Abstract
Hydroxycinnamic acid amides (HCAA) are the secondary metabolites ubiquitously exist in flowering plants, formed by condensation between hydroxycinnamates and mono or polyamines. HCAA species not only serve multiple functions in plant growth and development, but also exert significant positive effects on human health. In this study, we combined organic synthesis and UPHLC-TripleQ-MS/MS specifically targeting at HCAA species. The method was fully validated with respect to specificity, linearity, intra- and inter-day precision and accuracy, limit of detection (LOD), limit of quantification (LOQ), recovery, and reproducibility. We applied this method to identify and quantify HCAAs from the root barks and leaves of Lycium barbarum. HCAA species were reported in leaves for the first time, and 10 new HCAA species were further identified in root barks in addition to the ones reported in the literature. We also examine anti-inflammatory properties of identified HCAAs species. Seven HCAA compounds had a potent NO inhibitory effect with IC50 as low as 2.381 μM (trans-N-caffeoyl phenethylamine). Our developed method largely improved analytical sensitivity of HCAAs species that potentially contributes to plant metabolomics studies.
Keywords: Hydroxycinnamic acid amide, Lycium barbarum, UHPLC-MS/MS, Quantification, Anti-inflammatory
1. Introduction
Hydroxycinnamic acid amides (HCAAs) are secondary metabolites that exist abundantly in plants, forming upon conjugation between hydroxycinnamic acid and either mono- or polyamines [1]. They are derived from phenylalanine and tyrosine pathways which are composed of C3–C6 carbon skeleton with a series of hydroxylations and O-methylations on the aromatic ring, yielding distinct structural patterns [2]. HCAAs serve a wide array of functions in plant growth and development with presence in flowers, pollen grains, seeds, leaves and the roots of plants [3]. They participate in cell division, tuber formation, flowering regulation, defense mechanisms in response to biotic and abiotic stresses and cell wall cross linker [1,4–7]. However, the contribution of HCAAs to various plant developmental processes is still not fully clear and under debate that if HCAAs are storage molecules or actual bioactive species [8]. A great deal of effort is still needed in order to gain a detailed understanding of the biosynthesis, function and biotechnological applications of HCAA species in the plants. In particular, one of the difficulties from an analytical perspective of metabolomics studies is to precisely define the chemical and the physical compartmentation of the HCAAs, aiming to explore the molecular mechanism in relation to developmental and environmental effects. Thus, quantitative procedures with high sensitivity and specificity are required to overcome this challenge to progress the research in this area.
In addition, HCAAs are important class of antioxidants with potential application in prevention of human diseases [9,10]. The nature of amide species not only enhances the antioxidant activities of the molecules, but also improves the stability in physiological conditions and delivery applications [11]. Unlike esters that are readily hydrolyzed by the rich variety of hydrolase enzymes present in the human body, amides have the advantages of being more suitable for oral use [12]. HCAA family has been found to exhibit various biological activities, including anti-fungal [13], anti-inflammatory [14] and anti-cancer [15] properties. The large diversity and ubiquitous presence of HCAA species lays great potential to exploit the chemical diversity offered in them to obtain analogs with improved potency.
Advances in analytical techniques greatly facilitate biomedical researches by expediting identification and separation of compounds from complex matrices. Directly coupling high performance liquid chromatography with mass spectrometer (LC-MS) provides high sensitivity and enhances quantitative analysis accuracy. Nuclear magnetic resonance (NMR) is the widely used detection tool for structure elucidation of unknown compounds. In this paper, we developed an innovative approach that emerges organic synthesis and advances in chromatography, mass spectrometry and NMR technologies and to overcome the challenges encountered in both natural product chemistry and plant metabolomics studies, specifically targeting at HCAAs with high detection and quantification sensitivity. Our previous work identified a series of HCAA species from the fruits of L. barbarum; furthermore, HCAAs were also reported as major and characteristic chemical constituents of the root barks of the plant [16,17]. In our study, we developed the methodology that combined organic synthesis with NMR and UHPLC-TripleQ, aiming to improve sensitivity of HCAA family identification and quantification from plant tissues. We also validated the highly sensitive method by applying to profile HCAA family in the root barks and leaves of L. barbarum, exploring the chemical diversity of HCAA family in the plant as well as investigate the improved anti-inflammatory potency of the analogs by using in vitro models.
2. Material and method
2.1. Chemicals and reagents
Chemical synthesis: trans-caffeic acid, trans-ferulic acid, 3,4-dihydroxyhydrocinnamic acid, phenethylamine, tryptamine, tyramine, dopamine hydrochloride, and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride were purchased from Sigma–Aldrich (St. Louis, MO, USA). Triethylamine, 3,4-dimethoxyphenethylamine, LC-MS grade methanol, acetonitrile, water, and formic acid were purchased from Fisher Scientific (Fair Lawn, NJ, USA). Dimethylformamide (DMF), ethyl acetate, and hexane were purchased from Pharmco-AAPER (Brookfield, CT, USA).
In vitro studies: Gibco Dulbecco’s Modified Eagle Medium (DMEM), fetal bovine serum (FBS), and streptomycin/penicillin solution were purchased from Thermo Fisher Scientific (Hagerstown, MD, USA). Dimethyl sulfoxide-d6 was purchased from Cambridge Isotope Laboratories, Inc. (Tewksbury, MA, USA) sulfanilamide, naphthylethylenediamine dihydrochloride, molecular biology grade dimethyl sulfoxide (DMSO) and lipopolysaccharide (LPS) (Escherichia coli O127: E8) were purchased from Sigma–Aldrich (St. Louis, MO, USA).
2.2. Synthetic procedure
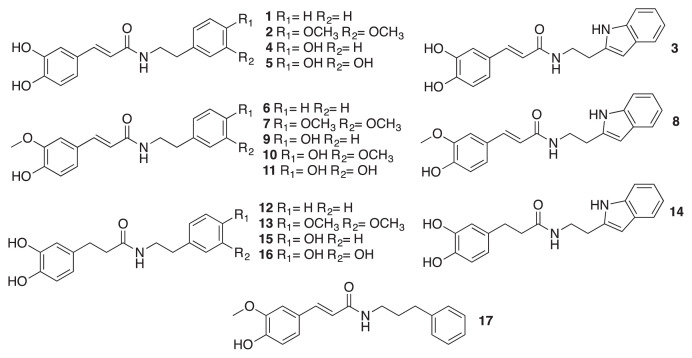
Five mM of hydroxycinnamic acid (trans-caffeic acid, trans-ferulic acid, and 3,4-dihydroxyhydrocinnamic acid) was mixed with 5 mmol triethylamine in DMF and placed on ice for 15 min 7.5 mM of amine (phenethylamine, tryptamine, tyramine, dopamine hydrochloride, 3,4-dimethoxyphenethylamine, or 3-phenylpropylamine) and 5 mM of N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride were added to DMF under nitrogen atmosphere at room temperature for 12 h. The reaction solution was then mixed with 100 mL distilled water and extracted by 100 mL ethyl acetate for three times. The ethyl acetate layer was washed with 0.2 M hydrochloric acid and brine, dried, evaporated and purified by column chromatography (eluted with ethyl acetate and hexane) and finally freeze dried resulting in final compounds. The purity of synthetic compounds was above 95%, which was checked by NMR and TLC. The NMR data could be found in our previous work [18]. The chemical structures of all HCAA compounds used in this study are shown in Fig. 1, and their corresponding numbers are indicated in Table 1.
