Abstract
Kluyveromyces marxianus protein hydrolysates were prepared by two different sonicated-enzymatic (trypsin and chymotrypsin) hydrolysis treatments to obtain antioxidant and ACE-inhibitory peptides. Trypsin and chymotrypsin hydrolysates obtained by 5 h, exhibited the highest antioxidant and ACE-inhibitory activities. After fractionation using ultra-filtration and reverse phase high performance liquid chromatography (RP-HPLC) techniques, two new peptides were identified. One fragment (LL-9, MW = 1180 Da) with the amino acid sequence of Leu-Pro-Glu-Ser-Val-His-Leu-Asp-Lys showed significant ACE inhibitory activity (IC50 = 22.88 μM) while another peptide fragment (VL-9, MW = 1118 Da) with the amino acid sequence of Val-Leu-Ser-Thr-Ser-Phe-Pro-Pro-Lys showed the highest antioxidant and ACE inhibitory properties (IC50 = 15.20 μM, 5568 μM TE/mg protein). The molecular docking studies revealed that the ACE inhibitory activities of VL-9 is due to interaction with the S2 (His513, His353, Glu281) and S’1 (Glu162) pockets of ACE and LL-9 can fit perfectly into the S1 (Thr345) and S2 (Tyr520, Lys511, Gln281) pockets of ACE.
Keywords: K. marxianus, Bioactive peptides, Antioxidant, ACE inhibitory, Protein hydrolysate
1. Introduction
During the last several years many investigators reported the presence of Kluyveromyces marxianus in the different dairy products [1–5]. Yeast biomass is generally regarded as safe (GRAS) [6] and can be used for large-scale production of single cell protein (SCP) [7]. The biomass of K. marxianus is used as animal feed and as yeast extract for use in food processing industry [8]. Also, with protein levels ranging from 50 to 70%, yeast extract provides an abundance source of amino acids and bioactive peptides.
Bioactive peptides contain 2–20 amino acids with possible bioactivities including antihypertensive, antioxidant, antimicrobial, anticancer, and opioid activity [9–11]. There has been a great interest in antihypertensive peptides for their efficiency in lowering blood pressure. The antihypertensive peptides are effective mainly due to inhibiting the angiotensin-converting enzyme (ACE) [9]. ACE has two different isoforms transcribed by the same gene; a larger with 1277 amino acids that is referred to as somatic ACE (sACE), and a smaller one which is composed of 701 amino acids and is referred to as testis ACE (tACE). Somatic ACE is composed of two homologous catalytic domains (N and C) but testis ACE exhibits just one domain which is identical to the C domain of sACE [12]. Some studies have shown that the C domain is more important for blood pressure regulation activity of ACE [13]. According to ACE’s catalytic mechanism and relevant experimental reports, ACE’s active site contains 19 amino acid residues: His353, Ala354, Ser355, Ala356, His383, Glu384, His387, Phe391, Pro407, His410, Glu411, Phe512, His513, Ser516, Ser517, Val518, Pro519, Arg522, and tyr523 [12].
Antioxidant peptides are also important because of their effect on inhibiting the deterioration of foods and their significant role in the treatment of different diseases such as arthrosclerosis, cancer, and diabetes [14].
Antioxidant and ACE inhibitory of bioactive peptides depends on their structure including size of peptides and their amino acid sequences, which are influenced by the source of protein and conditions of hydrolysis process [9].
To the best of our knowledge, there is no report on the biological activity of yeast extract and purified peptides obtained from K. marxianus. The possibility of using strain of K. marxianus as a source of yeast extract with antioxidant and ACE-inhibitory activity and isolation of the peptides with the most activity was the subject of this research. Ultrafiltration and reversed-phase high-performance liquid chromatography (RP-HPLC) were used to purify the antioxidant and ACE-inhibitory peptides. The sequence of the peptide was identified by MALDI/TOF/TOF technique. The binding interaction of purified peptides within the active site of ACE was also proposed according to the results of molecular docking experiments.
2. Materials and methods
2.1. Materials
Trypsin (EC 3.4.21.4), chymotrypsin (EC 3.4.21.1), furanacrylolyl tripeptide (FAPGG), ortho-phthalaldehyde (OPA), 2, 2, Diphenyl-1-Picryl hydrazyl (DPPH), 2, 2′–azinobis (3-ethyl-benzothiazoline-6-sulphonate) (ABTS), 6-hydroxy-2, 5, 7, 8-tetramethylchroman-2-carboxylic acid (trolox), trifluoroacetic acid (TFA), acetonitrile were obtained from Sigma–Aldrich Chemie GmbH (Munich, Germany). Ultrafiltration membranes with 3, 5 and 10 kDa cut-off were purchased from Sartorius Stedim (Biotech company, Geottingen, Germany).
2.2. Samples preparation
K. marxianus (PTCC 5195) was supplied by Persian Type Culture Collection (PTCC) of the Iranian Research Organization for Science and Technology (IROST). This strain was grown aerobically in yeast mold (YM) culture [Glucose (1%), yeast hydrolysate (0.3%), malt extract (0.3%), peptone (0.5%)]. The temperature was controlled at 28 °C in a shaking incubator (IRC-1-U) at 150 rpm. Cells were harvested in middle of logarithmic phase by centrifugation at 3000×g for 10 min. The yeast pellet was washed with distilled water three times and stored at −20 °C.
Sonicated-enzymatic hydrolysis treatment was carried out to produce yeast protein hydrolysates. Samples were heated at 85 °C for 15 min to terminate enzymes activity. Cell debris was removed by centrifugation at 11,500×g for 10 min. For sonicated-enzymatic hydrolysis, 50 ml of 2.5% dry yeast cells in distilled water was disrupted by a sonicator (Part NO. S-4000) at a fixed power of 600 W, amplitude of 50%, and frequency of 20 kHz. Total cycle time for ultrasonic treatment was 10 min. The cellular debris and particles were removed by centrifugation at 11,500×g for 10 min. The intrinsic yeast cell enzymes in the supernatant were thermally (85 °C for 15 min) inactivated. A solution of extracted protein (4 mg/ml) in phosphate buffer (50 mM, pH 7.8) was subjected to enzymatic hydrolysis using trypsin and chymotrypsin with an enzyme/substrate ratio of 1:10 at 37 °C for 5 h. The enzymatic hydrolysis was stopped by heating in water (85 °C, 15 min). One sample which contains just intrinsic yeast cell enzymes was also considered as control sample.
