 48 h, bacterial cultures were resuspended in 5 mM magnesium chloride (MgCl2) at the desired concentrations for in planta growth assays. The same growing and culture suspension method was followed for the adenylate cyclase (CyaA) fusion‐protein‐tagged variant of AvrRpt2 and type‐III secretion system (T3SS) mutant hrcC− , with the exception that NYGA plates contained 25 μg ml−1 rif and 10 μg ml−1 gentamycin (gen).
In planta bacterial growth assays
Pst harboring AvrRpt2, AvrPphB, AvrRpm1, and EV (control) inoculums were prepared at an optical density at 600 nm OD600 nm = 0.0005 (5 × 105 CFU ml−1), OD600 nm = 0.1 (108 CFU ml−1) and OD600 nm = 0.0075 (7.5 × 106 CFU ml−1) for growth curve, hypersensitive response (HR), and benzothiadiazole (BTH) assays, respectively. Bacterial inoculations were performed on multiple (n > 3) fully expanded leaves from 3‐wk‐old Arabidopsis plants grown at permissive (21°C) and elevated (29°C) temperatures. Plants were inoculated with Pst isolates using a needleless syringe. Three biological replicates were performed for each assay. For in planta bacterial growth curve analyses, 3 mm leaf disks from three plants (three leaves per sample) were collected at 3 d after inoculation (DAI). Harvested leaf discs were incubated in 5 mM MgCl2 + 0.1% Tween‐20 at 28°C, on a rotary platform shaker, for 1 h. After 1 h, each sample was serially diluted (10‐fold increments) and 5 μl of each dilution was plated on NGYA plates containing half‐strength antibiotics (i.e. rif and kan). After 2 d incubation at 28°C, bacterial CFUs were counted. For HR analysis, infected leaves (24 h post‐inoculation (hpi)) were collected, and phenotypes were recorded by digital photography.
RIN4 Western blot analysis
Two leaves from each plant genotype were hand infiltrated (OD600 nm = 0.1; c. 108 CFU ml−1) with Pst expressing either AvrRpt2 or EV. Infiltrated leaves were collected at the designated timepoints, placed into a sterile 2 ml centrifuge tube, and were flash frozen in liquid nitrogen (N2). Samples were ground in extraction buffer (20 mM Tris pH 7.5, 150 mM sodium chloride, 1 mM EDTA, % Triton X‐100, 0.1% sodium dodecyl sulfate (SDS), 5 mM dithiothreitol (DTT), 10× Sigma protease inhibitor mixture) and centrifuged at 20 000 g for 10 min at 4°C. After centrifugation, the supernatant was collected as the total protein extract. Total protein of 50 μg was equalized using 6× loading buffer (0.375 M Tris pH 6.8, 12% SDS, 60% glycerol, 0.6 M DTT, 0.06% bromophenol blue) as a dilutant. Samples were separated by SDS polyacrylamide gel electrophoresis using 4–12% NuPAGE gels (Invitrogen) followed by transfer onto nitrocellulose membranes (GVS North America, Sanford, ME, USA) for Western blot analysis.
Polyclonal rabbit anti‐RIN4 antibody was produced by Cocalico Biologicals Inc. (Stevens, PA, USA). The specificity of anti‐RIN4 antibody was confirmed by Western blot analysis using WT Col‐0 and rps2/rin4 mutant plant lines, as well as transient expression of RIN4 protein in Nicotiana benthamiana. Anti‐RIN4 sera was used at a concentration of 1 : 5000 in 1× TBST (1 M Tris pH 8.0, 1% Tween 20, 5% dehydrated milk).
Phytohormone analysis
Leaves were infiltrated with Pst suspended in 5 mM MgCl2 expressing AvrRpt2, AvrRpm1, or Pst harboring pVSP61 (EV) using a 1 ml needleless syringe at a concentration of OD600 nm = 0.0005. Mock inoculation controls were performed using 5 mM MgCl2. Quantification of phytohormones was performed as previously described (Velasquez et al., 2017), with minor modifications. For hormone extraction and quantitative evaluation, frozen tissue was ground using a TissueLyser II (Qiagen) and incubated on a rocking platform at 4°C for 24 h in extraction buffer (80 : 20 v/v HPLC‐grade methanol : water with 0.1% formic acid (v/v), 0.1 g l−1 butylated hydroxytoluene). Samples were centrifuged at 12 000 g for 10 min at 4°C, and the resultant supernatants were collected and filtered through a 0.2 mm polytetrafluoroethylene membrane (Millipore).
Abscisic acid (ABA)‐d 6 (Toronto Research Chemicals Inc., North York, ON, Canada) served as an internal standard. Injections of plant extracts (10 ml per injection) were separated on a Waters Acquity BEH‐C18 column (2.1 mm × 50 mm, 1.7 mm) installed in the column heater of an Acquity ultraperformance liquid chromatography system (Waters Corp.). A gradient of 0.1% aqueous formic acid (solvent A) and methanol (solvent B) was applied in a 5 min program with a mobile phase flow rate of 0.4 ml min−1 as follows: 0–0.5 min hold at 98% A and 2% B, transition to 70% B at 3 min, to 99% B at 4 min, hold at 99% B to 5 min, return to 98% A at 5.01 min and hold at 98% A to 6 min. The column was maintained at 40°C and interfaced to a Waters Xevo TQ‐XS mass spectrometer equipped with electrospray ionization and operated in negative‐ion mode with a capillary voltage of 1.00 kV. The flow rates of cone gas and desolvation gas were 150 and 800 l h−1, respectively. The source temperature was 150°C, and the desolvation temperature was 400°C. Collision energies and source cone potentials were optimized for each compound using QuanOptimize software (Waters Corp.). Peak areas were integrated, and the analytes were quantified based on standard curves generated from peak area ratios of analytes. Data acquisition and processing were performed using Masslynx 4.1 software (Waters Corp., Milford, MA, USA). Analytes were quantified by converting peak area to phytohormone concentration (nanomolar) per gram of DW of leaf tissue using a standard curve specific to each compound.
Adenylate cyclase assay
To monitor Pst type‐III effector delivery, a CyaA assay was performed as previously described (Fu et al., 2006; Chakravarthy et al., 2017), with slight modification. In brief, leaves from 4‐wk‐old Arabidopsis plants were infiltrated with Pst expressing AvrRpt2‐CyaA or the T3SS mutant hrcC− carrying AvrRpt2‐CyaA suspended in 5 mM MgCl2 at a concentration of OD600 nm = 0.005 (c. 5 × 106 CFU cm−1) using a 1 ml needleless syringe. Leaf samples were harvested from two plants (two leaves per sample) at 0, 6, and 10 hpi and snap frozen in liquid N2. Cyclic adenosine monophosphate (cAMP) levels were quantified using the direct cAMP ELISA kit (ADI‐900‐066; Enzo Life Sciences, Farmingdale, NY, USA).
RNA extraction, library preparation, and RNA sequencing
RNA sequencing (RNA‐seq) analyses were performed on Arabidopsis plants representing four genotypes: WT Col‐0, ndr1, ndr1/35S::NDR1, and ics1‐1. Plants were grown at permissive temperatures (i.e. 21°C) for 23 d and then moved to elevated (i.e. 29°C) temperatures. Upon moving to 29°C, two fully expanded leaves from four different plants (eight leaves) were harvested as a single biological replicate at 0, 6, and 24 h. Tissue isolations were collected from three independent experimental replications, each containing three biological replicates. Total RNA was extracted using the RNeasy Plant Mini kit (Qiagen). DNA was removed from the sample by using a TURBO DNA‐Free™ kit (Thermo‐Fisher, Waltham, MA, USA). RNA samples were quantified using a Nanodrop 2000 spectrophotometer (Thermo‐Fisher).
Construction of strand‐specific RNA‐sequencing libraries
Construction of the RNA‐seq libraries and sequencing on the Illumina NovaSeq 6000 were performed at the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana‐Champaign. Total RNAs were run on a fragment analyzer (Agilent, Santa Clara, CA, USA) to evaluate RNA integrity. RNA‐seq libraries were constructed with the TruSeq Stranded mRNAs Sample Prep kit (Illumina, San Diego, CA, USA). Polyadenylated mRNAs were enriched from 500 ng of high‐quality DNA‐free total RNA with oligo‐dT beads. The final libraries were quantitated using Qubit (Thermo‐Fisher), and the average library fragment length was determined on a fragment analyzer. The libraries were diluted to 10 nM and further quantitated by quantitative PCR (qPCR) on a CFX Connect Real‐Time qPCR system (Bio‐Rad) for accurate pooling of the barcoded libraries and maximization of number of clusters in the flow cell. A total of 90 RNA‐seq libraries were prepared from 1 μg of total RNA.
Sequencing of libraries on the NovaSeq instrument
The pooled barcoded RNA‐seq libraries were loaded on a NovaSeq S2 lane for cluster formation and sequencing. Sequencing was performed by the Roy J. Carver Biotechnology Center at the University of Illinois, Urbana‐Champaign. Libraries were sequenced from one end of the fragments for a total of 100 nt. The Fastq read files were generated and demultiplexed with the Bcl2fastq v.2.20 conversion software (Illumina).
