Summary
Indole‐3‐acetic acid (IAA) controls a plethora of developmental processes. Thus, regulation of its concentration is of great relevance for plant performance. Cellular IAA concentration depends on its transport, biosynthesis and the various pathways for IAA inactivation, including oxidation and conjugation.
Group II members of the GRETCHEN HAGEN 3 (GH3) gene family code for acyl acid amido synthetases catalysing the conjugation of IAA to amino acids. However, the high degree of functional redundancy among them has hampered thorough analysis of their roles in plant development.
In this work, we generated an Arabidopsis gh3.1,2,3,4,5,6,9,17 (gh3oct) mutant to knock out the group II GH3 pathway. The gh3oct plants had an elaborated root architecture, showed an increased tolerance to different osmotic stresses, including an IAA‐dependent tolerance to salinity, and were more tolerant to water deficit. Indole‐3‐acetic acid metabolite quantification in gh3oct plants suggested the existence of additional GH3‐like enzymes in IAA metabolism. Moreover, our data suggested that 2‐oxindole‐3‐acetic acid production depends, at least in part, on the GH3 pathway. Targeted stress‐hormone analysis further suggested involvement of abscisic acid in the differential response to salinity of gh3oct plants.
Taken together, our data provide new insights into the roles of group II GH3s in IAA metabolism and hormone‐regulated plant development.
Keywords: Arabidopsis, auxin, drought, GH3, salinity, stress tolerance
Introduction
Regulation of the concentrations and distribution of the major auxin indole‐3‐acetic acid (IAA) is of pivotal importance for plant development because it regulates multiple processes, including embryo, seed, root and leaf development, meristem maintenance, shoot branching and responses to environmental stresses (Casanova‐Saez & Voss, 2019; Leftley et al., 2021). Spatiotemporal fluctuations of IAA concentrations in response to external and internal stimuli drive plant developmental responses by triggering signalling cascades that result in modulation of cell division, differentiation and growth (Gallei et al., 2020).
Control of IAA concentrations and distribution within plant cells and tissues are achieved by intercellular and intracellular transport mechanisms (Band, 2021; Hammes et al., 2022) and spatiotemporally regulated IAA biosynthesis and inactivation (Zhao, 2018; Casanova‐Saez et al., 2021). IAA metabolic inactivation primarily involves oxidation processes and the formation of IAA‐sugar and IAA‐amino acid (IAA‐aa) conjugates (Casanova‐Saez et al., 2021). IAA oxidation is facilitated by the angiosperm‐related DIOXYGENASE FOR AUXIN OXIDATION (DAO) proteins (Zhao et al., 2013; Porco et al., 2016; Zhang et al., 2016; Takehara et al., 2020). The formation of 2‐oxindole‐3‐acetic acid (oxIAA) is an irreversible IAA inactivation process that operates to remove excess auxin, as inferred from higher oxIAA formation detected at sites of IAA maxima (Pencik et al., 2013). Formation of ester‐linked IAA conjugates with glucose (IAA‐glc) is a reversible reaction known to regulate IAA concentrations and homeostasis. Several members of the UDP‐glucosyltransferase superfamily have been shown to mediate the conjugation of both IAA and oxIAA to glucose in vivo (Mateo‐Bonmatí et al., 2021).
Indole‐3‐acetic acid is also inactivated by conjugation to amino acids via amide bonds, a reaction catalysed by members of the GRETCHEN HAGEN 3 (GH3) family of acyl acid amido synthetases (Staswick et al., 2005). Most of these conjugates, including IAA‐leucine, IAA‐alanine and IAA‐phenylalanine, can be hydrolysed to free IAA, and therefore serve as IAA storage forms (LeClere et al., 2002; Rampey et al., 2004). However, IAA irreversibly conjugates to aspartate (IAA‐Asp) and glutamate (IAA‐Glu) during catabolism (Ostin et al., 1998; Rampey et al., 2004). IAA‐Asp and IAA‐Glu catabolites have been found at higher concentrations compared with reversible IAA‐aa conjugates in different plant species (Kowalczyk & Sandberg, 2001; Kojima et al., 2009; Pencik et al., 2009). Several IAA‐Asp metabolites, including 6‐OH‐IAA‐Asp and oxIAA‐Asp, have been identified in the last few decades (Ostin et al., 1998; Kai et al., 2007). Recently, the IAA oxidase DAO1 was found to additionally operate in the GH3 pathway by mediating the formation of oxIAA‐Asp from IAA‐Asp in tobacco and Arabidopsis (Müller et al., 2021).
Based on sequence homology and substrate specificity studies, 19 Arabidopsis GH3 proteins have been classified into three functionally diverse groups, of which group II consists of eight GH3 members with the ability to mediate the conjugation of IAA to amino acids (i.e. GH3.1, GH3.2, GH3.3, GH3.4, GH3.5, GH3.6, GH3.9 and GH3.17) (Staswick et al., 2002, 2005). These group II GH3s are conserved across land plants (Terol et al., 2006).
Even though all Arabidopsis group II GH3s can conjugate IAA to different amino acids, substrate preferences exist among them. For instance, GH3.17 preferentially conjugates IAA to Glu, GH3.3 and GH3.4 preferentially conjugate IAA to Asp, and GH3.2, GH3.5 and GH3.6 show a similar predilection for Asp and Glu (Staswick et al., 2005; Brunoni et al., 2019). A range of amino acid substrate specificities has also been documented for GH3 orthologues in other plant species (e.g. Physcomitrella patens (Ludwig‐Muller et al., 2009), Vitis vinifera (Bottcher et al., 2011) and Picea abies (Brunoni et al., 2020)). Group II GH3s are also known to be promiscuous regarding the acyl acid substrate. GH3.3, GH3.5 and GH3.6 have been found to modulate jasmonic acid homeostasis by mediating its conjugation to amino acids (Gutierrez et al., 2012). GH3.5 has been further shown to mediate the conjugation of the auxin phenylacetic acid, and the benzoates salicylic acid and benzoic acid (Westfall et al., 2016).
Although specific roles have been reported for individual group II GH3 members (Khan & Stone, 2007; Park et al., 2007; Du et al., 2012; Zheng et al., 2016; Di Mambro et al., 2017; Kirungu et al., 2019), their overlapping expression domains and functions in IAA inactivation have probably masked additional roles of these GH3s in IAA metabolism and plant development. In the present study, we generated a group II GH3 octuple knockout to bypass the redundancy in GH3‐mediated IAA metabolism. The mutant plants showed pleiotropic high‐auxin‐related phenotypes, such as longer hypocotyls and petioles, as well as a more branched root system with no apparent penalty on primary root growth. Phenotypic, physiological and targeted‐hormonomic analyses further indicated that group II GH3s redundantly modulate IAA‐dependent salinity tolerance and that they collectively participate in the response to water deficit. We also obtained evidence supporting a connection between the GH3 pathway and oxIAA production. Finally, our data suggested that additional GH3 or GH3‐like genes might operate in IAA conjugation to amino acids.