2.3. Isolation of the antioxidant and ACE-inhibitory peptides
Ultrafiltration was performed with 3, 5 and 10 kDa membranes to purify antioxidant and ACE-inhibitory peptides. The resulting permeates were then lyophilized and stored at −20 °C until further analysis. The resulting filtrate with the highest antioxidant and ACE-inhibitory activities was dissolved in distilled water and further subjected to reverse phase-high performance liquid chromatography (RP-HPLC). Peptide solution (15 μl) was injected into an analytical C18 column (Perfect sil target, ODS-3, 250 × 4.6, 5 μm, 100 Å). The elusion was made in linear gradient mode from eluent A, which was composed of 0.1% TFA in distilled water to eluent B, which was composed of 0.1% TFA in acetonitrile at a flow rate of 0.5 ml/min for 45 min. The elution solution was determined at 215 nm using a UV detector, and collected for further analysis. The fractions exhibiting the highest ACE inhibitory and antioxidant activities were followed by identification of the amino acid sequence.
2.4. Protein assay
Protein concentration in supernatants was determined by modified Lowry method [5] as described by Hartree [15]. Bovine Serum Albumin (BSA) used as the protein standard.
2.5. Evaluation the degree of hydrolysis (DH)
A spectrophotometric assay using OPA was used for determination the extent of hydrolysis. The OPA reagent (1 ml), prepared by method described by Church, Swaisgood [16] and Sun, Luo [17] was mixed with an aliquot (usually 10–50 μl containing 5–100 μg protein) of sample. The solution was mixed briefly. After 2 min incubation at room temperature, the absorbance was read at 340 nm using a spectrophotometer (Jenway, Model, 6315). A standard curve (0–4 mg/ml) of l-leucine was used to create a standard curve and free amino groups were calculated using standard curve. DH was calculated using following formula.
L1: amount of free amino groups released after hydrolysis, L0: amount of free amino groups in the original yeast hydrolysate, Lmax: total amount of free amino groups in the original yeast hydrolysate obtained after acid hydrolysis (6 M HCl at 120 °C for 24 h).
2.6. Measurement of antioxidant activity
Antioxidant activity was measured based on 2, 2-diphenyl-1-picryl-hydrazyl (DPPH) and 2, 2′–azinobis (3-ethyl-benzothiazoline-6-sulphonate) (ABTS) radical-scavenging activity.
2.6.1. DPPH radical scavenging activity
The DPPH radical scavenging activity of peptide solutions was measured according the method described by Son and Lewis [18] with slight modifications. An aliquot of 1 ml peptide solution (or ethanol itself as control) was added to 1 ml of DPPH (0.2 mM) in 95% ethanol solution. After mixing vigorously for 10 s, the mixture was incubated for 30 min in the darkness at room temperature, and the absorbance of resulting solution was measured at 517 nm with a spectrophotometer. DPPH radical scavenging activity (%) was calculated as follows:
The trolox standard curve was used to determine trolox equivalent antioxidant capacity (TEAC).
2.6.2. ABTS-radical scavenging activity test
ABTS radical-scavenging activity was tested according to the method described by Re and others [19]. An aliquot of 25 μl sample was mixed with 1.0 ml ABTS free radical solution diluted with 5 mM phosphate buffer (pH 7.4). The initial absorbance at 734 nm was 0.7 ± 0.02. The absorbance of the resultant solution was recorded at 734 nm. The extent of radical scavenging activity was calculated as follows:
The percentage of radical scavenging activity was calculated and plotted as a function of concentration (0–25 μM) of trolox standard and sample solution to determine trolox equivalent antioxidant capacity (TEAC).
2.7. In vitro assay of ACE-inhibitory activity
ACE inhibitory activity of samples was measured based on the spectrophotometric assay described by Vermeirssen, Van Camp [20]. The rabbit lung powder was prepared following the procedure of Lossow et al. [21] with modifications [22]. Rabbit lung extract as the source of ACE was prepared by dissolving lung acetone powder (1 g) in 10 ml tris-HCl (50 mM, pH 8.3) and 5% v/v glycerol. The mixture was stored at 4 °C overnight and centrifuged at 14,000×g for 20 min. Substrate (5 mM FAPGG in 50 mM tris-HCl buffer (pH 8.3) containing 400 mM NaCl, pH 8.3) and peptide fractions or water (25 μl) were added to each well of ELISA plate and pre-incubated for 20 min at 37 °C before adding 10 μl of ACE extract. The absorbance at 340 nm was recorded every 30 s, for 30 min by an ELISA reader Expert 96 (Power wave xS2, Bioteck, Winooski, USA) for periods of 30 min at 37 °C. The concentration of sample need to inhibit ACE by 50% was defined as IC50 value.
2.8. Determination of amino acid sequence of the most active peptides
The sequences of the most ACE inhibitor and antioxidant peptides were determined by analysing the samples with MALDI/TOF/TOF mass spectrometer with a 5800 Proteomics Analyzer [Applied Biosystems at Proteomics International Pty Ltd., Nedlands, Western Australia]. MS/MS spectra was analysed using PEAKS Studio Version 4.5 SP2 [Bioinformatics Solutions] and manual interpretation.
2.9. Molecular docking of purified peptides on the ACE binding site
The crystal structure of human ACE complexed with inhibitor lisinopril (PDB: 1O8A) was derived from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do//) and used as the template for docking studies. Before the docking, all water molecules and the inhibitor lisinopril were removed whereas the cofactors zinc and chloride atoms were retained in the active site of ACE model. The polar hydrogens were then added to ACE model. Structure of the purified peptide was generated using Hyperchem program version and the geometry of the peptide was subsequently optimized to minimal energy using the same software. The HADDOCK software was selected for molecular docking studies.
