Abstract
Phytases catalyze the hydrolysis of phosphomonoester bonds of phytate (myo-inositol hexakisphosphate), thereby creating lower forms of myo-inositol phosphates and inorganic phosphate. In this study, cDNA expression libraries were constructed from four basidiomycete fungi (Peniophora lycii, Agrocybe pediades, a Ceriporia sp., and Trametes pubescens) and screened for phytase activity in yeast. One full-length phytase-encoding cDNA was isolated from each library, except for the Ceriporia sp. library where two different phytase-encoding cDNAs were found. All five phytases were expressed in Aspergillus oryzae, purified, and characterized. The phytases revealed temperature optima between 40 and 60°C and pH optima at 5.0 to 6.0, except for the P. lycii phytase, which has a pH optimum at 4.0 to 5.0. They exhibited specific activities in the range of 400 to 1,200 U · mg, of protein−1 and were capable of hydrolyzing phytate down to myo-inositol monophosphate. Surprisingly, 1H nuclear magnetic resonance analysis of the hydrolysis of phytate by all five basidiomycete phytases showed a preference for initial attack at the 6-phosphate group of phytic acid, a characteristic that was believed so far not to be seen with fungal phytases. Accordingly, the basidiomycete phytases described here should be grouped as 6-phytases (EC 3.1.3.26).
Phytases (myo-inositol hexakisphosphate phosphohydrolases) belong to the family of histidine acid phosphatases sharing the sequence consensus pattern [LIVM]-X-X-[LIVMA]-X-X-[LIVM]-X-R-H-[GN]-X-R-X-[PAS] (27; http://www.expasy.ch/cgi-bin/get-prodoc-entry?PDOC00538). They are capable of catalyzing the hydrolysis of phosphomonoester bonds of phytate (salts of myo-inositol hexakisphosphate or myo-inositol 1,2,3,4,5,6-hexakis dihydrogen phosphate), thereby creating lower forms of myo-inositol phosphates and inorganic phosphate (22, 24). Phytases are grouped according to the specific position of the phosphate ester group on the phytate molecule at which hydrolysis is initiated, i.e., as 3-phytases (EC 3.1.3.8) or as 6-phytases (EC 3.1.3.26).
Phytate is the primary source of inositol and the primary storage form of phosphate in plant seeds (23). Seeds, cereal grains, and legumes are important components of food and, in particular, of animal feed preparations. However, monogastric animals such as poultry and swine are incapable of utilizing the phosphorus bound in phytate due to low levels of phytase activity in the digestive tract. Furthermore, phytate acts as an antinutrient by chelating divalent cations and preventing the uptake of minerals, e.g., Zn (9). Thus, phytases are used as a cereal feed additive that enhances the phosphorus and mineral uptake in monogastric animals and reduces the level of phosphate output in their manure. Recently, there have been several reports on the cloning of fungal PhyA phytases from Aspergillus niger (16, 20, 28), Aspergillus fumigatus (19), Aspergillus terreus, Myceliophthora thermophila (14), Emericella nidulans, Talaromyces thermophilus (18), and Thermomyces lanuginosus (2). Based on their characteristics and on their sequence similarity, the PhyA phytases form a subclass within the histidine acid phosphatase family (14). So far, all the reported cloned filamentous fungal PhyA phytases were from the order Eurotiales or Sordariales within the phylum Ascomycota. Here, we report evidence for PhyA phytases being more widely distributed in the fungal kingdom. This was done by expression cloning, overexpression, and characterization of the first five PhyA phytases from four orders within the phylum Basidiomycota: Stereales, Agaricales, Phanerochaetales, and Coriolales.
MATERIALS AND METHODS
Materials.
All chemicals were obtained from Merck and were of analytical grade unless stated otherwise.
Fungal strains and growth conditions.
Peniophora lycii CBS 686.96, Agrocybe pediades CBS No. 900.96, Ceriporia sp. CBS 100231 (the CBS 100231 strain was previously designated Paxillus involutus), and Trametes pubescens CBS 100232 were cultivated in shake flasks containing 100 ml of growth medium (soya flour, 30 g/liter; maltodextrin, 15 g/liter; Bacto Peptone, 5 g/liter; and pluronic PE 6100, 0.2 g/liter). The P. lycii culture was incubated without agitation at 26°C for 15 days, and the A. pediades, Ceriporia sp., and T. pubescens cultures were incubated at 26°C for 5 days with agitation. Aspergillus oryzae A1560 transformants were grown in YP medium containing 35 g of maltodextrin per liter as described in reference 4.
RNA isolation and construction of directional cDNA libraries.
RNA isolation and the construction of the four directional cDNA libraries in the yeast expression vector pYES2 (Invitrogene) were carried out essentially as described in reference 13.
Screening of the cDNA library.
