Abstract
Beacon is a basic data discovery protocol issued by the Global Alliance for Genomics and Health (GA4GH). The main goal addressed by version 1 of the Beacon protocol was to test the feasibility of broadly sharing human genomic data, through providing simple “yes” or “no” responses to queries about the presence of a given variant in datasets hosted by Beacon providers. The popularity of this concept has fostered the design of a version 2, that better serves real‐world requirements and addresses the needs of clinical genomics research and healthcare, as assessed by several contributing projects and organizations. Particularly, rare disease genetics and cancer research will benefit from new case level and genomic variant level requests and the enabling of richer phenotype and clinical queries as well as support for fuzzy searches. Beacon is designed as a “lingua franca” to bridge data collections hosted in software solutions with different and rich interfaces. Beacon version 2 works alongside popular standards like Phenopackets, OMOP, or FHIR, allowing implementing consortia to return matches in beacon responses and provide a handover to their preferred data exchange format. The protocol is being explored by other research domains and is being tested in several international projects.
Keywords: Beacon, clinical genomics, data discovery, data sharing, GA4GH, REST API
Beacon is a basic data discovery protocol issued by the Global Alliance for Genomics and Health (GA4GH). Beacon version 2 serves real world requirements and addresses the needs of clinical genomics research and healthcare, allowing implementing consortia to return matches in beacon responses and provide a handover to their preferred data exchange format. Beacon is designed as a “lingua franca” to bridge data collections hosted in software solutions with different and rich interfaces. Beacon queries could be send to Beacon instances directly or via Beacon networks. The response could be yes/no, counts or details if the user is properly authorized.
1. INTRODUCTION
The Global Alliance for Genomics and Health (GA4GH) (e.g., Rehm et al., 2021) was created in 2013 on the core mission to create a federated ecosystem for sharing genomic data and associated clinical information within a human rights framework. At its foundational meeting, the concept of a genomics “Beacon” was presented by Jim Ostell, as a means to engage and connect genomic data providers and developers as well as researchers with interest to access genomic variation data. The concept of this Beacon system was purposefully simple: Design a data access API, which allows users to query genomic data collections for the existence of a specific genomic variation and responds with a “Yes” or “No” answer. The name “Beacon” referred to the hope that such a system would be simple enough to engage willing participants, thereby lighting up the so far dark landscape of genomic data sharing.
While kept simple to encourage broad participation, the Beacon concept was aimed to trigger potential issues—for example, related to institutional policies or general regulatory issues regarding genomic information—but also to demonstrate the power of such a simple data sharing concept especially if implemented through a federated model, distributing Beacon queries to a large number of international nodes and providing an aggregation of the individual responses. While it was clear that such a Beacon network could become more useful through complex queries and richer responses, such extensions were proposed for a “version 2,” following the successful establishment of a working implementation of the original concept.
With the Beacon Project becoming one of the original GA4GH Driver Projects, it was enthusiastically adopted by members of the GA4GH developer community and genomic resource providers alike. By 2016, more than 35 organizations from all over the world had “beaconized” over 90 genomics datasets, many of which were connected to the network aggregator provided by DNAstack (beacon‐network.org). At this point ELIXIR ‐ the European bioinformatics infrastructure organization ‐ joined the further development of the community project toward a standard specification, with improved usability and the goal of future use throughout biomedical genomics.
In 2018, the Beacon v1.0 protocol was accepted as official GA4GH standard (Fiume et al., 2019) following a formal review process. While this version and its updates introduced some improvements over the earlier editions, such as limited support for structural variant queries, some quantitative responses as well as the “handover” to external protocols, overall Beacon v1 stayed with the original “variant query and aggregate response” concept. However, at this point, it had become clear that further expansions of the protocol such as requested—especially for clinical applications in rare diseases and cancer genomics—required a re‐design of the Beacon protocol to serve a wide range of use cases and leading to initiation of the “Beacon v2” design process.
1.1. Designing a “clinical Beacon”
The initial concept of the Beacon protocol of returning a simple boolean response focuses more toward technical implementers and scientific researchers than clinicians. For broader use in clinical settings, each allele or mutation‐specific query should ideally offer options to query and retrieve associated phenotypic data, metadata (e.g., age at disease onset, genotypic sex), associated diagnoses, therapeutic interventions, pedigree information, and so on. The success of the Beacon v1 concept and its enthusiastic adoption by genomics and rare disease communities has provided a strong argument to expand its usability toward a more general use in healthcare environments, while building on its conceptual simplicity.