Materials and Methods
Plant material and culture conditions
Seeds from the Arabidopsis thaliana (L.) Heynh. accessions Columbia‐0 (Col‐0) and Landsberg erecta (Ler) as well as from the gh3 insertional lines (Supporting Information Table S1) were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info) and from the Arabidopsis Biological Resource Center (https://abrc.osu.edu). The presence and position of all insertions were confirmed by PCR amplification using gene‐specific primers together with the insertion‐specific primers Ds5′‐1 (for SGT lines), 3′‐dSpm (for SM lines) and LBb1.3 (for SALK lines) (see Tables S1, S2).
Arabidopsis seeds were surface‐sterilized with a bleach solution (40% v/v commercial bleach in dH2O and 0.002% Triton X‐100) for 8 min and then washed four times with sterile deionized water. Seeds were stratified for a minimum of 2 d and then sown under sterile conditions on square Petri dishes containing half‐strength Murashige & Skoog salt mixture (M0221; Duchefa Biochemie, Haarlem, the Netherlands), 1% sucrose, 0.05% MES hydrate (M2933; Sigma) and 0.8% plant agar (P1001; Duchefa Biochemie) with the pH adjusted to 5.7 with potassium hydroxide. Flowering plants were grown in pots containing a 3 : 1 mixture of organic soil and vermiculite. All plants were grown, unless otherwise stated, under long‐day conditions (16 h : 8 h, light : dark) at 22 ± 1°C under cool white fluorescent light (150 μmol photons m−2 s−1). Flowering time was recorded as the number of rosette leaves at bolting under long‐ and short‐day (8 h : 16 h, light : dark) conditions.
Chemical treatments and drought experiments
Salinity and osmotic treatments were performed on half‐strength Murashige & Skoog (MS) plates supplemented with sodium chloride, d‐sorbitol or d‐mannitol before autoclaving. The inhibitors of YUCCA‐mediated IAA biosynthesis, yucasin (5‐(4‐chlorophenyl)‐4H‐1,2,4‐triazole‐3‐thiol) and 4‐phenoxyphenylboronic acid (PPBo), kindly provided by Prof. Ken‐ichiro Hayashi, were prepared in dimethyl sulfoxide stock solutions. IAA treatments were performed on MS plates supplemented with IAA from a stock solution in absolute ethanol. Yucasin, PPBo and IAA were added to the autoclaved MS media before pouring into plates. To mimic drought conditions in vitro, we prepared half‐strength MS plates with a reduced water potential as described previously (Verslues et al., 2006). Solidified MS medium was perfused with an overlay solution containing poly(ethylene glycol) (PEG8000; P2139; Sigma). Control plates were perfused with an overlay MS solution lacking PEG8000. For water‐deficit treatment on soil‐grown plants, stratified seeds were planted in pots containing 40 g of soil mixture. Initially, 1 l of water was added to each tray. Irrigation was then withheld until both gh3oct and wild‐type plants displayed dryness symptoms. The weight of the pots was recorded to estimate and compare the water loss in the pots for each genotype.
Genome‐wide identification of T‐DNA and transposon insertions
Around 14 µg of nuclear‐enriched DNA was purified from 1 g of gh3oct seedlings using a previously described protocol (Hanania et al., 2004). Whole‐genome sequencing of the sample was performed at BGI Hong Kong using a BGISEQ‐500 sequencing platform. A total of 47 million 150‐bp‐long reads were obtained, reaching a 59× genome depth. Trimmed Fastq files were used to map the different insertions using Easymap software (Lup et al., 2021). Raw reads were deposited in the short read archive (SRA) with the code SRX8771538.
Gene expression analyses
For expression analyses, RNA was isolated using the Total RNA Purification Kit (Norgen, Thorold, ON, Canda). DNA was removed using the RNase‐Free DNase I Kit (Norgen). First‐strand cDNA synthesis was performed with the iScript cDNA Synthesis Kit (Bio‐Rad). ACTIN2 gene was used as an internal control for relative expression quantification. Four biological replicates (each being a pool of several plants) were analysed in triplicate. Quantitative PCR (qPCR) reactions were performed in 10 µl reactions containing 4 µl of LightCycler 480 SYBR Green I Master (Roche), 4 µl of PCR‐grade water (Roche), 1 µl of the corresponding primer pair (10 µM each) and 1 µl of the cDNA template. The primers used are listed in Table S2. Quantification of relative gene expression was performed using the comparative C T method (2−ΔΔCt) (Schmittgen & Livak, 2008) on a CFX384 Touch Real‐Time PCR Detection System (Bio‐Rad).
Quantification of IAA and IAA metabolites
Extraction and purification of the targeted compounds (IAA, oxIAA, IAA‐Asp, IAA‐Glu, IAA‐glc, oxIAA‐glc) were performed according to Novak et al. (2012). Briefly, 10 mg of frozen material per sample was homogenized using a bead mill (27 Hz, 10 min, 4°C; MixerMill, Retsch GmbH, Haan, Germany) and extracted in 1 ml of 50 mM sodium phosphate buffer containing 0.1% sodium diethyldithiocarbamate and a mixture of 13C6 isotopically labelled internal standards (Olchemim, Olomouc, Czech Republic). After centrifugation (20 000 g , 15 min, 4°C), the supernatant was transferred into new Eppendorf tubes. The pH was then adjusted to 2.5 with 1 M HCl and samples were immediately applied to preconditioned solid‐phase extraction columns (Oasis HLB, 30 mg of 1 ml; Waters Inc., Milford, MA, USA). After sample application, each column was rinsed with 2 ml 5% methanol. Compounds of interest were subsequently eluted with 2 ml of 80% methanol. Ultrahigh‐performance liquid chromatography (UHPLC)‐MS/MS analysis was performed according to the method described in Pěnčík et al. (2018), using an LC‐MS/MS system consisting of a 1290 Infinity Binary LC System coupled to a 6490 Triple Quad LC/MS System with Jet Stream and Dual Ion Funnel technologies (Agilent Technologies, Santa Clara, CA, USA). The quantification was carried out in Agilent MassHunter Workstation quantitative analysis software (Agilent Technologies).
Quantification of jasmonic acid (JA), JA‐isoleucine (JA‐Ile), salicylic acid (SA) and abscisic acid (ABA)
Samples were extracted, purified and analysed according to a method described in Simura et al. (2018). Briefly, c. 20 mg of frozen material per sample was homogenized and extracted in 1 ml of ice‐cold 50% aqueous acetonitrile (v/v) containing a mixture of [13C], [15N] or [2H] isotopically labelled internal standards using a bead mill (27 Hz, 10 min, 4°C; MixerMill, Retsch GmbH, Haan, Germany) and sonicator (3 min, 4°C; Ultrasonic bath P 310 H; Elma, Pforzheim, Germany). After centrifugation (20 000 g , 15 min, 4°C), the supernatant was purified as follows. A solid‐phase extraction column Oasis HLB (30 mg 1 cc; Waters) was conditioned with 1 ml of 100% methanol and 1 ml of deionized water (Milli‐Q; Merck Millipore, Burlington, MA, USA). After the conditioning steps, each sample was loaded onto a solid phase extraction column and the flow‐through fraction was collected as well as the 1 ml 30% aqueous acetonitrile (v/v) elution fraction. Samples were evaporated to dryness using a SpeedVac SPD111V (Thermo Scientific, Waltham, MA, USA). Prior to LC‐MS analysis, samples were dissolved in 40 µl of 30% acetonitrile (v/v) and transferred to insert‐equipped vials. Mass spectrometry analysis of targeted compounds was performed using an UHPLC‐electrospray ionization (ESI)‐MS/MS system comprising a 1290 Infinity Binary LC system coupled to a 6490 Triple Quad LC/MS system with Jet Stream and Dual Ion Funnel technologies (Agilent Technologies). The quantification was carried out in Agilent MassHunter Workstation software (Agilent Technologies).