The libraries were amplified in Escherichia coli strain DH10B (Life Technologies) and transformed into Saccharomyces cerevisiae W3124 (25) by electroporation. The S. cerevisiae transformants were plated on synthetic complete agar containing 2% glucose and incubated at 30°C for 3 days. The transformants were transferred to phytate replication plates (synthetic complete agar containing 2% galactose, 0.5% threonine, 20 mM CaCl2, 20 mM MgCl2, 20 mM Na-phytate, and 0.1% trace element solution [pH 6.5]) (trace element solution no. 141; Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH Catalogue of Strains) and incubated for 3 to 5 days at 30°C. LSB-agarose (1%; BioWhittaker Molecular Applications) containing 0.2 M CaCl2 was poured over the plates and after 1 to 4 days, the phytase-positive colonies were identified by a surrounding clearing zone. Isolation and rescuing of the phytase-encoding cDNA in pYES2 were carried out as previously described (5).
Nucleotide sequence analysis.
The cDNA sequences from both strands were determined with the Dye Terminator Cycle Sequencing kit (Perkin-Elmer) and an Applied Biosystems ABI PRISM 377 DNA sequencer according to the manufacturer's instructions.
Recombinant expression in A. oryzae.
The phytase-encoding cDNA inserts were subcloned into Aspergillus expression vector pHD414 (7) or, for the A. pediades insert, into the pHD423 expression vector (a pHD414 derivative with a KpnI site in the polylinker). Plasmid DNA was isolated and cotransformed into A. oryzae A1560 with an amdS+ plasmid (4). The amdS+ transformants were screened for phytase activity in culture, and phytase-producing transformants were isolated.
Purification.
The phytases were purified from the culture supernatants of phytase-producing A. oryzae transformants.
The A. pediades, P. lycii, and T. pubescens phytases were purified according to the following procedure. Filter aid was added to the culture broth, and the broth was first filtered through a filtration cloth and then through a Seitz depth filter plate. The filtrate was concentrated by ultrafiltration on 10-kDa cut-off polyethersulfone membranes, followed by diafiltration with distilled water to reduce the conductivity to less than 2 mS/cm. The concentrated enzyme was adjusted to a pH of 7.5 and applied to a Q-Sepharose FF anion-exchange column (Amersham Pharmacia Biotech) equilibrated in 20 mM Tris–CH3COOH, pH 7.5. Bound proteins were eluted with a linear NaCl gradient from 0 to 0.5 M. After addition of (NH4)2SO4 to a final concentration of 1.5 M, the phytase eluate was applied to a Phenyl Toyopearl 650S column (TosoHaas) equilibrated in 1.5 M (NH4)2SO4–10 mM succinic acid–NaOH, pH 6.0. The column was washed with the equilibration buffer, and bound protein was eluted with a linear (NH4)2SO4 gradient from 1.5 to 0 M. The phytase eluate was buffer exchanged on a Sephadex G25 column (Amersham Pharmacia Biotech) equilibrated in 20 mM HEPES–NaOH, pH 7.5, and applied to a SOURCE 30Q column (Amersham Pharmacia Biotech) equilibrated in the same buffer. The column was washed thoroughly with the equilibration buffer, and bound proteins were eluted with a linear NaCl gradient from 0 to 0.3 M. Fractions from the SOURCE 30Q column were evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), pure phytase fractions were pooled, and the pH was adjusted to 6.0.
The two phytases from the Ceriporia sp., PhyA1 and PhyA2, were purified as described above with the following modifications: after concentration by ultrafiltration, (NH4)2SO4 was added to a final concentration of 2.0 M and the sample was applied to a Phenyl Toyopearl 650S column equilibrated in 2.0 M (NH4)2SO4 and 10 mM succinic acid–NaOH, pH 6.0, followed by the elution of bound protein with a linear (NH4)2SO4 gradient from 2.0 to 0 M. The phytase eluate was buffer exchanged on a Sephadex G25 column equilibrated in 20 mM HEPES–NaOH, pH 7.5, and applied to a Q Sepharose FF column equilibrated in the same buffer, followed by the elution of bound protein as described above. The phytase eluate was dialyzed overnight against 20 mM HEPES–NaOH, pH 7.5, before being applied to a SOURCE 30Q column. The remaining steps of the procedure were performed as described above.
Tris-glycine SDS-PAGE, and IEF.
SDS-PAGE was performed with 8 to 16% Tris-glycine gradient gels and isoelectric focusing (IEF) on pH 3 to 7 or pH 3 to 10 IEF gels (Novex/Invitrogen). The gels were stained with colloidal Coomassie blue (Novex/Invitrogen) or semidry blotted onto Immobilon P polyvinylidene difluoride membranes (Millipore), followed by amido black (naphthol blue-black) staining.
Measurement of enzymatic activity.
Enzyme samples diluted in 0.1 M sodium acetate and 0.01% Tween-20, pH 5.5, were further diluted 26-fold into the substrate solution (5 mM sodium phytate [Sigma] in 0.1 M sodium acetate, and 0.01% Tween-20 [pH 5.5], preincubated at 37°C) to start the reaction. After 30 min at 37°C, the reaction was stopped by adding an equal volume of 10% trichloroacetic acid. Free inorganic phosphate was measured by the addition of an equal volume of molybdate reagent containing, in 100 ml, 7.3 g of FeSO4, 1.0 g of (NH4)6Mo7O24 · 4H2O, and 3.2 ml of H2SO4. Absorbance was measured at 750 nm (Vmax microtiter plate reader; Molecular Devices). 1 unit equals the amount of enzyme that releases 1 μmol of phosphate per min. pH activity profiles were obtained by running the assay with 0.1 M concentrations of the following buffers: glycine-HCl at pH 3.0 to 3.5, sodium acetate at pH 4.0 to 5.5, morpholincethanesulfonic acid (MES) at pH 6.0 to 6.5, and Tris-HCl at pH 7.0 to 9.0.