With these objectives in mind, the GA4GH Beacon group engaged with GA4GH Driver Projects and with ELIXIR partners to identify the consensus requirements for the next generation Beacon. The following Driver projects were interviewed: Autism Speaks (https://www.autismspeaks.org), BRCA Exchange (Cline et al., 2018), CanDIG (Rehm et al., 2021), EGA (Freeberg et al., 2022)/ENA (Harrison et al., 2021)/EVA (Cezard et al., 2022), EuCanCan (https://eucancan.com), European Joint Programme on Rare Diseases (EJP‐RD, https://www.ejprarediseases.org), H3Africa (e.g., Mulder et al., 2018), GEM Japan (https://www.amed.go.jp/en/aboutus/collaboration/ga4gh_gem_japan.html#anc-2), Genomics England (Koepfli et al., 2015), Matchmaker Exchange (Pilippakis et al., 2015), Swiss Variant Interpretation Platform (SVIP, https://svip.ch)/Swiss Personalized Health Network (SPHN, https://sphn.ch/fr/home/), and Variant Interpretation for Cancer Consortium (VICC). Some ELIXIR partners and communities such as Café Variome (Lancaster et al., 2015), hCNV community (https://elixir-europe.org/communities/hcnv), Fundación Progreso y Salud (https://www.clinbioinfosspa.es/), RD‐Connect (Thompson et al., 2014), CINECA (https://www.cineca-project.eu), and DisGeNET (Piñero et al., 2020), were also interviewed. In addition, members of the Beacon team detailed (a) requirements for specific diagnoses, including rare diseases, (b) a support for the clinical center to build their own Beacon, and (c) a procedure to gather them under clinical network of Beacon installations, via tight collaborations with hospitals in Catalunya, Spain, as well as Cancer Core Europe (https://cancercoreeurope.eu/) and Health‐RI (https://www.health-ri.nl/), Netherlands. Finally, to address specific aspects in the Beacon development, working groups ("Scouts") are working regularly on different aspects of the Beacon protocol such as security, filters, genomic variants, and protocol documentation.
1.2. Requirements for a clinical Beacon
The health sector is increasingly seeking to use genetic/genomic tests as an integral part of the diagnostic process or in the selection of therapeutic procedures. Typical genomic data generation and sharing in healthcare includes five types of roles: the patient, the clinicians, the genetic analysts, intramural contributing researchers, and external partners involved in associated research projects or technical aspects of diagnosis and data handling. Each contributor has a set of needs regarding data discovery. Citizens are both at the start and the end (as ultimate benefiters) of the data cycle: subject to different rules in different countries, they can consent to the use of their data for healthcare or secondary research use as long as their privacy and identity are protected. The general data protection regulation (GDPR) adds additional responsibilities on the operator of a Beacon to protect the privacy and rights of individuals whose data exists within a beacon, therefore data security (see section on security aspects) is therefore a key component to discovery tools such as Beacon. Finally, the new Beacon model should provide context for the genomic variant finding, including information about the biosample and molecular analysis procedures as well as observations and measurements describing the phenotypic state of individuals. Local researchers are responsible for structuring the data so it can be queried. As it is usually a manual process, different studies from the same institution often use different tools and select different information, making cross‐querying or reuse of data difficult. For this reason, the Beacon team is committed to train internal researchers on data structuring for Beacon. External partners in large‐scale European projects (e.g., CINECA or B1MG) are proponents of the Beacon v2. Understanding the partner needs has driven the addition of new features like cohorts, which are of high relevance for the scientific community. Projects also provide a platform to enable new Beacon features to be more visible (Fromont, 2021), and facilitate the use of these additions via training events. The ELIXIR Beacon project aims to disseminate Beacon and the Beacon network. In 2021, nine, preliminary version 2 and newly developed, Beacons were implemented across the ELIXIR Nodes in the ELIXIR Beacon network prototype (https://beacon-network.elixir-europe.org). Once the Beacon v2 specification stabilized, it was straightforward to write parsers to connect and translate to the various backends used in these implementations.
1.3. Rare diseases use case
Even though many candidate variants can be identified in rare disease patients, a reliable genetic diagnosis cannot be achieved in at least 50% of cases (Zurek et al., 2021). Pinpointing causal genetic variants in these cases can be greatly assisted by increasing sample size(s) and finding other patients with similar phenotype profiles: an approach called “matchmaking.”
“Matchmaking” cases—or patients with particular sets of disease phenotypes—are made difficult by the huge phenotypic and genotypic diversity of rare diseases patients, and the correspondingly large and variable way(s) in which rare diseases related data(sets) are collected into registries, biobanks, and sample catalogs. Each data set typically has its own access rules and gateways, making it difficult to connect data and deduce meaningful insights across resources for in depth genome analysis.
Previous projects such as matchmaker exchange (MME—Philippakis et al., 2015) and RD‐connect genome‐phenome analysis platform (GPAP—Matalonga et al., 2021) were successful in different ways in tackling these problems. MME is a small network of large databases that can interoperate to “matchmake” patients. However, the process requires the supply and transmission of patient profiles, with limited control over how a match is defined. This model comprises many data sharing policies, and therefore dissuades many potential users. The RD‐connect GPAP approach utilizes a combination of phenotype, genotype, and biobank data to allow users to find subjects of interest. However, it is based on a multi‐site centralized platform requiring the submission of data to the GPAP environment. Both systems also require authorized access, which further limits the availability of these data discovery solutions. Additionally, many smaller rare disease patient registries exist, along with sample catalogs and biobanks that operate their own systems. There is limited interoperability between any of these and the current larger solutions.