The best ranked docking pose of purified peptides in the active site of ACE was obtained according to the scores and binding energy values. Discovery studio 2016 software was also used to identify the hydrogen bonds as well as the hydrophobic, hydrophilic, electrostatic, and coordination interactions between residues located at the ACE active sites. The effect of peptides on Zn(II) tetrahedral geometry was obtained from the docking results using ligplot viewer.
2.10. Statistical analysis
All data were presented as mean ± standard deviation (SD) for three replications for each sample. The ANOVA test and independent sample T-test using the software of SPSS 20 were used to analyse the experiment data. P value < 0.05 was considered significant.
3. Results and discussion
3.1. Preparation of yeast protein hydrolysates from K. marxianus
In this study, K. marxianus protein hydrolysates was used as a possible source of ACE inhibitory and antioxidant peptides. Since enzymes have specific cleavage positions on poly-peptide chains, sonicated-enzymatic (trypsin and chymotrypsin) hydrolysis was used to produce peptides with different amino acid sequences and peptide lengths. The extent of protein degradation during enzymatic hydrolysis was estimated by assessing the DH value by the OPA method. As it was reported in our last paper [23], DH values at the end, were 18.52% and 21.59% for trypsin and chymotrypsin hydrolysis, respectively.
Antioxidant activity is expressed as TEAC and ACE inhibitory activity is reported, a concentration at which 50% of ACE activity is inhibited. Overall, sonication in conjunction with trypsin and chymotrypsin treatments was successful in releasing antioxidant and ACE inhibitory peptides. As it was reported previously, extensive studies demonstrated that trypsin and chymotrypsin are capable of producing bioactive peptides when applied to hydrolyze food proteins [24–28]. According to results (Table 1), there was no significant (P > 0.05) difference in antioxidant and ACE inhibitory activities of protein hydrolysates prepared by trypsin and chymotrypsin. So, trypsin and chymotrypsin hydrolysates were selected for further purification process.
Table 1.
ACE inhibitory and antioxidant activity of yeast protein hydrolysates.
| Samples | DH (%) | IC50 (mg/ml) | DPPH radical scavenging activity (μM TE/mg protein) | ABTS radical scavenging activity (μM TE/mg protein) |
|---|---|---|---|---|
| Control | 15.91a | 0.77 ± 0.01b | 91.85 ± 0b | 273.04 ± 8.63c |
| Physical-trypsin hydrolysis | 18.52b | 0.49 ± 0.009a | 118.53 ± 0a | 489.53 ± 0a |
| Physical-chymotrypsin hydrolysis | 21.59c | 0.51 ± 0.01a | 121.11 ± 0a | 482.03 ± 2.34a |
IC50 is the concentration (mg/ml) of an ACE inhibitor needed to inhibit 50% of ACE activity. The data of DPPH and ABTS radical-scavenging activity are expressed as trolox equivalent antioxidant capacity (μM TE/mg protein). The results are mean values of experiments carried out in triplicate. Different superscript letters in each column indicate significant differences (P < 0.05).
3.2. Purification of antioxidant and ACE-inhibitory peptides from yeast protein hydrolysates
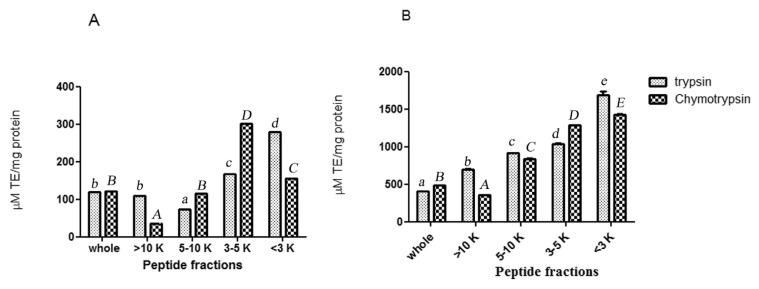
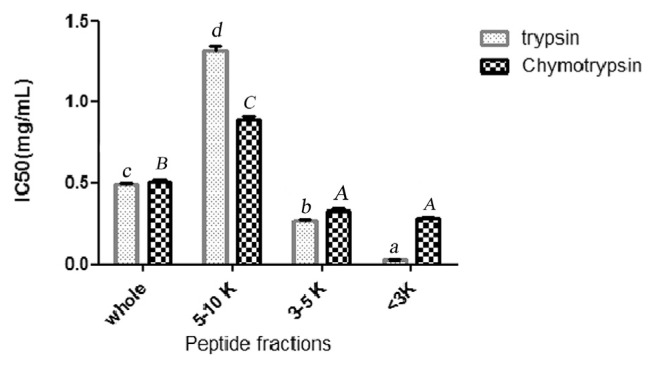
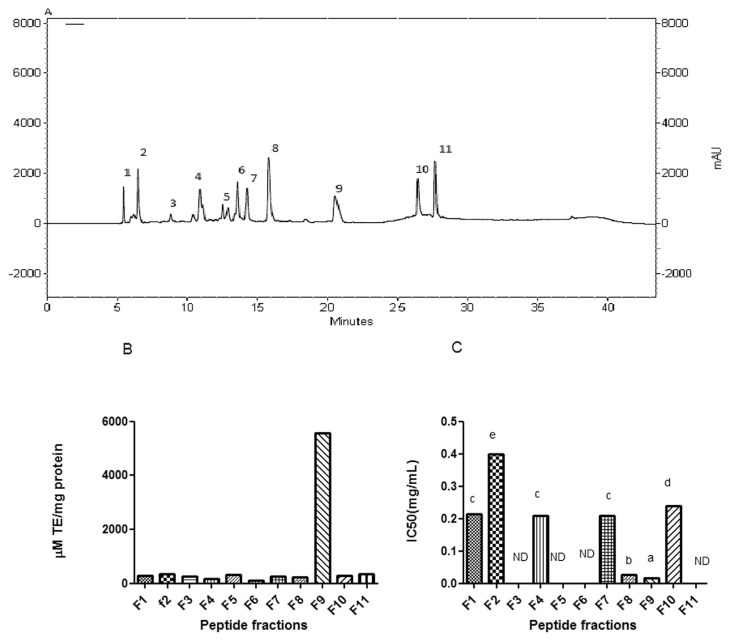
The ultrafiltration membranes were used to perform peptide separation according to their molecular weight. Trypsin and chymotrypsin hydrolysates were fractionated using membranes with molecular weight cut-off of 3, 5, and 10 kDa. The antioxidant and ACE inhibitory activities of trypsin and chymotrypsin hydrolysates and their UF-fractions were measured (Figs. 1 and 2). Considering both antioxidant and ACE inhibitory activities, trypsin MW < 3 kDa permeate peptide fraction was selected for further purification process and was applied on a RP-HPLC system using a RP-C18 column with gradient elution. The chromatographic profile shown in Fig. 3A, fraction is separated to 11 fractions according to their hydrophobicity. The fractions were numbered sequentially (F1–F11). All fractions were collected and lyophilized. Concentration of peptides, ACE inhibitory and antioxidant activity were determined. Results are presented in Fig. 3B and C. As it is shown in Fig. 3A, ABTS radical scavenging activity of fraction F9 was measured to be 5568 μM TE/mg protein and it was 3 folds higher than parent peptide fraction. It confirms 3 folds purification through RP-HPLC treatment. Among all of the fractions collected, fraction F9 also expressed the highest ACE inhibitory activity (IC50 = 15.20 μM) that was 1.76 folds higher than parent peptide fraction. Fraction F8 also showed ACE inhibitory activity (IC50 = 22.88 μM).