N-terminal sequencing.
Automated Edman degradation of purified phytases was done with a Perkin-Elmer ABI 494HT sequencer with online microbore phenylthiohydantoin-amino acid detection.
DSC.
The samples were desalted on a NAP-5 column (Amersham Pharmacia Biotech) equilibrated with 0.1 M sodium acetate, pH 5.5. Differential scanning calorimetry (DSC) was performed with the VP-DSC Micro Calorimeter (MicroCal) by applying a constant scan rate of 90°/h from 20 to 100°C.
Calculation of theoretical Mr and pI values.
Theoretical Mr and pI values were calculated from the cDNA deduced amino acid sequences corresponding to the mature proteins with the programs in Vector NTI (Molecular Biology software; InforMax, Inc.).
HPLC analysis of phytic acid degradation intermediates.
Samples of purified phytases were incubated at 37°C in 200 μl of an assay mixture containing 200 mM sodium acetate, pH 5.0, with a final concentration of 0.2 mM phytic acid which was radioactively labeled with 1 μCi of myo-[inositol-2-3H(N)]hexakisphosphate (NEN Research Products, DuPont) per ml. The incubation was stopped after 2.5, 5, 10, 15, 20, 25, 30, 40, 50, 60, or 90 min by the addition of an equal volume of acetonitrile and subsequent heating at 95°C for 2 min. After centrifugation for 10 min at 10,000 × g, a 150-μl aliquot of the supernatant was diluted with an equal volume of H2O. A 200-μl sample was assayed by high-performance liquid chromatography (HPLC) anion-exchange chromatography on a 4.6 by 250 mm Zorbax SAX (5-μm) column at a flow rate of 1 ml/min, according to reference 26.
1H NMR spectroscopic analysis of phytase-catalyzed hydrolysis of phytic acid.
Nuclear magnoic resonance (NMR) spectra were recorded at 27°C with a Bruker DRX400 instrument equipped with a 5-mm selective inverse probe head. A total of 16 scans preceded by 4 dummy scans were accumulated using a sweep width of 2,003 Hz (5 ppm) covered by 8,000 data points. Attenuation of the residual HOD resonance was achieved by a weak presaturation period of 3 seconds. The spectra were referenced to the HOD signal (δ 4.75).
Phytate samples for NMR analysis were prepared as follows: phytate (100 mg of phytic acid dipotassium salt; Sigma P-5681) was dissolved in deionized water (4.0 ml), and the pH was adjusted to 5.5 or 3.5 by the addition of NaOH (4 N). Deionized water was added to a total volume of 5 ml, 1-ml portions were transferred to screw-cap vials, and the solvent was evaporated (vacuum centrifuge). The dry samples were dissolved in deuterium oxide (2 ml, 99.5% D; Merck), evaporated to dryness, and stored at −18°C until use.
For NMR analysis, a phytate sample was dissolved in deuterium oxide (1.0 ml), thus yielding a phytate concentration of 27 mM. The solution was transferred to an NMR tube, and the 1H NMR spectrum was recorded. One unit of the phytase in question was added, followed by mixing and immediate initiation of recording of 1H NMR spectra. Subsequent spectra were acquired after 5, 10, 15, 20, 25, 30, 45, 60, 75, 90, 105, 120, 135, 150, 165, 180, 195, and 210 min (= 3.5 h) and 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 11.5, 13.5, 15.5, 17.5, 19.5, 21.5, and 23.5 h.
Nucleotide sequence accession numbers.
The cDNA nucleotide sequences of P. lycii phyA, A. pediades phyA, Ceriporia sp. phyA1 and phyA2, and T. pubescens phyA have been deposited in the EMBL database under accession no. AJ310696, AJ310697, AJ310698, AJ310699, and AJ310700, respectively.
RESULTS
Isolation and characterization of phytase-encoding cDNA clones.
Between 20,000 and 30,000 S. cerevisiae colonies from each of the four cDNA libraries were screened for phytase activity, and one to four phytase-producing S. cerevisiae clones were isolated from each cDNA library. The positive colonies corresponded to five different phytase genes, P. lycii phyA, A. pediades phyA, Ceriporia sp. phyA1 and phyA2, and T. pubescens phyA. Where more than one clone containing the same gene was isolated, the clone with the longest cDNA insert was chosen for further analysis. For details of the cDNA sequences, the reader is referred to the EMBL database (for accession numbers, see Materials and Methods).
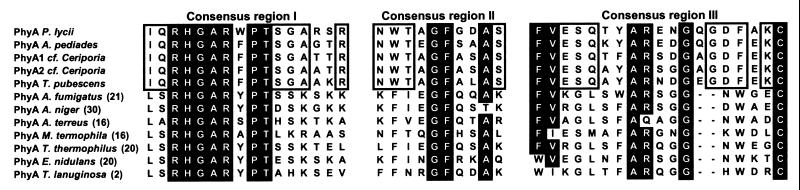
All five basidiomycete phytases contain the consensus pattern of histidine acid phosphatases and display some sequence similarity to the known PhyA phytases of the phylum Ascomycota, although there is a higher internal sequence similarity within the basidiomycete phytases. This suggests that these basidiomycete phytases form a distinct class of phytases, which is further supported by the observation of several amino acid sequence motifs conserved for the basidomycetes phytases (Fig. 1).