Phenotypic and statistical analyses
For root and hypocotyl phenotyping, vertically grown plates were imaged using Epson Perfection V600 photo scanners. For determination of petiole length, horizontally grown plates were photographed from above. Lengths were measured from scaled images using Fiji software (Schindelin et al., 2012).
Statistically significant differences between mean values from the wild‐type and mutants were analysed using a two‐tailed Student's t‐test. Multiple comparisons of genotypes, treatments and times were performed by one‐way ANOVA followed by Tukey's post hoc test to a 95% confidence level. Analyses were carried out in GraphPad Prism 6.01.
Accession numbers
These were as follows: ACTIN2 (At3g18780), GH3.1 (At2g14960), GH3.2 (At4g37390), GH3.3 (At2g23170), GH3.4 (At1g59500), GH3.5 (At4g27260), GH3.6 (At5g54510), GH3.9 (At2g47750), GH3.17 (At1g28130).
Results
Arabidopsis group II GH3s are required for vegetative and reproductive development
Group II GH3 enzymes (GH3.1, GH3.2, GH3.3, GH3.4, GH3.5, GH3.6, GH3.9 and GH3.17) are known to modulate IAA concentrations redundantly by catalysing the formation of IAA‐aa conjugates, which are inactive IAA forms (Staswick et al., 2005; Chen et al., 2010; Porco et al., 2016). Owing to functional redundancy, single gh3 mutants often show mild or no phenotypes (Staswick et al., 2005). To bypass such redundancy, we generated a gh3.1,2,3,4,5,6,9,17 octuple mutant (hereafter referred to as gh3oct) by crossing the individual insertional lines (Fig. S1a; Table S1). To verify that gh3oct was an octuple knockout, we performed reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis. We showed that expression of the full‐length GH3 transcript was completely abolished by the insertions in the gh3oct mutant, with just marginal expression of GH3.9 in reproductive tissues (Fig. S1b). Because the average number of T‐DNAs in insertional lines has been estimated to be 2.1 (Wilson‐Sánchez et al., 2014), the gh3oct mutant genome probably harboured several additional insertions that could have jeopardized our interpretations on the GH3‐related functions. To rule out this possibility, we sequenced the gh3oct genome and followed a tagged‐sequence strategy to map the positions of the insertions. We confirmed the presence of eight insertions within the corresponding GH3 coding sequences and found two extra insertions at intergenic regions (Fig. S2). Owing to their position, the latter were considered unlikely to contribute to the gh3oct phenotypes.
We then explored the developmental consequences of a group II GH3 knockout. We found that primary roots from gh3oct mutant seedlings were comparable in length to those of the wild‐type, whereas lateral root density was notably increased (Fig. 1a,b,j,k). Such a root phenotype was observed along the vegetative phase (Fig. 1j,k). The rosettes from gh3oct seedlings showed classical high‐auxin phenotypes, such as epinastic cotyledons (Fig. 1a,b) and elongated hypocotyls (Fig. 1a,b,l) and petioles (Fig. 1c,d,m). The gh3oct mutant also showed photoperiod‐independent early flowering (Figs 1e, S3). Most of the siliques in the gh3oct stems did not elongate properly (Fig. 1i), probably causing a delayed inflorescence arrest (Ware et al., 2020), and thus making gh3oct plants taller (Fig. 1h). We observed the formation of several aberrant flowers that developed into siliques with unfused valves in the gh3oct mutant (Fig. 1f,g), although this phenotype showed incomplete penetrance. Taken together, our data indicate that the gh3oct mutant is very likely an octuple GH3 knockout at the seedling stage and that functional group II GH3s redundantly control the progress and timing of several developmental processes throughout the plant’s life cycle.
Fig. 1.
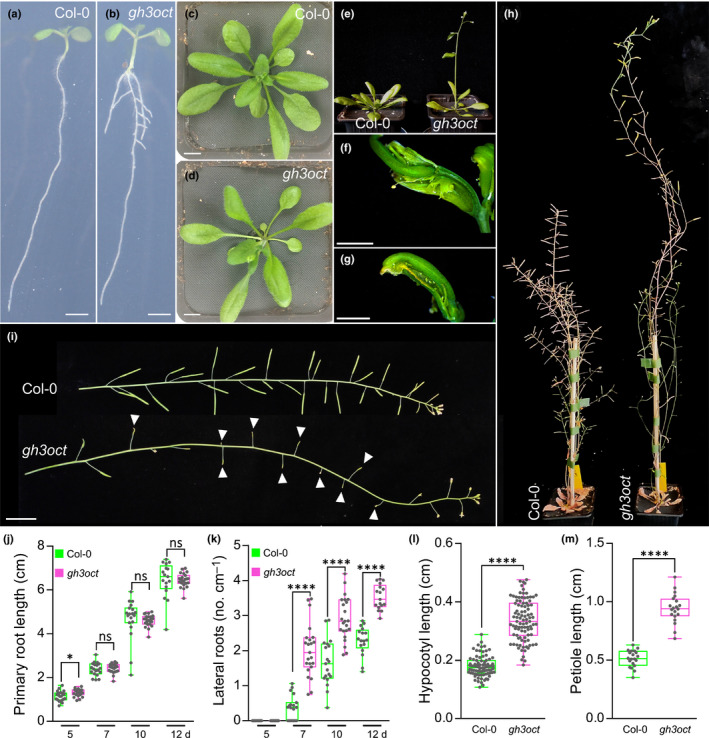
Phenotypes of the gh3oct mutant. (a–e) Arabidopsis thaliana Col‐0 and gh3oct plants at 7 (a, b), 20 (c, d) and 23 d (e) after stratification (das). (f, g) Aberrant reproductive structures occasionally formed by gh3oct mutant from latest inflorescences. (h) Plant height of Col‐0 and gh3oct flowering plants. (i) Stems from Col‐0 and the gh3oct mutant. White arrowheads indicate siliques that did not elongate further in the gh3oct mutant. (j, k) Primary root length (j) and lateral root density (k) determined at 5, 7, 10 and 12 das. (l) Hypocotyl length determined at 10 das. (m) Petiole length determined at 15 das. Box plots (j–m) showing the individual data points, the first and third quartiles (bottom and top of the box), the median (line in the box), and the highest (top whisker) and lowest (bottom whisker) values. Bars: (a, b) 2 mm; (c, d) 1 cm; (f, g) 2 mm; (i) 2 cm. Asterisks indicate values that are statistically different from Col‐0 in a two‐tailed Student’s t‐test (*, P < 0.05; ****, P < 0.0001; (j, k) n ≥ 17, (l) n ≥ 83, and (m) n = 20). ns, not statistically significant.
