Abstract
Sleep is regulated by two main processes: the circadian clock and sleep homeostasis. Circadian rhythms have been well studied at the molecular level. In the Drosophila circadian clock neurons, the core clock proteins are precisely regulated by post‐translational modifications and degraded via the ubiquitin‐proteasome system (UPS). Sleep homeostasis, however, is less understood; nevertheless, recent reports suggest that proteasome‐mediated degradation of core clock proteins or synaptic proteins contributes to the regulation of sleep amount. Here, we review the molecular mechanism of the UPS and summarize the role of protein degradation in the regulation of circadian clock and homeostatic sleep in Drosophila. Moreover, we discuss the potential interaction between circadian clock and homeostatic sleep regulation with a prime focus on E3 ubiquitin ligases.
Keywords: circadian clock, circadian rhythm, Drosophila, protein homeostasis, sleep, sleep homeostasis, ubiquitin proteasome system
Circadian clock regulates behavioral and physiological periodicity, while sleep homeostasis controls the sleep amount in response to the increased demand of sleep. Here, we review the molecular mechanism of the ubiquitin‐proteasome system (UPS) and summarize the role of protein degradation in the regulation of circadian clock and homeostatic sleep in Drosophila.
1. INTRODUCTION
Sleep is a conserved physiological phenomenon in the animal kingdom that is vital for survival, even though it increases the vulnerability to predators. Sleep is necessary for diverse body functions such as metabolic homeostasis (Maguire et al., 2015; Thimgan et al., 2015), learning and memory (Dissel et al., 2015; Gilestro et al., 2009), and clearance of reactive oxidative species (Hill et al., 2018; Vaccaro et al., 2020). Sleep deprivation has been documented to disrupt these functions, leading to premature death (Vaccaro et al., 2020). To ensure the timing and the amount of sleep, sleep is regulated by mainly two processes: circadian clock and sleep homeostasis (Deboer, 2018). Circadian clock regulates behavioral and physiological periodicity, while sleep homeostasis controls the sleep amount in response to the increased demand of sleep following the situations such as sleep deprivation or sickness, regardless of circadian rhythm (Dubowy & Sehgal, 2017; Toda et al., 2019).
Fruit fly (of the genus Drosophila) is a model organism that has been extensively used to investigate the mechanisms of circadian clock at the neural and molecular levels. The circadian rhythm of Drosophila is regulated by the transcriptional negative feedback loop of core clock genes including period (Konopka & Benzer, 1971), timeless (Sehgal et al., 1994), Clock (Allada et al., 1998), and cycle (Rutila et al., 1998). The core clock genes exhibit molecular oscillations in the clock neurons of the brain, which is the central pacemaker of the circadian rhythms. In mammals, the circadian clock‐mediated rest state in the activity rhythm has been comprehended as sleep (Hendricks, Sehgal, et al., 2000). In Drosophila, it has been demonstrated that the rest state is a sleep‐like state, characterized as behavioral quiescence that lasts for at least 5 min (Hendricks, Finn, et al., 2000; Shaw et al., 2000). During sleep, Drosophila exhibits an augmented arousal threshold, making it difficult to respond to external stimuli. However, the sleep state is reversible when the stimuli are stronger than the arousal threshold. After periods of sleep deprivation, fruit flies appear to prolong the duration of their sleep to compensate for the lack of sleep, which is a mechanism of sleep homeostasis. These observations are consistent with the features of sleep homeostasis in mammals (Huber et al., 2004). For the past 20 years, forward genetic screenings using Drosophila have identified various genes that regulate sleep. For instance, Shaker encodes a subunit of voltage‐gated potassium channels and plays a role in electrical excitability, while the mutation of the Shaker gene induces a reduction in sleep (Cirelli et al., 2005). Additionally, a mutant of the redeye gene encoding the nicotinic acetylcholine receptor α4 shows a similar short sleep phenotype. Redeye protein levels are upregulated in response to the sleep needs and are believed to be a sleep promoting factor under the homeostatic system (Shi et al., 2014). Likewise, a gene that encodes an antimicrobial peptide called nemuri has been identified using a gain‐of‐function screen for sleep induction (Toda et al., 2019). The overexpression of nemuri has been reported to prolong the sleep, implying a possible interaction between the immune system and sleep (Toda et al., 2019). Further, Argus, a transmembrane protein which is involved in the autophagosome degradation, is expressed in peptidergic neurons. Notably, argus mutants exhibit a short sleep phenotype, indicating the possibility that the degradation of waste products such as proteins in specific neurons could affect sleep (Bedont et al., 2021).
The ubiquitin‐proteasome system (UPS) is one of the protein degradation systems, which is highly conserved in eukaryotes and archaea. The UPS is involved in various biological events such as cell cycle progression, cell growth, cell proliferation, endocytosis, elimination of misfolded proteins, and response to environmental stimuli through the degradation of proteins (Nath & Shadan, 2009). The UPS also plays a role in the regulation of circadian clock by degrading core clock proteins (Vriend & Reiter, 2015). In addition, the UPS appears to have the potential to regulate homeostatic sleep in Drosophila (Li et al., 2017; Pfeiffenberger & Allada, 2012; Stavropoulos & Young, 2011). Here, we have reviewed the current knowledge of the UPS and its interaction with circadian clock and sleep homeostasis, specifically focusing on the E3 ubiquitin ligases, which have been identified in Drosophila (Table 1).
TABLE 1.
Ubiquitin enzymes and ubiquitin‐like protein involved in circadian clock and homeostatic sleep regulation
| Gene | Gene summary | Target | References | |
|---|---|---|---|---|
| Circadian clock | slmb | A substrate receptor of Cullin‐RING E3 ligase | Phosphorylated PER | (Chiu et al., 2008; Grima et al., 2002) |
| jet | A substrate receptor of Cullin‐RING E3 ligase | CRY‐bound TIM in light degradation pathway | (Koh et al., 2006; Ozturk et al., 2011; Vaidya et al., 2013) | |
| Cul3 | Cullin scaffold of Cullin‐RING E3 ligase | Hypophosphorylated TIM | (Grima et al., 2012) | |
| ctrip | HECT E3 ligase | CLK | (Lamaze et al., 2011) | |
| Homeostatic sleep regulation | stx | Ubiquitin‐like protein | PcG | (Du et al., 2016; Zhao et al., 2021) |
| inc | A substrate receptor of Cullin‐RING E3 ligase | Unknown | (Pfeiffenberger & Allada, 2012; Pirone et al., 2016; Stavropoulos & Young, 2011) |
2. UBIQUITIN PROTEASOME SYSTEM
The mechanism of UPS‐mediated protein degradation is divided into two major steps: first, tagging the target protein with ubiquitin, and second, degrading the ubiquitin‐tagged proteins via the 26S proteasome. Ubiquitin is a small protein consisting of 76 amino acids that is attached to the target proteins by the catalytic effects of three enzymes namely, the ubiquitin‐activating enzyme E1, ubiquitin‐conjugating enzyme E2, and ubiquitin ligase enzyme E3 (Bachiller et al., 2020; Ding & Shen, 2008; Leestemaker & Ovaa, 2017). E1 uses ATP to activate ubiquitin with high‐energy thioester covalent bonds to the C‐terminal glycine of ubiquitin. E2 forms another thioester bond with activated ubiquitin, which is then transferred to E2. E3 catalyzes the transfer of ubiquitin from E2 to the target protein. Subsequently, other activated ubiquitin moieties are added to ubiquitin conjugated target protein, resulting in the synthesizes of a polyubiquitin chain. This chain is then targeted by the 26S proteasome, resulting in target protein degradation (Glickman & Ciechanover, 2002; Leestemaker & Ovaa, 2017).