Expression and differential analysis
The adapter sequences and low‐quality bases (q < 10) were trimmed by Trimmomatic (Bolger et al., 2014). Resultant cleaned reads were mapped to the TAIR10 reference genome using Hisat2 (Kim et al., 2015). Mapped read counts for each gene were generated using the HTseq (Anders et al., 2015) command. The statistical analysis of the RNA‐seq data was performed in the R environment (v.4.0.5). Mitochondrial and chloroplast genes were excluded from analysis. Genes with mean read counts of fewer than 10 per library were also excluded from analysis. The resulting count data were subjected to trimmed‐mean of M‐values normalization using the function calcNormFactors in the package edger, followed by log‐transformation by the function voomWithQualityWeights in the package limma to yield log2 counts per million. To each gene, a linear model was applied using the lmFit function in the limma package with the following terms: Sgetr = GETget + ɛgetr, where S is the log2 expression value, GET is the (genotype : environment : time) interaction, r is the biological replicate, and ɛ is the residual. For variance shrinkage in the calculation of P‐values, the eBayes function in the limma package was used. Next, the resulting P‐values were corrected for multiple hypothesis testing by calculating the Storey q‐values using the function qvalue in the package qvalue. To extract genes with significant expression changes, the cutoff of q‐value < 0.01 and greater than two‐fold expression changes were applied. AgriGO was used for Gene Ontology (GO) enrichment analysis with default settings (Du et al., 2010). To create heatmaps, average linkage hierarchical clustering with uncentered Pearson correlation as a distance measure was carried out using Cluster 3.0 (Eisen et al., 1998), followed by visualization using Treeview (Eisen et al., 1998).
Coexpression network analysis was performed using the R package wgcna (Langfelder & Horvath, 2008). Genes with small expression variances (< 0.2) across the samples were excluded. Normalized and log2‐transformed read counts of the resulting 11 898 genes were used for constructing a signed hybrid network. The adjacency matrix was constructed using the adjacency function with the power of 14, and the topological overlap was then calculated from the adjacency matrix using the TOMsimilarity function. Average linkage hierarchical clustering was applied to the topological overlap for grouping genes with highly similar coexpression relationships. The Dynamic Hybrid Tree Cut algorithm was used to cut the hierarchal clustering tree, and 37 modules were defined as branches from the tree cutting. For the construction of the NDR1‐centered network, eigengene‐based gene connectivity, kME, was calculated using the signedKME function to select coexpression modules whose expression patterns are highly correlated to that of NDR1 (|kME| > 0.6). The relationships of these modules with NDR1 were visualized using Cytoscape (Shannon et al., 2003).
Results
Temporal dynamics of transcriptome responses to heat stress through NDR1‐dependent immune activation
The loss of NDR1 has a profound impact on pathogen defense signaling and disease resistance in plants (Century et al., 1995). Previous results suggest that one mechanism underpinning this activity may intersect with broader stress response processes, including those associated with plant hormone‐based signaling and the maintenance of cellular integrity (Knepper et al., 2011). To define how NDR1 influences plant response to abiotic stress response, we first conducted a comprehensive RNA‐seq analysis over a 24 h period following permissive temperature (i.e. 21°C) and heat stress (i.e. 29°C) exposure in WT Col‐0, the ndr1 mutant, a previously characterized ndr1/35S::NDR1‐overexpression line (Coppinger et al., 2004), and the SA‐deficient mutant ics1‐2. The impetus for this was to determine the rapid transcriptional responses required for signaling in response to pathogen infection and elevated temperature, as well as to define the potential priming of immune responses and their relationship to heat tolerance.
Hierarchical clustering analysis of 11 245 differentially expressed genes (DEGs) revealed gene expression changes over the 24 h time course across all genotypes (Fig. 1a; Supporting Information Table S1). Further analysis identified two significant gene clusters that showed significant response(s) to heat stress. The first, cluster 1, contains 800 genotype‐independent temperature‐responsive genes (Figs 1a, S1; Tables S2, S3). As revealed by GO enrichment analysis, this cluster contains a large number of genes involved in mitochondrial RNA editing, suggesting the role of mitochondrial RNA editing in acclimation to high temperature. This is supported by a recent paper reporting that an Arabidopsis mutant lacking the mitochondrial RNA editing enzyme GEND1 is hypersensitive to high temperature (Guo et al., 2021). Notably, cluster 2, which is comprised of 2151 genes, is enriched in transcripts that were highly expressed in the NDR1‐overexpression line (ndr1/35S::NDR1) and are related to defense response and immune system function based on GO enrichment analysis (Fig. 1c; Tables S4, S5). Interestingly, following 24 h exposure to elevated temperature, NDR1‐overexpressing plants had the greatest number of DEGs, up or downregulated, compared with ndr1 and ics1‐2 (with WT Col‐0 as a baseline) (Fig. 1b; Tables S6, S7).
Fig. 1.
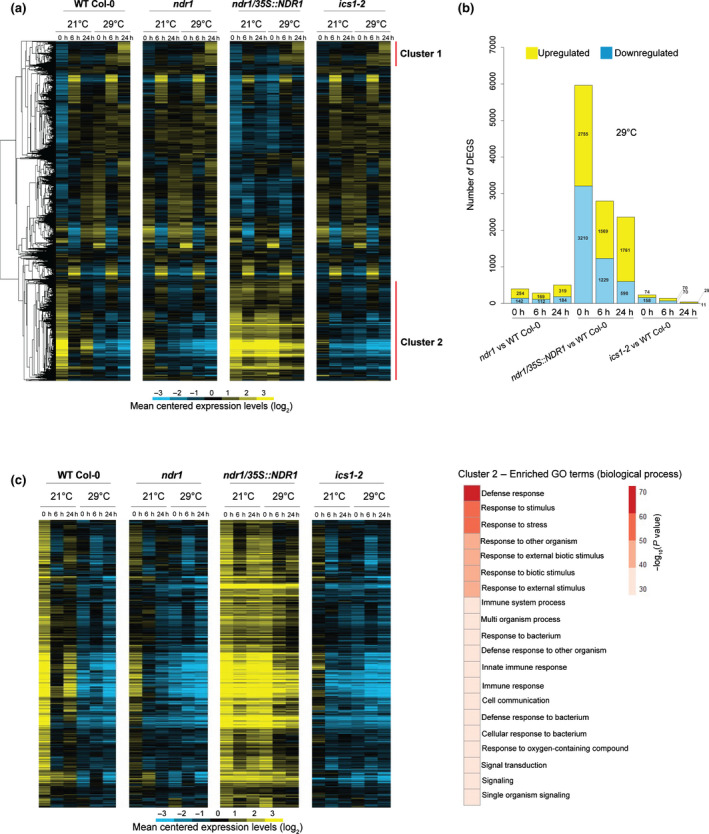
Temporal dynamics of transcriptome responses to heat stress through Non‐Race‐Specific Disease Resistant1 (NDR1)‐dependent immune activation in Arabidopsis. (a) Heat map showing log2‐fold gene expression changes over the 24 h heat stress. (b) Number of differentially expressed genes (DEGs) at 29°C over a 24 h time course in the ndr1, NDR1‐overexpression, and ics1‐2 mutant plants. (c) Heat map showing log2‐fold gene expression changes in cluster 2 genes that are highly expressed in ndr1/35S::NDR1 plants and are related to defense response and immune system based on Gene Ontology (GO) enrichment analysis. Blue indicates negative values, yellow indicates positive values, and black indicates zero.
To further evaluate how the NDR1 overexpression line maintains enhanced immune responses at elevated temperature, we next examined NDR1‐dependent and independent DEGs under heat stress. NDR1‐dependent genes were selected as DEGs that were upregulated (NDR1‐up) or downregulated (NDR1‐down) in ndr1/35S::NDR1 plants compared with WT Col‐0 after the exposure to elevated temperature at 29°C (q < 0.01 and or < −1). Heat‐responsive genes were selected as DEGs that were induced (heat‐induced) or suppressed (heat‐suppressed) in WT Col‐0 exposed to elevated temperature at 29°C compared with WT Col‐0 grown at 21°C (q < 0.01 and or < −1). As shown in Fig. S2, a large portion of heat‐suppressed genes overlap NDR1‐up genes at both 6 and 24 h. Similarly, there is a substantial overlap in the gene lists between NDR1‐down and heat‐induced genes. These results further support our claim that NDR1 overexpression maintains the expression of genes that are otherwise vulnerable to heat. Not surprisingly, little overlaps were found between NDR1‐up and heat‐induced genes and between NDR1‐down and heat‐suppressed genes, suggesting that NDR1 overexpression has little effect on the regulation of heat‐responsive genes (Tables S8–S11). Furthermore, GO terms ‘defense response’ and ‘response to salicylic acid’ are found in genes that are upregulated in NDR1‐overexpression line but suppressed by heat stress at both 6 and 24 h (Tables S12–S15). This further supports that NDR1 overexpression protects defense‐related genes from perturbation by heat. Genes that are suppressed in NDR1‐overexpression line but induced by heat at 6 h are associated with ‘response to water/ABA’. Taken together, overexpression of NDR1 imparts a preemptive activation of immunity by heat stress.