IAA metabolism in the absence of functional group II GH3s
Owing to the involvement of group II GH3s in IAA metabolism, we explored the concentrations of IAA, the irreversible IAA conjugates IAA‐Asp and IAA‐Glu, and the IAA inactive forms oxIAA, oxIAA‐glc and IAA‐glc in shoots and roots from Col‐0 and the gh3oct mutant. According to the high‐auxin phenotypes displayed by gh3oct plants (Fig. 1), IAA contents were found to be increased in gh3oct seedling shoots (Fig. 2). However, total IAA in root tissues was found at wild‐type concentrations (Fig. 2). Despite these modest differences in IAA contents, the roots of gh3oct seedlings were hypersensitive to exogenous IAA (Fig. S4). As expected for a total knockout of the IAA‐amido synthetase functions, no detectable IAA‐Asp was found in gh3oct plants (Fig. 2). Surprisingly, decreased yet still detectable amounts of IAA‐Glu were found in shoots, and unchanged IAA‐Glu concentrations were detected in roots from gh3oct seedlings (Fig. 2). The contents of the oxidative catabolites oxIAA and oxIAA‐glc were diminished in gh3oct shoots and roots (Fig. 2), whereas no differences in the content of IAA‐glc were found (Fig. 2). Overall, our data suggested that gh3oct plants have a reduced ability to inactivate IAA and that IAA inactivation, by means of conjugation to Glu, could still occur in the absence of functional group II GH3 members. Our data additionally indicated a connection between the production of IAA‐aa conjugates and IAA oxidative catabolites.
Fig. 2.
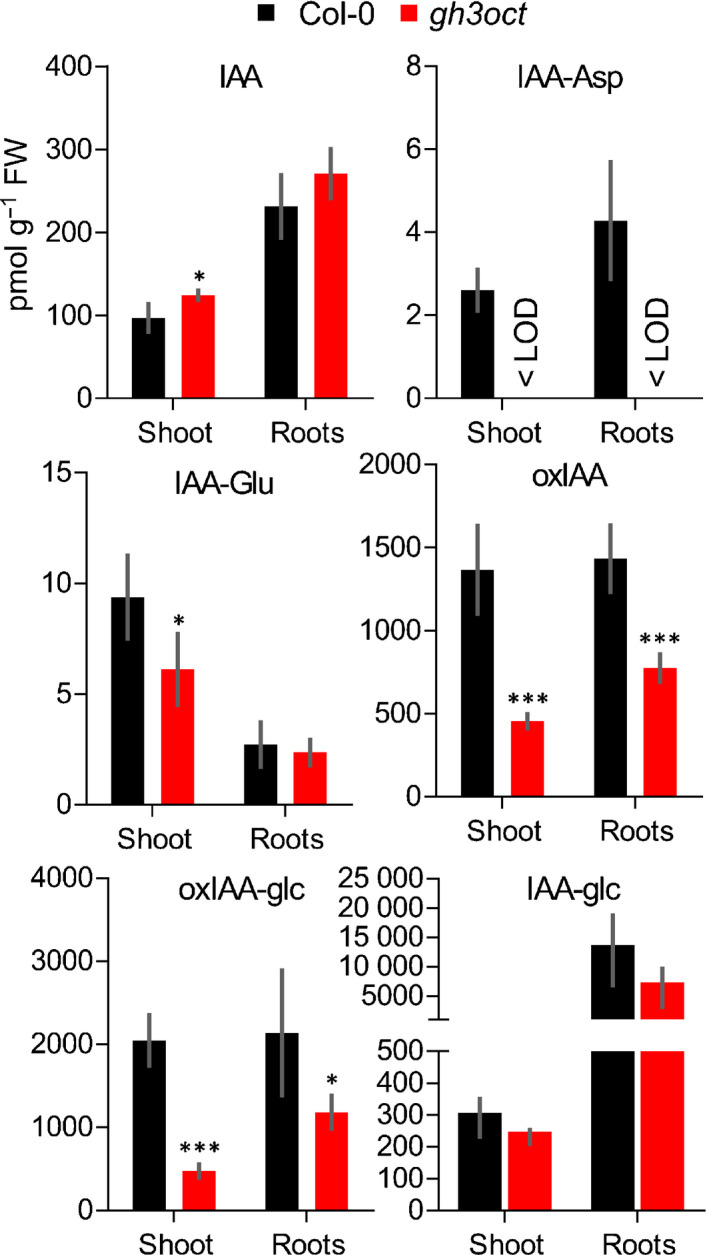
Concentrations of indole‐3‐acetic acid (IAA) and IAA metabolites in shoots and roots of the gh3oct mutant. Concentration of IAA, indole‐3‐acetyl‐aspartate (IAA‐Asp), indole‐3‐acetyl‐glutamate (IAA‐Glu), 2‐oxindole‐3‐acetic acid (oxIAA), 1‐O‐(2‐oxindole‐3‐ylacetyl)‐β‐d‐glucose (oxIAA‐glc) and 1‐O‐indole‐3‐ylacetyl‐β‐d‐glucose (IAA‐glc) in shoots and roots from 7‐d‐old Arabidopsis thaliana Col‐0 (black bars) and gh3oct (red bars) seedlings. Error bars indicate standard deviation. Asterisks indicate values significantly different from Col‐0 in a two‐tailed Student's t‐test (*, P < 0.05; ***, P < 0.001; n = 5). < LOD, below the limit of detection.
gh3oct mutant tolerance to salinity and osmotic stress
GRETCHEN HAGEN 3‐mediated IAA metabolism has previously been suggested to modulate abiotic stress responses in plants (Park et al., 2007; Du et al., 2012; Kirungu et al., 2019). Therefore, we focused on the involvement of group II GH3s in the response to salinity. When examining transcriptomic datasets from Arabidopsis plants exposed to NaCl, we found that Arabidopsis group II GH3s are responsive to salt treatments (Fig. S5). To corroborate this response under our experimental conditions, we used RT‐qPCR to determine the relative expression of each GH3 gene to NaCl in seedling roots. We found all root‐expressed group II GH3s to be upregulated upon NaCl application (Fig. 3a), suggesting that these enzymes might play a role in the response to salinity.
Fig. 3.