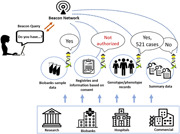
To improve on the above there needs to be a standardized way to interconnect diverse resources to provide safe, federated, flexible but powerful discovery queries, to (see Figure 1):
Find a suitable registry based on summary data—response could be a yes/no;
Find a suitable registry based on genotypes/phenotypes on a record level—response could be yes/no, counts;
Find a biobank based on the sample data—response could be yes/no or counts of samples;
Find a collection of subjects across many registries—response could be counts per registry;
Find registries and biobanks based on consent and use conditions that apply to the assets in those resources.
Figure 1.
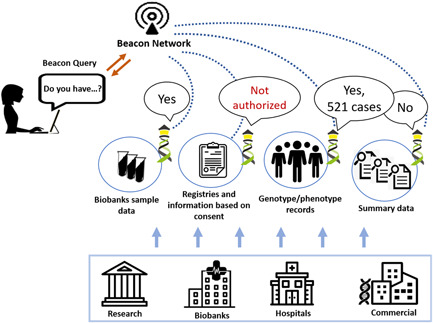
Beacon queries could be sent to Beacon instances directly or via Beacon networks. The response could be yes/no, counts or details if the user is properly authorized
The Beacon v2 API can help with these goals, as it is separated from the software and data models employed at any potential query target. The API provides a standardized way to send a query itself, and receive a standardized response containing yes/no, counts or data. The Beacon 2 models provide a description and attributes of the target type, an initial set of models covering query targets such as “individual” or “data set”are part of the API, however, this is extensible and new models can be made that fit a particular use case.
1.4. Scope of Beacon v2 model
The original Beacon protocol did not specify an explicit data model but rather limited itself to reference genome mapped genome variations and simple, boolean responses. In contrast, and driven by the requirements detailed above, the Beacon v2 protocol allows for an extensible data model on top of its flexible framework (see our website https://beacon-project.io/ for details). The Beacon v2 provides default support for a data model serving the needs of biomedical genomics but also accommodating simple boolean responses.
The complete v2 data model (Figure 2) implements default entities such as “individual,” “biosample,” “observed genome variation,” and “variant information” and their logical relations, as well as additional technical concepts. Here, the Beacon model follows common concepts such as those established through the GA4GH Data Working Group and compatible with, for example, the Phenopackets standard (Jacobsen et al., 2021) and the variant representation standard (VRS; Wagner et al., 2021). For example, Beacons can support queries combining phenotype parameters of an individual with genomic variation parameters (“are there individuals with phenotype X and variant Y”); or retrieve information from cancer samples of a certain histology that contain a mutation in a specified oncogene. Additionally, the "Variant Annotation" schema type can provide rich information about matched variants, but also can serve as the core of genomic knowledge resources for aggregated data about clinically actionable variants. Also, the Beacon v2 model supports the use of grouping concepts such as "data set" and "cohort," for example, to query data particular to a certain resource within a larger Beacon instance or a set of individuals from a given study cohort.
Figure 2.
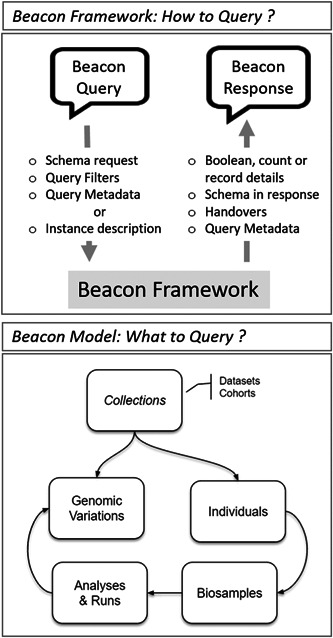
Beacon v2 is composed of two parts: the Beacon Framework and the Beacon Model. The former describes the request and response protocol, the latter describes the common entities in the clinical research domain although other models could be used
In summary, where Beacon v1 by design was limited to positional requests for genomic variations in specified datasets, v2 leverages common biomedical entity models for query and response. While default models and examples support the simple alignment across implementations and thereby empower federated Beacon queries, the extensibility of the model allows to tailor specific solutions for example, in the healthcare context.
1.5. Implementing Beacon over existing solutions
Beacon v2 is organized in two main blocks: the Beacon Framework and the Beacon Models. The Framework is agnostic to the knowledge domain and includes the features related to Beacon instance description (metadata), query requests, query responses, filters, handovers, and so on. The Beacon Models describe the domain entities and the relationships between them. To obtain interoperability between Beacon instances, Beacon v2 includes a recommended model for clinical genomics diagnoses and research, as it is described in a previous section. Separating the Framework from the Model allows other disciplines to adopt the Beacon concept without departing from the standard itself and without any servitude of implementing a model that is not relevant to their domain.