Overexpression of NDR1 results in sustained accumulation of RPS2 messenger RNA
The data described so far herein support a role for transcriptional induction of defense responses in the NDR1‐overexpression line at elevated temperature. This is exciting, as it points to a possible intersection between immunity and elevated temperature response through NDR1, a key regulator of ETI‐based immune activation and signaling. To gain insight into the role of NDR1 at the intersection of immunity and high‐temperature response, we performed a coexpression network analysis using the R package wgcna. This approach led to the identification of 37 modules with distinct expression patterns, as indicated by module eigengenes (MEs), which summarized the expression levels of the corresponding modules (Fig. S3). Using this, we calculated correlations of the expression pattern of NDR1 and those of MEs. From this, we selected correlated modules (|correlation coefficient| > 0.6) to construct an NDR1‐centered coexpression network (Fig. 2a). Within this network, NDR1 showed positive and negative correlations with modules 1 and 6 and modules 2 and 4, respectively. Modules 1 and 6 showed upregulation in the NDR1‐overexpression line, and this upregulation was maintained at elevated temperature (Fig. 2b). Further, these modules were enriched for genes associated with immunity‐related GO terms, such as ‘defense response’ and ‘innate immune response’ (Fig. 2a; Tables S16, S17). By contrast, modules 2 and 4 showed heat‐resistant downregulation in the NDR1‐overexpression line and were enriched for genes associated with photosynthesis and growth‐related GO terms (Fig. 2a; Tables S16, S17). The output of this analysis revealed that NDR1 overexpression activates defense‐associated gene expression and protects these expression networks from perturbation by elevated temperature.
Fig. 2.
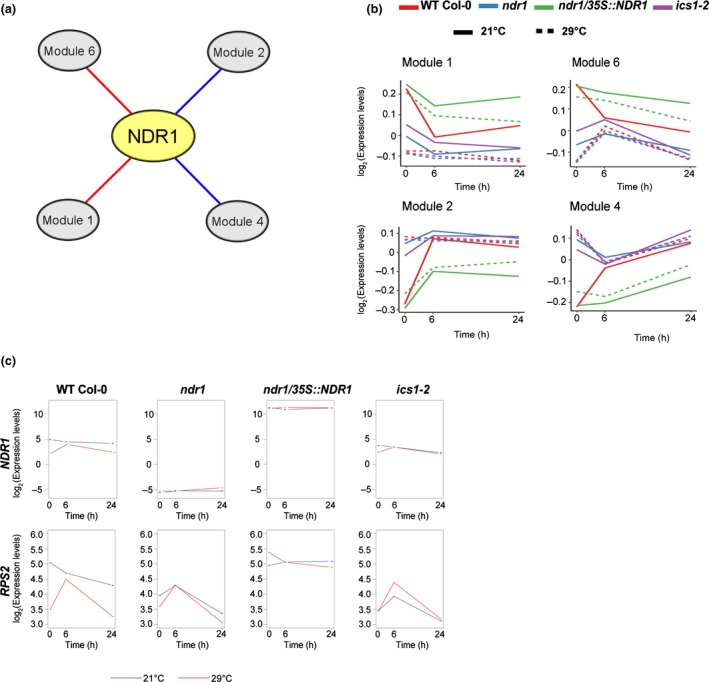
NON‐RACE‐SPECIFIC DISEASE RESISTANCE1 (NDR1) overexpression in Arabidopsis induces expression of immunity genes and protects it from perturbation by elevated temperature. (a) An NDR1‐centered coexpression network reveals modules whose expression levels are correlated at elevated temperature in the NDR1‐overexpression line. Red and blue edges indicate positive and negative correlation, respectively. (b) Averaged expression levels of genes in the modules summarized by module eigengenes. (c) Overexpression of NDR1 results in sustained accumulation of RESISTANCE TO PSEUDOMONAS SYRINGE‐2 (RPS2) messenger RNA at both 21°C and 29°C.
NDR1 is required for the activation of ETI through a defined set of NB‐LRR R‐proteins (e.g. RPS2, RPM1) (Day et al., 2006; van Wersch et al., 2020). Previous studies showed that RPS2 is required for Psm ES4326 AvrRpt2 and Pto DC3000 AvrRpt2‐induced SA accumulation and the induction of immune‐associated transcripts (Liu et al., 2016; Mine et al., 2018). Interestingly, we found that RPS2 is included in module 6 (i.e. heat‐resistant upregulation) in the NDR1‐overexpression line. Based on this, we further evaluated the mRNA accumulation of RPS2 and other key defense‐associated genes at both permissive (21°C) and elevated (29°C) temperatures (Figs 2c, S4). In contrast to the other genotypes, the downregulation of the R proteins RPS2, RPM1, and RPS1 together with NDR1 (control) at 24 h was not observed in the NDR1‐overexpression line at elevated temperature (Fig. 2c; Table S1). Expression of the key genes in SA response, ICS1, CALMODULIN BINDING PROTEIN 60g (CBP60g), and PATHOGENESIS RELATED GENE 1 (PR1) was reduced at elevated temperature, but still higher in the NDR1‐overexpression line than in the other genotypes (Fig. S4). The gene expression level changes observed at T 0 in all genotypes at 21°C and 29°C is likely due to occur through a combination of factors, such as the function of the genes themselves, changes occurring in response to the transfer of plants from permissive to elevated temperature chamber, and/or due to wounding during sampling.
To further define NDR1’s role as a regulator of general stress response signaling in Arabidopsis, we next asked if NDR1 is required for disease resistance signaling at elevated temperature. To do this, we first evaluated the activation of immune signaling in response to simultaneous exposure of elevated temperature and pathogen infection. Consistent with the requirement for NDR1 in the activation of RPS2‐mediated ETI, WT Col‐0 and ndr1/35S::NDR1, but not ndr1, responded to Pst‐AvrRpt2 with rapid induction of the HR at both permissive and elevated temperatures (Fig. 3a, top two panels). This result was consistent with the absence of disease symptoms in WT Col‐0 and the NDR1‐overexpression line, and the development of disease symptoms (e.g. chlorosis) (Fig. 3a, lower two panels) in both the ndr1 and ics1‐2 mutants. As a further confirmation of this interaction, we also evaluated the in planta bacterial growth at 3 DAI to examine the level of host resistance and/or susceptibility against Pst DC3000 (Pst) and Pst‐AvrRpt2. As shown, and consistent with the results of the HR assay, we observed enhanced susceptibility in plants lacking NDR1 (ndr1) and SA (ics1‐2), whereas WT Col‐0 and ndr1/35S::NDR1 showed resistance at elevated temperature (Fig. 3b). In planta bacterial growth at 0 hpi was also quantified to capture any population‐dependent growth rate differences (Fig. S5). Collectively, these results demonstrate that key regulators of SA, as well as the expression of NDR1‐dependent resistance signaling (e.g. RPS2), are enhanced in the NDR1‐overexpression line at elevated temperature.
Fig. 3.
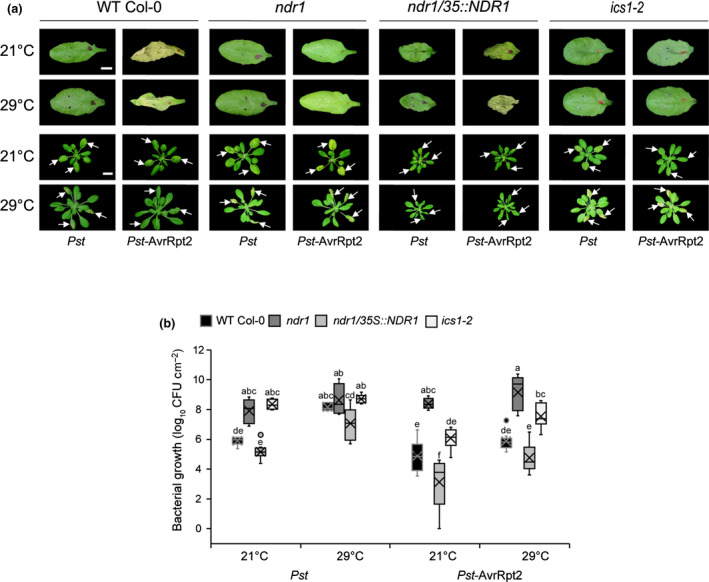
Disease resistance at elevated temperature is linked to stable RESISTANCE TO PSEUDOMONAS SYRINGE‐2 (RPS2) messenger RNA expression in ndr1/35S::NDR1 Arabidopsis plants. (a) Hypersensitive response at 24 h post‐infection (OD600 nm = 0.1) (upper panel) and disease symptoms (OD600 nm = 0.0005) at 3 d post‐inoculation (lower panel) at 21°C and 29°C. White arrows indicate leaves infiltrated. (b) Bacterial growth at 3 d after syringe‐infiltration with Pseudomonas syringae pv tomato (Pst) and Pst‐AvrRpt2 (OD600 nm = 0.0005) in wild‐type (Col‐0) and mutant plants at 21°C and 29°C. n represents the total number of leaves from three independent biological repeats (n = 9). Values are plotted as boxplots split by the median, and the whiskers show the range of data. Different letters represent a significant difference at P < 0.05 with Tukey’s honest significant difference (HSD) test. Bar, 0.5 cm. All data are representative of three independent experiments.
Pst‐AvrRpt2 promotes effector‐triggered immunity‐induced salicylic acid accumulation at elevated temperatures
To determine if the observed disease resistance phenotype in the NDR1‐overexpression line following challenge with avirulent Pst‐AvrRpt2 is mediated by SA at elevated temperatures, we quantified the level of SA in plants hand‐infiltrated with Pst and Pst‐AvrRpt2 at 24 hpi. Consistent with previous reports, we observed a decreased SA accumulation at elevated temperature, compared with those at permissive temperature (21°C), following mock and Pst treatment (Fig. 4a,b) (Huot et al., 2017). Intriguingly, we found that ETI triggered by Pst‐AvrRpt2 led to a significant increase in the levels of SA in WT Col‐0 (Fig. 4c). In addition, the SA levels in the ndr1/35S::NDR1‐overexpression line remained stable, in comparison with plants inoculated with the virulent pathogen Pst at elevated temperature (Fig. 4c). The low levels of SA in the ndr1 and ics1‐2 mutant plants are consistent with the observed susceptibility to Pst‐AvrRpt2 at both permissive and elevated temperatures (Fig. 3b).