Approximately 2 E1s, 40 E2s, and more than 600 E3s have been documented to be encoded in the human genome (Leestemaker & Ovaa, 2017; Morreale & Walden, 2016), while there are 8 E1s, 34 E2s, and 207 E3s genes that have been identified in Drosophila (Du et al., 2011). Moreover, the E3 enzymes are more diverse than the E1 and E2 enzymes, indicating that E3 ubiquitin ligases determine the specificity of the ubiquitin target proteins (Bachiller et al., 2020; Ding & Shen, 2008). E3 ubiquitin ligases are divided into two major groups: the R eally I nteresting N ew G ene (RING) E3, which has a zinc‐binding domain called RING or U‐box domain; and the Homologous to E6AP Carboxyl Terminus (HECT) E3, which has two lobes; one of which interacts with an E2 enzyme while the other interacts with the ubiquitin C terminus (Morreale & Walden, 2016; Rotin & Kumar, 2009). About 95% of E3 ligases belong to RING family (Li et al., 2008).Moreover, the cullin‐RING E3 ligases form the largest class of RING family ligases and contribute to approximately 20% of the total ubiquitination in a cell (Petroski & Deshaies, 2005; Soucy et al., 2009). RING E3 catalyzes the direct transfer of ubiquitin from E2 to the target protein (Morreale & Walden, 2016; Figure 1a). In contrast, the HECT family consists of only 28 E3 ligases, constituting about 5% of all the known human E3s (Rotin & Kumar, 2009). Mechanistically, the HECT E3 ligase has been shown to first catalyze the self‐transfer of ubiquitin, followed by the transfer of the ubiquitin to the target protein (Morreale & Walden, 2016; Figure 1b).
FIGURE 1.
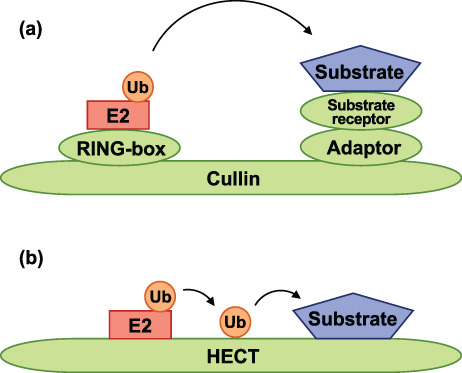
Molecular components of E3 ubiquitin ligases. (a) A schematic diagram of Cullin‐RING E3 ligases. Cullin‐RING E3 ligases (green) directly transfer ubiquitin (orange) from E2 ligase (red) to the substrate (blue). They are composed of multiple subunits (RING‐box protein, adaptor protein, and substrate receptor protein), assembled on a Cullin scaffold. The RING‐box protein binds to the N‐terminus of Cullin, whereas the adaptor protein and substrate receptor protein bind to the C‐terminus of Cullin. The RING‐box protein binds to the ubiquitin‐bound E2, after which ubiquitin is transferred to the substrate bound to the substrate receptor protein. (b) A schematic representation of HECT E3 ligases. The HECT E3 ligases (green) transfer ubiquitin (orange) to the substrate (blue) through a two‐step reaction. Ubiquitin is first transferred from E2 ligase (red) to the catalytic site of HECT and subsequently to the substrate. The N‐terminal lobe of HECT binds to the ubiquitin‐bound E2. The C‐terminal lobe of HECT functions as the catalyst for ubiquitin transfer
3. CIRCADIAN CLOCK AND POST‐TRANSLATIONAL MODIFICATION IN DROSOPHILA
The circadian rhythms are approximately 24 h cycles, which endogenously regulate the behavior and physiological phenomena in organisms. As the earth takes 24 h to complete one rotation on its axis, the “light and warm period” alternates with the “dark and cold period.” Organisms anticipate and adapt to the daily altering environments, and these adaptations are regulated by the clock genes (Dubowy & Sehgal, 2017; Patke et al., 2020). The clock genes are expressed rhythmically, in sync with the environmental changes that affect behavioral and physiological changes throughout a day. The rhythmic expression of clock genes is self‐sustained only in the clock cells, also known as circadian pacemakers (Herzog, 2007). The circadian pacemakers regulate the periodical physiology of other cells that do not express the clock genes. For instance, in Drosophila, although the lateral horn leucokinin neurons (LHLKs) do not produce the clock proteins, TIMELESS (TIM) and PERIOD (PER), the LHLKs excitability is rhythmically regulated by clock cells, which innervate the LHLKs (Cavey et al., 2016). Therefore, clock cells provide circadian information to other cells called the “clock output cells” and finally induce the behavioral and physiological oscillations.
Drosophila have been documented to exhibit rhythmic activities. In the laboratory conditions, flies anticipate the transition from dark to light and from light to dark for a 12 h light:12 h dark (LD) cycle, which seems to align with the light/dark transition (Axelrod et al., 2015; Figure 2a). Approximately 150 clock cells, also known as “clock neurons,” in the Drosophila brain have been demonstrated to regulate this rhythmic activity. The Drosophila clock neurons are divided into two groups: lateral neurons, which includes the large and small ventral lateral neurons (lLNvs and sLNvs), dorsal lateral neurons (LNds), lateral posterior neurons (LPNs), and dorsal neurons, including DN1, 2, and 3 (Figure 2b). Nearly all LNvs, except for the 5th sLNvs, express the neuropeptide pigment‐dispersing factor (PDF), which is essential for the inter‐clock‐neuron coordination (Helfrich‐Förster, 1997; Kaneko et al., 1997; Rieger et al., 2006). Moreover, the PDF‐positive LNvs are required for the morning anticipation of the fly activity (Dubowy & Sehgal, 2017; Renn, Park, et al., 1999). The LNds and PDF‐negative LNvs are required for the evening anticipation (Dubowy & Sehgal, 2017; Grima et al., 2004; Stoleru et al., 2004). Likewise, the DN1 play a role in relaying a molecular rhythm, as they receive inputs from the PDF neurons (Helfrich‐Förster, 2003; L. Zhang et al., 2010; Y. Zhang et al., 2010) and send projections to the pars intercerebralis (PI), which is crucial for converting the molecular rhythms into activity rhythms (Cavanaugh et al., 2014).
FIGURE 2.