Fig. 1.
Chemical structure of synthetic HCAA standards.
Table 1.
Synthetic HCAA standard numbers and corresponding compound names.
| Compound no. | Compound name |
|---|---|
| 1 | N-trans-caffeoyl phenethylamine |
| 2 | N-trans-caffeoyl 3,4-dimethoxyphenethylamine |
| 3 | N-trans-caffeoyl tryptamine |
| 4 | N-trans-caffeoyl tyramine |
| 5 | N-trans-caffeoyl dopamine |
| 6 | N-trans-feruloyl phenethylamine |
| 7 | N-trans-feruloyl 3,4-dimethoxyphenethylamine |
| 8 | N-trans-feruloyl tryptamine |
| 9 | N-trans-feruloyl tyramine |
| 10 | N-trans-feruloyl 3-methoxytyramine |
| 11 | N-trans-feruloyl dopamine |
| 12 | N-3,4-Dihydroxyhydrocinnamoyl phenethylamine |
| 13 | N-3,4-Dihydroxyhydrocinnamoyl 3,4-dimethoxyphenthylamine |
| 14 | N-3,4-Dihydroxyhydrocinnamoyl tryptamine |
| 15 | N-3,4-Dihydroxyhydrocinnamoyl tyramine |
| 16 | N-3,4-Dihydroxyhydrocinnamoyl dopamine |
| 17 | N-trans-feruloyl 3-phenylpropylamine |
2.3. Preparation of standard solutions and samples
The individual stock solutions were prepared by dissolving each compound in a solution of dimethyl sulfoxide-methanol (1:1 or 1:4) at a concentration of 1 mg/mL. The standard mixtures were prepared by mixing and diluting each stock solution with methanol. The stock solutions and standard mixtures were stored at −20 °C. The internal standard (compound 17) was dissolved in dimethyl sulfoxide-methanol (1:4) at a concentration of 1 mg/mL, and diluted in proper concentration with methanol. The concentration ranges of calibration standards were 0.05–20 ng/mL (0.05, 0.2, 1, 4, 10 and 20 ng/mL) for compound 1, 0.1–40 ng/mL (0.1, 0.4, 2, 8, 20 and 40 ng/mL) for compounds 6 and 8, 0.25–100 ng/mL (0.25, 1, 5, 20, 50, and 100 ng/mL) for compounds 7, 12, 13 and 14, and 0.5–200 ng/mL (0.5, 2, 10, 40, 100 and 200 ng/mL) for the others. Each calibration sample contained internal standard at a level of 10 ng/mL. Calibration curves were plotted by the ratios of analyte signal to internal standard signal versus concentration of each analyte.
The dried leaves and roots of L. barbarum were purchased from Anguo Mayway Herb Company Ltd. (Anguo, Hebei, China) at 20th July 2015. Plants were originated from Zhongning, Ningxia, China. Ten to one hundred milligrams of leaves and roots were extracted with 5 mL methanol containing internal standard (N-trans-feruloyl 3-phenylpropylamine, 17) by ultrasonic assisted extraction for 40 min followed by agitation for 60 min. After filtered with a 0.22 μm filter, samples were optionally concentrated by evaporation with nitrogen gas or directly injected into UHPLC system.
2.4. UHPLC-MS/MS analysis
UHPLC analyses were carried out with an Ultimate 3000 ultra high performance liquid chromatography (UHPLC) system from Dionex (Sunnyvale, CA, USA). The instrument was equipped with a XRS Open autosampler, a binary RS pump and a RS column compartment. Separations were conducted with a Phenomenex (Torrance, CA, USA) Synergi Fusion-RP column (2.0 mm × 100 mm, 2.5 μm particle size). The mobile phases were prepared by adding 0.1% formic acid to water (mobile phase A) and to acetonitrile (mobile phase B), respectively. Mobile phase B was linearly increased from 20 to 100% for 5 min, and maintained for 2 min before re-equilibration to the initial condition. The flow rate was 0.4 mL/min, and the injection volume was 10 μL. Column temperature was set at 25 °C.
The UHPLC system was hyphenated with a TSQ Quantiva (Thermo Fisher Scientific, San Jose, CA, USA) triple quadrupole mass spectrometer equipped with a heated-electrospray ionization (HESI) source. Electrospray ionization was operated in positive or negative polarity mode depending on the analytes, and spectra were acquired through selected reaction monitoring (SRM) measurements. Nitrogen gas was employed as sheath and auxiliary gases, and argon gas was used as collision gas. The source parameters were as follows: The positive spray voltage was set to 3500 V and the negative spray voltage was 2500 V. The sheath, aux and sweep gases were set to 45, 15 and 1 Arb, respectively. The ion transfer tube and vaporizer temperatures were both set at 350 °C. The following MS/MS parameters were used: The collision gas pressure was set to 2 mTorr, the dwell time was adjusted to 100 ms, and the chrom filter was set to 3 s. Data collection and processing were performed using Xcaliber software (Ver. 3.0).
2.5. Cell culture
RAW264.7 murine macrophages were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in high-glucose Dulbecco’s Modified Eagle’s medium, supplemented with 100 IU/mL penicillin/streptomycin, 1 mM sodium pyruvate and 10% fetal bovine serum. Cells were incubated in 10 cm culture Petri dishes in 5% CO2 with 70% humidity at 37 °C.
2.6. Cell viability assay
Cell viability was tested by MTT assay. Specifically, 4 × 105 cells/mL were seeded into 96-well plates and incubated for 12 h before treatment. Either the DMSO solution of compounds of interest or a vehicle (0.01% v/v DMSO) was added to the medium and then incubated for 24 h. Phenol red-free medium with 5 mg/mL of 3-(4,5-c)-2,5-diphenyltetrazolium bromide was added to cells and then incubated at 37 °C for 4 h. After removing the supernatant, formazan crystals were dissolved in 150 μL of DMSO. Optical densities were measured at 570 nm.
2.7. Nitrite assay
Cells (4 × 105 cells/mL) were treated with E. coli LPS (100 ng/mL) in either the presence of the compound of interest or 0.01% dimethyl sulfoxide (DMSO) as a vehicle in phenol red free medium for 24 h. After the incubation period, 50 μL of conditional supernatant was mixed with an equal volume of Griess reagent (1% sulfanilamide in 5% phosphoric acid and 0.1% naphthylethylenediamine dihydrochloride in water), and incubated at room temperature for 10 min. Production of nitrite was measured by an absorbance of 550 nm.