We can also refer to the architecture of the Beacon design and to the architecture of the Beacon implementations or instances. Implementations of the previous version of Beacon (v1) existed in two different flavors: (1) solutions developed from scratch, that is by "beaconizing" a pure variant collection without existing data interface; and (2) Beacons created on top of existing solutions, for example: Cafe Variome (Lancaster et al., 2015), OpenCGA (http://opencb.org/), Progenetix (Huang et al., 2021), or RD‐Connect GPAP. Beacon v1 was designed to be minimalistic, with a minimal implementation effort. Given that Beacon v2 has a broader scope and the effort to build a solution from scratch is significantly higher, Beacon v2 has been designed as a REST API façade for existing solutions. These could be professional and popular solutions or homemade, basic ones; in any case, the design principle was to make the implementation simple and as less intrusive as possible, lowering the barrier for implementation for existing solutions.
The Beacon designers envisioned a scenario in which genomics solution providers would be able to implement a Beacon on top of existing solutions, with a minimum of development effort and resources. To not be intrusive, the Beacon suggests an harmonization approach at query time. For example, a given backend could store the “male gender” concept as a “M” in the column “gender” of the “Person” table, while another Beacon could represent it as “male” in the column “sex” of the “cohort‐members” table. Beacon v2 suggests using an ontology term for “male,” for example, PATO:0000384, at query time. The Beacon instances will receive that term as part of the query and they would need to translate it, map it, to the corresponding internal representation to solve the query. The results are mapped to the model suggested by Beacon, obtaining, therefore, an harmonization at query and at response time. Importantly, such concepts can be implemented gradually, without the need to change the underlying data model ab initio.
Many Beacon instances will be part of networks, although a Beacon can be instantiated as a stand‐alone solution. The Beacon design includes several features aimed to be consumed by Beacon network aggregators. For example, a Beacon endpoint declares which entities are implemented in that particular instance, which are the ontology terms supported or the URL endpoints where different elements could be found.
In summary, Beacon v2 has been designed to be domain agnostic, but suggesting a model for clinical genomics as well as to act as an interoperability interface on top of existing resources, to enable their utilization as part of networked and federated data discovery solutions.
1.6. How does it relate to other clinical research standards?
An ecosystem of clinical data standards enables healthcare and research systems to interoperate to unambiguously describe, store, exchange, and analyze health data on an international scale. Beacon works alongside established standards to provide a flexible data discovery solution with optional clinical applications. Beacon supports the use of semantic standards used to describe clinical concepts and it is compatible with open syntactic standards for harmonized clinical data storage and communication.
Controlled vocabularies and ontologies standardize the labeling of concepts for various biomedical domains, for example, SNOMED for diagnoses, HPO for phenotypic abnormalities, and LOINC for laboratory results. Beacon v2 “filters” support the discovery of patients and biosamples using ontology terms. Beacon does not limit which systems may participate by being agnostic to the semantic standards used by a data source. Beacons which use controlled vocabularies and ontologies declare this by providing an informational filters endpoint that is defined in the Beacon framework. Beacon reuses Phenopackets v2 specifications for describing ontologies and representing ontology classes. Phenopackets is a GA4GH approved standard for sharing disease and phenotypic information and has been adopted by the rare diseases research community for consistent characterization and representation of disease manifestations (Rubinstein et al., 2020). Beacon individual and biosample schemas compatible with Phenopackets v2 architecture are available to help streamline implementations of Beacon discovery when Phenopackets are used by a data provider.
The openEHR, HL7 fast healthcare interoperability resources (FHIR), and Observational Medical Outcomes Partnership Common Data Model (OMOP CDM) standards are concerned with the storage of clinical data for healthcare, clinical data exchange, and the storage of clinical data for research, respectively. The Beacon architecture supports patient discovery across these three standards. The Beacon “individuals” model can be tailored and mapped to the components of these standards that store patient health information.
The openEHR specification defines the structure and function of electronic health record (EHR) systems. Archetypes are core concepts of the openEHR specification and are comprehensive, machine‐interpretable, and reusable discrete models of health information, such as observations of body mass index and arterial blood pressure (Bosca et al., 2015). The complementary FHIR standard is used by the healthcare industry to exchange EHR data. FHIR uses components called resources to access and perform operations on patient data. The resources define generic common health care concepts with clearly defined scope such as observation and condition (Ayaz et al., 2021). Dedicated Beacon model schemas can be used to accommodate implementations of openEHR archetypes and serializations of FHIR resources. Patients can be discovered by filtering on the patient characteristics captured by the clinical coding used by implementations, with the Beacon response accommodating a handover to the native data exchange format.
The OMOP CDM supports research by harmonizing healthcare data from diverse sources in a consistent and standardized way (Hripcsak et al., 2015). The CDM can be adapted to accommodate specialized medical research use cases, for example, the storage and analysis of rare disease patient data are enabled by including dedicated rare disease terminologies within the CDM (Zoch et al., 2021). The OMOP CDM consists of a collection of table schemas where each schema represents a specific OMOP domain such as observation and measurement. A community effort organized through Biohackathon Europe (described below) has mapped the Beacon model to OMOP table schemas to demonstrate the Beacon discovery of patients using OMOP vocabularies.