Fig. 4.
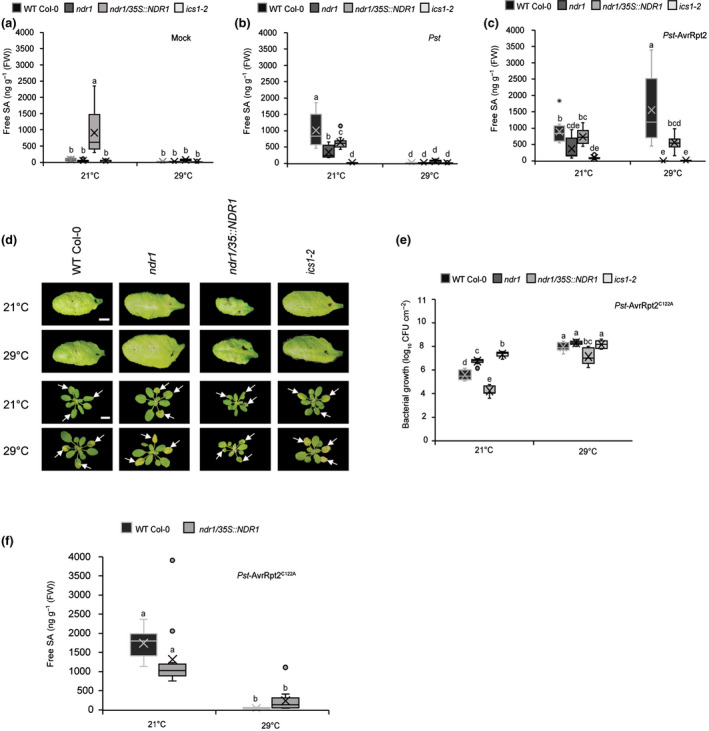
Pseudomonas syringae pv tomato (Pst)‐AvrRpt2 promotes effector‐triggered‐immunity‐induced salicylic acid (SA) synthesis at in Arabidopsis at elevated temperature. (a) Basal accumulation of total SA in wild‐type (Col‐0) and mutant plants. Leaves of mock infiltrated plants were harvested at 24 h post‐infection and evaluated for SA content. (b) Pathogen‐induced SA levels in wild‐type (Col‐0) and mutant plants treated with Pst and (c) Pst‐AvrRpt2 (OD600 nm = 0.0005). Leaves of pathogen‐infiltrated plants were harvested at 24 h post‐infection for SA quantification. (d) Hypersensitive response at 24 h post‐inoculation (OD600 nm = 0.1) (upper panel) and disease symptoms (OD600 nm = 0.0005) at 3 d post‐inoculation (lower panel) after syringe‐infiltration with Pst‐AvrRpt2C122A in wild‐type (Col‐0) and mutant plants. White arrows indicate leaves infiltrated. (e) Bacterial growth at 3 d after syringe‐infiltration with Pst‐AvrRpt2 C122A (OD600 = 0.0005) in wild‐type (Col‐0) and mutant plants. (f) Levels of free SA in wild‐type (WT) Col‐0 and ndr1/35S::NDR1 plants treated with Pst‐AvrRpt2C122A (OD600 = 0.0005). n represent total number of leaves from three independent biological repeats (for hormone quantification and disease assays, n = 12 and 9, respectively). Measures are plotted as boxplots split by the median, and the whiskers show the range of data. Different letters represent a significant difference at P < 0.05 with Tukey’s honest significant difference (HSD) test. Bar, 0.5 cm. All data are representative of three independent experiments.
To determine if overexpression of NDR1, and/or Pst‐AvrRpt2 infection, is responsible for the induction of SA at elevated temperature, we first evaluated the in planta bacterial growth in plants infiltrated with Pst‐AvrRpt2 and the AvrRpt2 cysteine protease mutant AvrRpt2C122A (Kim et al., 2005). As expected, in the absence of the cysteine protease activity of AvrRpt2 (e.g. Pst alone or AvrRpt2C122A), we observed the absence of HR elicitation and the development of pronounced disease phenotypes in all plant lines at both permissive and elevated temperatures (Fig. 4d,e). Next, to define the link between the cysteine protease activity of AvrRpt2 and the induced accumulation of SA, we quantified the level of SA in WT Col‐0 and the NDR1‐overexpression plants at 24 hpi with Pst‐AvrRpt2C122A. In contrast to elevated levels of SA in plants following Pst‐AvrRpt2 infection, we observed low levels of SA at elevated temperature in WT Col‐0 and NDR1‐overexpression plants following AvrRpt2C122A inoculation (Fig. 4f). Coupled with the aforementioned results (Fig. 4c), these data support a role for Pst‐AvrRpt2‐associated temperature‐independent SA levels in WT Col‐0 or the NDR1‐overexpression plants grown at elevated temperatures. Based on this, we hypothesize that the observed resistance at elevated temperature is mediated by Pst‐AvrRpt2‐induced SA production/stabilization, as well as through NDR1 overexpression.
Overexpression of NDR1 leads to enhanced stability of RIN4 in the presence of Pseudomonas syringae expressing Pst‐AvrRpt2
To further define the mechanism(s) underpinning the observation of pathogen‐induced SA and the sustained accumulation of SA at elevated temperatures in the NDR1 overexpression line, we first investigated the activation of ETI through the NDR1–RIN4 signaling node. We first evaluated the activity of the T3E cysteine protease AvrRpt2, by examining its ability to cleave RIN4 (Axtell et al., 2003; Chisholm et al., 2005). To begin, we quantified the RIN4 protein levels of the untreated plants at both permissive and elevated temperatures. The RIN4 protein levels were similar at both temperatures, as shown in Fig. S4(a). Next, we observed a decrease in RIN4 protein stability over time in the presence of Pst‐AvrRpt2 at both permissive and elevated temperatures in WT Col‐0, as well as in the ndr1 and ics1‐2 mutants (Fig. 5a). Interestingly, in the ndr1/35S::NDR1‐overexpression line, we did not observe a reduction in RIN4 following infection with Pst‐AvrRpt2, suggesting that overexpression of NDR1 may protect RIN4 from cleavage. As expected, we observed no RIN4 disappearance following Pst inoculation over the same time frame (Fig. S6; Table S18).
Fig. 5.
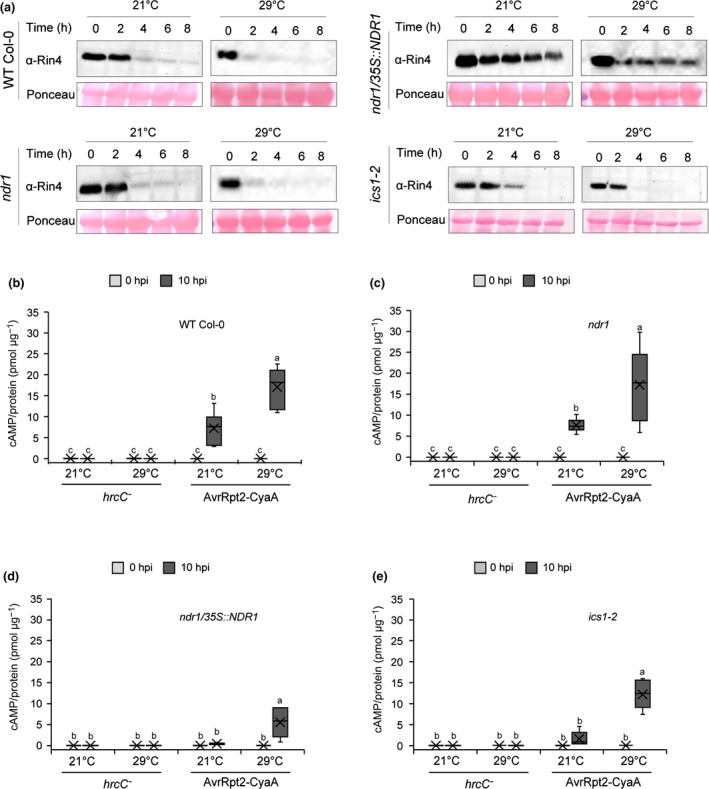
Pseudomonas syringae pv tomato (Overexpression of NON‐RACE‐SPECIFIC DISEASE RESISTANCE1 (NDR1) in Arabidopsis results in enhanced RPM1‐interacting protein 4 (RIN4) stability in the presence of Pst)‐AvrRpt2. (a) Detection of RIN4 at 0, 2, 4, 6, and 8 h after syringe‐infiltration with Pst‐AvrRpt2 (OD600 nm = 0.1) in wild‐type (Col‐0) and mutant plants. The total protein extracts were subjected to ⍺‐RIN4 Western blot. Equal loading of protein was verified by ponceau S staining of the membrane after protein transfer. (b–e) Effector translocation in ndr1, ndr1/35S::NDR1, and ics1‐2, respectively, following syringe‐infiltration with Pst hrcC− or Pst‐expressing AvrRpt2‐CyaA (OD600 = 0.005). Tissue was collected at 0 and 10 h post‐inoculation (hpi) for quantification of cyclic adenosine monophosphate (cAMP) which was normalized by total protein. Higher levels of cAMP indicate more translocation of bacterial effectors. n represents the total number of leaves from three independent biological repeats (n = 6). Values are plotted as boxplots split by the median, and the whiskers show the range of data. Different letters represent a significant difference at P < 0.05 with Tukey’s honest significant difference (HSD) test. All data are representative of three independent experiments.