2.8. Statistical analysis
Results are shown as mean ± standard deviation. Statistical analysis for IC50 was performed using Prism 7 by non-linear regression. IC50 values are shown as 95% confidence interval. All experimental data were obtained independently and replicated a total of three times. Significant differences were determined as p < 0.05.
3. Results and discussion
3.1. Synthesis design
The biosynthesis of HCAA compounds involves phenyl-propanoid pathway that contributes to the formation of hydroxycinnamate moiety of the amides through deamination of l-phenylalanine into trans-cinnamic acid or cinnamate [1,2]. Serial modifications, including hydroxylation and further methylation on the hydroxyl sites, yield more elaborate substitution patterns on the cinnamate moiety. The condensation is catalyzed by hydroxycinnamoyl-CoA:tyramine N-(hydroxycinnamoyl) transferase (THT) by joining two chemical backbones such as hydroxycinnamoyl-CoA thioester and hydroxyphenylethylamines [1]. Hydroxycinnamoyl-CoA thioester is formed through sequential modification of cinnamic acid through series of enzymatic catalysis. A wide array of chemical species could serve as substrates of THT, including various hydroxycinnamic acid and phenolic amine derivatives, which leads to a potential large of chemical diversity of amide species [19].
We designed the synthesis consisting three hydroxycinnamic acids and six different phenolic amines, including trans-caffeic acid, trans-ferulic acid, 3,4-dihydroxyhydrocinnamic acid, phenethylamine, 3,4-dimethoxyphenethylamine, tryptamine, tyramine, 3-methoxytyramine and dopamine. The synthesis design yields a series of analogous HCAA compounds, which would a comprehensive investigation of this family by our method from the plant tissue.
3.2. Optimization of UHPLC-MS/MS
Although UHPLC-TripleQ provides very limited structural information of analytes, especially those from extracted from plants containing multiple structural isomers and analogs, this drawback could be overcome by targeting synthesis and NMR with structure elucidation and confirmation of each synthetic standard.
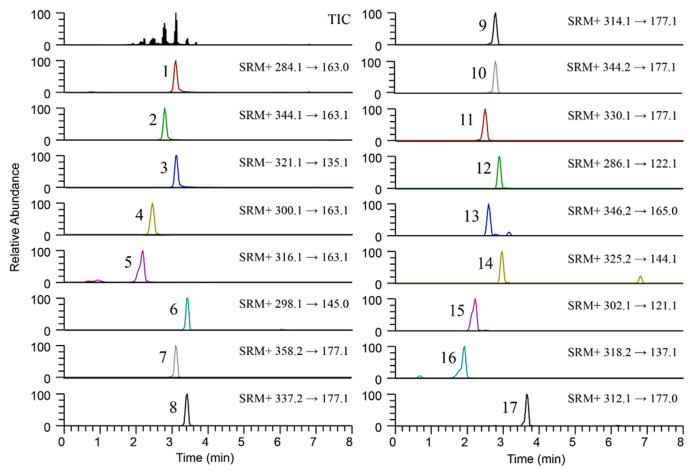
0.1% formic acid was added in both water (mobile phase A) and acetonitrile (mobile phase B) as a mobile phase modifier to obtain desirable peak shapes and adequate retention times of analytes. Gradient elution was employed to decrease retention time as well as to further improve peak sharpness. All compounds were eluted within 5 min (Fig. 2), which means that the method is suitable for large-scale application with large number of samples.
Fig. 2.
UHPLC-MS/MS chromatograms of standard mixture containing 100 ng/mL N-trans-caffeoyl phenethylamine (1), 200 ng/mL N-trans-caffeoyl 3,4-dimethoxyphenethylamine (2), 200 ng/mL N-trans-caffeoyl tryptamine (3), 200 ng/mL N-trans-caffeoyl tyramine (4), 200 ng/mL N-trans-caffeoyl dopamine (5), 40 ng/mL N-trans-feruloyl phenethylamine (6), 100 ng/mL N-trans-feruloyl 3,4-dimethoxyphenethylamine (7), 40 ng/mL N-trans-feruloyl tryptamine (8), 200 ng/mL N-trans-feruloyl tyramine (9), 200 ng/mL N-trans-feruloyl 3-methoxytyramine (10), 200 ng/mL N-trans-feruloyl dopamine (11), 100 ng/mL N-3,4-Dihydroxyhydrocinnamoyl phenethylamine (12), 100 ng/mL N-3,4-Dihydroxyhydrocinnamoyl 3,4-dimethoxyphenthylamine (13), 100 ng/mL N-3,4-Dihydroxyhydrocinnamoyl tryptamine (14), 200 ng/mL N-3,4-Dihydroxyhydrocinnamoyl tyramine (15), 200 ng/mL N-3,4-Dihydroxyhydrocinnamoyl dopamine (16), and 10 ng/mL N-trans-feruloyl 3-phenylpropylamine (internal standard) (17).
Each synthetic analyte was infused into the mass spectrometer, and the precursor ions and at two product ions were preliminarily selected in both positive ion and negative ion modes. Positive ion mode showed better signal response than negative ion mode. Only compound 3 had higher intensity in negative mode. Parameters such as spray voltage, sheath gas, aux gas, ion transfer tube temperature, vaporizer temperature, collision gas pressure and dwell time were evaluated to obtain suitable signal for the precursor and product ions of the analytes. The precursor ion, product ion, collision energy and RF lens voltage of the analytes were individually optimized by direct flow infusion of each standard, and the optimum values are summarized in Supplementary Table 1.
3.3. Method validation
The developed UHPLC-MS/MS method was validated in terms of specificity, linearity, intra- and inter-day precision and accuracy, limit of detection (LOD), limit of quantification (LOQ), recovery, and reproducibility according to the guideline established by the International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use Validation of analytical procedures: Text and methodology, 2005 [20].
The specificity was determined by a chromatogram of target compounds (Fig. 2). The retention times of compounds 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 and 17 (internal standard) are 3.11, 2.80, 3.13, 2.45, 2.19, 3.44, 3.10, 3.42, 2.79, 2.80, 2.50, 2.90, 2.59, 2.97, 2.22, 1.93 and 3.65 min, respectively, and the analytes were detected without any apparent interference. The LC-MS/MS chromatograms of each compound identified from root barks and leaves extract could be found in Supplemental material Fig. 1. Calibration curves were constructed by plotting the ratio of the analyte peak area to the internal standard peak area against the respective analyte concentrations. As presented in Table 2, the calibration curves of analytes were linear over the concentration ranges with high correlation of determination (r2) values above 0.996. The limit of detection (LOD) and limit of quantification (LOQ) were set to three times and ten times the signal to noise ratio by analyzing serial diluted standards, respectively. All LODs and LOQs were estimated at picogram (pg/mL) levels, which means that the method was proven to be highly sensitive to determine trace amount of compounds (Table 2).
Table 2.