Fig. 1.
DPPH (A) and ABTS (B) radical-scavenging activity of whole yeast protein hydrolysates (physical-trypsin hydrolysate, physical-chymotrypsin hydrolysate) and their peptide fractions. The data are expressed as trolox equivalent antioxidant capacity (μM TE/mg protein). The results are mean values of experiments carried out in triplicate. Values with different small letters (trypsin treatments) and capital letters (chymotrypsin treatments) are statistically different at P < 0.05.
Fig. 2.
The size dependent ACE – inhibitory activity of trypsin and chymotrypsin hydrolysates and its peptide fractions. IC50 is the concentration (mg/ml) of an ACE inhibitor needed to inhibit 50% of ACE activity. (P > 0.05). Values with different small letters (trypsin treatments) and capital letters (chymotrypsin treatments) are statistically different at P < 0.05.
Fig. 3.
(A) RP-HPLC chromatogram of the prepared yeast protein hydrolysate (physical-trypsin hydrolysis) with MW < 3 kDa. (B) ABTS radical-scavenging activity of purified peptide fractions. The data are expressed as trolox equivalent antioxidant capacity (μM TE/mg protein). (C) ACE inhibitory activity of purified peptide fractions. IC50 is the concentration (mg/ml) of an ACE inhibitor needed to inhibit 50% of ACE activity. The results are mean values of experiments carried out in triplicate.
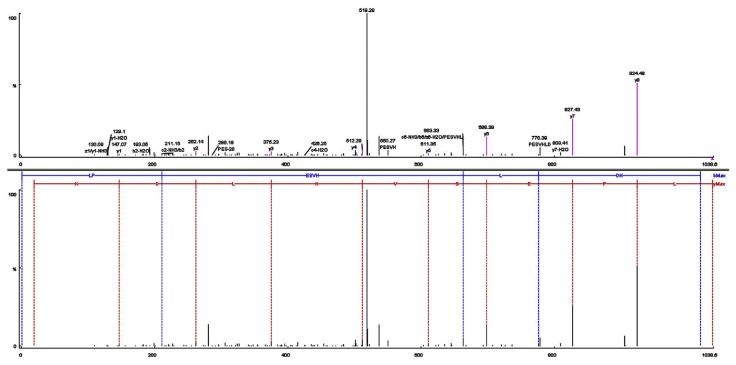
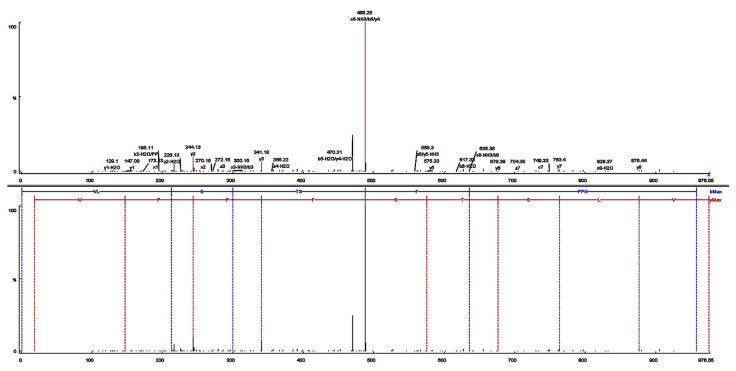
3.3. Sequences and molecular mass of purified peptide
Fractions F8 and F9 were analysed for amino acid sequences (Figs. 4 and 5). The amino acid sequences of the F8 and F9 peptides were identified respectively, as Leu-Pro-Glu-Ser-Val-His-Leu-Asp-Lys (LL-9) and Val-Leu-Ser-Thr-Ser-Phe-Pro-Pro-Lys (VL-9). Many potent ACE-inhibitory and antioxidant peptides have been isolated and identified from various food proteins but to the best of our knowledge, this report is the first that introduces new ACE-inhibitory and antioxidant peptides from K. marxianus protein hydrolysates. De Leo F, Panarese S [29] reported IC50 values between 0.21 and 1682 μM for potent ACE inhibitory peptide sequences derived from milk and plant protein sources. Among them, IPP and VPP are well characterized ACE-inhibitory peptides from fermented milk with IC50 values of 5 μM and 9 μM, respectively [30]. Ni, Li [31] purified and identified a hexapeptide, Thr-Pro-Thr-Gln-Gln-Ser (660 Da) with ACE inhibitory activity (IC50 = 73.25 μg/ml) from water-soluble protein extracted of Saccharomyces cerevisiae. Also, peptide with amino acids sequence of Tyr-Gly-Lys-Pro-Val-Ala-Val-Pro-Ala-Arg (IC50 = 397.26 μM, 26.25 μM TE/μgr protein) was purified from S. cerevisiae protein hydrolysates [22]. ACE Inhibitory activities are often compared with that of captopril, the most widely used antihypertensive drug. The IC50 of captopril is approximately 0.0015 g/ml (0.0071 μM) [32,33]. Therefore, the inhibition potential of LL-9 and VL-9 is less than captopril.