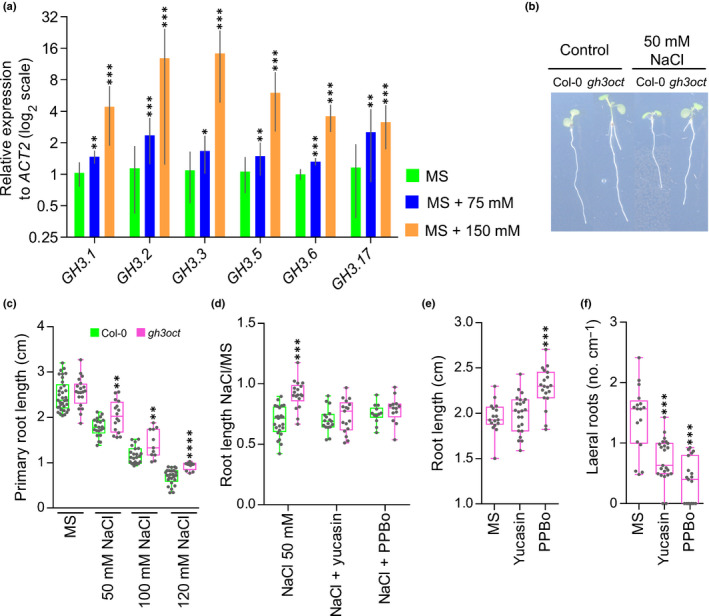
Arabidopsis thaliana group II GH3s are upregulated upon salt treatment and the gh3oct mutant showed indole‐3‐acetic acid (IAA)‐dependent tolerance to salinity. (a) Transcriptional response of group II GH3 genes from 10‐d‐old Col‐0 roots determined by reverse transcription‐quantitative polymerase chain reaction 6 h after transferring to MS media supplemented with 0, 75 or 150 mM NaCl. No expression of GH3.4 and GH3.9 was detected in any sample. Error bars indicate SD. (b) Seven‐day‐old Col‐0 and gh3oct seedlings transferred at day 5 to control MS plates or MS plates supplemented with 50 mM NaCl. (c) Primary root length of Col‐0 and gh3oct seedlings grown for 7 d at different concentrations of NaCl. (d) Root growth ratio of 4‐d‐old seedlings transferred for another 3 d to media plates containing 50 mM NaCl, 50 mM NaCl + 20 μM of the YUCCA inhibitor yucasin or 50 mM NaCl + 1 μM of the YUCCA inhibitor 4‐phenoxyphenylboronic acid (PPBo). To calculate the ratio, the length of the root from seedlings grown on NaCl, NaCl + yucasin and NaCl + PPBo plates was divided by the mean root length of seedlings grown on control MS, MS + yucasin and MS + PPBo plates, respectively. (e, f) Root length (e) and lateral root density (f) in 7‐d‐old gh3oct mutant seedlings grown on control MS plates, and on plates supplemented with 20 μM of yucasin or 1 μM of PPBo. Box plots (c–f) showing the individual data points, the first and third quartiles (bottom and top of the box), the median (line in the box), and the highest (top whisker) and lowest (bottom whisker) values. Asterisks indicate ΔCT values significantly different from MS (n = 4) (a), values significantly different from Col‐0 (n ≥ 11) (c, d) and values significantly different from MS (n ≥ 17) (e, f) in a two‐tailed Student's t‐test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Salinity is well known to cause general arrest in plant growth (Zhao et al., 2020), and a decreased length of the primary root in salt‐stressed Arabidopsis plants is documented (Smolko et al., 2021). Thus, we used the growth of the primary root as an indicator of the tolerance to salinity stress. Compared with the Col‐0 wild‐type, gh3oct plants were more tolerant to 50 mM NaCl (Fig. 3b,c). The gh3oct mutant also grew better than Col‐0 at higher NaCl concentrations (Fig. 3c). Besides NaCl, we found that the gh3oct mutant exhibited higher tolerance to other osmotic stressors, such as sorbitol and mannitol, over the vegetative phase (Fig. S6). Thus, the data suggested that genetic disruption of the group II GH3s confers salinity tolerance, probably as part of a general osmotolerant mechanism.
Tolerance to salinity in the gh3oct mutant is related to endogenous IAA content
To investigate the relevance of the elevated endogenous IAA content in gh3oct plants for the tolerance to salinity, we co‐treated gh3oct seedlings with both NaCl and IAA synthesis inhibitors. As shown in Fig. 3(d), the higher tolerance to NaCl of the gh3oct mutant, in terms of a higher root growth ratio, was abolished by addition of either yucasin or PPBo, which are two inhibitors of YUCCA enzymes that cause a decrease in plant IAA concentrations by competing with the IAA precursor IPyA (Nishimura et al., 2014; Kakei et al., 2015). To rule out the possibility that the decreased root growth ratio was caused by the effect of the IAA synthesis inhibitors on root growth rather than the tolerance to salinity, we inspected the effects of yucasin and PPBo on the root growth of gh3oct seedlings in the absence of NaCl. We found that, at the employed concentrations, none of the inhibitors caused a decrease in the length of the root, whereas both decreased the lateral root density of the gh3oct mutant (Fig. 3e,f).
We next explored the contents of IAA and IAA metabolites and the tolerance to salinity of the gh3 single mutants. IAA concentrations were found to be comparable to the wild‐type in all gh3 mutant shoots and roots, except for gh3.1 (which showed decreased IAA concentrations in roots) and gh3.2 (which showed increased IAA concentrations in shoots but decreased concentrations in roots) (Fig. S7). The IAA‐Asp concentration was slightly decreased in gh3.3 shoots and gh3.6 roots, while a reduced concentration of IAA‐Glu was detected in gh3.1 and gh3.2 roots (Fig. S7). Concentrations of oxIAA were decreased in gh3.2 shoots but were comparable to the wild‐type in the other gh3 mutant shoots and roots (Fig. S7). Under our experimental conditions, only gh3.5 plants showed salt‐tolerant root growth (Fig. S8a,b). However, different from the gh3oct mutant, the tolerance to salinity of gh3.5 plants was not suppressed by decreasing endogenous IAA concentrations (Fig. S8c). Taken together, the results indicated that the tolerance to salinity stress of the gh3oct mutant is related to their higher endogenous IAA contents.
gh3oct mutant tolerance to water‐deficit stress
Salinity and water deficit are initially perceived by the plant roots as an osmotic stress. Therefore, it is not uncommon for salt‐tolerant genotypes also to exhibit a degree of tolerance to drought stress and vice versa (Uddin et al., 2016; Lamers et al., 2020). To test whether gh3oct plants tolerated water deficit better, we first grew Col‐0 and gh3oct seedlings on media plates with a reduced water potential by using polyethylene glycol (PEG) perfusion (Verslues et al., 2006). The root growth ratio of gh3oct plants grown on PEG‐perfused plates was higher than that of the wild‐type (Fig. 4a), suggesting that the gh3oct mutant was more tolerant to water‐deficit stress when grown in vitro. Because gh3.5 plants also showed more tolerant growth under salinity, we investigated the growth ratio of gh3.5 roots in PEG‐perfused media. In contrast to the gh3oct mutant, gh3.5 plants were as sensitive as the wild‐type to PEG‐mediated reduction of the water potential (Fig. S8d).
Fig. 4.