FIG. 1.
Amino acid sequence alignment of three regions where basidiomycete phytases share a high degree of amino acid conservation. In P. lycii PhyA, consensus region I corresponds to residues 68 to 83, consensus region II corresponds to residues 162 to 171, and consensus region III corresponds to residues 415 to 433. Identical residues in at least 10 of the sequences are indicated by a black box; a white box indicates identical residues for the basidiomycete phytases.
Enzyme characterization.
Molecular properties and characteristics of the recombinantly expressed and purified basidiomycete phytases are presented in Tables 1 and 2. SDS-PAGE analysis showed the pure preparations for the recombinantly expressed phytases as a single broad band, and furthermore the Mrs determined were significantly higher than the Mrs calculated from the corresponding mature amino acid sequences. This is probably due, as shown previously for several ascomycete phytases (29), to glycosylation of the secreted protein as all the phytase sequences contain several potential N-glycosylation sites (Table 1).
TABLE 1.
Molecular properties of the recombinantly expressed basidiomycete PhyA phytases
| Phytase | Length (aa)a | Predicted cleavage siteb between: | N-terminal sequencec | N-gly sitesd | Mr
|
pI
|
||
|---|---|---|---|---|---|---|---|---|
| Calculated | SDS-PAGE | Calculatede | IEF | |||||
| P. lycii PhyA | 439 | A29 and Q30 | L31PIPAQXTXN | 10 | 44,583 | 72,000 | 4.35 | 3.61 ± 0.01 |
| A. pediades PhyA | 453 | G19 and G20 | F31PPQIQDSXAAY | 6 | 46,781 | 59,000 | 5.00 | 4.15 ± 0.01 |
| 4.66 ± 0.01 | ||||||||
| 4.78 ± 0.01 | ||||||||
| 4.86 ± 0.01 | ||||||||
| Ceriporia sp. PhyA1 | 442 | A19 and T20 | S21VPKNTAPXFX | 7 | 45,942 | 59,000 | 6.40 | 7.36 ± 0.01 |
| 8.01 ± 0.03 | ||||||||
| Ceriporia sp. PhyA2 | 442 | A19 and A20 | N25IAPKFSIP | 8 | 44,955 | 54,000 | 4.64 | 3.94 ± 0.01 |
| 4.00 ± 0.01 | ||||||||
| 4.06 ± 0.01 | ||||||||
| 4.18 ± 0.01 | ||||||||
| T. pubescens PhyA | 443 | A19 and V20 | S31AXLDVTRDV | 9 | 44,531 | 62,000 | 4.40 | 3.58 ± 0.02 |
Including the signal sequence. aa, amino acids.
Signal peptide cleavage site predicted with the program SignalP, version 1.1 (17).
Determined from the purified recombinantly expressed phytases. The subscript number indicates the position of the first amino acid determined by Edman sequencing.
Number of potential N-glycosylation (N-gly) sites.
Calculated by using amino acid sequence of the mature protein (i.e., without signal sequence).
TABLE 2.
Characteristics of purified, recombinantly expressed phytases
| Phytase | Sp act (U/mg) | pH optimum | Temp optimum (°C) | Tm (°C) | % Residual activitya |
|---|---|---|---|---|---|
| P. lycii PhyA | 1,080 ± 110 | 4.0–4.5 | 50–55 | 60 | 62 |
| A. pediades PhyA | 400 | 5.0–6.0 | 50 | 58 | 47 |
| Ceriporia sp. PhyA1 | 700 ± 80 | 5.5–6.0 | 55–60 | 59 | 38 |
| Ceriporia sp. PhyA2 | 1,040 ± 310 | 5.0–6.0 | 40–45 | 48 | 22 |
| T. pubescens PhyA | 1,210 ± 30 | 5.0–5.5 | 50 | 55 | 15 |
| A. niger PhyA | 100b | 2.5–5.5b | 50b | 57 | 52 |
Residual activities were measured after preincubation of the enzymes for 60 min at 80°C in 0.1 M sodium acetate, pH 5.5.
R. F. M. van Gorcom, W. van Hartingsveldt, P. A. van Paridon, A. E. Veenstra, R. G. M. Luiten, and G. C. M. Selten, 3 April 1991, European Patent Application EP-0420358-A1.
The temperature optima obtained from temperature-activity profiles fully agree with the thermal stabilities measured by DSC; even at the relatively high enzyme concentrations used in DSC, the phytases showed a significant degree of refolding after thermal unfolding. This confirms the findings in reference 30 that the previously reported heat stability of fungal PhyAs (20) is due to reversible thermal unfolding rather than to intrinsic thermostability.