Fig. 4.
Identification of the molecular mass and amino acid sequence of the F8 peptide using MALDL-TOF-TOF mass-spectrometer. (A) MS/MS spectra of the F8 peptide, and (B) the interpretation of the obtained spectra.
Fig. 5.
Identification of the molecular mass and amino acid sequence of the F9 peptide using MALDL-TOF-TOF mass-spectrometer. (A) MS/MS spectra of the F9 peptide, and (B) the interpretation of the obtained spectra.
The amino acid composition of purified peptides consist of hydrophobic amino acids (Leu, Pro, Val, Leu, Phe) with a total percentage of, respectively, 44.4% and 55.5% for LL-9 and VL-9 peptides. The hydrophilic-hydrophobic ratio in the peptide sequence is a critical factor in ACE inhibitory activity [34,35]. Because hydrophilic amino acid residues could disrupt the access of peptide to the active site of ACE. Many studies have shown that ACE prefers to have substrate or competitive inhibitory that contain aromatic residues such as tryptophan, phenylalanine, tyrosine or proline at their C-terminal tripeptide sequence and branched aliphatic amino acids such as glycine, valine, leucine, and isoleucine at the N-terminal position [34,36–38]. Therefore, presence of two proline residue at C-terminus and valine and leucine at N-terminus of VL-9 and presence of leucine at N-terminus of LL-9 may contribute to their ACE inhibitory activity. Also, phenylalanine may contribute to ACE inhibitory activity of VL-9 as Pripp [39] reported that highly active ACE inhibitory peptide should be composed of large, hydrophobic, aromatic amino acid. Positively charged ɛ-amine group in lysine at the C-terminus of both peptides and the presence of aspartic and glutamic acids in the sequence of LL-9 may contribute to ACE inhibitory activity [40]. The capability of glutamic acid to chelate zinc, as a component of the ACE active centre was reported previously [40]. Also, hydrophobic amino acids (55.5% of the total amino acid residues) in VL-9 enhance the solubility of peptide in lipid and facilitates accessibility to hydrophobic radical species, thereby increase the antioxidant activity of peptide [41–43]. Chen, Muramoto [44] reported the presence of valine and leucine at the N-terminal and proline in the sequence of peptide contribute to its antioxidant activity. Qian, Jung [42] reported high reactivity of aliphatic groups in valine and leucine to hydrophobic poly unsaturated fatty acids. Suetsuna and Chen [45] reported the presence of alanine and leucine at the N-terminus, glutamine and proline residues in the sequences of antioxidant peptide purified from gluten. The presence of lysine is reported to be important factor in the antioxidant activities of the peptides, especially due to their ability to reduce Fe3+ to Fe2+ and to chelate Fe2+ and Cu2+ ions [46–48]. Also, aromatic amino acids phenylalanine act as proton donors to electron deficient radicals and efficiently scavenge them [49].
3.4. Insight into molecular docking simulation
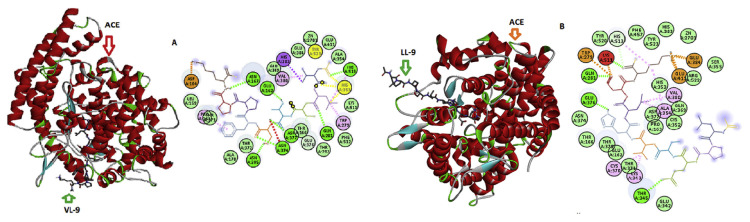
In order to explore the molecular mechanism of interactions between LL-9 and VL-9 and ACE, the docking simulation was studied using the flexible docking tool of HADDOCK software. The docking study of the peptides VL-9 and LL-9 at the ACE active site, in the presence of Zn(II), showed a best pose (Fig. 6A and B) with Z scores of −2.1 and −2.9, respectively. Researches have shown that the inhibitor combined with ACE residues through the interaction forces of hydrogen bonds, hydrophobic, van der waals and electrostatic interactions that exist between amino acids residues of ACE and those of each peptides within a distance of 3.5 Å [50,51]. Among them, hydrogen bonds interaction force plays the most important role for stabilizing the docking complex and enzyme catalytic reactions [52]. As shown in Fig. 6, the peptide pose was stabilized by hydrogen bonds. Seven and four hydrogen bonds were formed, respectively, between VL-9, LL-9 and ACE, suggesting that the peptides effectively interacted with the ACE active site, and maybe explain stronger inhibition activity of VL-9 [52].
Fig. 6.
The docking results for the of VL-9 (A) and LL-9 (B) with ACE (PDB:1O8A). 3D and 2D structures of VL(9)-ACE (A) and LL(9)-ACE (B). Van der Waals bonds are represented in blue, salt bridge in orange, conventional hydrogen bond in green dotted lines, while other residues and the zinc atom are represented automatically. Image obtained with Accelrys DS Visualizer software.
As reported by Wu et al. [53], ACE contained three main active site pockets (S1, S2, and S’1). S1 pocket included Ala354, Glu384 and Tyr523 residues, and S2 pocket included Glu281, His353, Lys511, His513 and Tyr520 residues, while S’1 contained Glu162 residue. The molecular docking study revealed that VL-9 establishes hydrogen bonds with the S2 (His513, His353, Glu281) and S’1 (Glu162) pockets mostly due to Val1, Leu2, Thr4, Ser5, and Lys9 (Fig. 6A). The peptide LL-9 also established hydrogen bonds with S1(Thr345) and S2 (Tyr520, Lys511, Gln281) pockets of ACE due to Glu3, His6, Asp8, and Lys9. The results suggested that the peptides could effectively interact with the active site of ACE and inhibit its activity [52]. These are also key residues at ACE active site which interact with ACE inhibitor lisinopril and is consistence with the competitive inhibition model [54,55].