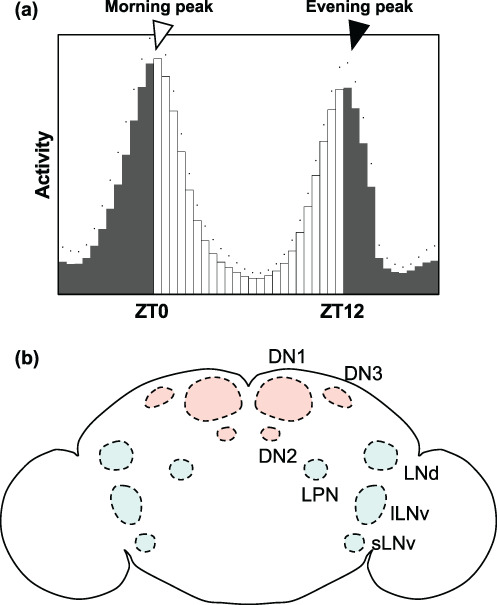
Activity rhythm and related neurons in Drosophila. (a) Activity of wild‐type male Drosophila flies in a 12 h:12 h light:dark cycle at 25°C. Light is turned on at Zeitgeber Time (ZT) 0 (morning) turned off at ZT12 (evening). The flies' activity increased during morning and the evening, thus indicating that flies can anticipate the light‐on and light‐off timings, respectively. (b) The cell body regions of clock neurons in an adult fly's brain. The dorsal neurons (DNs) and lateral neurons (LNs) are shown in red and blue circles, respectively. All LNvs, except for the 5th sLNv, express PDF
The well‐known clock genes in Drosophila are period (per) and timeless (tim) (Konopka & Benzer, 1971; Sehgal et al., 1994). These genes are expressed rhythmically, thereby driving the circadian rhythm. PER and TIM proteins have been reported to suppress their own transcription and are degraded during the day. Additionally, the per and tim mRNA have been documented to be elevated during the day and reach their highest levels in the early evening. In contrast, during the night, the PER and TIM proteins accumulate in the nucleus, which reduces the expression of per and tim mRNAs. Moreover, the PER and TIM regulate their own transcription through the inhibition of CLOCK (CLK) and CYCLE (CYC) heterodimers, which bind to the enhancer regions of the per and tim genes. The CLK/CYC heterodimers also bind to the enhancer of clock output genes, which results in the rhythmic and broader physiological changes (Dubowy & Sehgal, 2017; Sharma et al., 2020). Therefore, the cyclic expression of clock genes leads to rhythmic physiological changes.
The CYC protein has been reported to be stabilized by the CLK protein (Liu et al., 2017). Moreover, the cyc mRNA expression does not oscillate (Rutila et al., 1998) and the CLK protein is constitutively expressed (Houl et al., 2006); therefore, the CLK/CYC complex persists constantly. To sustain the CLK/CYC‐regulated rhythmic expression of per and tim, the transcriptional capability of CLK/CYC should be accurately regulated by the PER and TIM proteins, which inhibit the CLK/CYC binding to DNA in the nucleus (Lee et al., 1999). Therefore, the entry of PER/TIM into the nucleus is significant and mostly depends on post‐translational modifications such as protein phosphorylation. The alpha subunit of casein kinase 2 (CK2α) phosphorylates the Ser149‐151‐153 of PER and promotes the nuclear entry of PER/TIM (Lin et al., 2002; Lin et al., 2005; Top et al., 2016). While the nuclear entry of PER/TIM is an essential step in inhibiting the expression of clock output genes, the degradation of accumulated PER/TIM is imperative for resuming the CLK/CYC‐dependent transcription. It has been reported that PER phosphorylation is the initiation signal for the degradation of PER/TIM. DOUBLE‐TIME (DBT), the fly ortholog of mammalian casein kinase 1 (CK1) δ and ε, phosphorylates Ser47 of PER and promotes the degradation of PER (Chiu et al., 2008; Kloss et al., 1998; Price et al., 1998). Additionally, the degradation of PER/TIM after phosphorylation is dependent on the UPS. Therefore, phosphorylation at the differential sites of PER can induce the nuclear entry and degradation of PER/TIM, which serves as the essential mechanism governing the core of the circadian clock.
4. THE ROLE OF E3 UBIQUITIN LIGASES IN THE DROSOPHILA CIRCADIAN CLOCK
The periodic accumulation and degradation of clock proteins largely rely on the UPS‐dependent degradation (Stojkovic et al., 2014; Szabó et al., 2018). In the UPS, the specificity of the substrate has been reported to be dependent on the E3 ligases. Here, we describe the E3 ligases that affect the rhythmic behavior and degradation of core clock proteins in Drosophila.
Supernumerary limb (slmb) gene encodes a Cullin‐RING E3 ubiquitin ligase component that functions as a substrate receptor (Figure 1a). Grima et al. have demonstrated that the slmb mutant does not show the anticipation of light‐off transition under the light/dark (LD) conditions and cannot sustain rhythmic locomotor activity under the constant darkness (DD) conditions (Grima et al., 2002). Moreover, the Ser47 of PER has been shown to be phosphorylated by DBT, which induces an efficient interaction between PER and SLMB (Chiu et al., 2008). SLMB has been reported to target the phosphorylated PER for proteasomal degradation (Grima et al., 2002; Figure 3). In addition, both the levels of SLMB protein and slmb mRNA have been shown to not oscillate (Grima et al., 2002), suggesting that slmb is not subjected to a circadian regulation. Additionally, the expression of slmb only in PDF‐positive LNvs, one of the clock neuron group, can rescue the activity rhythm phenotype of the slmb mutant (Grima et al., 2002). Overall, these findings suggest that slmb is essential for the regulation of clock gene expression in PDF‐positive LNvs, a subset of clock neurons.
FIGURE 3.
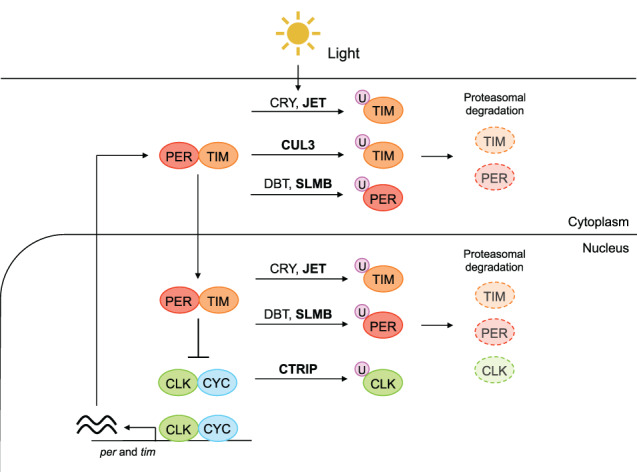
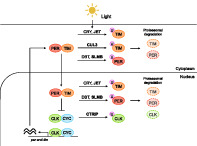
E3 ubiquitin ligases and core clock components in Drosophila. A scheme of UPS in Drosophila circadian clock regulation. When the heterodimer of PER (red) and TIM (orange) is not degraded, the PER/TIM complex accumulates in the nucleus and represses the CLK (green) and CYC (blue) complex. When the PER/TIM is degraded, the CLK/CYC complex promotes the expression of per and tim (the left side of the scheme). Core clock proteins are degraded by UPS (the right side of the scheme). Light exposure facilitates the binding of CRY to TIM, which allows JET to induce the binding of ubiquitin (pink) to TIM. CUL3 binds to hypophosphorylated TIM and induces ubiquitination of TIM. PER is phosphorylated by DBT in the absence of TIM and subsequently subjected to ubiquitination by SLMB. The transcription factor CLK is ubiquitinated by CTRIP. The ubiquitinated proteins are degraded by 26S proteasome. The boldface shows E3 ligase. The localization of E3 ligases are based on Flybase. It is known that the clock protein is degraded both in cytoplasm and nucleus in mammals (Yoo et al., 2013)
JETLAG (JET) is also a substrate receptor for the Cullin‐RING E3 ubiquitin ligase (Figure 1a). Meanwhile, the jet mutants have been reported to show normal activity rhythms under LD, DD, and constant light (LL) conditions (Koh et al., 2006). Under LL conditions, it is known that the wild‐type fly shows arrhythmic behavior due to the constant degradation of TIM by light. The degradation of TIM requires light and CRY, the blue light photoreceptor cryptochrome (Ceriani et al., 1999). Studies have shown that light induces conformational changes in CRY, which then binds to TIM, allowing the JET protein to bind to TIM to induce ubiquitin‐dependent TIM degradation (Lamba et al., 2014; Ozturk et al., 2011; Vaidya et al., 2013; Figure 3). Additionally, the JET protein contains leucine‐rich repeats (LRRs), which is a protein–protein interaction domain (Koh et al., 2006). Therefore, it is speculated that the LRRs of JET bind to the CRY‐bound TIM.