The annual Biohackathon Europe event hosted by ELIXIR brings together bioinformaticians, software engineers, data providers, and consumers to work on life sciences data challenges. During Beacon‐focused projects at the event over consecutive years (in 2020 and 2021), we aimed to demonstrate the reality of Beacon discovery alongside existing clinical data standards. In 2020 we devised a proof of concept (POC) Beacon implementation to enable patient discovery of individuals described in the OMOP CDM. Serializations of SNOMED‐coded synthetic FHIR resources were transformed and loaded into the OMOP CDM, and the POC Beacon was mapped to OMOP table schemas to enable individuals to be discovered using SNOMED ontology filters. This capability has been extended in 2021 to discover synthetic patients from EHRs and our ambition is to deliver a POC Beacon to demonstrate and support Beacon adoption alongside, and complementary to, established health information systems implementing these open standards.
1.7. Security aspects
The Beacon uses a 3‐tiered access model ‐ anonymous, registered, and controlled access. A Beacon that supports anonymous access responds to queries irrespective of the source of the query. For a Beacon to respond to a query at the registered tier, the user must identify themselves to the Beacon, for example by using an ELIXIR identity. ELIXIR identities are controlled by the ELIXIR Authorization and Authentication Infrastructure (ELIXIR AAI; https://elixir-europe.org/services/compute/aai). The ELIXIR Authentication and Authorization Infrastructure (AAI) enables researchers to use their home organization credentials, community, or commercial identities (e.g., ORCID, LinkedIn) to sign in and access data and the services that they need. For a Beacon to respond to a controlled‐access query, the user must have applied for and been granted access to, the Beacon (or data derived from one or more individuals within the Beacon) before sending the query. Note that a Beacon may contain datasets (or collections of individuals) whose data is only accessible at specified tiers within the Beacon. This tiered access model allows the owner of a Beacon to determine which responses are returned to whom depending on the query itself and the user who is making the request, for example, to ensure the response respects the consent or legal basis under which the data were collected, or to support requirements in different legal jurisdictions, for example, the data minimization or purpose limitation principles within GDPR (European Parliament and Council, 2016). As an example, the ELIXIR Beacon Network supports Beacons which respond at different tiers, for example, only Beacons which have a response for anonymous queries need respond to an anonymous request. A security document (ELIXIR, 2021) has been written to describe security best practice for users interested in deploying or running a Beacon or users who govern data hosted within a Beacon, and the requirements for adding the Beacon to the ELIXIR Beacon Network. Additionally, as Beacon implements a GA4GH approved standard it must go through the GA4GH approval process, which means the standard must be approved by both the Regulatory and Ethics, and Data Security foundational workstreams. As the Beacon standard extends in V2 toward supporting phenotype and range queries, the tiered access model becomes more important to ensure the Beacon response is appropriate to the underlying data.
All the measures described should allow a Beacon administrator to configure the access to the hosted data according to their sensitivity, ranging from total openness for allele frequencies in population studies to fully protected in particular diseases, therefore minimizing the risk of undesired re‐identification (Bernier et al., 2022).
As a Beacon is designed to support data discoverability of controlled access datasets, it is recommended that synthetic or artificial data is used for testing and initial deployment of Beacon instances. The use of synthetic data for testing is important in that it ensures that the full functionality of a Beacon can be tested and/or demonstrated without risk of exposing data from individuals. In addition to testing or demonstrating a deployment, synthetic data should be used for development, for example adding new features. An example data set that contains chromosome specific vcf files is hosted at EGA under data set accession EGAD00001006673. This data set is accessible via EGA's test user and does not require obtaining separate credentials.
1.8. Toward an “Internet of genomics”?
Since the inception of Beacon v2 idea in October 2018, many projects and initiatives have shown interest in the Beacon concept and its possibilities. The scope of projects is broad: resource discovery (e.g., biobanks or registries), cohort discovery and description, proteomics, viral genomics (e.g., SARS‐CoV‐2 in Viral Beacon ‐ https://covid19beacon.crg.eu/), plants, and so on. Some flagship projects like the European Joint Program on Rare Diseases (EJP‐RD), the 1+ Million Genomes initiative (European Commission, 2021) and its supporting Beyond One Million Genomes project, or Horizon 2020 funded projects like BY‐COVID, CINECA, CONVERGE, or EUCANCan.
The goals of these projects are diverse, ranging from sharing data in domains where there are no established standard solutions, to allow total control on the granularity of sensitive data sharing (from boolean answers to complete details, depending on the level of trust and if the audience is intramural or external). All of them share the vision that the future of sensitive data is federated discovery, query, and analysis and that only pragmatic approaches would make that possible. These pragmatic approaches translate into control, flexibility, simplicity, and capability to deal with heterogeneity. All of them are attributes that Beacon v2 has included in their design, therefore, these projects have looked at it as a solution to observe. Several of the mentioned projects have implemented Beacon v1 instances and tested the preliminary versions of Beacon v2.