Parameters linear range, regression equation, correlation coefficient (r2), retention time (tr), limit of quantification (LOQ) and limit of detection (LOD) for HCAA compounds.
| Compound | Linear range (ng/mL) | Regression equation | r | tr (min) | LOQ (ng/mL) | LOD (ng/mL) |
|---|---|---|---|---|---|---|
| 1 | 0.05–20 | y = 0.0566x − 0.0007 | 0.9999 | 3.1 | 0.05 | 0.02 |
| 2 | 0.5–200 | y = 0.0169x + 0.0290 | 0.9989 | 2.8 | 0.1 | 0.025 |
| 3 | 0.5–200 | y = 0.0092x − 0.0097 | 0.9996 | 3.1 | 0.1 | 0.02 |
| 4 | 0.5–200 | y = 0.0243x + 0.0136 | 0.9999 | 2.5 | 0.05 | 0.01 |
| 5 | 0.5–200 | y = 0.0083x − 0.0287 | 0.9964 | 2.2 | 0.25 | 0.1 |
| 6 | 0.1–40 | y = 0.0619x + 0.0038 | 0.9999 | 3.4 | 0.02 | 0.005 |
| 7 | 0.25–100 | y = 0.0830x + 0.0113 | 0.9999 | 3.1 | 0.05 | 0.01 |
| 8 | 0.1–40 | y = 0.0558x + 0.0011 | 0.9985 | 3.4 | 0.02 | 0.008 |
| 9 | 0.5–200 | y = 0.0591x + 0.1257 | 0.9992 | 2.8 | 0.05 | 0.01 |
| 10 | 0.5–200 | y = 0.0329x + 0.0324 | 0.9998 | 2.8 | 0.1 | 0.025 |
| 11 | 0.5–200 | y = 0.0201x + 0.0128 | 0.9995 | 2.5 | 0.1 | 0.025 |
| 12 | 0.25–100 | y = 0.0332x + 0.0089 | 0.9995 | 2.9 | 0.1 | 0.025 |
| 13 | 0.25–100 | y = 0.0174x + 0.0020 | 0.9988 | 2.6 | 0.05 | 0.01 |
| 14 | 0.25–100 | y = 0.0512x + 0.0038 | 0.9995 | 3.0 | 0.05 | 0.02 |
| 15 | 0.5–200 | y = 0.0228x + 0.0007 | 0.9983 | 2.2 | 0.1 | 0.025 |
| 16 | 0.5–200 | y = 0.0050x − 0.0114 | 0.9982 | 1.9 | 0.1 | 0.04 |
y means peak area ratio and x means concentration (ng/mL).
The intra- and inter-day precision and accuracy were estimated at three different concentrations (0.05, 4 and 20 ng/mL for compound 1, 0.1, 8 and 40 ng/mL for compounds 6 and 8, 0.25, 20 and 100 ng/mL for compounds 7, 12, 13 and 14, and 0.5, 40 and 200 ng/mL for the others) in the range of the calibration curve. Three replicate analyses were conducted on the same day and on three consecutive days. The precision was defined as the relative standard deviation (RSD, %), and the accuracy was expressed as the observed concentration relative to the nominal concentration. The intra-day precision was measured from 0.1 to 10.9%, and the inter-day precision was from 0.7 to 11.8%. The intra-day accuracy ranged from 90.5 to 111.6%, and the inter-day accuracy ranged from 93.8 to 112.8% (Table 3).
Table 3.
Intra- and inter-day precision, accuracy (n =3) and reproducibility (n =6) of HCAA compounds.
| Compound | Precisiona | Accuracya | Reproducibility | ||
|---|---|---|---|---|---|
|
|
|
|
|||
| Intra-day (RSD, %) | Inter-day (RSD, %) | Intra-day (%) | Inter-day (%) | RSD (%) | |
| 1 | 2.4–8.9 | 4.2–8.4 | 96.8–111.6 | 96.6–108.7 | 1.3 |
| 2 | 0.8–5.1 | 4.0–6.3 | 94.7–104.3 | 93.8–100.3 | 3.0 |
| 3 | 1.4–3.6 | 1.6–9.1 | 96.5–106.6 | 96.3–110.6 | 2.2 |
| 4 | 0.8–2.3 | 1.7–4.7 | 97.3–102.8 | 97.4–103.5 | 2.2 |
| 5 | 0.8–4.5 | 2.2–5.7 | 95.0–106.3 | 100.0–106.5 | 3.3 |
| 6 | 1.2–8.5 | 3.1–8.9 | 101.4–111.0 | 96.3–102.6 | 2.4 |
| 7 | 1.1–4.0 | 0.8–4.8 | 93.7–103.6 | 99.1–102.2 | 2.6 |
| 8 | 1.6–2.5 | 1.2–8.2 | 97.5–109.2 | 96.0–102.7 | 2.3 |
| 9 | 5.1–6.9 | 0.7–9.9 | 90.5–96.6 | 99.0–101.6 | 2.1 |
| 10 | 4.6–10.9 | 4.9–7.6 | 93.4–107.8 | 100.3–106.5 | 2.4 |
| 11 | 0.1–5.3 | 0.7–9.1 | 98.8–109.0 | 97.0–105.3 | 2.5 |
| 12 | 1.7–4.5 | 2.9–11.8 | 99.0–103.5 | 98.7–112.8 | 2.2 |
| 13 | 1.1–4.6 | 2.4–3.7 | 99.0–103.9 | 101.5–103.1 | 2.1 |
| 14 | 0.5–5.8 | 2.6–11.3 | 97.4–100.2 | 99.1–110.1 | 2.7 |
| 15 | 1.7–4.2 | 2.1–6.8 | 96.0–100.4 | 97.0–101.6 | 2.4 |
| 16 | 1.4–2.0 | 3.2–6.0 | 97.2–101.2 | 100.8–103.9 | 4.7 |
Three different concentration levels (n = 3).
The recovery was determined by a standard addition method using low and high amounts (40 ng/mL and 100 ng/mL) of analytes. Recoveries from root barks and leaves were evaluated, and each experiment was performed in five times (n = 5). The mean recovery was in the range of 94.8–105.1% and the RSD was less than 15% (Table 4). Lastly, the reproducibility was examined by six replicate analyses (n = 6) of a standard solution (10 ng/mL for compound 1, 20 ng/mL for compounds 6 and 8, 50 ng/mL for compounds 7, 12, 13 and 14, and 100 ng/mL for the others). The resulting RSD was below 5% for all compounds (Table 3).
Table 4.