CULLIN‐3 (CUL3), a member of the CULLIN family, is a scaffold of the Cullin‐RING E3 ubiquitin ligase involved in the function of the circadian clock (Figure 1a). It has been reported that the knockdown of Cul3 E3 in a limited number of clock neurons induces defects in morning anticipation under LD conditions and behavioral arrhythmicity in DD conditions, whereas the knockdown of Cul3 in all clock neurons results in 100% lethality (Grima et al., 2012). Likewise, CUL3 binds to hypophosphorylated TIM and controls the TIM oscillation (Grima et al., 2012; Guo et al., 2014); however, it is unclear which substrate receptor protein binds to CUL3 and bears the specificity for TIM (Figure 3).
Besides the Cullin‐RING E3 ubiquitin ligase, the HECT E3 ubiquitin ligase, circadian trip (ctrip), has also been reported as a clock control gene (Lamaze et al., 2011; Figure 1b). CTRIP has been shown to contain a WWE protein–protein interaction domain, which is a putative binding site for the target protein, CLK. The expression of ctrip is strictly restricted to the PDF‐positive LNvs (Lamaze et al., 2011), indicating that ctrip contributes to the degradation of the core clock gene (Figure 3). Additionally, the knockdown of ctrip leads to high levels of CLK protein at all times and extends the period length of endogenous rhythm in the absence of external stimuli, such as light (Lamaze et al., 2011).
Together, the E3 ubiquitin ligases, which bind to clock proteins, affect the behavior and molecular cycling associated with circadian clock. Notably, the E3 ligases mentioned above do not exhibit oscillation in their expression, suggesting that ubiquitin‐dependent degradation is regulated by other systems such as protein phosphorylation, which are important for the precise cycling of clock proteins (Stojkovic et al., 2014). However, the absence of E3 ligases does not result in the abolishment of the core clock protein functions. Therefore, the E3 ligases play an essential role in the degradation of core clock proteins.
5. PROTEASOMAL DEGRADATION IN DROSOPHILA SLEEP HOMEOSTASIS
Circadian clock induces sleep after the beginning of the dark period, and to wake up in advance of the light period, whereas the homeostatic regulation of sleep determines the amount of sleep to meet the body's sleep requirements. Sleep homeostasis in Drosophila shares the features of mammalian sleep homeostasis, with the intensity and duration of sleep rebound being dependent on the prior awakening time or sleep deprivation (Huber et al., 2004). When sleep‐deprived, the duration of sleep in flies is less fragmented and is accompanied by high arousal thresholds (Figure 4a). Moreover, the sleep homeostatic system depends on the R5 neurons (previously referred to as R2 neurons) in the ellipsoid body (EB), which is a doughnut‐shaped synaptic neuropil domain in the adult fly brain. The R5 neurons have been reported to activate the dorsal fan‐shaped body (dFB), thus integrating various sleep signals to promote sleep (Dubowy & Sehgal, 2017; Figure 4b). Moreover, the activation of the R5 neurons has been shown to strongly induce sleep (Liu et al., 2016). Slow‐wave oscillation, which is characteristic of deep sleep in the vertebrate brain and is believed to be the synchronization of neural activity, has been discovered in R5 neurons (Raccuglia et al., 2019). Therefore, while the homeostatic regulation of sleep has been revealed in neural circuits, it is poorly understood at the molecular level. For instance, it is not known what molecule is specifically responsible for inducing the homeostatic sleep drive.
FIGURE 4.
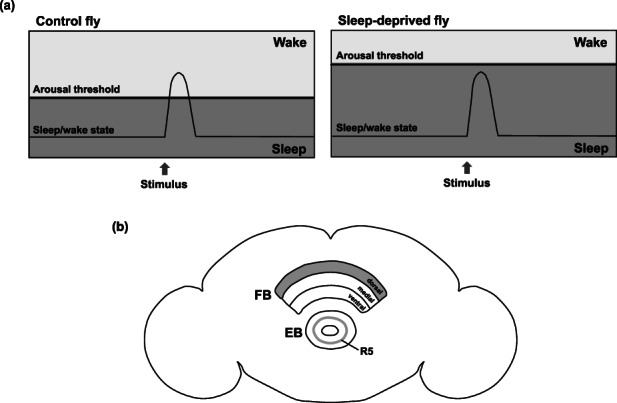
Sleep homeostasis and related neurons in Drosophila. (a) Schematics showing arousal threshold and sleep/wake states. A control fly is aroused when subjected to a stimulus that exceeds the arousal threshold (left panel). Moreover, the arousal threshold is higher in a sleep‐deprived fly than in the control fly. Therefore, the sleep‐deprived fly is not aroused even when subjected to intense stimuli (right panel). (b) A diagram showing the regions in the adult fly brain involved in the homeostatic sleep regulation. The fan‐shaped body (FB) comprises three domains: a dorsal domain, a medial domain, and a ventral domain. Only the dorsal domain of FB (dFB) regulates sleep by the activation of R5 neurons, a type of R neurons arborizing as a ring in the ellipsoid body (EB) (Omoto et al., 2017; Renn, Armstrong, et al., 1999), thereby driving the regulation of sleep homeostasis
One of the vital functions of sleep is the clearance of inessential byproducts from the brain. In the mammalian brain, sleep modulates interstitial space size and induces cerebrospinal fluid influx, resulting in the clearance of β‐amyloids, a toxic metabolic byproduct, from the brain (Xie et al., 2013). Aggregation of such degradation byproducts leads to neural injury and ultimately the Alzheimer's disease (Mattson, 1994). Moreover, Varshavsky hypothesized that the fragmented proteins cleaved by proteases accumulate when the organism is awake and are degraded by the proteasome system during the night (Varshavsky, 2012, 2019b). Additionally, the calpains and caspases are present at post‐synaptic densities (PSDs) and cleave post‐synaptic proteins (Varshavsky, 2019b). The resultant cleaved proteins generate two fragments: a protein containing a new N‐terminus and a protein containing a new C‐terminus (Varshavsky, 2019b). The new N‐terminus and C‐terminus function as degrons, which are the targets of ubiquitin‐dependent degradation (Varshavsky, 2019a). In summary, the UPS is likely required to degrade the accumulated protein fragments during sleep. Therefore, we have hereby deliberated on two proteasome‐related molecules that have the potential to reveal the regulatory mechanism of sleep homeostasis at the molecular level in Drosophila.
Forward genetic screens have identified insomniac (inc), which a mutant exhibiting a significant reduction in sleep (Stavropoulos & Young, 2011). Moreover, inc is an ortholog of the potassium channel tetramerization domain (KCTD) subfamily in vertebrates (Stavropoulos & Young, 2011). Several KCTD subfamilies function as substrate receptors for CUL3 (Cho et al., 2020; He et al., 2018; Rutz et al., 2015; Stavropoulos & Young, 2011). Therefore, INC is a component of SCF E3 ligase that might be involved in the induction of proteasomal degradation. Moreover, Cul3 knockdown reduces sleep, suggesting that the INC/CUL3 complex plays a vital role in regulating sleep and maintaining the sleep homeostatic system (Pfeiffenberger & Allada, 2012; Pirone et al., 2016; Stavropoulos & Young, 2011). The inc gene is expressed in sleep regulatory neurons, including EB, FB, and mushroom bodies (MBs); the INC protein is localized in the cytosol, or the pre‐ and post‐synaptic compartments of neurons (Li et al., 2017). These findings imply that the INC/CUL3 complex regulates sleep by degrading its targets that are localized at the synapse. According to the hypothesis that the accumulated protein fragments are vital for the sleep/wake cycle, the INC/CUL3 complex may potentially be involved in the degradation of the fragments derived from synapse proteins cleaved by proteases. Protein structure analysis and in vitro isothermal calorimetry revealed that Drosophila Golgi reassembly stacking protein (dGRASP) is thought to be a potential target of INC/CUL3 complex (Pirone et al., 2016). However, the genetic interaction between INC and dGRASP in vivo requires further study. Therefore, the identification of the INC/CUL3 complex target may lead to the elucidation of the molecular regulation mechanism of sleep homeostasis.