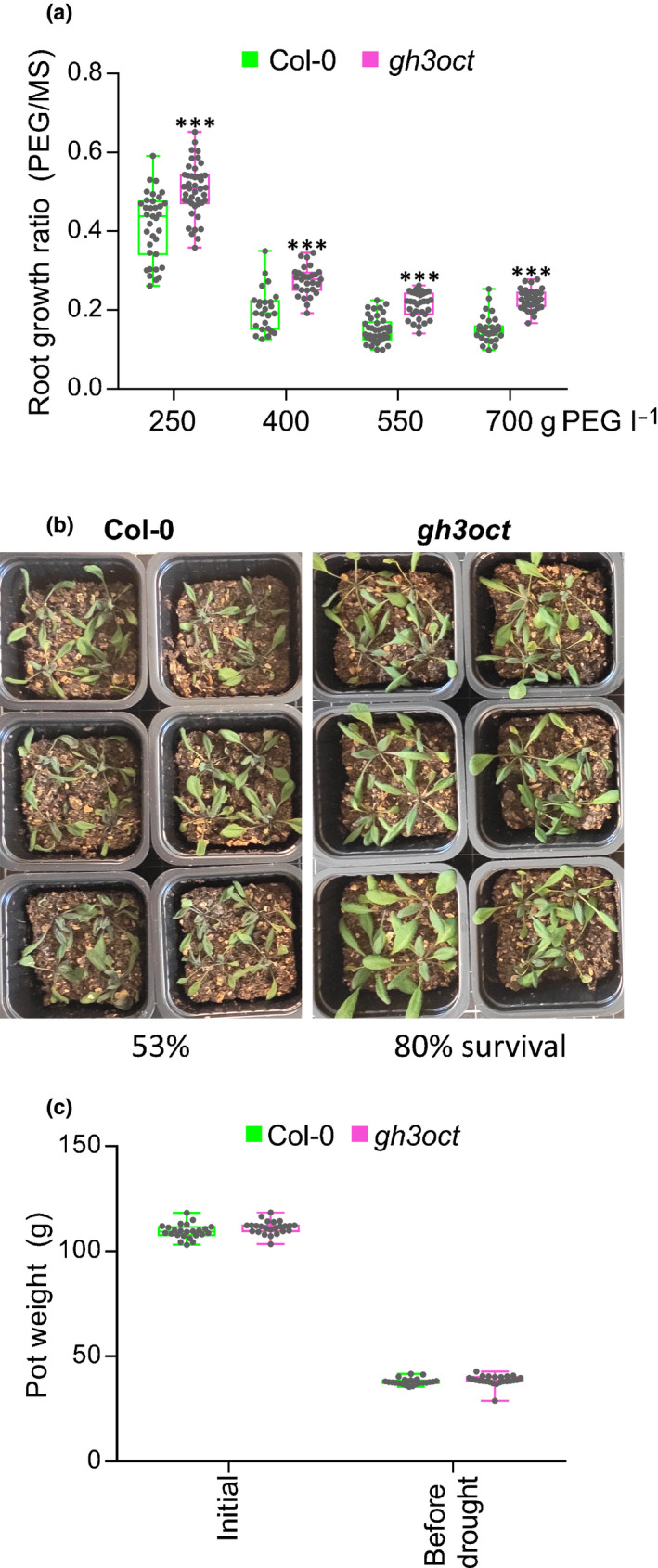
gh3oct plants are tolerant to water deficit. (a) Root growth ratio of Arabidopsis thaliana Col‐0 and gh3oct seedlings in media with lowered water potential. Four‐day‐old seedlings were transferred to plates perfused with a solution of PEG8000 at the indicated concentration and grown for another 6 d. To calculate the ratio, the root length from seedlings in polyethylene glycol (PEG)‐containing plates was divided by the mean root length of seedlings grown on control plates (perfused with liquid MS). Asterisks indicate statistically significantly different growth ratios from Col‐0 in a two‐tailed Student's t‐test (***, P < 0.001; n ≥ 25). (b) Tolerance of Col‐0 and gh3oct plants to drought stress on soil. The image corresponds to 3‐wk‐old plants on dry soil. Survival rate after rewatering is indicated at the bottom and represents the mean percentage from three independent experiments with 27, 36 and 23 plants per genotype. (c) Soil pot weight in the drought experiment. Pots were prepared with 40 g of soil mixture, watered and weighed at the beginning of the experiment (initial). Pots were reweighed when the soil was noticeably dry and before plants started showing drought symptoms (before drought). No significant differences were found between Col‐0 and gh3oct pots in a two‐tailed Student's t‐test. Box plots (a, c) showing the individual data points, the first and third quartiles (bottom and top of the box), the median (line in the box), and the highest (top whisker) and lowest (bottom whisker) values.
Finally, we explored whether the tolerance to water deficit of the gh3oct mutant could be reproduced when grown on soil. We planted stratified Col‐0 and gh3oct seeds in well‐watered soil pots and then withheld irrigation until the plants manifested drought symptoms. As shown in Fig. 4(b), gh3oct mutant plants showed milder drought symptoms and a higher survival rate after rewatering (80% in gh3oct vs 53% in Col‐0). To rule out the possibility that pots containing gh3oct plants could retain more water, we compared the weights of well watered and dry pots (before drought symptoms appeared) for both genotypes. As shown in Fig. 4(c), we found no differences between Col‐0 and gh3oct pots. Thus, our results further supported the contention that the gh3oct mutant was also more tolerant to water deficit on soil.
Hormonal landscape in gh3oct plants under salinity
In addition to IAA, group II GH3 members have been shown to conjugate amino acids to other phytohormones, such as jasmonic acid (JA) and salicylic acid (SA) (Gutierrez et al., 2012; Westfall et al., 2016). Given the involvement of GH3s in the response to salinity, we quantified the endogenous contents of these phytohormones in aerial and root tissues from wild‐type and gh3oct plants grown in control and NaCl‐supplemented media. The IAA concentration was higher in gh3oct plants grown at 0 and 75 mM NaCl, whereas it became comparable to Col‐0 at 150 mM NaCl (Fig. 5a). The increased IAA content in gh3oct plants was related to aerial tissues (Fig. 5a). The content of JA increased markedly in both Col‐0 and gh3oct aerial tissues at 150 mM NaCl, although this increase was less pronounced in the gh3oct mutant (Fig. 5b). The JA bioactive form, JA‐Ile, was found to be increased in gh3oct shoots grown at 75 mM NaCl, but comparable to Col‐0 in control conditions and at 150 mM NaCl (Fig. 5c). Concentrations of jasmonates were very low in wild‐type and gh3oct roots under both control and salinity conditions (Fig. 5b,c). Increased SA concentrations were specifically found in gh3oct roots grown at 75 mM NaCl, while similar concentrations were quantified from Col‐0 and gh3oct shoots under control and NaCl‐stress conditions (Fig. 5d). Because NaCl stress is known to trigger an increase in concentrations of ABA (Simura et al., 2018), we also measured the concentrations of this phytohormone. The ABA content was higher in gh3oct plants grown under control and salinity conditions, and this genotype‐related increase was observed in aerial but not root tissues (Fig. 5e).
Fig. 5.
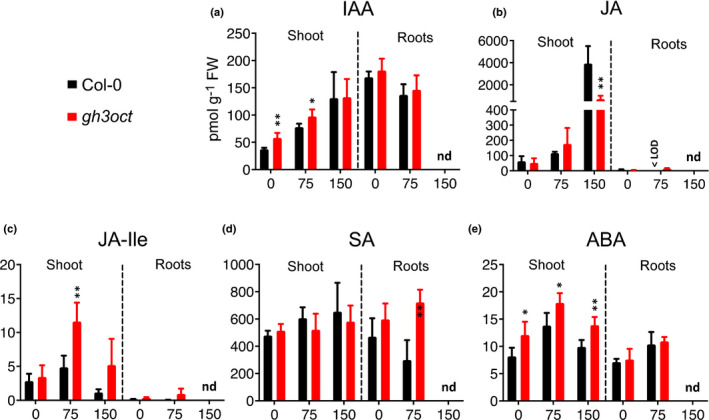
Phytohormone concentrations in shoots and roots from Arabidopsis thaliana Col‐0 and gh3oct plants grown under different NaCl concentrations. (a–e) Concentrations of indole‐3‐acetic acid (IAA) (a), jasmonic acid (JA) (b), jasmonic acid‐isoleucine (JA‐Ile) (c), salicylic acid (SA) (d) and abscisic acid (ABA) (e) in Col‐0 and gh3oct shoots and roots were determined at 7 d after germination on plates containing 0, 75 or 150 mM NaCl. Error bars indicate SD. Asterisks indicate values significantly different from the corresponding values of Col‐0 in a two‐tailed Student's t‐test (*, P < 0.05; **, P < 0.01; n = 4–6). < LOD, below the limit of detection; nd, not determined (roots from plants on 150 mM NaCl were extremely short and samples could not be harvested to meet our LC‐MS/MS protocol requirements).