To gain a comprehensive understanding of the role of NDR1 in ETI at elevated temperature, we further conducted disease assays in WT Col‐0, ndr1, ndr1/35S::NDR1, and ics1‐2 plants following infection with Pst expressing, individually, the T3Es AvrRpm1 or AvrPphB. As shown, we observed comparable disease resistance to Pst‐AvrRpt2 following both T3E infections in the NDR1‐overexpression line at elevated temperature (Figs S7, S8). Based on these data, we surmise that enhanced resistance in ndr1/35S::NDR1 against T3Es may be the result of their respective R protein interactions – both genetic and potentially physical – with NDR1, as well as via a yet to be defined function for NDR1 in SA‐dependent signaling cascades.
To determine how overexpression of NDR1 and the associated increase in SA might function in the activation of resistance, we next monitored the release of the negative regulation of immunity via the cleavage of RIN4 by the T3E cysteine protease AvrRpt2 (Axtell et al., 2003). To do this, we first evaluated the translocation of AvrRpt2 into plant cells via the T3SS at 0, 6, and 10 h via the infection of plants with Pst‐expressing AvrRpt2 fused to a CyaA reporter (i.e. AvrRpt2‐CyaA). As a negative control for these experiments, we employed a T3SS mutant hrcC− carrying AvrRpt2‐CyaA to ensure that detected in planta levels of CyaA arose via the action of a functional T3SS (Li et al., 2017). Additionally, in planta bacterial levels at each time point were quantified to eliminate any population‐dependent translocation rate errors (Fig. S9). Consistent with previous studies that evaluated the impact of elevated temperatures on T3E translocation (Huot et al., 2017), we observed increased levels of cAMP at elevated temperature in all four plant genotypes compared with plants grown at permissive temperatures (i.e. 21°C; Fig. 5b–e). However, at 10 h, we observed the lowest levels of cAMP at elevated temperature in the NDR1‐overexpression line compared with WT Col‐0, followed by ics1‐2 and ndr1 mutants (Fig. 5b–e). These data are consistent with the increased stability of RIN4 in the NDR1‐overexpression line (Fig. 5a) and support a role for NDR1 in protecting RIN4 in the presence of Pst‐AvrRpt2.
Previous work demonstrated that the plant defense inducer BTH induces pathogen resistance in an SA‐dependent manner (Huot et al., 2017; Kouzai et al., 2018). To further uncouple the role of NDR1 and SA as a function of pathogen T3E activity, we first evaluated the effect of BTH on AvrRpt2‐CyaA effector translocation in plants grown at both permissive and elevated temperatures. At neither temperature did we observe a significant change in the level of cAMP following BTH treatment, with the exception for in the ics1‐2 line; we hypothesize that this is due to the low cAMP amount detected with mock treatment (Fig. S10a,b), which is likely due to subtle differences in buffer content (e.g. dimethyl sulfoxide). Indeed, the lower levels of cAMP observed in BTH‐treated ics1‐2 plants is consistent with our observations presented in Fig. 5, wherein the amount of effector translocation was reduced in the NDR1‐overexpression line, which also has increased levels of SA. Thesr data agree with the results shown in Fig. 4(a), wherein mock‐treated NDR1‐overexpression lines also had elevated levels of SA. As a control for these assays, we also enumerated in planta bacterial levels at 0 hpi to eliminate any population‐dependent translocation rate errors (Fig. S11a,b). Overall, this result suggests that lower translocation rates observed in the NDR1‐overexpression line (Fig. 5d) could be a consequence of the elevated levels of SA in this line.
Having demonstrated the impact of SA on bacterial T3E translocation into the host cell, we next queried the role of SA on RIN4 cleavage by Pst‐AvrRpt2, a function required for the robust activation of R‐protein (e.g. RPS2)‐mediated ETI (Axtell et al., 2003) following T3E (i.e. AvrRpt2) delivery and recognition. At the onset of this line of investigation, our working hypothesis was that, given the enhanced level of resistance in the NDR1‐overexpression line (Coppinger et al., 2004), cleavage of RIN4 by AvrRpt2 occurs more rapidly in this line, thereby leading to the release of negative regulation on the R‐proteins RPS2 (Axtell et al., 2003) and RPM1 (Mackey et al., 2003) and the robust activation of ETI. To test this, we first monitored the levels of RIN4 protein in BTH‐treated plant lines hand‐infiltrated with Pst‐AvrRpt2. As shown in Fig. S10(c,d), exogenous application of BTH did not protect RIN4 from cleavage by AvrRpt2 in WT Col‐0, nor in the ndr1 or ics1‐2 mutants. However, similar to results observed in Fig. 5(a), overexpression of NDR1 did result in enhanced protection of RIN4 from cleavage by Pst‐AvrRpt2. Based on this result, we conclude BTH‐induced SA does not protect RIN4 under Pst‐AvrRpt2 treatment. Thus, the inability of SA to protect RIN4 from cleavage, coupled with the observed RIN4 protection in the NDR1‐overexpression line, is likely due to the physical interaction, and stoichiometry of this association, between RIN4 and NDR1.
Discussion
Plant immune signaling during heat stress response has been described since the early 1900s, wherein it was demonstrated that the spread of tobacco mosaic virus necrotic lesions in Nicotiana glutinosa‐infected leaves was more prevalent at elevated temperatures (Samuel, 1931). More than 75 yr after this discovery, similar correlations have been described as they relate to the impact of elevated temperature on plant growth (Penfield, 2008), reproduction (McClung & Davis, 2010), and hormone signaling (Sakata et al., 2010). More recent work has shown that plant resistance to pathogens is reduced under conditions of elevated temperature, a phenomenon that is hypothesized to be associated with the downregulation of SA signaling (Li et al., 2010; Huot et al., 2017). Collectively, these studies have provided foundational support for the ‘growth–defense’ paradigm (Guo et al., 2018). In the current study, to expand our understanding of the mechanisms that function at the nexus of heat stress response and immune signaling activation, we focused on the activation of a well‐defined and genetically tractable immune signaling cascade, ETI.
Previous studies have shown that plant response(s) to both biotic and abiotic stimuli are initiated by rapid, highly specific, changes in the transcriptional landscape; notably, the induction of genes associated with plant defense (Hu et al., 2012), and the attenuation of those required for growth and reproduction (Lee et al., 2014; Quint et al., 2016). These observations have led to the development of models that describe an important role for the co‐regulation of processes that function antagonistically during simultaneous exposure to abiotic and biotic stressors (Hossain et al., 2018; Kim et al., 2020; Lu et al., 2021). To dissect the role of elevated temperature on the activation of ETI, we generated 69 transcriptomes from four plant lines that have reported varied responses related to pathogen infection and hormone signaling, under permissive and elevated temperature (Century et al., 1997; Tao et al., 2003; Strawn et al., 2007; Catinot et al., 2008; Li et al., 2021). Through this approach, we identified two main clusters of DEGs that segregated based on genotype‐independent expression, as well as those that were regulated in a genotype‐specific manner under elevated temperature. In the case of the latter, this cluster was comprised of highly expressed genes in the NDR1‐overexpression line, many of which were related to GO terms including defense response and immune system function. Interestingly, our analysis revealed the stability of RPS2 mRNA at elevated temperatures in the NDR1‐overexpression line, a phenomenon we hypothesize is possibly attributable to downstream, preemptive, transcriptional activation of defense, and/or a consequence of elevated SA levels in the NDR1 overexpressor. In either case, we posit that such a response would prime the immune system for protection during simultaneous biotic and abiotic stress exposure.
Phytohormones are an indispensable component of the plant immune system, required for the robust activation of both PTI and ETI (Miller et al., 2017; Yuan et al., 2021). Interestingly, SA production is also affected when plants are exposed to both low (P. Li et al., 2020; Z. Li et al., 2020) and elevated (Huot et al., 2017) temperatures. As immune signaling modulators, previous work showed that RPS2 is required not only for the production of SA, but also for the generation of pathogen‐induced jasmonic acid and ABA production, supporting the hypothesis that, to some degree, defense hormone production is ETI dependent (Liu et al., 2016). In contrast to published data showing loss of virulent Pst‐induced SA biosynthesis at elevated temperature, we observed that avirulent Pst‐AvrRpt2 promotes SA synthesis in a temperature‐independent manner in WT Col‐0 and the NDR1‐overexpression line at 29°C (Huot et al., 2017). Here, we observed that SA production was compromised in the ndr1 mutant, similar to that in the SA‐deficient ics1‐2 mutant line at elevated temperature. This is interesting; and with observed stability of RPS2 mRNA at elevated temperature, high SA levels following Pst‐AvrRpt2 treatment in the NDR1‐overexpression line may be an underlying mechanism that contributes to reduced effector translocation.