The ubiquitin‐like protein Stuxnet (stx) promotes the degradation of an epigenetic repressor polycomb group (PcG) by the proteasome, independently of ubiquitination (Du et al., 2016). The stx mutant increases sleep duration and reduces sleep latency, indicating that sleep pressure is elevated in the mutants (Zhao et al., 2021). Stx protein is localized in the EB, which is an important region for homeostatic sleep regulation. Moreover, it has been reported to be downregulated in response to octopamine and reduces the expression of the sleep‐inhibiting octopamine β2 receptor through epigenetic regulation in EB (Zhao et al., 2021).
6. CONCLUSION AND FUTURE PERSPECTIVE
Sleep behavior is poorly understood at the molecular level. Contrary to the neural circuit of sleep regulation, the molecular agents responsible for sleep regulation have not been well elucidated. The INC/CUL3 complex is predicted to play a role in proteasomal degradation (Cho et al., 2020; He et al., 2018; Li et al., 2017; Pirone et al., 2016; Rutz et al., 2015; Stavropoulos & Young, 2011) and the substrate degraded by INC/CUL3‐dependent UPS may be a key factor in regulating the sleep. Thus, the INC/CUL3 complex is considered to be significant in elucidating the molecular mechanisms underlying sleep regulation.
CUL3 plays crucial role in the regulation of circadian clock through the degradation of core clock protein, TIM (Grima et al., 2012), and the homeostatic sleep regulation (Stavropoulos & Young, 2011). Moreover, CUL3 in clock neurons is essential for TIM degradation in the circadian clock, whereas the CUL3/INC complex in sleep regulatory neurons is important for unknown‐target degradation (Pfeiffenberger & Allada, 2012; Stavropoulos & Young, 2011). Besides, INC is localized in the PDF‐positive neurons (Li et al., 2017). The substrate receptor protein of CUL3 for degradation of TIM has not been reported yet. However, the difference in the substrate receptor protein of CUL3 possibly contributes to both the circadian clock and homeostatic sleep regulation (Freeman et al., 2013). Consequently, identifying the substrate receptor protein of CUL3 within the circadian clock may be useful in elucidating the interaction between the circadian clock and homeostatic sleep at the molecular level.
It is hypothesized that synaptic properties contribute significantly to the regulation and function of sleep. For instance, sleep loss induces an increased accumulation of the synaptic proteins (Gilestro et al., 2009) in the presynaptic active zone, which is responsible for the occurrence of high sleep pressure in Drosophila (Huang et al., 2020). These facts imply that the elevated levels of synaptic proteins induce sleep to eliminate the accumulated synaptic protein, which may be one of the primary functions of sleep. Varshavsky hypothesized that another function of sleep is to eliminate the fragmented proteins at the synapse accumulated while awake (Varshavsky, 2012, 2019b). According to this hypothesis, fragmented proteins are eliminated by the UPS. In summary, the UPS is predicted to contribute to the function of sleep through the degradation of synaptic proteins. Moreover, the localization of INC in pre‐ and post‐synaptic compartments (Li et al., 2017) implies a potential relationship between sleep and the degradation of synaptic proteins.
CONFLICT OF INTEREST
The authors declare no potential conflict of interests.
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI (20K15903) to Misako Okumura and (21H02479) to Takahiro Chihara, AMED (JP21gm6310003) and RIKEN‐Hiroshima University Science & Technology Hub Collaborative Research Program to Misako Okumura, the Toray Science Foundation, the Astellas Foundation for Research on Metabolic Disorders, the Naito Foundation, the Frontier Development Program for Genome Editing, and the Core Research for Organelle Diseases at Hiroshima University, Japan (the MEXT program for promoting the enhancement of research universities, Japan). We would like to thank Editage (www.editage.jp) for the English language editing.
Ukita, Y. , Okumura, M. , & Chihara, T. (2022). Ubiquitin proteasome system in circadian rhythm and sleep homeostasis: Lessons from Drosophila . Genes to Cells, 27(6), 381–391. 10.1111/gtc.12935
Communicated by: Eisuke Nishida
Funding information Astellas Foundation for Research on Metabolic Disorders; Core Research for Organelle Diseases at Hiroshima University; Japan Agency for Medical Research and Development, Grant/Award Number: JP21gm6310003; Japan Society for the Promotion of Science, Grant/Award Numbers: 20K15903, 21H02479; Naito Foundation; RIKEN‐Hiroshima University Science & Technology Hub Collaborative Research Program; Japanese Toray Science Foundation
REFERENCES
- Allada, R. , White, N. E. , So, W. V. , Hall, J. C. , & Rosbash, M. (1998). A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell, 93(5), 791–804. 10.1016/s0092-8674(00)81440-3 [DOI] [PubMed] [Google Scholar]
- Axelrod, S. , Saez, L. , & Young, M. W. (2015). Studying circadian rhythm and sleep using genetic screens in Drosophila. Methods in Enzymology, 551, 3–27. 10.1016/bs.mie.2014.10.026 [DOI] [PubMed] [Google Scholar]
- Bachiller, S. , Alonso‐Bellido, I. M. , Real, L. M. , Pérez‐Villegas, E. M. , Venero, J. L. , Deierborg, T. , Armengol, J. Á. , & Ruiz, R. (2020). The ubiquitin proteasome system in neuromuscular disorders: Moving beyond movement. International Journal of Molecular Sciences, 21(17), 6429. 10.3390/ijms21176429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont, J. L. , Toda, H. , Shi, M. , Park, C. H. , Quake, C. , Stein, C. , Kolesnik, A. , & Sehgal, A. (2021). Short and long sleeping mutants reveal links between sleep and macroautophagy. eLife, 10, e64140. 10.7554/eLife.64140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh, D. J. , Geratowski, J. D. , Wooltorton, J. R. , Spaethling, J. M. , Hector, C. E. , Zheng, X. , Johnson, E. C. , Eberwine, J. H. , & Sehgal, A. (2014). Identification of a circadian output circuit for rest: Activity rhythms in Drosophila. Cell, 157(3), 689–701. 10.1016/j.cell.2014.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey, M. , Collins, B. , Bertet, C. , & Blau, J. (2016). Circadian rhythms in neuronal activity propagate through output circuits. Nature Neuroscience, 19(4), 587–595. 10.1038/nn.4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani, M. F. , Darlington, T. K. , Staknis, D. , Más, P. , Petti, A. A. , Weitz, C. J. , & Kay, S. A. (1999). Light‐dependent sequestration of TIMELESS by CRYPTOCHROME. Science (New York, N.Y.), 285(5427), 553–556. 10.1126/science.285.5427.553 [DOI] [PubMed] [Google Scholar]