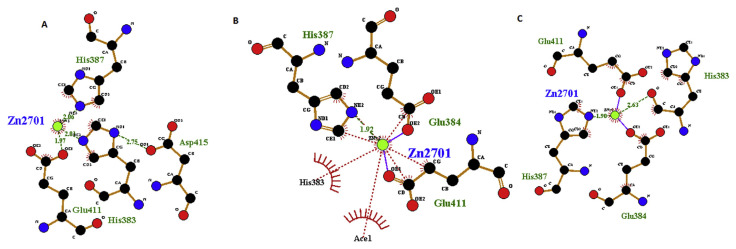
Zn(II) at the ACE active site usually plays a significant role and tetra-coordinated with three ACE residues (His383, His387, Glu411) [56] The values of bond length are shown in Fig. 7A. Although VL-9 and LL-9 not directly interact with Zn(II), after docking the initial values of bond length made some differences and some hydrogen bonds were lost (Fig. 7B and C). This result suggests that the peptides may have the ability to coordinate with Zn(II) of ACE and distort the tetrahedral geometry and cause ACE inhibitory activities. This phenomena is in agreement with the finding of Jia et al. [57] who reported the changes in the distance between the Zn(II) and its surrounding atoms after interaction of Lys-His-Val with ACE. Pan et al. [58] also reported the distorted Zn(II) tetrahedral geometry in the interaction of ACE with Leu-Leu. Many studies had shown that lisinopril directly interacted with the Zn(II) of ACE [12,59] and it can explain higher ACE inhibitory activity of lisinopril compare to purified peptides.
Fig. 7.
Details of Zn(II) (green) coordination with ACE residues before docking (A), after docking with VL-9 (B), and LL-9 (C). Green dotted, Blue and brown lines indicate respectively, hydrogen, ligand and non ligand bonds formation. Image obtained with Ligplot version v.1.4.5 software.
Acknowledgements
The present study has been supported by Iranian Research Organization for Science and Technology (IROST) (1012195004). The FP7 WeNMR (project# 261572) and H2020 West-Life (project# 675858) European e-Infrastructure projects are acknowledged for the use of their web portals, which make use of the EGI infrastructure and DIRAC4EGI service with the dedicated support of CESNET-MetaCloud, INFN-PADOVA, NCG-INGRID-PT, RAL-LCG2, TW-NCHC, SURFsara and NIKHEF, and the additional support of the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, South Africa, Malaysia, Taiwan and the US Open Science Grid. The authors would like to thank Dr. Marzieh Dehghan for her help in computational techniques.
Funding Statement
The present study has been supported by Iranian Research Organization for Science and Technology (IROST) (1012195004).
References
- 1. Lagneau PE, Lebtahi K, Swinne D. Isolation of yeasts from bovine milk in Belgium. Mycopathologia. 1996;135(2):99–102. doi: 10.1007/BF00436458. [DOI] [PubMed] [Google Scholar]
- 2. Corbo MR, Lanciotti R, Albenzio M, Sinigaglia M. Occurrence and characterization of yeasts isolated from milks and dairy products of Apulia region. Int J Food Microbiol. 2001;69(1–2):147–52. doi: 10.1016/s0168-1605(01)00585-2. [DOI] [PubMed] [Google Scholar]
- 3. Romano P, Ricciardi A, Salzano G, Suzzi G. Yeasts from water buffalo mozzarella, a traditional cheese of the Mediterranean area. Int J Food Microbiol. 2001;69(1–2):45–51. doi: 10.1016/s0168-1605(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 4. Simova E, Beshkova D, Angelov A, Hristozova T, Frengova G, Spasov Z. Lactic acid bacteria and yeasts in kefir grains and kefir made from them. J Ind Microbiol Biotechnol. 2002;28(1):1–6. doi: 10.1038/sj/jim/7000186. [DOI] [PubMed] [Google Scholar]
- 5. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–75. [PubMed] [Google Scholar]
- 6. Caballero R, Olguín P, Cruz-Guerrero A, Gallardo F, García-Garibay M, Gómez-Ruiz L. Evaluation of Kluyveromyces marxianus as baker’s yeast. Food Res Int. 1995;28(1):37–41. [Google Scholar]
- 7. Pas M, Piskur B, Sustaric M, Raspor P. Iron enriched yeast biomass–a promising mineral feed supplement. Bioresour Technol. 2007;98(8):1622–8. doi: 10.1016/j.biortech.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 8. Lane MM, Morrissey JP. Kluyveromyces marxianus: a yeast emerging from its sister’s shadow. Fungal Biol Rev. 2010;24(1–2):17–26. [Google Scholar]
- 9. Shahidi F, Zhong Y. Bioactive peptides. J AOAC Int. 2008;91(4):914–31. [PubMed] [Google Scholar]
- 10. Homayouni-Tabrizi M, Asoodeh A, Soltani M. Cytotoxic and antioxidant capacity of camel milk peptides: effects of isolated peptide on superoxide dismutase and catalase gene expression. J Food Drug Anal. 2016;25(3):567–75. doi: 10.1016/j.jfda.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suwanmanon K, Hsieh P-C. Effect of γ-aminobutyric acid and nattokinase-enriched fermented beans on the blood pressure of spontaneously hypertensive and normotensive Wistar–Kyoto rats. J Food Drug Anal. 2014;22(4):485–91. doi: 10.1016/j.jfda.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan D, Guo H, Zhao B, Cao J. The molecular mechanisms of interactions between bioactive peptides and angiotensin-converting enzyme. Bioorg Med Chem Lett. 2011;21(13):3898–904. doi: 10.1016/j.bmcl.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 13. Ni H, Li L, Liu G, Hu SQ. Inhibition mechanism and model of an angiotensin I-converting enzyme (ACE)-inhibitory hexapeptide from yeast (Saccharomyces cerevisiae) PLoS One. 2012;7(5):e37077. doi: 10.1371/journal.pone.0037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sarmadi BH, Ismail A. Antioxidative peptides from food proteins: a review. Peptides. 2010;31(10):1949–56. doi: 10.1016/j.peptides.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 15. Hartree EF. Determination of protein: a modification of the lowry method that gives a linear photometric response. Anal Biochem. 1972;48(2):422–7. doi: 10.1016/0003-2697(72)90094-2. [DOI] [PubMed] [Google Scholar]