Next, we explored changes in endogenous concentrations of IAA, JA, JA‐Ile, SA and ABA in a time‐course salinity stress experiment. For that, we transferred Col‐0 and gh3oct seedlings to media plates supplemented with 150 mM NaCl and quantified the phytohormone concentrations at 0, 6 and 24 h upon exposure to NaCl. Concentrations of IAA were higher in gh3oct seedlings at all inspected times under control conditions, but they became comparable to those of Col‐0 in response to 150 mM NaCl (Fig. S9a). This result is in accordance with the similar IAA concentrations in Col‐0 and gh3oct shoots grown at 150 mM NaCl (Fig. 5a). Concentrations of JA and JA‐Ile were not affected by either genotype or salinity (Fig. S9b,c). Concentrations of SA were significantly lower in gh3oct seedlings after 6 h of exposure to salinity, although this difference vanished after 24 h (Fig. S9d). Endogenous concentrations of ABA prominently increased in both genotypes in response to salinity. However, the ABA content was higher in the gh3oct mutant after 24 h of exposure to NaCl (Fig. S9e). Because higher ABA concentrations were also found in gh3oct aerial tissues grown under control and salinity stress conditions (Fig. 5e), and as ABA is known to modulate the response to osmotic stresses by mediating stomatal closure (Hedrich & Shabala, 2018), we hypothesized a higher ability of gh3oct plants to deal with water loss. However, our observations indicated that the water loss rate in gh3oct detached rosettes was indistinguishable from that of the wild‐type (Fig. S10). In summary, we found altered concentrations of IAA and ABA under control conditions, and of SA and ABA in response to salinity in gh3oct mutant plants.
Discussion
IAA metabolism and plant development without functional group II GH3s
Conjugation of IAA to amino acids is an important metabolic pathway regulating concentrations of this bioactive auxin and is performed by group II GH3 proteins (Staswick et al., 2005). Studies on single gh3 mutants have demonstrated the relevance of IAA conjugation for plant developmental processes, such as root meristem size control (Di Mambro et al., 2017) and hypocotyl elongation (Zheng et al., 2016). To identify additional roles of the group II GH3s in auxin metabolism and plant development, we generated an octuple group II GH3 mutant. Our phenotypical analyses of gh3oct mutant plants supported redundant involvement of group II GH3s in lateral root, shoot and fruit development, as well as in vegetative‐to‐reproductive transition. Although our data were not sufficient to support a direct causality between IAA metabolism and early flowering or aberrant fruit development in gh3oct plants, the observed increased lateral root density and longer hypocotyls and petioles are well‐known high‐auxin phenotypes (Sugawara et al., 2009; Gao et al., 2020). Our analyses found that the IAA content was only slightly increased in gh3oct shoots but was at wild‐type concentrations in roots. This suggests that the high‐auxin phenotypes displayed by gh3oct plants might arise from highly spatiotemporally localized IAA increases which do not alter the total IAA content remarkably, such as those required to form lateral roots (Du & Scheres, 2018).
We previously reported a sextuple GH3 knockout mutant in which six group II GH3s (GH3.1‐6) were disrupted (Porco et al., 2016). No detectable concentrations of IAA‐Asp were found in the gh3 sextuple mutant, whereas conjugation of IAA to Glu was upregulated. This was probably attributable to the activity of GH3.17, which is highly specific for the formation of IAA‐Glu (Staswick et al., 2005; Brunoni et al., 2019). Therefore, one might expect that formation of both IAA‐Asp and IAA‐Glu would be abolished after knocking out all group II GH3s. However, the presence of detectable IAA‐Glu content in gh3oct seedlings, which displayed null expression of group II GH3 genes, strongly suggests that additional GH3 or GH3‐like enzymes work in IAA inactivation by means of conjugation to glutamate in Arabidopsis.
DIOXYGENASE FOR AUXIN OXIDATION proteins have been shown to regulate IAA concentrations by mediating its metabolic inactivation through oxidation (Zhao et al., 2013; Porco et al., 2016; Zhang et al., 2016). Concentrations of the oxidative IAA catabolite oxIAA are known to rapidly increase in response to endogenous or exogenous IAA increases (Kubes et al., 2012; Pencik et al., 2013). However, despite showing locally increased IAA concentrations, as inferred from their multiple high‐auxin phenotypes and hypersensitivity to IAA, reduced concentrations of oxIAA and oxIAA‐glc were observed in gh3oct plants. DAOs and GH3s have been suggested to establish parallel and redundant pathways for IAA inactivation, with the GH3 pathway playing a compensatory function, as supported by increased and decreased conjugate concentrations in the DAO1 mutant and overexpressing plants, respectively (Porco et al., 2016; Zhang et al., 2016). This model has recently been challenged by the finding that IAA‐Asp is an in vivo substrate for the DAO1 oxidase, which additionally accounts for the formation of oxIAA‐Asp conjugates (Müller et al., 2021). In a context where DAOs function downstream of group II GH3s, the reduced concentrations of oxIAA and oxIAA‐glc in gh3oct plants found in the present work suggest that a significant fraction of the plant oxIAA pool derives from oxIAA‐aa conjugates. This is indeed supported by a newly proposed pathway for IAA inactivation (Hayashi et al., 2021).
Cooperative involvement of group II GH3s in response to salinity and drought
Salinity and drought are major constraints on plant growth, and therefore represent serious threats to agriculture and forestry. Data from several studies over the past decade support a link between auxin and plant responses to salinity (Wang et al., 2009; Liu et al., 2015; Korver et al., 2018; Fu et al., 2019) and drought stress (Zhang et al., 2009, 2012, 2020; Shi et al., 2014; Jung et al., 2015). Spatiotemporal auxin distribution and auxin sensing modulate adaptive growth responses to NaCl stress (Wang et al., 2009; Liu et al., 2015). Recently, Aux/IAA auxin co‐receptors were shown to be required for plant tolerance to water deficit, thus suggesting a central role for auxin in integrating drought signals to elaborate genetic and physiological responses (Shani et al., 2017). Although the pathways that mediate auxin‐mediated responses to salinity and drought are only just being elucidated, it appears that auxin may mediate tolerance to water deprivation by regulating several adaptive responses leading to stress avoidance. These include adaptive root growth (Leftley et al., 2021), reactive oxygen species scavenging (Fu et al., 2019), regulation of stomatal apertures (Salehin et al., 2019) and crosstalk with ABA and other hormones involved in the responses to salinity and drought (Sun & Li, 2014; Yu et al., 2020; Salvi et al., 2021).