- Chiu, J. C. , Vanselow, J. T. , Kramer, A. , & Edery, I. (2008). The phospho‐occupancy of an atypical SLIMB‐binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes & Development, 22(13), 1758–1772. 10.1101/gad.1682708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. J. , Ryu, K. J. , Baek, K. E. , Lim, J. , Kim, T. , Song, C. Y. , Yoo, J. , & Lee, H. G. (2020). Cullin 3/KCTD5 promotes the ubiqutination of rho guanine nucleotide dissociation inhibitor 1 and regulates its stability. Journal of Microbiology and Biotechnology, 30(10), 1488–1494. 10.4014/jmb.2007.07033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli, C. , Bushey, D. , Hill, S. , Huber, R. , Kreber, R. , Ganetzky, B. , & Tononi, G. (2005). Reduced sleep in Drosophila Shaker mutants. Nature, 434(7037), 1087–1092. 10.1038/nature03486 [DOI] [PubMed] [Google Scholar]
- Deboer, T. (2018). Sleep homeostasis and the circadian clock: Do the circadian pacemaker and the sleep homeostat influence each other's functioning? Neurobiology of Sleep and Circadian Rhythms, 5, 68–77. 10.1016/j.nbscr.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, M. , & Shen, K. (2008). The role of the ubiquitin proteasome system in synapse remodeling and neurodegenerative diseases. BioEssays, 30(11–12), 1075–1083. 10.1002/bies.20843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel, S. , Melnattur, K. , & Shaw, P. J. (2015). Sleep, performance, and memory in flies. Current Sleep Medicine Reports, 1(1), 47–54. 10.1007/s40675-014-0006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J. , Zhang, J. , He, T. , Li, Y. , Su, Y. , Tie, F. , Liu, M. , Harte, P. J. , & Zhu, A. J. (2016). Stuxnet facilitates the degradation of polycomb protein during development. Developmental Cell, 37(6), 507–519. 10.1016/j.devcel.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J. , Zhang, J. , Su, Y. , Liu, M. , Ospina, J. K. , Yang, S. , & Zhu, A. J. (2011). In vivo RNAi screen reveals neddylation genes as novel regulators of hedgehog signaling. PLoS One, 6(9), e24168. 10.1371/journal.pone.0024168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy, C. , & Sehgal, A. (2017). Circadian rhythms and sleep in Drosophila melanogaster . Genetics, 205(4), 1373–1397. 10.1534/genetics.115.185157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, A. A. , Mandilaras, K. , Missirlis, F. , & Sanyal, S. (2013). An emerging role for Cullin‐3 mediated ubiquitination in sleep and circadian rhythm: Insights from Drosophila . Fly, 7(1), 39–43. 10.4161/fly.23506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilestro, G. F. , Tononi, G. , & Cirelli, C. (2009). Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila . Science (New York, N.Y.), 324(5923), 109–112. 10.1126/science.1166673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M. H. , & Ciechanover, A. (2002). The ubiquitin‐proteasome proteolytic pathway: Destruction for the sake of construction. Physiological Reviews, 82(2), 373–428. 10.1152/physrev.00027.2001 [DOI] [PubMed] [Google Scholar]
- Grima, B. , Chélot, E. , Xia, R. , & Rouyer, F. (2004). Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature, 431(7010), 869–873. 10.1038/nature02935 [DOI] [PubMed] [Google Scholar]
- Grima, B. , Dognon, A. , Lamouroux, A. , Chélot, E. , & Rouyer, F. (2012). CULLIN‐3 controls TIMELESS oscillations in the Drosophila circadian clock. PLoS Biology, 10(8), e1001367. 10.1371/journal.pbio.1001367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima, B. , Lamouroux, A. , Chélot, E. , Papin, C. , Limbourg‐Bouchon, B. , & Rouyer, F. (2002). The F‐box protein slimb controls the levels of clock proteins period and timeless. Nature, 420(6912), 178–182. 10.1038/nature01122 [DOI] [PubMed] [Google Scholar]
- Guo, F. , Cerullo, I. , Chen, X. , & Rosbash, M. (2014). PDF neuron firing phase‐shifts key circadian activity neurons in Drosophila . eLife, 3, e02780. 10.7554/eLife.02780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, H. , Peng, Y. , Fan, S. , Chen, Y. , Zheng, X. , & Li, C. (2018). Cullin3/KCTD5 induces monoubiquitination of ΔNp63α and impairs its activity. FEBS Letters, 592(13), 2334–2340. 10.1002/1873-3468.13104 [DOI] [PubMed] [Google Scholar]
- Helfrich‐Förster, C. (1997). Development of pigment‐dispersing hormone‐immunoreactive neurons in the nervous system of Drosophila melanogaster. The Journal of Comparative Neurology, 380(3), 335–354. [DOI] [PubMed] [Google Scholar]
- Helfrich‐Förster, C. (2003). The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microscopy Research and Technique, 62(2), 94–102. 10.1002/jemt.10357 [DOI] [PubMed] [Google Scholar]
- Hendricks, J. C. , Finn, S. M. , Panckeri, K. A. , Chavkin, J. , Williams, J. A. , Sehgal, A. , & Pack, A. I. (2000). Rest in Drosophila is a sleep‐like state. Neuron, 25(1), 129–138. 10.1016/s0896-6273(00)80877-6 [DOI] [PubMed] [Google Scholar]
- Hendricks, J. C. , Sehgal, A. , & Pack, A. I. (2000). The need for a simple animal model to understand sleep. Progress in Neurobiology, 61(4), 339–351. 10.1016/s0301-0082(99)00048-9 [DOI] [PubMed] [Google Scholar]
- Herzog, E. D. (2007). Neurons and networks in daily rhythms. Nature Reviews. Neuroscience, 8(10), 790–802. 10.1038/nrn2215 [DOI] [PubMed] [Google Scholar]
- Hill, V. M. , O'Connor, R. M. , Sissoko, G. B. , Irobunda, I. S. , Leong, S. , Canman, J. C. , Stavropoulos, N. , & Shirasu‐Hiza, M. (2018). A bidirectional relationship between sleep and oxidative stress in Drosophila . PLoS Biology, 16(7), e2005206. 10.1371/journal.pbio.2005206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houl, J. H. , Yu, W. , Dudek, S. M. , & Hardin, P. E. (2006). Drosophila CLOCK is constitutively expressed in circadian oscillator and non‐oscillator cells. Journal of Biological Rhythms, 21(2), 93–103. 10.1177/0748730405283697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. , Piao, C. , Beuschel, C. B. , Götz, T. , & Sigrist, S. J. (2020). Presynaptic active zone plasticity encodes sleep need in Drosophila . Current Biology, 30(6), 1077–1091.e5. 10.1016/j.cub.2020.01.019 [DOI] [PubMed] [Google Scholar]
- Huber, R. , Hill, S. L. , Holladay, C. , Biesiadecki, M. , Tononi, G. , & Cirelli, C. (2004). Sleep homeostasis in Drosophila melanogaster. Sleep, 27(4), 628–639. 10.1093/sleep/27.4.628 [DOI] [PubMed] [Google Scholar]
- Kaneko, M. , Helfrich‐Förster, C. , & Hall, J. C. (1997). Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: Newly identified pacemaker candidates and novel features of clock gene product cycling. The Journal of Neuroscience, 17(17), 6745–6760. 10.1523/JNEUROSCI.17-17-06745.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss, B. , Price, J. L. , Saez, L. , Blau, J. , Rothenfluh, A. , Wesley, C. S. , & Young, M. W. (1998). The Drosophila clock gene double‐time encodes a protein closely related to human casein kinase Iε. Cell, 94(1), 97–107. 10.1016/s0092-8674(00)81225-8 [DOI] [PubMed] [Google Scholar]