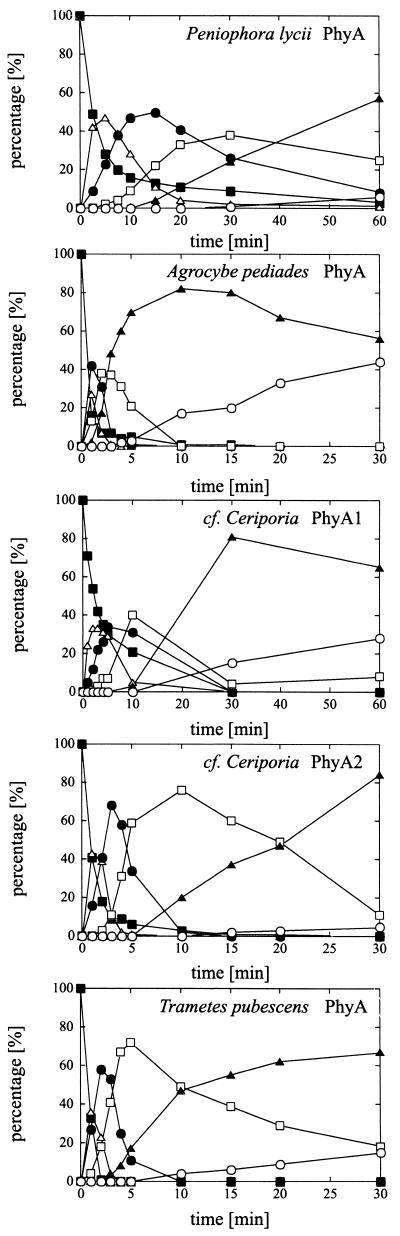
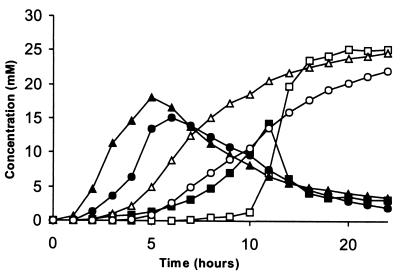
The pattern of phytic acid degradation intermediates obtained with the different phytases was studied by HPLC using a limited amount of phytate (0.2 mM) as the substrate at pH 5.0 (Fig. 2). A rapid decline of the myo-inositol hexakisphosphate is observed for all the phytases, as is the rapid buildup and decline of the myo-inositol pentakisphosphate. After prolonged incubation, all the phytases tended to accumulate relatively high levels of myo-inositol bisphosphate, whereas myo-inositol monophosphate was accumulated only slowly. This indicates that the more highly phosphorylated inositols are better substrates for the phytases.
FIG. 2.
HPLC analysis of phytate degradation patterns for recombinant basidiomycete phytases. The fraction of the different degrada tion intermediates out of the total amount of myo-inositol compounds is plotted as the percentage versus time. ■, myo-Inositol hexakisphosphate; ▵, myo-inositol pentakisphosphate; ●, myo-inositol tetrakisphosphate; □, myo-inositol trisphosphate; ▴, myo-inositol bisphosphate; ○, myo-inositol monophosphate. Concentrations: P. lycii PhyA, 0.1 U/ml; A. pediades PhyA, 0.2 U/ml; Ceriporia sp. PhyA1, 0.25 U/ml; Ceriporia sp. PhyA2, 0.1 U/ml; T. pubescens PhyA, 0.1 U/ml.
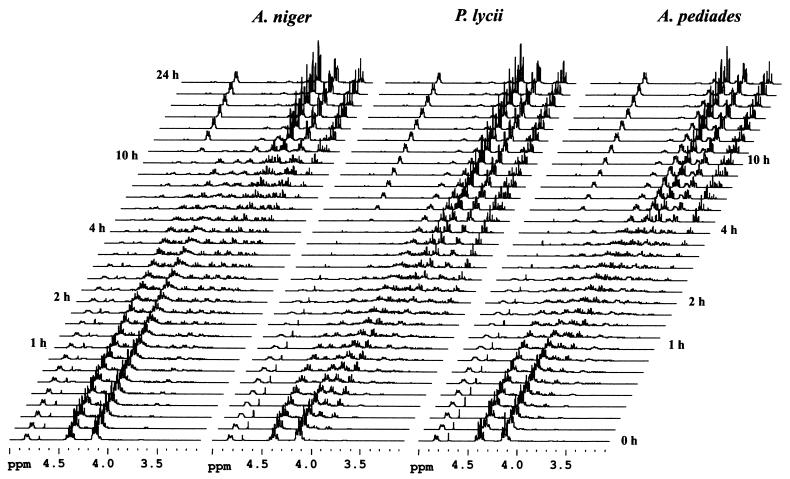
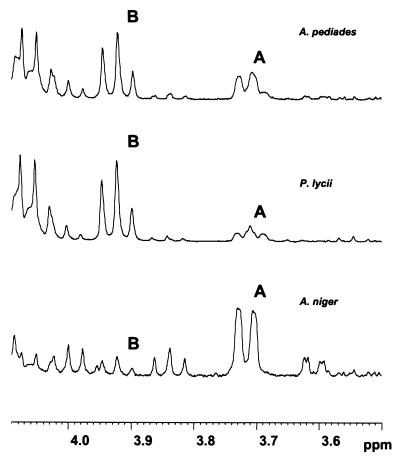
Figure 3 displays stacked plots of 1H NMR spectra recorded over a period of 24 h during the hydrolysis of 27 mM phytate by the A. niger, P. lycii, and A. pediades phytases (27°C, pH 5.5). Concomitant with the disappearance of the phytate-derived signals, resonances shifted up field as a result of dephosphorylation upgrowth. Distinctive differences, particularly with respect to the positional preference for the initial attack by the enzyme on the substrate, are reflected in the initial product profiles (Fig. 4). For the A. niger phytase, the dominant pentakisphosphate was identified as Ins(1,2,4,5,6)P5, corresponding to removal of the phosphate group in the 3-position, as diagnosed by the intense doublet signal at δ 3.72 attributable to H-3. For the P. lycii phytase, the triplet signal at δ 3.92 was assigned to H-6 in Ins(1,2,3,4,5)P5, formed by hydrolysis of the 6-phosphate ester linkage. Accordingly, the P. lycii phytase was characterized as a 6-phytase. Finally, for the A. pediades phytase, Ins(1,2,3,4,5)P5 was identified as the most abundant pentaphosphate, accompanied by significant amounts of Ins(1,2,4,5,6)P5. Thus, the A. pediades phytase attacks both the 3- and 6-positions, but with a preference for the 6-position. A similar mixed 3- or 6-position specificity was observed for the Ceriporia sp. and T. pubescens phytases (data not shown).