Beacon v2 is designed to be an interface on top of existing solutions, however, the clinical genomics research facilities are, in many cases, facing a more basic issue: the lack of a solution to manage the genomic data and its relationship with the clinical care associated data (phenotypes and clinical journey). This need has led to the concept of the EGA Community Platform. The European Genome‐phenome Archive (EGA) Community Platform is a proposal to combine existing solutions for genomic and metadata data management, with existing analysis solutions, all topped with a Beacon v2 interface. The clinical research facility could choose among the already tested solutions or add any of their preference, the only requirement is that it must implement a Beacon v2 interface. The aim of this concept is to facilitate the reuse of existing data, initially inside the institution, while paving the way for sharing with the community the generated knowledge in a safe and controlled way.
Beacons lighted independently, through EGA Community packages or by any other means, could be integrated in Beacon networks. Beacon networks could be internal to a hospital campus, a consortium, a region or country, or be organized by topic, one example being the ELIXIR Beacon Network (https://beacon-network.elixir-europe.org/), whose goal is to trigger the discovery of Beacons and to showcase the utility of such networks.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
The Beacon v2 project has received continued support from both GA4GH and ELIXIR. As a prominent GA4GH Discovery Work Stream product, the Beacon protocol and associated team has benefitted from administrative support in product submission and meeting management. We would like to acknowledge the support and contribution from the GA4GH Driver projects and other partners mentioned in the text. And last, but not least all the volunteers of the Beacon Scouts and the enthusiastic Beacon v2 implementers. This study was funded by ELIXIR, the research infrastructure for life‐science data and also by LaCaixa Foundation (grant number 004745/008034). Tim Beck was supported by a UKRI Innovation Fellowship at Health Data Research UK (MR/S003703/1). Anthony J. Brookes and Jordi Rambla were supported, in part, by the European Union's Horizon 2020 research and innovation program under the EJP RD COFUND‐EJP #825575. Michael Baudis acknowledges funding under the BioMedIT Network project of Swiss Institute of Bioinformatics (SIB) and Swiss Personalized Health Network (SPHN).
Rambla, J. , Baudis, M. , Ariosa, R. , Beck, T. , Fromont, L. A. , Navarro, A. , Paloots, R. , Rueda, M. , Saunders, G. , Singh, B. , Spalding, J. D. , Törnroos, J. , Vasallo, C. , Veal, C. D. , & Brookes, A. J. (2022). Beacon v2 and Beacon networks: A “lingua franca” for federated data discovery in biomedical genomics, and beyond. Human Mutation, 43, 791–799. 10.1002/humu.24369
Contributor Information
Jordi Rambla, Email: jordi.rambla@crg.eu.
Michael Baudis, Email: michael.baudis@uzh.ch.
REFERENCES
- Ayaz, M. , Pasha, M. F. , Alzahrani, M. Y. , Budiarto, R. , & Stiawan, D. (2021). The fast health interoperability resources (FHIR) standard: Systematic literature review of implementations, applications, challenges and opportunities. JMIR Medical Informatics, 9(7), e21929. 10.2196/21929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier, A. , Liu, H. , & Knoppers, B. M. (2022). Computational tools for genomic data de‐identification: Facilitating data protection law compliance. Nature Communications, 13, 391. 10.1038/s41467-021-27890-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosca, D. , Moner, D. , Maldonado, J. A. , & Robles, M. (2015). Combining archetypes with fast health interoperability resources in future‐proof health information systems. Studies in Health Technology and Informatics, 210, 180–184. [PubMed] [Google Scholar]
- Cezard, T. , Cunningham, F. , Hunt, S. E. , Koylass, B. , Kumar, N. , Saunders, G. , Shen, A. , Silva, A. F. , Tsukanov, K. , Venkataraman, S. , Flicek, P. , Parkinson, H. , & Keane, T. M. (2022). The European Variation Archive: A FAIR resource of genomic variation for all species. Nucleic Acids Research, 50, gkab960. 10.1093/nar/gkab960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, M. S. , Liao, R. G. , Parsons, M. T. , Paten, B. , Alquaddoomi, F. , Antoniou, A. , Baxter, S. , Brody, L. , Cook‐Deegan, R. , Coffin, A. , Couch, F. J. , Craft, B. , Currie, R. , Dlott, C. C. , Dolman, L. , den Dunnen, J. T. , Dyke, S. , Domchek, S. M. , Easton, D. , … Spurdle, A. B. (2018). BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genetics, 14, e1007752. 10.1371/journal.pgen.1007752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELIXIR . (2021). ELIXIR Beacon 2019–21 Deliverable D3.3. https://docs.google.com/document/d/1q7XuUB-Z4A_DogWT1AVrvkp_qHWWtbbICxokHup_tts/edit [Google Scholar]