- Koh, K. , Zheng, X. , & Sehgal, A. (2006). JETLAG resets the Drosophila circadian clock by promoting light‐induced degradation of TIMELESS. Science (New York, N.Y.), 312(5781), 1809–1812. 10.1126/science.1124951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka, R. J. , & Benzer, S. (1971). Clock mutants of Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America, 68(9), 2112–2116. 10.1073/pnas.68.9.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaze, A. , Lamouroux, A. , Vias, C. , Hung, H. C. , Weber, F. , & Rouyer, F. (2011). The E3 ubiquitin ligase CTRIP controls CLOCK levels and PERIOD oscillations in Drosophila . EMBO Reports, 12(6), 549–557. 10.1038/embor.2011.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba, P. , Bilodeau‐Wentworth, D. , Emery, P. , & Zhang, Y. (2014). Morning and evening oscillators cooperate to reset circadian behavior in response to light input. Cell Reports, 7(3), 601–608. 10.1016/j.celrep.2014.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. , Bae, K. , & Edery, I. (1999). PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK‐CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: A basis for circadian transcription. Molecular and Cellular Biology, 19(8), 5316–5325. 10.1128/MCB.19.8.5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leestemaker, Y. , & Ovaa, H. (2017). Tools to investigate the ubiquitin proteasome system. Drug Discovery Today: Technologies, 26, 25–31. 10.1016/j.ddtec.2017.11.006 [DOI] [PubMed] [Google Scholar]
- Li, Q. , Kellner, D. A. , Hatch, H. , Yumita, T. , Sanchez, S. , Machold, R. P. , Frank, C. A. , & Stavropoulos, N. (2017). Conserved properties of Drosophila insomniac link sleep regulation and synaptic function. PLoS Genetics, 13(5), e1006815. 10.1371/journal.pgen.1006815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Bengtson, M. H. , Ulbrich, A. , Matsuda, A. , Reddy, V. A. , Orth, A. , Chanda, S. K. , Batalov, S. , & Joazeiro, C. A. (2008). Genome‐wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One, 3(1), e1487. 10.1371/journal.pone.0001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. M. , Kilman, V. L. , Keegan, K. , Paddock, B. , Emery‐Le, M. , Rosbash, M. , & Allada, R. (2002). A role for casein kinase 2alpha in the Drosophila circadian clock. Nature, 420(6917), 816–820. 10.1038/nature01235 [DOI] [PubMed] [Google Scholar]
- Lin, J. M. , Schroeder, A. , & Allada, R. (2005). In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila period. The Journal of Neuroscience, 25(48), 11175–11183. 10.1523/JNEUROSCI.2159-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Liu, Q. , Tabuchi, M. , & Wu, M. N. (2016). Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell, 165(6), 1347–1360. 10.1016/j.cell.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Mahesh, G. , Yu, W. , & Hardin, P. E. (2017). CLOCK stabilizes CYCLE to initiate clock function in Drosophila . Proceedings of the National Academy of Sciences of the United States of America, 114(41), 10972–10977. 10.1073/pnas.1707143114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire, S. E. , Rhoades, S. , Chen, W. F. , Sengupta, A. , Yue, Z. , Lim, J. C. , Mitchell, C. H. , Weljie, A. M. , & Sehgal, A. (2015). Independent effects of γ‐aminobutyric acid transaminase (GABAT) on metabolic and sleep homeostasis. The Journal of Biological Chemistry, 290(33), 20407–20416. 10.1074/jbc.M114.602276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson, M. P. (1994). Calcium and neuronal injury in Alzheimer's disease. Contributions of beta‐amyloid precursor protein mismetabolism, free radicals, and metabolic compromise. Annals of the New York Academy of Sciences, 747, 50–76. [PubMed] [Google Scholar]
- Morreale, F. E. , & Walden, H. (2016). Types of ubiquitin ligases. Cell, 165(1), 248–248.e1. 10.1016/j.cell.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Nath, D. , & Shadan, S. (2009). The ubiquitin system. Nature, 458(7237), 421. 10.1038/458421a [DOI] [PubMed] [Google Scholar]
- Omoto, J. J. , Keleş, M. F. , Nguyen, B. M. , Bolanos, C. , Lovick, J. K. , Frye, M. A. , & Hartenstein, V. (2017). Visual input to the Drosophila central complex by developmentally and functionally distinct neuronal populations. Current Biology, 27(8), 1098–1110. 10.1016/j.cub.2017.02.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk, N. , Selby, C. P. , Annayev, Y. , Zhong, D. , & Sancar, A. (2011). Reaction mechanism of Drosophila cryptochrome. Proceedings of the National Academy of Sciences of the United States of America, 108(2), 516–521. 10.1073/pnas.1017093108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patke, A. , Young, M. W. , & Axelrod, S. (2020). Molecular mechanisms and physiological importance of circadian rhythms. Nature Reviews. Molecular Cell Biology, 21(2), 67–84. 10.1038/s41580-019-0179-2 [DOI] [PubMed] [Google Scholar]
- Petroski, M. D. , & Deshaies, R. J. (2005). Function and regulation of cullin‐RING ubiquitin ligases. Nature Reviews. Molecular Cell Biology, 6(1), 9–20. 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Pfeiffenberger, C. , & Allada, R. (2012). Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila . PLoS Genetics, 8(10), e1003003. 10.1371/journal.pgen.1003003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirone, L. , Smaldone, G. , Esposito, C. , Balasco, N. , Petoukhov, M. V. , Spilotros, A. , Svergun, D. I. , Di Gaetano, S. , Vitagliano, L. , & Pedone, E. M. (2016). Proteins involved in sleep homeostasis: Biophysical characterization of INC and its partners. Biochimie, 131, 106–114. 10.1016/j.biochi.2016.09.013 [DOI] [PubMed] [Google Scholar]
- Price, J. L. , Blau, J. , Rothenfluh, A. , Abodeely, M. , Kloss, B. , & Young, M. W. (1998). Double‐time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell, 94(1), 83–95. 10.1016/s0092-8674(00)81224-6 [DOI] [PubMed] [Google Scholar]
- Raccuglia, D. , Huang, S. , Ender, A. , Heim, M. M. , Laber, D. , Suárez‐Grimalt, R. , Liotta, A. , Sigrist, S. J. , Geiger, J. , & Owald, D. (2019). Network‐specific synchronization of electrical slow‐wave oscillations regulates sleep drive in Drosophila . Current Biology, 29(21), 3611–3621. 10.1016/j.cub.2019.08.070 [DOI] [PubMed] [Google Scholar]
- Renn, S. C. , Armstrong, J. D. , Yang, M. , Wang, Z. , An, X. , Kaiser, K. , & Taghert, P. H. (1999). Genetic analysis of the Drosophila ellipsoid body neuropil: Organization and development of the central complex. Journal of Neurobiology, 41(2), 189–207. [PubMed] [Google Scholar]
- Renn, S. C. , Park, J. H. , Rosbash, M. , Hall, J. C. , & Taghert, P. H. (1999). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila . Cell, 99(7), 791–802. 10.1016/s0092-8674(00)81676-1 [DOI] [PubMed] [Google Scholar]
- Rieger, D. , Shafer, O. T. , Tomioka, K. , & Helfrich‐Förster, C. (2006). Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. The Journal of Neuroscience, 26(9), 2531–2543. 10.1523/JNEUROSCI.1234-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin, D. , & Kumar, S. (2009). Physiological functions of the HECT family of ubiquitin ligases. Nature Reviews. Molecular Cell Biology, 10(6), 398–409. 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- Rutila, J. E. , Suri, V. , Le, M. , So, W. V. , Rosbash, M. , & Hall, J. C. (1998). CYCLE is a second bHLH‐PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell, 93(5), 805–814. 10.1016/s0092-8674(00)81441-5 [DOI] [PubMed] [Google Scholar]