Recovery of HCAA compounds in root barks and leaves of Lycium barbarum (n =5).
| Compound | Spiked concentration (ng/mL) | Recovery | |||
|---|---|---|---|---|---|
|
| |||||
| Root barks | Leaves | ||||
|
|
|
||||
| Mean (%) | RSD (%) | Mean (%) | RSD (%) | ||
| 1 | 40 | 99.7 | 2.3 | 97.3 | 7.3 |
| 100 | 96.8 | 2.5 | 97.8 | 1.2 | |
| 2 | 40 | 96.8 | 1.3 | 97.3 | 6.8 |
| 100 | 99.0 | 1.3 | 98.1 | 6.6 | |
| 3 | 40 | 98.6 | 2.5 | 98.1 | 1.8 |
| 100 | 99.6 | 1.3 | 101.0 | 0.6 | |
| 4 | 40 | 96.2 | 2.2 | 96.1 | 4.2 |
| 100 | 94.8 | 4.0 | 97.5 | 2.8 | |
| 5 | 40 | 99.5 | 2.4 | 98.7 | 3.7 |
| 100 | 98.4 | 3.5 | 99.5 | 2.6 | |
| 6 | 40 | 99.9 | 2.2 | 101.0 | 6.6 |
| 100 | 98.0 | 4.0 | 96.0 | 4.5 | |
| 7 | 40 | 100.1 | 1.5 | 100.7 | 1.4 |
| 100 | 99.8 | 1.4 | 101.0 | 2.6 | |
| 8 | 40 | 98.7 | 2.9 | 98.2 | 3.6 |
| 100 | 100.1 | 4.8 | 97.9 | 4.4 | |
| 9 | 40 | 104.5 | 6.4 | 98.0 | 11.3 |
| 100 | 102.3 | 6.1 | 99.4 | 8.6 | |
| 10 | 40 | 98.7 | 1.8 | 105.1 | 6.5 |
| 100 | 97.7 | 1.4 | 98.5 | 4.0 | |
| 11 | 40 | 97.0 | 7.3 | 97.6 | 1.1 |
| 100 | 98.3 | 1.3 | 101.3 | 4.7 | |
| 12 | 40 | 99.1 | 1.9 | 98.7 | 6.9 |
| 100 | 98.2 | 1.3 | 98.9 | 2.4 | |
| 13 | 40 | 96.5 | 1.4 | 102.0 | 1.6 |
| 100 | 96.6 | 1.8 | 98.3 | 3.0 | |
| 14 | 40 | 100.0 | 1.2 | 98.4 | 7.9 |
| 100 | 99.8 | 1.5 | 99.6 | 2.1 | |
| 15 | 40 | 95.2 | 0.9 | 100.3 | 5.7 |
| 100 | 98.8 | 2.8 | 96.9 | 4.0 | |
| 16 | 40 | 104.7 | 3.7 | 102.1 | 3.6 |
| 100 | 100.8 | 2.7 | 101.8 | 3.1 | |
3.4. Quantification of HCAAs from the plant tissue
In order to further validate our developed method, we applied it to comprehensively profile HCAA from different plant tissues, which were the root barks and leaves of L. barbarum, aiming to explore the chemical diversity of the HCAA family as well as systematically quantify them at different locations of the plant. The established UHPLC-MS/MS method was applied to quantitative evaluation of target compounds in samples of leaves and root barks. The analysis was performed in triplicate (n = 3). HCAA compounds were found in various quantities in the root bark and leaves of L. barbarum. Table 5 lists the quantification results of HCAA compounds in the leaves and root barks. Fourteen HCAA compounds were identified from the root barks and 10 from the leaves, which indicated that root barks had more diverse HCAA species compared to the leaves and fruits, compared to our previous work [18]. Compounds 4, 9, 11 and 15 presented at highest concentration in the root bark as 26446.0 ± 154.8, 10600.0 ± 509.2, 5392.1 ± 236.3 and 4864.0 ± 74.9 ng/g, respectively. Compounds 9 and 11 were trans-feruloyltyr-amine and its derivative trans-feruloyl 3-methoxytyramine, which were found to ubiquitously present in plants, serving as major metabolic constituents of cell wall alterations that play a role in plant defense response to pathogen challenge [21]. Compounds 4 and 15 also had tyramine moiety but conjugated with trans-caffeic acid and 3,4-dihydroxyhydrocinnamic acid, which may biosynthesized from similar precursors so that existed at higher concentration. Similar trend was also observed in quantification results of leaves. Compound 4 was demonstrated to have promising anti-inflammatory properties in previous studies [14], which promoted us to investigate the anti-inflammatory activities of its structural analogs in the later sections. Following the tyramine conjugated species, compounds 5, 11 and 16 were also found as major HCAA components in both root barks and leaves. They had dopamine extensions and conjugated by trans-caffeic acid, trans-ferulic acid and 3,4-dihydroxyhydrocinnamic acid, respectively. They may result from the hydroxylation of tyramine, which was the decarboxylated product of tyrosine [4]. Interestingly, compound 6 was found minor in the root barks (3.9 ± 0.1 ng/g) but had much higher quantity in the leaves (182.9 ± 6.8 ng/g).
Table 5.
Mean concentrations of HCAA compounds in root barks and leaves of Lycium barbarum (n =3).
| Compound | Mean ± SDa (ng/g) | |
|---|---|---|
|
| ||
| Root barks | Leaves | |
| 1 | 0.6 ± 0.1 | ND |
| 2 | NDb | ND |
| 3 | 49.6 ± 2.8 | ND |
| 4 | 26446.0 ± 154.8 | 2143.2 ± 53.0 |
| 5 | 317.6 ± 21.9 | 523.7 ± 39.7 |
| 6 | 3.9 ± 0.1 | 182.9 ± 6.8 |
| 7 | 6.4 ± 0.2 | 25.3 ± 0.3 |
| 8 | 2.1 ± 0.1 | 2.6 ± 0.1 |
| 9 | 10600.0 ± 509.2 | 20762.2 ± 1304.2 |
| 10 | 5392.1 ± 236.3 | 42200.3 ± 1692.6 |
| 11 | 611.3 ± 10.6 | 688.6 ± 34.9 |
| 12 | 5.2 ± 0.2 | ND |
| 13 | ND | ND |
| 14 | 2.7 ± 0.1 | ND |
| 15 | 4864.0 ± 74.9 | 1694.2 ± 64.5 |
| 16 | 324.3 ± 8.0 | 508.0 ± 23.2 |
Standard deviation.
Not detected.
As far as we know, HCAA compounds were first time reported in leaves of the plant. Previous studies only detected several major HCAA compounds in the root bark but not in the leaves [16,22], and 10 HCAA compounds were newly detected in the root bark. The developed method combined organic synthesis and UHPLC-TripleQ specifically targeting at HCAA species identification and quantification largely improved sensitivity with respect to quantification and detection limit. The improvement potentially leads to targeting compartmental simultaneous analysis with fractionation and extraction method.
3.5. Nitric oxide inhibition and potential anti-inflammatory properties of HCAAs
Over past few decades, many studies reveal that chronic inflammation is a critical component in many human diseases and conditions, including obesity, cardiovascular diseases, neurodegenerative diseases, diabetes and cancers [23–25], which motivates much research on discover anti-inflammatory agent from natural sources and investigate their pharmacological mechanisms. Traditional purification and bioassay-guided method caused great loss of minor chemical constituents that potentially exhibited potent activities. Our developed analytical method targeting at HCAA species compensated this drawback and identified novel structural analogs from the plant. The findings drove us to further examine their bioactivities by using in vitro model in order to support our method with improved sensitivity overcoming the challenges remained in traditional purification and bioactivities guided techniques.