Recent studies aimed at identifying the molecular–genetic mechanisms controlling immune signaling stability at elevated temperature have uncovered a relationship between an increase in temperature and the sustainable activity of host R‐proteins (Venkatesh & Kang, 2019). For example, Arabidopsis plants subjected to a long‐term (c. 10 d) temperature acclimation at 28°C resulted in an approximate eight‐fold increase in the in planta growth at 3 DAI of the virulent pathogen Pst compared with the plants grown at 22°C after 3 DAI. Furthermore, the same study also demonstrated that Pst expressing the T3Es AvrRpt2 and AvrRpm1 showed 10 times more bacterial growth at 28°C compared with 22°C, indicating the plant defense responses mediated by R‐genes are likely suppressed at higher temperatures (Wang et al., 2009), and/or are affected by the virulence activity of these effectors. In support of the former, it has been demonstrated that R‐protein stability is linked to the presence of SA, which, as already noted, plays an indispensable role in the plant defense response to bacterial pathogens. For example, the R gene‐like toll/interleukin‐1 receptor (TIR)‐NB‐LRR‐type gene SUPPRESSOR OF NPR1‐1 CONSTITIUTIVE1 (SNC1) has emerged as a case‐study for SA‐dependent resistance signaling and a model defining the crosstalk between the R genes and hormones (Zhang et al., 2003; Yang & Hua, 2004). Interestingly, SNC1 protein accumulation is reduced at elevated temperatures, a phenomenon that is coincident with the reduction of SA at elevated temperatures (Zhu et al., 2003, 2010; Huot et al., 2017). Using a mutagenesis‐based approach, 102snc1‐1 was identified, which showed pathogen resistance at both basal and elevated temperatures. This temperature‐insensitive immune response in the 102snc1‐1 was further attributed to the high expression level of PR1, further supporting the involvement of SA (Zhu et al., 2003, 2010). Similarly, we observed sustained PR1 mRNA accumulation/gene expression under elevated temperature in the NDR1‐overexpression line.
Mounting evidence suggests that both nuclear localized TIR‐NB‐LRRs and plasma‐membrane‐localized CC‐NB‐LRR receptor‐mediated signaling pathways in a temperature‐sensitive manner (Mang et al., 2012; Cheng et al., 2013). Unexpectedly, in contrast to the reported suppression of RPS2 mediated ETI signaling at elevated temperatures, we found that the NDR1‐overexpression line remained resistant at elevated temperature. Previous work demonstrated that temperature acclimation at 32°C for 6 h (i.e. short‐term) did not impact the mRNA accumulation of key NB‐LRR signaling components (e.g. RPM1, RPS2, RIN4, and NDR1) in the absence of pathogen infection (Cheng et al., 2013). Here, our data revealed the stable expression level of RPS2 gene in the NDR1‐overexpression line in the absence of pathogen infection under long‐term (24 h) heat stress, an observation that suggests the early establishment of defense gene expression could be a preemptive strategy to defend against pathogen infection under conditions of environmental stress.
Previous results showed that RIN4 cleavage by AvrRpt2 occurs within c. 8 hpi at both 23°C and 32°C (Cheng et al., 2013). Consistent with this, we showed RIN4 degradation in WT Col‐0, ndr1, and ics1‐2 at basal and elevated temperatures. However, in the NDR1‐overexpression line, RIN4 remained protected from cleavage by AvrRpt2, suggesting a role of RIN4 protection conferred by the overexpression of NDR1. Here, we demonstrate that BTH‐induced SA production was not sufficient to protect RIN4 from cleavage in WT Col‐0, ndr1, and ics1‐2, thereby eliminating the possibility of the involvement of the high levels of SA in the NDR1‐overexpression line in the protection of RIN4. The simplest explanation for this is that overexpression of NDR1, and the interaction of NDR1 with RIN4, protects RIN4 from cleavage. What remains unclear is how NDR1 overexpression results in reduced effector translocation, an observation previously observed in ics1‐2 at both basal and elevated temperatures (van Dijk et al., 1999; Huot et al., 2017). Moreover, reduced effector translocation in the NDR1 overexpressor would appear to be in conflict with enhanced resistance in this line (Coppinger et al., 2004). The simplest explanation is that only a small amount of RIN4 cleavage is required for full activation of ETI, a mechanism that ensures the robust activation of resistance following release of RIN4 negative regulation. In total, we propose that the rescue of ETI in the absence of RIN4 degradation in the NDR1‐overexpression line is potentially due to a complex interaction involving the stability of RPS2 mRNA expression and a decrease in T3E translocation rates at elevated temperature (Fig. 6). Though much work remains towards fully defining the role of NDR1 at the nexus of biotic and abiotic signaling, the data herein provide insight into a role for NDR1 as a stabilizing component, and potential scaffolding mechanism, required for the maintenance of homeostasis during abiotic and biotic stress response signaling.
Fig. 6.
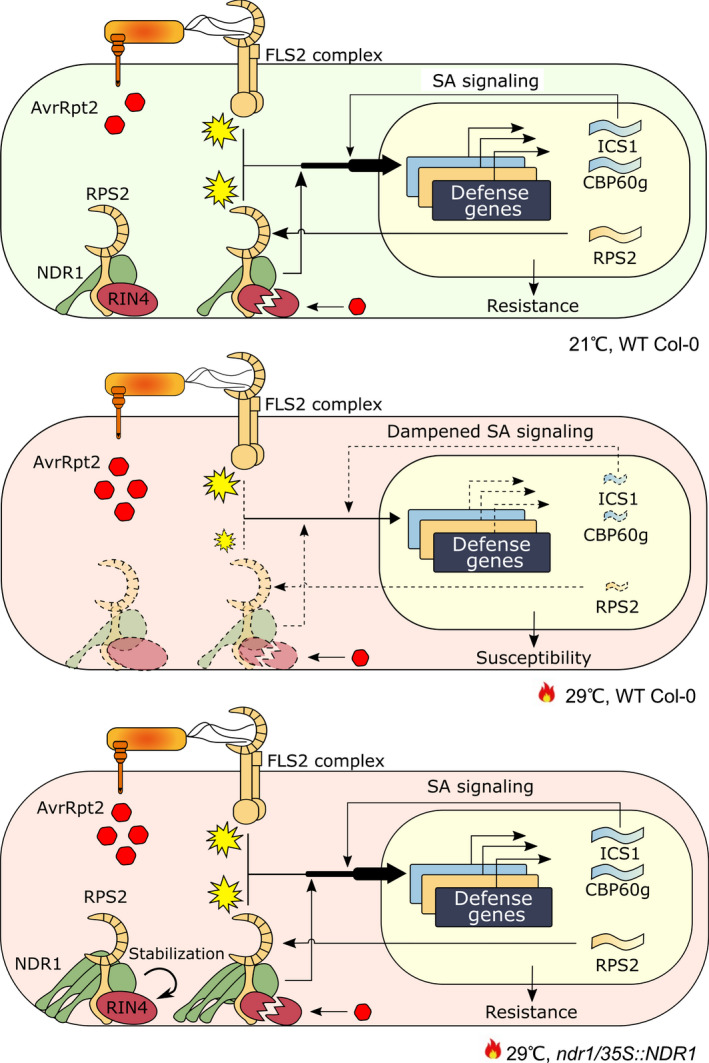
The schematic diagram of the mechanism of rescued effector‐triggered immunity (ETI) in elevated temperature by overexpressed NON‐RACE‐SPECIFIC DISEASE RESISTANCE1 (NDR1) in Arabidopsis. In plant immunity, NDR1 genetically interacts with Resistance to Pseudomonas Syringae‐2 (RPS2) and RPM1‐interacting protein 4 (RIN4) to facilitate ETI in response to AvrRpt2. Concomitantly, NDR1 contributes to a robust pro‐immune transcriptome, including the upregulation of RPS2 and genes involved in salicylic acid (SA) signaling. At elevated temperature (29°C), transcription of these defense genes is inhibited, rendering plants susceptible to bacteria pathogen infection. In the ndr1/35S::NDR1 overexpression line, increased levels of NDR1 rescue the transcription of ISOCHRISMATE SYNTHASE 1 (ICS1) and CALMODULIN BINDING PROTEIN 60g (CBP60g) under elevated temperatures, thus sustaining the production of SA and its signaling pathway. In parallel, NDR1 overexpression enhances RPS2 messenger RNA accumulation and stabilizes RIN4, the guardee, to sustain the function of the nucleotide‐binding leucine‐rich repeat complex. Dashed lines indicate dampened signaling pathway. Yellow explosion symbols indicate immune activation.
Author contributions
SPS and BD planned and designed the research. SPS, Y‐JL and YK performed experiments. SPS, HC, Y‐JL, PL, YK, AM, KT and BD analyzed data. SPS and BD wrote the paper with input from all co‐authors.
Supporting information
Fig. S1 Temporal dynamics of transcriptome responses to heat stress.
Fig. S2 Venn diagrams illustrating the overlap between NDR1‐dependent genes and heat‐responsive genes.
Fig. S3 mRNA accumulation of key defense response genes.
Fig. S4 Expression patterns of coexpression modules.
Fig. S5 Disease assays at 0 hpi after avirulent phytopathogen bacteria treatment.
Fig. S6 Temporal detection of RIN4 after virulent Pst treatment.
Fig. S7 Hypersensitive response and disease phenotypes after treatment with Pst‐AvrRpm1 and Pst‐AvrPphB.
Fig. S8 Disease assays at 3 dpi after avirulent phytopathogen bacteria treatment.
Fig. S9 Bacterial growth after syringe‐infiltration with Pst‐expressing hrcC− and AvRpt2‐CyaA in wild‐type (Col‐0) and mutant plants.
Fig. S10 Benzothiadiazole (BTH)‐induced SA does not protect RIN4 under Pst‐AvrRpt2 treatment.
Fig. S11 Bacterial growth at 0 hpi in BTH‐untreated wild‐type (Col‐0) and mutant plants after syringe‐infiltration with mock and Pst‐expressing AvrRpt2‐Cya.
Table S1 Mean centered expression levels (log2) of differentially expressed genes.
Table S2 Differentially expressed genes in cluster 1.
Table S3 Gene ontology enrichment of cluster 1 containing genotype‐independent temperature‐responsive genes (FDR < 0.05).