- Rutz, N. , Heilbronn, R. , & Weger, S. (2015). Interactions of cullin3/KCTD5 complexes with both cytoplasmic and nuclear proteins: Evidence for a role in protein stabilization. Biochemical and Biophysical Research Communications, 464(3), 922–928. 10.1016/j.bbrc.2015.07.069 [DOI] [PubMed] [Google Scholar]
- Sehgal, A. , Price, J. L. , Man, B. , & Young, M. W. (1994). Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science (New York, N.Y.), 263(5153), 1603–1606. 10.1126/science.8128246 [DOI] [PubMed] [Google Scholar]
- Sharma, A. , Lee, S. , Kim, H. , Yoon, H. , Ha, S. , & Kang, S. U. (2020). Molecular crosstalk between circadian rhythmicity and the development of neurodegenerative disorders. Frontiers in Neuroscience, 14, 844. 10.3389/fnins.2020.00844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, P. J. , Cirelli, C. , Greenspan, R. J. , & Tononi, G. (2000). Correlates of sleep and waking in Drosophila melanogaster. Science (New York, N.Y.), 287(5459), 1834–1837. 10.1126/science.287.5459.1834 [DOI] [PubMed] [Google Scholar]
- Shi, M. , Yue, Z. , Kuryatov, A. , Lindstrom, J. M. , & Sehgal, A. (2014). Identification of Redeye, a new sleep‐regulating protein whose expression is modulated by sleep amount. eLife, 3, e01473. 10.7554/eLife.01473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy, T. A. , Smith, P. G. , Milhollen, M. A. , Berger, A. J. , Gavin, J. M. , Adhikari, S. , Brownell, J. E. , Burke, K. E. , Cardin, D. P. , Critchley, S. , Cullis, C. A. , Doucette, A. , Garnsey, J. J. , Gaulin, J. L. , Gershman, R. E. , Lublinsky, A. R. , McDonald, A. , Mizutani, H. , Narayanan, U. , … Langston, S. P. (2009). An inhibitor of NEDD8‐activating enzyme as a new approach to treat cancer. Nature, 458(7239), 732–736. 10.1038/nature07884 [DOI] [PubMed] [Google Scholar]
- Stavropoulos, N. , & Young, M. W. (2011). Insomniac and Cullin‐3 regulate sleep and wakefulness in Drosophila . Neuron, 72(6), 964–976. 10.1016/j.neuron.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojkovic, K. , Wing, S. S. , & Cermakian, N. (2014). A central role for ubiquitination within a circadian clock protein modification code. Frontiers in Molecular Neuroscience, 7, 69. 10.3389/fnmol.2014.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru, D. , Peng, Y. , Agosto, J. , & Rosbash, M. (2004). Coupled oscillators control morning and evening locomotor behaviour of Drosophila . Nature, 431(7010), 862–868. 10.1038/nature02926 [DOI] [PubMed] [Google Scholar]
- Szabó, Á. , Papin, C. , Cornu, D. , Chélot, E. , Lipinszki, Z. , Udvardy, A. , Redeker, V. , Mayor, U. , & Rouyer, F. (2018). Ubiquitylation dynamics of the clock cell proteome and TIMELESS during a circadian cycle. Cell Reports, 23(8), 2273–2282. 10.1016/j.celrep.2018.04.064 [DOI] [PubMed] [Google Scholar]
- Thimgan, M. S. , Seugnet, L. , Turk, J. , & Shaw, P. J. (2015). Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila . Sleep, 38(5), 801–814. 10.5665/sleep.4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, H. , Williams, J. A. , Gulledge, M. , & Sehgal, A. (2019). A sleep‐inducing gene, nemuri, links sleep and immune function in Drosophila . Science (New York, N.Y.), 363(6426), 509–515. 10.1126/science.aat1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top, D. , Harms, E. , Syed, S. , Adams, E. L. , & Saez, L. (2016). GSK‐3 and CK2 kinases converge on timeless to regulate the master clock. Cell Reports, 16(2), 357–367. 10.1016/j.celrep.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro, A. , Kaplan Dor, Y. , Nambara, K. , Pollina, E. A. , Lin, C. , Greenberg, M. E. , & Rogulja, D. (2020). Sleep loss can cause death through accumulation of reactive oxygen species in the gut. Cell, 181(6), 1307–1328.e15. 10.1016/j.cell.2020.04.049 [DOI] [PubMed] [Google Scholar]
- Vaidya, A. T. , Top, D. , Manahan, C. C. , Tokuda, J. M. , Zhang, S. , Pollack, L. , Young, M. W. , & Crane, B. R. (2013). Flavin reduction activates Drosophila cryptochrome. Proceedings of the National Academy of Sciences of the United States of America, 110(51), 20455–20460. 10.1073/pnas.1313336110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky, A. (2012). Augmented generation of protein fragments during wakefulness as the molecular cause of sleep: A hypothesis. Protein Science, 21(11), 1634–1661. 10.1002/pro.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky, A. (2019a). N‐degron and C‐degron pathways of protein degradation. Proceedings of the National Academy of Sciences of the United States of America, 116(2), 358–366. 10.1073/pnas.1816596116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky, A. (2019b). On the cause of sleep: Protein fragments, the concept of sentinels, and links to epilepsy. Proceedings of the National Academy of Sciences of the United States of America, 116(22), 10773–10782. 10.1073/pnas.1904709116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend, J. , & Reiter, R. J. (2015). Melatonin feedback on clock genes: A theory involving the proteasome. Journal of Pineal Research, 58(1), 1–11. 10.1111/jpi.12189 [DOI] [PubMed] [Google Scholar]
- Xie, L. , Kang, H. , Xu, Q. , Chen, M. J. , Liao, Y. , Thiyagarajan, M. , O'Donnell, J. , Christensen, D. J. , Nicholson, C. , Iliff, J. J. , Takano, T. , Deane, R. , & Nedergaard, M. (2013). Sleep drives metabolite clearance from the adult brain. Science (New York, N.Y.), 342(6156), 373–377. 10.1126/science.1241224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S. H. , Mohawk, J. A. , Siepka, S. M. , Shan, Y. , Huh, S. K. , Hong, H. K. , Kornblum, I. , Kumar, V. , Koike, N. , Xu, M. , Nussbaum, J. , Liu, X. , Chen, Z. , Chen, Z. J. , Green, C. B. , & Takahashi, J. S. (2013). Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell, 152(5), 1091–1105. 10.1016/j.cell.2013.01.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Chung, B. Y. , Lear, B. C. , Kilman, V. L. , Liu, Y. , Mahesh, G. , Meissner, R. A. , Hardin, P. E. , & Allada, R. (2010). DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila . Current Biology, 20(7), 591–599. 10.1016/j.cub.2010.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, Y. , Bilodeau‐Wentworth, D. , Hardin, P. E. , & Emery, P. (2010). Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Current Biology, 20(7), 600–605. 10.1016/j.cub.2010.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z. , Zhao, X. , He, T. , Wu, X. , Lv, P. , Zhu, A. J. , & Du, J. (2021). Epigenetic regulator Stuxnet modulates octopamine effect on sleep through a Stuxnet‐Polycomb‐Octβ2R cascade. EMBO Reports, 22(2), e47910. 10.15252/embr.201947910 [DOI] [PMC free article] [PubMed] [Google Scholar]