- European Commission . (2021). 1+ Million genomes. Shaping Europe's digital future. https://digital-strategy.ec.europa.eu/en/policies/1-million-genomes
- European Parliament and Council . (2016). General Data Protection Regulations (GDPR): Regulation (EU) 2016/679; Article 5. https://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:32016R0679%26from=EN#d1e1807-1-1
- Fiume, M. , Cupak, M. , Keenan, S. , Rambla, J. , de la Torre, S. , Dyke, S. , Brookes, A. J. , Carey, K. , Lloyd, D. , Goodhand, P. , Haeussler, M. , Baudis, M. , Stockinger, H. , Dolman, L. , Lappalainen, I. , Törnroos, J. , Linden, M. , Spalding, J. D. , Ur‐Rehman, S. , … Scollen, S. (2019). Federated discovery and sharing of genomic data using Beacons. Nature Biotechnology, 37(3), 220–224. 10.1038/s41587-019-0046-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeberg, M. A. , Fromont, L. A. , D'altri, T. , Romero, A. F. , Ciges, J. I. , Jene, A. , Kerry, G. , Moldes, M. , Ariosa, R. , Bahena, S. , Barrowdale, D. , Barbero, M. C. , Fernandez‐Orth, D. , Garcia‐Linares, C. , Garcia‐Rios, E. , Haziza, F. , Juhasz, B. , Llobet, O. M. , Milla, G. , … Rambla, J. (2022). The European Genome‐phenome Archive in 2021. Nucleic Acids Research, 50, gkab1059. 10.1093/nar/gkab1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont, L. A. (2021). Beacon cohorts: A model for cohort discovery in CINECA and beyond—CINECA—Common Infrastructure for National Cohorts in Europe, Canada, and Africa. CINECA. https://www.cineca-project.eu/blog-all/beacon-cohorts-a-model-for-cohort-discovery-in-cineca-and-beyond
- Harrison, P. W. , Ahamed, A. , Aslam, R. , Alako, B. T. F. , Burgin, J. , Buso, N. , Courtot, M. , Fan, J. , Gupta, D. , Haseeb, M. , Holt, S. , Ibrahim, T. , Ivanov, E. , Jayathilaka, S. , Balavenkataraman Kadhirvelu, V. , Kumar, M. , Lopez, R. , Kay, S. , Leinonen, R. , … Cochrane, G. (2021). The European Nucleotide Archive in 2020. Nucleic Acids Research, 49(D1), D82–D85. 10.1093/nar/gkaa1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hripcsak, G. , Duke, J. D. , Shah, N. H. , Reich, C. G. , Huser, V. , Schuemie, M. J. , & Ryan, P. B. (2015). Observational health data sciences and informatics (OHDSI): Opportunities for observational researchers. Studies in Health Technology and Informatics, 216, 574–578. [PMC free article] [PubMed] [Google Scholar]
- Huang, Q. , Carrio‐Cordo, P. , Gao, B. , Paloots, R. , & Baudis, M. (2021). The Progenetix oncogenomic resource in 2021. Database: The Journal of Biological Databases and Curation, 2021, baab043. 10.1093/database/baab043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, J. O. B. , Baudis, M. , Baynam, G. S. , Beckmann, J. S. , Beltran, S. , Callahan, T. J. , Chute, C. G. , Courtot, M. , Danis, D. , Elemento, O. , Freimuth, R. R. , Gargano, M. A. , Groza, T. , Hamosh, A. , Harris, N. L. , Kaliyaperumal, R. , Khalifa, A. , Krawitz, P. M. , Köhler, S. , … Robinson, P. N. (2021). The GA4GH Phenopacket schema: A computable representation of clinical data for precision medicine. medRxiv, 11(27), 21266944. 10.1101/2021.11.27.21266944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli, K. P. , Paten, B. , Genome 10K Community of Scientists O'Brien, S. J . (2015). The Genome 10K Project: A way forward. Annual Review of Animal Biosciences, 3, 57–111. 10.1146/annurev-animal-090414-014900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster, O. , Beck, T. , Atlan, D. , Swertz, M. , Thangavelu, D. , Veal, C. , Dalgleish, R. , & Brookes, A. J. (2015). Cafe Variome: General‐purpose software for making genotype‐phenotype data discoverable in restricted or open access contexts. Human Mutation, 36(10), 957–964. 10.1002/humu.22841 [DOI] [PubMed] [Google Scholar]