FIG. 3.
1H NMR profiles of phytate degradation by A. niger, P. lycii, and A. pediades phytases (27 mM phytate, pH 5.5, 27°C) recorded over a period of 24 h.
FIG. 4.
The 4.1- to 3.5-ppm region of the 1H NMR profiles recorded after 20 min of hydrolysis with A. niger, P. lycii, and A. pediades phytases (27 mM phytate, pH 5.5, 27°C) exhibiting the characteristic signals corresponding to H-3 in Ins(1,2,4,5,6)P5 (A) and H-6 in Ins(1,2,3,4,5)P5 (B).
The positional specificities were likewise studied at pH 3.5. The results were similar to those obtained at pH 5.5, except for a slightly decreased 6-position selectivity of the P. lycii phytase. H-5 in Ins(1,2)P2 and Ins(2)P displays characteristic triplet signals in the 1H NMR spectrum at δ 3.22 and δ 3.29, respectively. The transient accumulation of Ins(1,2)P2 and its subsequent transformation into Ins(2)P for the A. niger, P. lycii, and A. pediades phytases are shown in Fig. 5. For the P. lycii phytase, the Ins(1,2)P2 level rises steeply and peaks after 5 h where it represents approximately two-thirds of total inositol; whereas in the case of A. niger phytase, the build-up of Ins(1,2)P2 occurs more slowly. In contrast, the subsequent hydrolysis to the end product, Ins(2)P, proceeded more rapidly with the A. niger phytase than with the P. lycii phytase. Except for the initial specificity differences between the P. lycii and A. pediades phytase, the overall picture, including their Ins(2)P and Ins(1,2)P2 concentration profiles, appears very similar and distinctively different from that of A. niger phytase.
FIG. 5.
Concentration profiles of Ins(1,2)P2 and Ins(2)P observed by 1H NMR spectroscopy during enzymatic digestion of phytate by A. niger phytase [■, Ins(1,2)P2; □, Ins(2)P], P. lycii phytase [▴, Ins(1,2)P2; ▵, Ins(2)P], and A. pediades phytase [●, Ins(1,2)P2; ○, Ins(2)P] (27 mM phytate, pH 5.5, 27°C).
DISCUSSION
We have cloned and characterized five full-length cDNAs encoding phytase, phyA, from four different basidiomycete fungi. The deduced amino acid sequences show the characteristics of extracellular fungal enzymes with a cleavable signal sequence (Table 1).
The N-terminal sequences determined for the mature proteins did not correspond to the predicted signal peptide cleavage site (17) for any of the five recombinantly expressed basidiomycete phytases, a phenomenon also seen with the recombinantly expressed ascomycete phytases studied in reference 29. It has been proposed that the difference of 10 amino acids between the predicted and observed N-terminal amino acid sequences for the T. lanuginosus phytase is due to a propeptide, a notion that is supported by the observation of a Kex2 site (2). It is not possible to apply this hypothesis in general to the phytases here reported, since a Kex2 site (Pro-Arg) (15) was found only for Ceriporia sp. PhyA2. It is more likely that the N terminus was exposed to aminopeptidase activity after the signal sequence had been cleaved off. This notion is further supported by the recent characterization of secreted aminopeptidase (3), dipeptidyl peptidases (8), and tripeptidyl peptidase (K. Holm, G. Rasmussen, T. Halkaier, and J. Lehmbeck, 8 November 1995, world patent application WO 96/14404) in A. oryzae, of which at least the aminopeptidase and the tripeptidyl peptidase are non-specific.