- 16. Church FC, Swaisgood HE, Porter DH, Catignani GL. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J Dairy Sci. 1983;66(6):1219–27. [Google Scholar]
- 17. Sun Q, Luo Y, Shen H, Hu XIN. Effects of pH, temperature and enzyme to substrate ratio on the antioxidant activity of porcine hemoglobin hydrolysate prepared with pepsin. J Food Biochem. 2011;35(1):44–61. [Google Scholar]
- 18. Son S, Lewis BA. Free radical scavenging and antioxidative activity of caffeic acid amide and ester analogues: structure-activity relationship. J Agric Food Chem. 2002;50(3):468–72. doi: 10.1021/jf010830b. [DOI] [PubMed] [Google Scholar]
- 19. Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med. 1999;26(9–10):1231–7. doi: 10.1016/s0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- 20. Vermeirssen V, Van Camp J, Verstraete W. Optimisation and validation of an angiotensin-converting enzyme inhibition assay for the screening of bioactive peptides. J Biochem Biophys Methods. 2002;51(1):75–87. doi: 10.1016/s0165-022x(02)00006-4. [DOI] [PubMed] [Google Scholar]
- 21. Lossow WJ, Migiorini RH, Brot N, Chaikoff IL. Effect of total exclusion of the exocrine pancreas in the rat upon in vitro esterification of C14-Labeled cholesterol by the intestine and upon lymphatic absorption of C14-Labeled cholesterol. J Lipid Res. 1964;5:198–202. [PubMed] [Google Scholar]
- 22. Mirzaei M, Mirdamadi S, Ehsani MR, Aminlari M, Hosseini E. Purification and identification of antioxidant and ACE-inhibitory peptide from Saccharomyces cerevisiae protein hydrolysate. J Funct Foods. 2015;19(Part A):259–68. [Google Scholar]
- 23. Mirzaei M, Mirdamadi S, Ehsani MR, Aminlari M. Antioxidant, ACE-inhibitory and antimicrobial activities of Kluyveromyces marxianus protein hydrolysates and their peptide fractions. FFHD. 2016;6(7):428–39. [Google Scholar]
- 24. Ko JY, Lee JH, Samarakoon K, Kim JS, Jeon YJ. Purification and determination of two novel antioxidant peptides from flounder fish (Paralichthys olivaceus) using digestive proteases. Food Chem Toxicol. 2013;52:113–20. doi: 10.1016/j.fct.2012.10.058. [DOI] [PubMed] [Google Scholar]
- 25. Jun S-Y, Park P-J, Jung W-K, Kim S-K. Purification and characterization of an antioxidative peptide from enzymatic hydrolysate of yellowfin sole (Limanda aspera) frame protein. Euro Food Res Technol. 2004;219(1):20–6. [Google Scholar]
- 26. Karaki H, Doi K, Sugano S, Uchiwa H, Sugai R, Murakami U, et al. Antihypertensive effect of tryptic hydrolysate of milk casein in spontaneously hypertensive rats. Comp Biochem Physiol C. 1990;96(2):367–71. [PubMed] [Google Scholar]
- 27. Fan J, He J, Zhuang Y, Sun L. Purification and identification of antioxidant peptides from enzymatic hydrolysates of Tilapia (Oreochromis niloticus) frame protein. Molecules. 2012;17(11):12836–50. doi: 10.3390/molecules171112836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qian Z-J, Je J-Y, Kim S-K. Antihypertensive effect of angiotensin I converting enzyme-inhibitory peptide from hydrolysates of bigeye tuna dark muscle, Thunnus obesus. J Agric Food Chem. 2007;55(21):8398–403. doi: 10.1021/jf0710635. [DOI] [PubMed] [Google Scholar]
- 29. De Leo F, Panarese S, Gallerani R, Ceci LR. Angiotensin converting enzyme (ACE) inhibitory peptides: production and implementation of functional food. Curr Pharm Des. 2005;15(31):3622–43. doi: 10.2174/138161209789271834. [DOI] [PubMed] [Google Scholar]
- 30. Yun Li, Faizan A, Sadi Q, Fu L, Zhu H, Zhong M, et al. Identification of angiotensin I-converting enzyme inhibitory peptides derived from enzymatic hydrolysates of razor clam sinonovacula constricta. Mar drugs. 2016;14(110) doi: 10.3390/md14060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ni H, Li L, Guo S-S, Li H-H, Jiang R, Hu S-Q. Isolation and identification of an Angiotensin-I converting enzyme inhibitory peptide from yeast (Saccharomyces cerevisiae) Curr Anal Chem. 2012;8(1):180–5. [Google Scholar]
- 32. Tsai J-S, Chen T-J, Pan BS, Gong S-D, Chung M-Y. Antihypertensive effect of bioactive peptides produced by protease-facilitated lactic acid fermentation of milk. Food Chem. 2008;106(2):552–8. [Google Scholar]
- 33. Pihlanto-Leppala A, Rokka T, Korhonen H. Angiotensin I converting enzyme inhibitory peptides derived from bovine milk proteins. Int Dairy J. 1998;8(4):325–31. [Google Scholar]
- 34. Li GH, Le GW, Shi YH, Shrestha S. Angiotensin I–converting enzyme inhibitory peptides derived from food proteins and their physiological and pharmacological effects. Nutr Res. 2004;24(7):469–86. [Google Scholar]
- 35. Asoodeh A, Homayouni-Tabrizi M, Shabestarian H, Emtenani S, Emtenani S. Biochemical characterization of a novel antioxidant and angiotensin I-converting enzyme inhibitory peptide from Struthio camelus egg white protein hydrolysis. J Food Drug Anal. 2016;24(2):332–42. doi: 10.1016/j.jfda.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kapel R, Rahhou E, Lecouturier D, Guillochon D, Dhulster P. Characterization of an antihypertensive peptide from an alfalfa white protein hydrolysate produced by a continuous enzymatic membrane reactor. Process Biochem. 2006;41:1961–6. [Google Scholar]
- 37. Ondetti MA, Cushman DW. Angiotensin-converting enzyme inhibitors: biochemical properties and biological actions. CRC Crit Rev Biochem. 1984;16:381–411. doi: 10.3109/10409238409108720. [DOI] [PubMed] [Google Scholar]