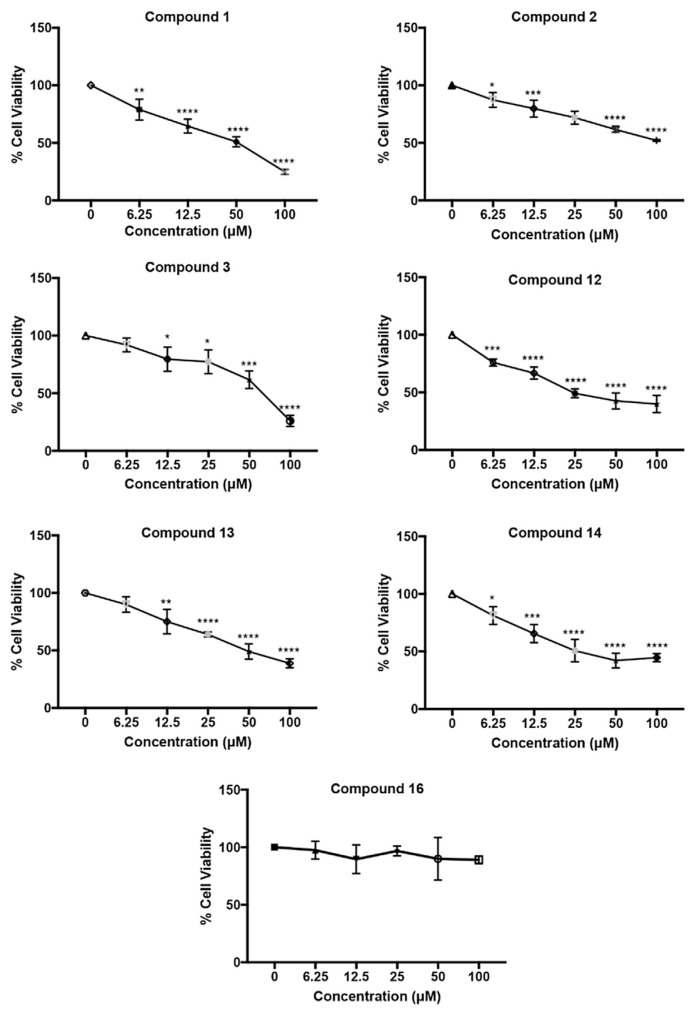
In order to screen the improved anti-inflammatory properties obtained from newly identified structural analogs, MTT assays were firstly performed in order to select proper testing concentration as well as investigate their potential cytotoxicity effects. Among the IC50 values shown in Table 6, including the results from our previous work in order to compare, 10 of the 16 compounds possess IC50 values larger than 100 μM, indicating low cytotoxicity of these compounds. The compounds exhibiting higher cytotoxicity contain large of amounts of trans-caffeic acid or 3,4-dihydroxyhydrocinnamic acid moiety, and were compounds 1, 3 and 12–14, which each contain a catechol structure (Fig. 3). These results led to the hypothesis that the ortho bis-hydroxylation structure is crucial for cytotoxicity. Similar findings were observed by studying the anti-cancer properties of trans-caffeic acid derivatives due to hydroxyl groups in the molecule that enhances the affinity for proteins and nucleic acids through potential hydrogen donating and accepting [26–28]. We confirmed that methylation of the hydroxyl group did reduce cytotoxicity by comparing compounds 1, 3 and 12–14 with compounds 5–10 that had lost the catechol structure of trans-ferulic acid moiety (see Fig. 4).
Table 6.
Cytotoxicity IC50 value and NO production inhibition IC50 value of HCAA compound 1–16 to RAW 264.7.
| Compound No. | Cytotoxicity IC50, Mean ± STDa (μM) | NO production IC50, Mean ± STD (μM) | ||
|---|---|---|---|---|
| trans-Caffeic acid | 1 | 35.25 ± 3.148 | 2.381 ± 0.1497 | |
| 2 | >100 | 5.575 ± 0.3469 | ||
| 3 | 72.71 ± 7.811 | 4.227 ± 0.3854 | ||
| 4 | >100 | 12.76 ± 1.611 | [18] | |
| 5 | >100 | 39.05 ± 3.527 | [18] | |
| trans-Ferulic acid | 6 | >100 | 14.38 ± 2.099 | [18] |
| 7 | >100 | >10 | [18] | |
| 8 | >100 | >50 | [18] | |
| 9 | >100 | >50 | [18] | |
| 10 | >100 | >50 | [18] | |
| 11 | >100 | 15.08 ± 0.8049 | [18] | |
| 3,4-Dihydroxyhydrocinnamic acid | 12 | 35.53 ± 4.066 | >5 | |
| 13 | 52.00 ± 4.849 | >10 | ||
| 14 | 40.34 ± 6.697 | >5 | ||
| 15 | >100 | 40.36 ± 4.648 | [18] | |
| 16 | >100 | >20 |
Standard error.
Fig. 3.
Cytotoxicity of HCAA compounds on RAW264.7 cells, measured by MTT assay. The cells were incubated with compounds of interest (6.25, 12.5, 25, 50, 100 μM) or vehicle control (0.01% DMSO v/v) for 24 h. Asterisks indicate significant differences from the control (*p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001).
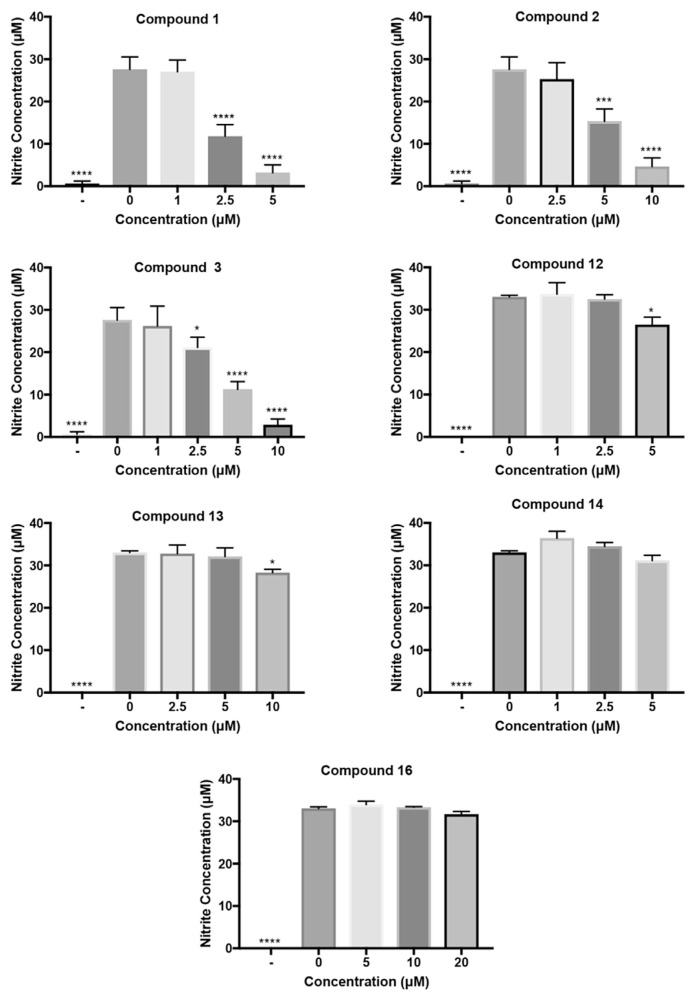
Fig. 4.