Most of the known phytases are either 3- or 6-phytases (EC 3.1.3.8 or EC 3.1.3.26, respectively), grouped according to the specific position of the phosphomonoester group on the phytate molecule at which hydrolysis is initiated. One exception is an alkaline phytase from lily pollen that initially hydrolyzes the 5-phosphate group (1). Previously, phytases of microbial origin such as A. niger (11), Neurospora crassa (12), and Pseudomonas phytase (6) were generally considered to belong to the 3-phytases, while 6-phytases such as wheat bran phytase (21) were believed to be restricted mainly to the plant kingdom. Exceptions are the 6-phytases from Paramecium (26) and E. coli (10) and now the 6-phytases reported here. Thus, general rules about the evolutionary distribution of 3- and 6-phytases appear to be of limited relevance. Furthermore, the distinction between 3- and 6-specific phytases is not clear cut. The E. coli phytase was shown not to be strictly 6-specific but also to initiate phytate hydrolysis at the 3-phosphate group, and vice versa for the 3-phytase from A. niger. A notable example of an enzyme displaying such a mixed-type behavior is the A. pediades phytase.
The 3- and 6-phytase classification and the NMR experiments do not address all aspects of stereoisomerism; namely, no distinction is made between enantiomers. Therefore, the group of 3-phytases may comprise enzymes with preference for the D-3 (= L-1) or L-3 (= D-1) positions, and the group of 6-phytases enzymes may comprise those with preference for the D-6 (= L-4) or L-6 (= D-4) positions. Since the pentakisphosphates formed by cleaving the D-6 and L-6 phosphomonoester bonds are enantiomers (mirror-image isomers), they are indistinguishable by most techniques, including NMR spectroscopy, and more elaborate procedures have to be applied to determine the exact stereospecificity.
REFERENCES
- 1.Barrientos L, Scott J J, Murthy P P N. Specificity of hydrolysis of phytic acid by alkaline phytase from lily pollen. Plant Physiol. 1994;106:1489–1495. doi: 10.1104/pp.106.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berka R M, Rey M W, Brown K M, Byun T, Klotz A V. Molecular characterization and expression of a phytase gene from the thermophilic fungus Thermomyces lanuginosus. Appl Environ Microbiol. 1998;64:4423–4427. doi: 10.1128/aem.64.11.4423-4427.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blinkovsky A M, Byun T, Brown M K, Golightly E J, Klotz A V. A non-specific aminopeptidase from Aspergillus. Biochim Biophys Acta. 2000;1480:171–181. doi: 10.1016/s0167-4838(00)00064-9. [DOI] [PubMed] [Google Scholar]
- 4.Christensen T, Woeldike H, Boel E, Mortensen S B, Hjortshoej K, Thim L, Hansen M T. High level of expression of recombinant genes in Aspergillus oryzae. Bio/Technology. 1988;6:1419–1422. [Google Scholar]
- 5.Christgau S, Sandal T, Kofod L V, Dalbøge H. Expression cloning, purification and characterization of a β-1, 4-galactanase from Aspergillus aculeatus. Curr Genet. 1995;27:135–141. doi: 10.1007/BF00313427. [DOI] [PubMed] [Google Scholar]
- 6.Cosgrove D J. Inositol phosphate phosphatases of microbial origin. Inositol phosphate intermediates in the dephosphorylation of the hexaphosphates of myo-inositol, scyllo-inositol, and d-chiro-inositol by bacterial (Pseudomonas sp.) phytase. Aust J Biol Sci. 1970;23:1207–1220. doi: 10.1071/bi9701207. [DOI] [PubMed] [Google Scholar]
- 7.Dalbøge H, Heldt-Hansen H P. A novel method for efficient cloning of fungal enzyme genes. Mol Gen Genet. 1994;243:253–260. doi: 10.1007/BF00301060. [DOI] [PubMed] [Google Scholar]
- 8.Doumas A, Van den Broek P, Affolter M, Monod M. Characterization of the prolyl dipeptidyl peptidase gene (dppIV) from the koji mold Aspergillus oryzae. Appl Environ Microbiol. 1998;64:4809–4815. doi: 10.1128/aem.64.12.4809-4815.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdman J W., Jr Oilseed phytase: nutritional implications. J Am Oil Chem Soc. 1979;56:736–741. [Google Scholar]
- 10.Greiner R, Konietzny U, Jany K-D. Purification and characterization of two phytases from Escherichia coli. Arch Biochem Biophys. 1993;260:215–221. doi: 10.1006/abbi.1993.1261. [DOI] [PubMed] [Google Scholar]
- 11.Irving G C J, Cosgrove D J. Inositol phophate phosphatases of microbiological origin: the inositol pentaphosphate products of Aspergillus ficuum phytases. J Bacteriol. 1972;112:434–438. doi: 10.1128/jb.112.1.434-438.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson L F, Tate M E. The structure of myo-inositol pentaphosphates. Ann NY Acad Sci. 1969;165:526–551. [PubMed] [Google Scholar]
- 13.Kofod L V, Kauppinen S K, Christgau S, Andersen L N, Heldt-Hansen H P, Dörreich K, Dalbøge H. Cloning and characterization of two structurally and functionally divergent rhamnogalacturonases from Aspergillus aculeatus. J Biol Chem. 1994;269:29182–29189. [PubMed] [Google Scholar]