Table S4 Differentially expressed genes in cluster 2.
Table S5 Gene Ontology enrichment of cluster 2 containing genotype‐specific temperature‐responsive genes (FDR < 0.05).
Table S6 The number of upregulated genes between genotypes.
Table S7 The number of downregulated genes between genotypes.
Table S8 NDR1‐upregulated genes, heat‐suppressed genes, and their intersections at 6 h.
Table S9 NDR1‐udownregulated genes, heat‐induced genes, and their intersections at 6 h.
Table S10 NDR1‐upregulated genes, heat‐suppressed genes, and their intersections at 24 h.
Table S11 NDR1‐downregulated genes, heat‐induced genes, and their intersections at 24 h.
Table S12 Gene Ontology enrichment of NDR1‐upregulated genes, heat‐suppressed genes at 6 h.
Table S13 Gene Ontology enrichment of NDR1‐downregulated genes, heat‐induced genes at 6 h.
Table S14 Gene Ontology enrichment of NDR1‐upregulated genes, heat‐suppressed genes at 24 h.
Table S15 Gene Ontology enrichment of NDR1‐downregulated genes, heat‐induced genes at 24 h.
Table S18 Quantification of RIN4 after virulent Pst treatment.
Table S16 Gene Ontology enrichment analysis of the genes in each coexpression module (FDR < 0.05).
Table S17 Description of the genes in each coexpression module.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We would like to acknowledge the support of the MSU Plant Resilience Institute for providing funding to the laboratory of BD. Research in the laboratory of BD is supported by the National Institutes of General Medical Sciences (1R01GM125743) and the National Science Foundation‐National Institute of Food and Agriculture (USDA) joint Plant–Biotic Interactions Program (IOS‐1146128). Research in the laboratory of AM is supported by JST PRESTO (JPMJPR17Q6), Grant‐in‐Aid for Scientific Research (B) (19H02960), and Grant‐in‐Aid for Scientific Research on Innovative Areas (B) (21H05151). Research in the laboratory of KT is supported by the Fundamental Research Funds for the Central Universities (program no. 2662020ZKPY009), the Huazhong Agricultural University Scientific & Technological Self‐innovation Foundation, and Joint Funding of Huazhong Agricultural University and Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences (SZYJY2021007). Special thanks to Alvaro Hernandez, Director of DNA Services at the Roy J. Carver Biotechnology Center, The University of Illinois, Urbana‐Champaign, for providing consultation in advance of sequencing. The authors acknowledge the services of the staff at the MSU Research Technology Support Facility for conducting the LC–MS analysis, Masaki Shimono for experimental assistance, Sheng Yang He for providing the ics1‐2 seeds, and Noel Day for editorial comments. The authors declare no competing financial interests.
Data availability
The RNA‐seq data generated herein are contained within the National Center for Biotechnology Information Short Read Archive . The Illumina RNA‐seq reads were deposited in BioProject under project ID PRJNA778239.
References
- Anders S, Pyl PT, Huber W. 2015. HTseq – a Python framework to work with high‐throughput sequencing data. Bioinform 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Chishol ST, Dahlbeck D, Staskawicz BJ. 2003. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Molecular Microbiology 49: 1537–1546. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. 2003. Initiation of RPS2‐specified disease resistance in Arabidopsis is coupled to the AvrRpt2‐directed elimination of RIN4. Cell 112: 369–377. [DOI] [PubMed] [Google Scholar]
- Bahuguna RN, Jagadish KSV. 2015. Temperature regulation of plant phenological development. Environmental and Experimental Botany 111: 83–90. [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. 2006. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genetics 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Song W‐M, Pan J, Jiang C‐M, Srivastava R, Li B, Zhu L‐Y, Su H‐Y, Gao X‐S, Liu H et al. 2016. Overexpression of the NDR1/HIN1‐Like Gene NHL6 modifies seed germination in response to abscisic acid and abiotic stresses in Arabidopsis . Plos ONE 11: e0148572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens ML, Wolinska KW, Spaepen S, Ziegler J, Nobori T, Nair A, Kruler V, Winkelmuller TM, Wang Y, Mine A et al. 2019. Balancing trade‐offs between biotic and abiotic stress responses through leaf age‐dependent variation in stress hormone cross‐talk. Proceedings of the National Academy of Sciences, USA 116: 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catinot J, Buchala A, Abou‐Mansour E, Métraux JP. 2008. Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana . FEBS Letters 582: 473–478. [DOI] [PubMed] [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ. 1995. NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proceedings of the National Academy of Sciences, USA 92: 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. 1997. NDR1, a pathogen‐induced component required for Arabidopsis disease resistance. Science 278: 1963–1965. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Huot B, Kvitko BH. 2017. Effector translocation: Cya reporter assay. In: L. Journet and E. Cascales, eds. Bacterial protein secretion systems: methods and protocols. New York, NY, USA: Springer, 473–487. [Google Scholar]
- Cheng C, Gao X, Feng B, Sheen J, Shan L, He P. 2013. Plant immune response to pathogens differs with changing temperatures. Nature Communications 4: 2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. 2005. Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proceedings of the National Academy of Sciences, USA 102: 2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppinger P, Repetti PP, Day B, Dahlbeck D, Mehlert A, Staskawicz BJ. 2004. Overexpression of the plasma membrane‐localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana . The Plant Journal 40: 225–237. [DOI] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Staskawicz BJ. 2006. NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis . Plant Cell 18: 2782–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Xu J, Hofte M. 2014. Making sense of hormone‐mediated defense networking: from rice to Arabidopsis . Frontiers in Plant Science 5: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk K, Fouts DE, Rehm AH, Hill AR, Collmer A, Alfano JR. 1999. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature‐ and pH‐sensitive manner. Journal of Bacteriology 181: 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome‐wide expression patterns. Proceedings of the National Academy of Sciences, USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JM, Lin ZJ, Coaker G. 2011. Plant NB‐LRR signaling: upstreams and downstreams. Current Opinion in Plant Biology 14: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Alfano JR. 2006. Pseudomonas syringae HrpJ is a type III secreted protein that is required for plant pathogenesis, injection of effectors, and secretion of the HrpZ1 harpin. Journal of Bacteriology 188: 6060–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez E, Salinas M, Manzano‐Agugliaro F. 2018. Worldwide research on plant defense against biotic stresses as improvement for sustainable agriculture. Sustainability 10: 391. [Google Scholar]
- Guo Q, Major IT, Howe GA. 2018. Resolution of growth‐defense conflict: mechanistic insights from jasmonate signaling. Current Opinion in Plant Biology 44: 72–81. [DOI] [PubMed] [Google Scholar]
- Guo Z, Wang X, Hu Z, Wu C, Shen Z. 2021. The pentatricopeptide repeat protein GEND1 is required for root development and high temperature tolerance in Arabidopsis thaliana . Biochemical and Biophysical Research Communications 578: 63–69. [DOI] [PubMed] [Google Scholar]
- Havko NE, Kapali G, Das MR, Howe GA. 2020. Stimulation of insect herbivory by elevated temperature outweighs protection by the jasmonate pathway. Plants 9: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Li ZG, Hoque TS, Burritt DJ, Fujita M, Munne‐Bosch S. 2018. Heat or cold priming‐induced cross‐tolerance to abiotic stresses in plants: key regulators and possible mechanisms. Protoplasma 255: 399–412. [DOI] [PubMed] [Google Scholar]
- Hu Y, Dong Q, Yu D. 2012. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae . Plant Science 185–186: 288–297. [DOI] [PubMed] [Google Scholar]
- Huot B, Castroverde CDM, Velasquez AC, Hubbard E, Pulman JA, Yao J, Childs KL, Tsuda K, Montgomery BL, He SY. 2017. Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis . Nature Communications 8: 1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Z, Iqbal MS, Hashem A, Abd Allah EF, Ansari MI. 2021. Plant defense responses to biotic stress and its interplay with fluctuating dark/light conditions. Frontiers in Plant Science 12: 631810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. Hisat: a fast spliced aligner with low memory requirements. Nature Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. 2005. The Pseudomonas syringae effector AvrRpt2 cleaves its c‐terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proceedings of the National Academy of Sciences, USA 102: 6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Gilmour SJ, Chao L, Park S, Thomashow MF. 2020. Arabidopsis CAMTA transcription factors regulate pipecolic acid biosynthesis and priming of immunity genes. Molecular Plant 13: 157–168. [DOI] [PubMed] [Google Scholar]
- Knepper C, Savory EA, Day B. 2011. Arabidopsis NDR1 is an integrin‐like protein with a role in fluid loss and plasma membrane‐cell wall adhesion. Plant Physiology 156: 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. 2009. High temperature‐mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Current Biology 19: 408–413. [DOI] [PubMed] [Google Scholar]
- Kouzai Y, Noutoshi Y, Inoue K, Shimizu M, Onda Y, Mochida K. 2018. Benzothiadiazole, a plant defense inducer, negatively regulates sheath blight resistance in Brachypodium distachyon . Scientific Reports 8: 17358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. 1993. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2 . Plant Cell 5: 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. wgcna: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Jung JH, Cortes Llorca L, Kim SG, Lee S, Baldwin IT, Park CM. 2014. FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis . Nature Communications 5: 5473. [DOI] [PubMed] [Google Scholar]