- 38. Sharma S, Singh R, Rana S. Bioactive peptides: a review. Int J Bioautomation. 2011;15:223–50. [Google Scholar]
- 39. Pripp A. Initial proteolysis of milk proteins and its effect on formation of ACE-inhibitory peptides during gastrointestinal proteolysis: a bioinformatic, in silico, approach. Eur Food Res Technol. 2005;221(5):712–6. [Google Scholar]
- 40. FitzGerald RJ, Murray BA, Walsh DJ. Hypotensive peptides from milk proteins. J Nutr. 2004;134(4):980S–8S. doi: 10.1093/jn/134.4.980S. [DOI] [PubMed] [Google Scholar]
- 41. Chen H-M, Muramoto K, Yamauchi F, Nokihara K. Antioxidant activity of designed peptides based on the antioxidative peptide isolated from digests of a soybean protein. J Agric Food Chem. 1996;44(9):2619–23. [Google Scholar]
- 42. Qian Z-J, Jung W-K, Kim S-K. Free radical scavenging activity of a novel antioxidative peptide purified from hydrolysate of bullfrog skin, Rana catesbeiana Shaw. Bioresour Technol. 2008;99(6):1690–8. doi: 10.1016/j.biortech.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 43. Suetsuna K, Ukeda H, Ochi H. Isolation and characterization of free radical scavenging activities peptides derived from casein. J Nutr Biochem. 2000;11(3):128–31. doi: 10.1016/s0955-2863(99)00083-2. [DOI] [PubMed] [Google Scholar]
- 44. Chen H-M, Muramoto K, Yamauchi F. Structural analysis of antioxidative peptides from soybean.beta.-conglycinin. J Agric Food Chem. 1995;43(3):574–8. [Google Scholar]
- 45. Suetsuna K, Chen JR. Isolation and characterization of peptides with antioxidant activity derived from wheat gluten. Food Sci Technol Res. 2002;8(3):227–30. [Google Scholar]
- 46. Carrasco-Castilla J, Hernández-Álvarez AJ, Jiménez-Martínez C, Jacinto-Hernández C, Alaiz M, Girón-Calle J, et al. Antioxidant and metal chelating activities of Phaseolus vulgaris L. var. Jamapa protein isolates, phaseolin and lectin hydrolysates. Food Chem. 2012;131(4):1157–64. doi: 10.1016/j.foodchem.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 47. Huang S-M, Chen K-N, Chen Y-P, Hong W-S, Chen M-J. Immunomodulatory properties of the milk whey products obtained by enzymatic and microbial hydrolysis. Int J Food Sci Technol. 2010;45(5):1061–7. [Google Scholar]
- 48. Wang W, De Mejia EG. A new frontier in soy bioactive peptides that may prevent age-related chronic diseases. Compr Rev Food Sci Food Saf. 2005;4(4):63–78. doi: 10.1111/j.1541-4337.2005.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 49. Duan X, Ocen D, Wu F, Li M, Yang N, Xu J, et al. Purification and characterization of a natural antioxidant peptide from fertilized eggs. Food Res Int. 2014;56(0):18–24. [Google Scholar]
- 50. Li P, Jia J, Fang M, Zhang L, Guo M, Xie J, et al. In vitro and in vivo ACE inhibitory of pistachio hydrolysates and in silico mechanism of identified peptide binding with ACE. Process Biochem. 2014;49(5):898–904. [Google Scholar]
- 51. Girgih AT, Fau He R, Aluko RE, Aluko RE. Kinetics and molecular docking studies of the inhibitions of angiotensin converting enzyme and renin activities by hemp seed (Cannabis sativa L.) peptides. doi: 10.1021/jf5002606. (1520 5118 (Electronic)) [DOI] [PubMed] [Google Scholar]
- 52. Chaudhary S, Vats ID, Chopra M, Biswas P, Pasha S. Effect of varying chain length between P1 and P1′ position of tripeptidomimics on activity of angiotensin-converting enzyme inhibitors. Bioorg Med Chem Lett. 2009;19(15):4364–6. doi: 10.1016/j.bmcl.2009.05.079. [DOI] [PubMed] [Google Scholar]
- 53. Wu Q, Jia J, Yan H, Du J, Gui Z. A novel angiotensin-capital I, Ukrainian converting enzyme (ACE) inhibitory peptide from gastrointestinal protease hydrolysate of silkworm pupa (Bombyx mori) protein: biochemical characterization and molecular docking study. Peptides. 2015;68:17–24. doi: 10.1016/j.peptides.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 54. Rawendra RD, Aisha, Chang C-I, Aulanni’am, Chen H-H, Huang T-C, Hsu JL. A novel angiotensin converting enzyme inhibitory peptide derived from proteolytic digest of Chinese soft-shelled turtle egg white proteins. J Proteomics. 2013;94:359–69. doi: 10.1016/j.jprot.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 55. Natesh R, Schwager SL, Evans HR, Sturrock ED, Acharya KR. Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochem. 2004;43(27):8717–24. doi: 10.1021/bi049480n. [DOI] [PubMed] [Google Scholar]
- 56. Jalkute CB, Barage SH, Dhanavade MJ, Sonawane KD. Molecular dynamics simulation and molecular docking studies of Angiotensin converting enzyme with inhibitor lisinopril and amyloid beta peptide. Protein J. 2013;32(5):356–64. doi: 10.1007/s10930-013-9492-3. [DOI] [PubMed] [Google Scholar]
- 57. Jia J, Wu Q, Yan H, Gui Z. Purification and molecular docking study of a novel angiotensin-I converting enzyme (ACE) inhibitory peptide from alcalase hydrolysate of ultrasonic-pretreated silkworm pupa (Bombyx mori) protein. Process Biochem. 2015;50(5):876–83. [Google Scholar]
- 58. Pan D, Cao J, Guo H, Zhao B. Studies on purification and the molecular mechanism of a novel ACE inhibitory peptide from whey protein hydrolysate. Food Chem. 2012;130(1):121–6. [Google Scholar]
- 59. Jimsheena VK, Gowda LR. Arachin derived peptides as selective angiotensin I-converting enzyme (ACE) inhibitors: structure-activity relationship. Peptides. 2010;31(6):1165–76. doi: 10.1016/j.peptides.2010.02.022. [DOI] [PubMed] [Google Scholar]