- Matalonga, L. , Hernández‐Ferrer, C. , Piscia, D. , Solve‐RD SNV‐indel working, g , Schüle, R. , Synofzik, M. , Töpf, A. , Vissers, L. , de Voer, R. , Solve‐Rd, D. , Solve‐Rd, D. , Solve‐Rd, D. , Solve‐Rd, D. , Tonda, R. , Laurie, S. , Fernandez‐Callejo, M. , Picó, D. , Garcia‐Linares, C. , Papakonstantinou, A. , … Solve‐RD, C. (2021). Solving patients with rare diseases through programmatic reanalysis of genome‐phenome data. European Journal of Human Genetics, 29(9), 1337–1347. 10.1038/s41431-021-00852-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder, N. , Abimiku, A. , Adebamowo, S. N. , de Vries, J. , Matimba, A. , Olowoyo, P. , Ramsay, M. , Skelton, M. , & Stein, D. J. (2018). H3Africa: Current perspectives. Pharmacogenomics and Personalized Medicine, 11, 59–66. 10.2147/PGPM.S141546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis, A. A. , Azzariti, D. R. , Beltran, S. , Brookes, A. J. , Brownstein, C. A. , Brudno, M. , Brunner, H. G. , Buske, O. J. , Carey, K. , Doll, C. , Dumitriu, S. , Dyke, S. O. , den Dunnen, J. T. , Firth, H. V. , Gibbs, R. A. , Girdea, M. , Gonzalez, M. , Haendel, M. A. , Hamosh, A. , … Rehm, H. L. (2015). The Matchmaker Exchange: A platform for rare disease gene discovery. Human Mutation, 36(10), 915–921. 10.1002/humu.22858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero, J. , Ramírez‐Anguita, J. M. , Saüch‐Pitarch, J. , Ronzano, F. , Centeno, E. , Sanz, F. , & Furlong, L. I. (2020). The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Research, 48(D1), D845–D855. 10.1093/nar/gkz1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm, H. L. , Page, A. J. H. , Smith, L. , Adams, J. B. , Alterovitz, G. , Babb, L. J. , Barkley, M. P. , Baudis, M. , Beauvais, M. , Beck, T. , Beckmann, J. S. , Beltran, S. , Bernick, D. , Bernier, A. , Bonfield, J. K. , Boughtwood, T. F. , Bourque, G. , Bowers, S. R. , Brookes, A. J. , … Rodarmer, K. W. (2021a). CanDIG: Federated network across Canada for multi‐omic and health data discovery and analysis. Cell Genomics, 1(2), 100033. 10.1016/j.xgen.2021.100033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm, H. L. , Page, A. J. H. , Smith, L. , Adams, J. B. , Alterovitz, G. , Babb, L. J. , Barkley, M. P. , Baudis, M. , Beauvais, M. J. S. , Beck, T. , Beckmann, J. S. , Beltran, S. , Bernick, D. , Bernier, A. , Bonfield, J. K. , Boughtwood, T. F. , Bourque, G. , Bowers, S. R. , Brookes, A. J. , … Rodarmer, K. W. (2021b). GA4GH: International policies and standards for data sharing across genomic research and healthcare. Cell Genomics, 1(2), 100029. 10.1016/j.xgen.2021.100029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein, Y. R. , Robinson, P. N. , Gahl, W. A. , Avillach, P. , Baynam, G. , Cederroth, H. , Goodwin, R. M. , Groft, S. C. , Hansson, M. G. , Harris, N. L. , Huser, V. , Mascalzoni, D. , McMurry, J. A. , Might, M. , Nellaker, C. , Mons, B. , Paltoo, D. N. , Pevsner, J. , Posada, M. , … Haendel, M. A. (2020). The case for open science: Rare diseases. JAMIA Open, 3(3), 472–486. 10.1093/jamiaopen/ooaa030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, R. , Johnston, L. , Taruscio, D. , Monaco, L. , Béroud, C. , Gut, I. G. , Hansson, M. G. , t Hoen, P.‐B. A. , Patrinos, G. P. , Dawkins, H. , Ensini, M. , Zatloukal, K. , Koubi, D. , Heslop, E. , Paschall, J. E. , Posada, M. , Robinson, P. N. , Bushby, K. , & Lochmüller, H. (2014). RD‐Connect: An integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. Journal of General Internal Medicine, 29, 780–787. 10.1007/s11606-014-2908-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, A. H. , Babb, L. , Alterovitz, G. , Baudis, M. , Brush, M. , Cameron, D. L. , Cline, M. , Griffith, M. , Griffith, O. L. , Hunt, S. E. , Kreda, D. , Lee, J. M. , Li, S. , Lopez, J. , Moyer, E. , Nelson, T. , Patel, R. Y. , Riehle, K. , Robinson, P. N. , … Hart, R. K. (2021). The GA4GH Variation Representation Specification: A computational framework for variation representation and federated identification. Cell Genomics, 1(2), 100027. 10.1016/j.xgen.2021.100027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoch, M. , Gierschner, C. , Peng, Y. , Gruhl, M. , Leutner, L. A. , Sedlmayr, M. , & Bathelt, F. (2021). Adaption of the OMOP CDM for rare diseases. Studies in Health Technology and Informatics, 281, 138–142. 10.3233/SHTI210136 [DOI] [PubMed] [Google Scholar]
- Zurek, B. , Ellwanger, K. , Vissers, L. E. L. M. , Schüle, R. , Synofzik, M. , Töpf, A. , de Voer, R. M. , Laurie, S. , Matalonga, L. , Gilissen, C. , Ossowski, S. , 't Hoen, P. , Vitobello, A. , Schulze‐Hentrich, J. M. , Riess, O. , Brunner, H. G. , Brookes, A. J. , Rath, A. , Bonne, G. , … Solve‐RD, c (2021). Solve‐RD: Systematic pan‐European data sharing and collaborative analysis to solve rare diseases. European Journal of Human Genetics, 29, 1325–1331. 10.1038/s41431-021-00859-0 [DOI] [PMC free article] [PubMed] [Google Scholar]