Inhibitive effects of HCAA compounds on NO production from LPS-activated RAW624.7 cell, measured by Griess reagent. Compounds of interested or vehicle control (0.01% DMSO v/v) were co-incubated with 100 ng/mL LPS for 24 h.
A total of 8 HCAA compounds exhibited NO inhibitory properties, among these, 7 were identified in the plant. Furthermore, compound 1 (2.381 μM) and compound 3 (4.227 μM) showed potent NO inhibitory activity with a low NO IC50 value, and compound 4, which was abundantly present (26446.0 ± 154.8 ng/g) also effectively inhibited NO production with a 50% inhibition concentration of 12.760 μM.
According to our results, the compounds with a trans-caffeic acid moiety exhibited the most significant inhibition of NO accumulation. All five trans-caffeic acid derivatives (compounds 1–5) have a 50% inhibition concentration (<50 μM) of NO production from macrophages. Among trans-ferulic acid derivatives, two compounds, 6 and 11, exhibited inhibitory effects of NO production, which implies a reduced NO suppression effect. Moreover, only compound 15 in the 3,4-dihydroxyhydrocinnamic acid derivatives group was found to significantly decrease NO levels with a 50% inhibition concentration of 40.360 μM. The reduction of effectiveness on NO production by macrophages suggests that catechol and tethered conjugated double bond structures are essential for inhibitory potency, and the conjugated double bond structure may be more important for inhibitory effects. The inhibitory activity of HCAAs containing the trans-caffeic acid moiety decreased in following order: 5 < 4 < 2 < 3 < 1. Furthermore, the inhibitory properties of NO accumulation in the cell medium are not fully related to the free radical scavenging effects of these compounds. A free radical scavenging study of caffeoyl amide compounds shows effectiveness decreases in the order: 1 < 4 < 5 [29], which is not consistent with our results, and indicates these compounds exhibited NO inhibitory activities through cellular mechanisms. This sequence suggested that the effectiveness of NO inhibition is proportional to hydrophobicity resulting from a decrease in the number of hydroxyl and methoxyl groups. The difference in NO inhibition between 3 and 1 may be due to the size of the side chain of the amine moiety. Thus, the effectiveness to inhibit NO production may also require an optimal bulkiness of the side chain of amine moieties. Similar trends were observed in studies of anti-inflammatory properties of caffeic acid alkyl ester derivatives to suppress NO production [30,31]. The anti-inflammatory mechanism of caffeoyl phenthylester has been intensively reviewed by Murtaza et al. [32], but due to the lack of physiological stability and short elimination half-life, the amide derivatives reveal great potential to exert anti-inflammatory potency in biological systems. However, in our study, this trend was only observed in the caffeic acid derivative group, not the other two phenolic acid derivatives. The detailed mechanism underlying the structure-activity relationship remains unclear.
4. Conclusion
In this study, we developed and validated a highly sensitive method UHPLC-MS/MS combined with synthetic standards, targeting at profiling HCAA species from plant tissues. We further applied our method to determine of potential existing HCAA species from the leaves and root barks of L. barbarum. We successfully identified 10 HCAA species in leaves, which was reported first time from the leaves of the plants. A total of 14 HCAA compounds were found in root barks, and among them, 10 HCAA compounds were newly reported, which broadened the diversity of the HCAA species in this plant. Furthermore, we screened the potential anti-inflammatory activities of target HCAA compounds, and found that some compounds exhibited a promising inhibitory effect on NO production, which is more potent than any HCAA compounds reported before. We expect the developed method to be applicable to comprehensive analysis of plant metabolites from whole-cell level analysis to subcellular metabolite distributions, as well as the quantitative analysis of wide varieties of HCAA compounds in plant tissues. The developed methodology also facilitates natural product studies that can be expanded to identification of other bioactive components from the plants, in order to explore the chemical diversity of targeting metabolites as their potential improved potency [7].
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jfda.2017.06.002.
References
- 1. Facchini PJ, Hagel J, Zulak KG. Hydroxycinnamic acid amide metabolism: physiology and biochemistry. Can J Bot. 2002;80:577–89. [Google Scholar]
- 2. Macoy DM, Kim WY, Lee SY, Kim MG. Biosynthesis, physiology, and functions of hydroxycinnamic acid amides in plants. Plant Biotechnol Rep. 2015;9:269–78. [Google Scholar]
- 3. Ly D, Kang K, Choi JY, Ishihara A, Back K, Lee SG. HPLC analysis of serotonin, tryptamine, tyramine, and the hydroxycinnamic acid amides of serotonin and tyramine in food vegetables. J Med Food. 2008;11:385–9. doi: 10.1089/jmf.2007.514. [DOI] [PubMed] [Google Scholar]
- 4. Bassard JE, Ullmann P, Bernier F, Werck-Reichhart D. Phenolamides: bridging polyamines to the phenolic metabolism. Phytochemistry. 2010;71:1808–24. doi: 10.1016/j.phytochem.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 5. Martin-Tanguy J, Cabanne F, Perdrizet E, Martin C. The distribution of hydroxycinnamic acid amides in flowering plants. Phytochemistry. 1978;17:1927–8. [Google Scholar]
- 6. Campos L, Lisón P, López-Gresa MP, Rodrigo I, Zacarés L, Conejero V, et al. Transgenic tomato plants overexpressing tyramine N-hydroxycinnamoyltransferase exhibit elevated hydroxycinnamic acid amide levels and enhanced resistance to Pseudomonas syringae. Mol Plant Microbe Interact. 2014;27:1159–69. doi: 10.1094/MPMI-04-14-0104-R. [DOI] [PubMed] [Google Scholar]
- 7. Ishihara A, Hashimoto Y, Tanaka C, Dubouzet JG, Nakao T, Matsuda F, et al. The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. Plant J. 2008;54:481–95. doi: 10.1111/j.1365-313X.2008.03441.x. [DOI] [PubMed] [Google Scholar]