- 14.Mitchell D B, Vogel K, Weimann B J, Pasamontes L, van Loon A P G M. The phytase subfamily of histidine acid phosphatases: isolation of genes for two novel phytases from the fungi Aspergillus terreus and Myceliophthora thermophila. Microbiology. 1997;143:245–252. doi: 10.1099/00221287-143-1-245. [DOI] [PubMed] [Google Scholar]
- 15.Mizuno K, Nakamura T, Ohshima T, Tanaka S, Matsuo H. Characterization of Kex2-encoded endopeptidase from yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1989;159:305–311. doi: 10.1016/0006-291x(89)92438-8. [DOI] [PubMed] [Google Scholar]
- 16.Mullaney E J, Gibson D M, Ullah A H J. Positive identification of a lambda gt11 clone containing a region of fungal phytase gene by immunoprobe and sequnce verification. Appl Microbiol Biotechnol. 1991;35:611–614. doi: 10.1007/BF00169625. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Pasamontes L, Haiker M, Henriquez-Huecas M, Mitchell D B, van Loon A P G M. Cloning of the phytases from Emiricella nidulans and the thermophilic fungus Talaromyces thermophilus. Biochim Biophys Acta. 1997;1353:217–223. doi: 10.1016/s0167-4781(97)00107-3. [DOI] [PubMed] [Google Scholar]
- 19.Pasamontes L, Haiker M, Wyss M, Tessier M, van Loon A P G M. Gene cloning, purification, and characterization of a heat-stable phytase from fungus Aspergillus fumigatus. Appl Environ Microbiol. 1997;63:1696–1700. doi: 10.1128/aem.63.5.1696-1700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piddington C S, Houston C S, Paloheimo M, Cantrell M, Miettinen-Oinonen A, Nevalainen H, Rambosek J. The cloning and sequencing of the genes encoding phytase (phy) and pH 2.5-optimum acid phosphatase (aph) from Aspergillus niger var. awamori. Gene. 1993;133:55–62. doi: 10.1016/0378-1119(93)90224-q. [DOI] [PubMed] [Google Scholar]
- 21.Thomlinson R V, Ballou C E. myo-Inositol polyphosphate intermediates in the dephosphorylation of phytic acid by phytase. Biochemistry. 1962;1:166–171. doi: 10.1021/bi00907a025. [DOI] [PubMed] [Google Scholar]
- 22.Ullah A H J. Aspergillus ficuum phytase: partial primary structure, substrate selectivity, and kinetic characterization. Prep Biochem. 1988;18:459–471. doi: 10.1080/00327488808062544. [DOI] [PubMed] [Google Scholar]
- 23.Ullah A H J, Gibson D M. Extracellular phytase (E.C. 3.1.3.8) from Aspergillus ficuum NRRL 3135: purification and characterization. Prep Biochem. 1987;17:63–91. doi: 10.1080/00327488708062477. [DOI] [PubMed] [Google Scholar]
- 24.Ullah A H J, Phillippy B Q. Substrate selectivity in Aspergillus ficuum phytase and acid phosphatases using myo-inositol phosphates. J Agric Food Chem. 1994;42:423–425. [Google Scholar]
- 25.van den Hazel H B, Kielland-Brandt M C, Winther J R. Autoactivation of proteinase A initiates activation of yeast vacuolar zymogens. Eur J Biochem. 1992;207:277–283. doi: 10.1111/j.1432-1033.1992.tb17048.x. [DOI] [PubMed] [Google Scholar]
- 26.Van der Kaay J, Van Haastert P J. Stereospecificity of inositol hexakisphosphate dephosphorylation by Paramecium phytase. Biochem J. 1995;312:907–910. [PMC free article] [PubMed] [Google Scholar]
- 27.van Etten R L, Davidson R, Panayiotis E S, MacArthur H, Moore D L. Covalent structure, disulfide bonding, and identification of reactive surface and active site residues of human prostatic acid phosphatase. J Biol Chem. 1991;266:2313–2319. [PubMed] [Google Scholar]
- 28.van Hartingsveldt W, van Zeijl C M J, Harteveld G M, Gouka R J, Suykerbuyk M E G, Luiten R G M, van Paridon P A, Selten G C M, Veenstra A E, van Gorcom R F M, van den Hondel C A M J J. Cloning, characterization and overexpression of the phytase-encoding gene (phyA) of Aspergillus niger. Gene. 1993;127:87–94. doi: 10.1016/0378-1119(93)90620-i. [DOI] [PubMed] [Google Scholar]
- 29.Wyss M, Pasamontes L, Friedlein A, Rémy R, Tessier M, Kronenberger A, Middendorf A, Lehmann M, Schnoebelen L, Röthlisberger U, Kusznir E, Wahl G, Müller F, Lahm H-W, Vogel K, van Loon A P G M. Biophysical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): molecular size, glycosylation pattern, and engineering of proteolytic resistance. Appl Environ Microbiol. 1999;65:359–366. doi: 10.1128/aem.65.2.359-366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyss M, Pasamontes L, Rémy R, Kohler J, Kusznir E, Gadient M, Müller F, van Loon A P G M. Comparison of the thermostability properties of three acid phosphatases from molds: Aspergillus fumigatus phytase, A. niger phytase, and A. niger pH 2.5 acid phosphatase. Appl Environ Microbiol. 1998;64:4446–4451. doi: 10.1128/aem.64.11.4446-4451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]