- Li L, Li RF, Ming ZH, Lu GT, Tang JL. 2017. Identification of a novel type III secretion‐associated outer membrane‐bound protein from Xanthomonas campestris pv. campestris . Scientific Reports 7: 42724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L‐S, Ying J, Li E, Ma T, Li M, Gong L‐M, Wei G, Zhang Y, Li S. 2021. Arabidopsis CBP60b is a central transcriptional activator of immunity. Plant Physiology 186: 1645–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Day B. 2019. Battlefield cytoskeleton: turning the tide on plant immunity. Molecular Plant–Microbe Interactions 32: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Lu Y‐J, Chen H, Day B. 2020. Lifecycle of the plant immune response. Critical Reviews in Plant Sciences 39: 72–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhou X, Chen L, Huang W, Yu D. 2010. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Molecules and Cells 29: 475–483. [DOI] [PubMed] [Google Scholar]
- Li Z, Liu H, Ding Z, Yan J, Yu H, Pan R, Hu J, Guan Y, Hua J. 2020. Low temperature enhances plant immunity via salicylic acid pathway genes that are repressed by ethylene. Plant Physiology 182: 626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Sonbol FM, Huot B, Gu Y, Withers J, Mwimba M, Yao J, He SY, Dong X. 2016. Salicylic acid receptors activate jasmonic acid signalling through a non‐canonical pathway to promote effector‐triggered immunity. Nature Communications 7: 13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. 2009. Dissection of salicylic acid‐mediated defense signaling networks. Plant Signaling & Behavior 4: 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zhang C, Albrecht U, Shimizu R, Wang G, Bowman KD. 2013. Overexpression of a citrus NDR1 ortholog increases disease resistance in Arabidopsis. Frontiers in Plant Science 4: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y‐J, Chen H, Corrion A, Buyuk I, Li P, Samaradivakara S, Wai CM, Sakamoto H, Santos P, VanBuren R et al. 2021. NDR1 and the Arabidopsis plasma membrane ATPase AHA5 are required for processes that converge on drought tolerance and immunity. bioRxiv. doi: 10.1101/2021.06.10.445978. [DOI] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. 2003. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2‐mediated resistance. Cell 112: 379–389. [DOI] [PubMed] [Google Scholar]
- Maier BA, Kiefer P, Field CM, Hemmerle L, Bortfeld‐Miller M, Emmenegger B, Schafer M, Pfeilmeier S, Sunagawa S, Vogel CM et al. 2021. A general non‐self response as part of plant immunity. Nature Plants 7: 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang HG, Qian W, Zhu Y, Qian J, Kang HG, Klessig DF, Hua J. 2012. Abscisic acid deficiency antagonizes high‐temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis . Plant Cell 24: 1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR, Davis SJ. 2010. Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Current Biology 20: R1086–R1092. [DOI] [PubMed] [Google Scholar]
- Miller RN, Costa Alves GS, Van Sluys MA. 2017. Plant immunity: unravelling the complexity of plant responses to biotic stresses. Annals of Botany 119: 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine A, Seyfferth C, Kracher B, Berens ML, Becker D, Tsuda K. 2018. The defense phytohormone signaling network enables rapid, high‐amplitude transcriptional reprogramming during effector‐triggered immunity. Plant Cell 30: 1199–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB. 2005. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annual Review of Plant Biology 56: 509–531. [DOI] [PubMed] [Google Scholar]
- Nejat N, Mantri N. 2017. Plant immune system: crosstalk between responses to biotic and abiotic stresses the missing link in understanding plant defence. Current Issues in Molecular Biology 23: 1–16. [DOI] [PubMed] [Google Scholar]
- Penfield S. 2008. Temperature perception and signal transduction in plants. New Phytologist 179: 615–628. [DOI] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M. 2016. Molecular and genetic control of plant thermomorphogenesis. Nature Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Loo EP. 2020. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytologist 225: 87–104. [DOI] [PubMed] [Google Scholar]
- Sakata T, Oshino T, Miura S, Tomabechi M, Tsunaga Y, Higashitani N, Miyazawa Y, Takahashi H, Watanabe M, Higashitani A. 2010. Auxins reverse plant male sterility caused by high temperatures. Proceedings of the National Academy of Sciences, USA 107: 8569–8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel G. 1931. Some experiments on inoculating methods with plant viruses, and on local lesions. The Annals of Applied Biology 18: 494–507. [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn MA, Marr SK, Inoue K, Inada N, Zubieta C, Wildermuth MC. 2007. Arabidopsis isochorismate synthase functional in pathogen‐induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. Journal of Biological Chemistry 282: 5919–5933. [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang H‐S, Han B, Zhu T, Zou G, Katagiri F. 2003. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae . Plant Cell 15: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. 2009. Network properties of robust immunity in plants. PLoS Genetics 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Somssich IE. 2015. Transcriptional networks in plant immunity. New Phytologist 206: 932–947. [DOI] [PubMed] [Google Scholar]
- Velasquez AC, Oney M, Huot B, Xu S, He SY. 2017. Diverse mechanisms of resistance to Pseudomonas syringae in a thousand natural accessions of Arabidopsis thaliana . New Phytologist 214: 1673–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh J, Kang BC. 2019. Current views on temperature‐modulated R gene‐mediated plant defense responses and tradeoffs between plant growth and immunity. Current Opinion in Plant Biology 50: 9–17. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bao Z, Zhu Y, Hua J. 2009. Analysis of temperature modulation of plant defense against biotrophic microbes. Molecular Plant–Microbe Interactions 22: 498–506. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hua J. 2009. A moderate decrease in temperature induces COR15a expression through the CBF signaling cascade and enhances freezing tolerance. The Plant Journal 60: 340–349. [DOI] [PubMed] [Google Scholar]
- van Wersch S, Tian L, Hoy R, Li X. 2020. Plant NLRs: the whistleblowers of plant immunity. Plant Communications 1: 100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Hua J. 2004. A haplotype‐specific resistance gene regulated by BONZAI1 mediates temperature‐dependent growth control in Arabidopsis . Plant Cell 16: 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Ngou BPM, Ding P, Xin XF. 2021. PTI‐ETI crosstalk: an integrative view of plant immunity. Current Opinion in Plant Biology 62: 102030. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhu J, Gong Z, Zhu JK. 2022. Abiotic stress responses in plants. Nature Reviews Genetics 23: 104–119. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X. 2003. A gain‐of‐function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1‐1, constitutive 1. Plant Cell 15: 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Lu Z, Wang L, Jin B. 2020. Plant responses to heat stress: physiology, transcription, noncoding RNAs, and epigenetics. International Journal of Molecular Sciences 22: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qian W, Hua J. 2010. Temperature modulates plant defense responses through NB‐LRR proteins. PLoS Pathogens 6: e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Temporal dynamics of transcriptome responses to heat stress.
Fig. S2 Venn diagrams illustrating the overlap between NDR1‐dependent genes and heat‐responsive genes.
Fig. S3 mRNA accumulation of key defense response genes.
Fig. S4 Expression patterns of coexpression modules.
Fig. S5 Disease assays at 0 hpi after avirulent phytopathogen bacteria treatment.
Fig. S6 Temporal detection of RIN4 after virulent Pst treatment.
Fig. S7 Hypersensitive response and disease phenotypes after treatment with Pst‐AvrRpm1 and Pst‐AvrPphB.
Fig. S8 Disease assays at 3 dpi after avirulent phytopathogen bacteria treatment.
Fig. S9 Bacterial growth after syringe‐infiltration with Pst‐expressing hrcC− and AvRpt2‐CyaA in wild‐type (Col‐0) and mutant plants.
Fig. S10 Benzothiadiazole (BTH)‐induced SA does not protect RIN4 under Pst‐AvrRpt2 treatment.
Fig. S11 Bacterial growth at 0 hpi in BTH‐untreated wild‐type (Col‐0) and mutant plants after syringe‐infiltration with mock and Pst‐expressing AvrRpt2‐Cya.
Table S1 Mean centered expression levels (log2) of differentially expressed genes.
Table S2 Differentially expressed genes in cluster 1.
Table S3 Gene ontology enrichment of cluster 1 containing genotype‐independent temperature‐responsive genes (FDR < 0.05).
Table S4 Differentially expressed genes in cluster 2.
Table S5 Gene Ontology enrichment of cluster 2 containing genotype‐specific temperature‐responsive genes (FDR < 0.05).
Table S6 The number of upregulated genes between genotypes.
Table S7 The number of downregulated genes between genotypes.
Table S8 NDR1‐upregulated genes, heat‐suppressed genes, and their intersections at 6 h.
Table S9 NDR1‐udownregulated genes, heat‐induced genes, and their intersections at 6 h.
Table S10 NDR1‐upregulated genes, heat‐suppressed genes, and their intersections at 24 h.
Table S11 NDR1‐downregulated genes, heat‐induced genes, and their intersections at 24 h.
Table S12 Gene Ontology enrichment of NDR1‐upregulated genes, heat‐suppressed genes at 6 h.
Table S13 Gene Ontology enrichment of NDR1‐downregulated genes, heat‐induced genes at 6 h.
Table S14 Gene Ontology enrichment of NDR1‐upregulated genes, heat‐suppressed genes at 24 h.
Table S15 Gene Ontology enrichment of NDR1‐downregulated genes, heat‐induced genes at 24 h.
Table S18 Quantification of RIN4 after virulent Pst treatment.
Table S16 Gene Ontology enrichment analysis of the genes in each coexpression module (FDR < 0.05).
Table S17 Description of the genes in each coexpression module.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The RNA‐seq data generated herein are contained within the National Center for Biotechnology Information Short Read Archive . The Illumina RNA‐seq reads were deposited in BioProject under project ID PRJNA778239.