- 8. Martin-Tangtiy J. Conjugated poiyamines and reproductive deveiopment: biochemicai, moiecuiar and physioiogical approaches. Physiol Plant. 1997;100:675–88. [Google Scholar]
- 9. Cos P, Rajan P, Vedernikova I, Calomme M, Pieters L, Vlietinck AJ, et al. In vitro antioxidant profile of phenolic acid derivatives. Free Radic Res. 2009;36:711–6. doi: 10.1080/10715760290029182. [DOI] [PubMed] [Google Scholar]
- 10. Negri G, Teixeira EW, Alves ML, Moreti AC, Otsuk IP, Borguini RG, et al. Hydroxycinnamic acid amide derivatives, phenolic compounds and antioxidant activities of extracts of pollen samples from Southeast Brazil. J Agric Food Chem. 2011;59:5516–22. doi: 10.1021/jf200602k. [DOI] [PubMed] [Google Scholar]
- 11. Marinova E, Georgiev L, Totseva I, Seizova K, Milkova T. Antioxidant activity and mechanism of action of some synthesised phenolic acid amides of aromatic amines. Czech J Food Sci. 2013;31:5–13. [Google Scholar]
- 12. Razzaghi-Asl N, Garrido J, Khazraei H, Borges F, Firuzi O. Antioxidant properties of hydroxycinnamic acids: a review of structure-activity relationships. Curr Med Chem. 2013;20:4436–50. doi: 10.2174/09298673113209990141. [DOI] [PubMed] [Google Scholar]
- 13. Lee DG, Park Y, Kim MR, Jung HJ, Seu YB, Hahm KS, et al. Anti-fungal effects of phenolic amides isolated from the root bark of Lycium chinense. Biotechnol Lett. 2004;26:1125–30. doi: 10.1023/B:BILE.0000035483.85790.f7. [DOI] [PubMed] [Google Scholar]
- 14. Xie LW, Atanasov AG, Guo DA, Malainer C, Zhang JX, Zehl M, et al. Activity-guided isolation of NF-kappaB inhibitors and PPARgamma agonists from the root bark of Lycium chinense Miller. J Ethnopharmacol. 2014;152:470–7. doi: 10.1016/j.jep.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 15. Park JB, Schoene N. N-Caffeoyltyramine arrests growth of U937 and Jurkat cells by inhibiting protein tyrosine phosphorylation and inducing caspase-3. Cancer Lett. 2003;202:161–71. doi: 10.1016/j.canlet.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 16. Zhang J, Guan S, Sun J, Liu T, Chen P, Feng R, et al. Characterization and profiling of phenolic amides from Cortex Lycii by ultra-high performance liquid chromatography coupled with LTQ-Orbitrap mass spectrometry. Anal Bioanal Chem. 2015;07:581–95. doi: 10.1007/s00216-014-8296-4. [DOI] [PubMed] [Google Scholar]
- 17. Zhang JX, Guan SH, Yang M, Feng RH, Wang Y, Zhang YB, et al. Simultaneous determination of 24 constituents in Cortex Lycii using high-performance liquid chromatography-triple quadrupole mass spectrometry. J Pharm Biomed Anal. 2013;77:63–70. doi: 10.1016/j.jpba.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 18. Wang S, Suh JH, Zheng X, Wang Y, Ho CT. Identification and quantification of potential anti-inflammatory hydroxycinnamic acid amides from wolfberry. J Agric Food Chem. 2017;65:364–72. doi: 10.1021/acs.jafc.6b05136. [DOI] [PubMed] [Google Scholar]
- 19. Jonathan Nergel CM. The biosynthesis of feruloyltyramine in Nicotiana tabacum. Phytochemistry. 1984;23:2797–801. [Google Scholar]
- 20. IHT guideline. Validation of analytical procedures: text and methodology, Q2,1. 2005 [Google Scholar]
- 21. McLusky SR, Bennett MH, Beale MH, Lewis MJ, Gaskin P, Mansfield JW. Cell wall alterations and localized accumulation of feruloyl-3′-methoxytyramine in onion epidermis at sites of attempted penetration by Botrytis allii are associated with actin polarisation, peroxidase activity and suppression of flavonoid biosynthesis. Plant J. 1999;17:523–34. [Google Scholar]
- 22. Dong JZ, Lu DY, Wang Y. Analysis of flavonoids from leaves of cultivated Lycium barbarum L. Plant Foods Hum Nutr. 2009;64:199–204. doi: 10.1007/s11130-009-0128-x. [DOI] [PubMed] [Google Scholar]
- 23. Pan MH, Lai CS, Ho CT. Anti-inflammatory activity of natural dietary flavonoids. Food Funct. 2010;1:15–31. doi: 10.1039/c0fo00103a. [DOI] [PubMed] [Google Scholar]
- 24. Coussens LM. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Monteiro R, Azevedo I. Chronic inflammation in obesity and the metabolic syndrome. Mediators Inflamm. 2010;2010:1–10. doi: 10.1155/2010/289645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ali A, Bansal D, Kaushik NK, Kaushik N, Choi EH, Gupta R. Syntheses, characterization, and anti-cancer activities of pyridine-amide based compounds containing appended phenol or catechol groups. J Chem Sci. 2014;126:1091–105. [Google Scholar]
- 27. Ketabforoosh SHE, Amini M, Vosooghi M, Shafiee A, Azizi E, Kobarfard F. Synthesis, evaluation of anticancer activity and QSAR study of heterocyclic esters of caffeic acid. Iran. J Pharm Res. 2013;12:705–19. [PMC free article] [PubMed] [Google Scholar]
- 28. Lee YJ, Liao PH, Chen Wk, Yang CY. Preferential cytotoxicity of caffeic acid phenethyl ester analogues on oral cancer cells. Cancer Lett. 2000;153(1–2):51–6. doi: 10.1016/s0304-3835(00)00389-x. [DOI] [PubMed] [Google Scholar]
- 29. Son S, Lewis BA. Free radical scavenging and antioxidative activity of caffeic acid amide and ester analogues: structure-activity relationship. J Agric Food Chem. 2002;50:468–72. doi: 10.1021/jf010830b. [DOI] [PubMed] [Google Scholar]
- 30. Uwai K, Osanai Y, Imaizumi T, Kanno S, Takeshita M, Ishikawa M. Inhibitory effect of the alkyl side chain of caffeic acid analogues on lipopolysaccharide-induced nitric oxide production in RAW264. 7 macrophages. Bioorg Med Chem Lett. 2008;16:7795–803. doi: 10.1016/j.bmc.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 31. da Cunha FM, Duma D, Assreuy J, Buzzi FC, Niero R, Campos MM, et al. Caffeic acid derivatives: in vitro and in vivo anti-inflammatory properties. Free Radic Res. 2004;38:1241–53. doi: 10.1080/10715760400016139. [DOI] [PubMed] [Google Scholar]
- 32. Murtaza G, Sajjad A, Mehmood Z, Shah SH, Siddiqi AR. Possible molecular targets for therapeutic applications of caffeic acid phenethyl ester in inflammation and cancer. J Food Drug Anal. 2015;23:11–8. doi: 10.1016/j.jfda.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]