Our work revealed that gh3oct mutant plants were more tolerant to salinity, and that this tolerance was related to endogenous IAA concentrations. Tolerance to salinity was also higher in gh3.5 mutant plants, although this effect appeared to be unaffected by endogenous IAA concentrations. GH3.5 is well known to mediate the conjugation of other plant hormones, including SA and JA (Gutierrez et al., 2012; Westfall et al., 2016), suggesting that the salt‐tolerant growth of gh3.5 roots may rather be related to the SA or JA pathways. The increased root branching of gh3oct plants without penalty for primary root growth produced a more robust root system, which might contribute to the better performance of gh3oct plants under water stress conditions.
In addition to root system architecture, the increased tolerance of gh3oct plants to salinity and water deficit might be driven by the increased ABA content. Even though it remains unclear whether ABA increase is a direct or indirect consequence of the lack of group II GH3s, ABA is a stress‐related hormone whose concentrations increase very rapidly upon salt and drought stress sensing and is well known for its direct involvement in the response to osmotic stresses. This response is primarily based on promoting stomatal closure (Hedrich & Shabala, 2018), although other stomata‐independent mechanisms have also been reported (Thalmann et al., 2016). The increased ABA content in gh3oct plants grown in control conditions, but with a water loss rate similar to the wild‐type, goes against the idea that gh3oct plants have a predisposition to tolerate water deficit based on lower transpiration, and suggests that other ABA‐mediated mechanisms might help gh3oct plants to cope with this stress.
Conclusions
In this work, we present a group II GH3 knockout as a useful genetic tool to research metabolic pathways regulating concentrations of the main auxin IAA. It has long been accepted that group II GH3 members carry out conjugation of IAA to amino acids. Our metabolic analyses in gh3oct plants indicated that additional GH3s or GH3‐like proteins might modulate IAA concentrations by means of conjugation to glutamate. Additionally, our data suggested that the major IAA catabolite oxIAA is, at least in part, produced from the GH3 pathway.
Examination of mutant gh3oct plants also revealed that plant tolerance to water deprivation stresses is modulated by redundant auxin conjugation. Identifying as many components as possible of the plant's responses to salinity and drought is key to determining the best targets for chemical and/or gene‐editing approaches directed at enhancing stress tolerance in plants. While the mechanisms behind the auxin‐related tolerance to salinity and drought remain to be elucidated, the present work supports strategies based on salinity‐ and drought‐inducible disruption of group II GH3s as a profitable tool to engineer plants that better tolerate water stress.
Author contributions
EM‐B, RC‐S and KL conceived and designed the research; EM‐B and RC‐S performed most of the experiments; JŠ performed the hormone analyses; AP and ON quantified IAA metabolites from the gh3 single mutant; PS made the gh3oct mutant; RC‐S prepared the manuscript draft; EM‐B and RC‐S wrote the manuscript with input from all authors. RC‐S and EM‐B contributed equally to this work. This research was supported by funds to KL.
Supporting information
Fig. S1 The gh3oct mutant is an octuple knockout.
Fig. S2 The gh3oct mutant harbours two extra T‐DNA insertions at intergenic regions.
Fig. S3 The gh3oct is a photoperiod‐independent early‐flowering mutant.
Fig. S4 Root growth in the gh3oct mutant is hypersensitive to IAA.
Fig. S5 Transcriptional response of Arabidopsis thaliana group II GH3 genes to salinity according to published datasets.
Fig. S6 Root growth in the gh3oct mutant is tolerant to different osmolytes.
Fig. S7 Concentrations of IAA and IAA metabolites in shoots and roots from Arabidopsis thaliana group II gh3 single mutants.
Fig. S8 Tolerance to salinity and water deficit of the Arabidopsis thaliana gh3 single mutants.
Fig. S9 Concentrations of IAA and stress‐related phytohormones in response to salinity in the gh3oct mutant.
Fig. S10 Col‐0 and gh3oct plants lose water at the same rate.
Table S1 Insertion lines combined to obtain the gh3oct mutant.
Table S2 Primer sets used in this work.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
Research in KL’s laboratory is supported by grants from the Swedish Foundation for Strategic Research (Vinnova), the Knut and Alice Wallenberg Foundation (KAW), the Swedish research councils VR and Formas. EM‐B (JCK‐1811) held a postdoctoral fellowship from Kempestiftelserna. AP and ON were financially supported by the Ministry of Education, Youth and Sports of the Czech Republic (European Regional Development Fund‐Project ‘Plants as a tool for sustainable global development’ no. CZ.02.1.01/0.0/0.0/16_019/0000827). We also acknowledge the Swedish Metabolomics Centre (http://www.swedishmetabolomicscentre.se/) for access to instrumentation. We thank Ken‐ichiro Hayashi for the generous sharing of the auxin synthesis inhibitors.
Contributor Information
Rubén Casanova‐Sáez, Email: ruben.casanova.saez@slu.se, Email: karin.ljung@slu.se.
Karin Ljung, Email: karin.ljung@slu.se.
Data availability
The sequencing data generated in this study are openly available at the Short Read Archive (SRA) with the code SRX8771538.
References
- Band LR. 2021. Auxin fluxes through plasmodesmata. New Phytologist 231: 1686–1692. [DOI] [PubMed] [Google Scholar]
- Bottcher C, Boss PK, Davies C. 2011. Acyl substrate preferences of an IAA‐amido synthetase account for variations in grape (Vitis vinifera L.) berry ripening caused by different auxinic compounds indicating the importance of auxin conjugation in plant development. Journal of Experimental Botany 62: 4267–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoni F, Collani S, Simura J, Schmid M, Bellini C, Ljung K. 2019. A bacterial assay for rapid screening of IAA catabolic enzymes. Plant Methods 15: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoni F, Collani S, Casanova‐Sáez R, Šimura J, Karady M, Schmid M, Ljung K, Bellini C. 2020. Conifers exhibit a characteristic inactivation of auxin to maintain tissue homeostasis. New Phytologist 226: 1753–1765. [DOI] [PubMed] [Google Scholar]
- Casanova‐Saez R, Mateo‐Bonmati E, Ljung K. 2021. Auxin metabolism in plants. Cold Spring Harbor Perspectives in Biology 13: a039867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova‐Saez R, Voss U. 2019. Auxin metabolism controls developmental decisions in land plants. Trends in Plant Science 24: 741–754. [DOI] [PubMed] [Google Scholar]
- Chen Q, Westfall CS, Hicks LM, Wang S, Jez JM. 2010. Kinetic basis for the conjugation of auxin by a GH3 family indole‐acetic acid‐amido synthetase. Journal of Biological Chemistry 285: 29780–29786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Wu N, Fu J, Wang S, Li X, Xiao J, Xiong L. 2012. A GH3 family member, OsGH3‐2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. Journal of Experimental Botany 63: 6467–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Scheres B. 2018. Lateral root formation and the multiple roles of auxin. Journal of Experimental Botany 69: 155–167. [DOI] [PubMed] [Google Scholar]
- Fu Y, Yang Y, Chen S, Ning N, Hu H. 2019. Arabidopsis IAR4 modulates primary root growth under salt stress through ROS‐mediated modulation of auxin distribution. Frontiers in Plant Science 10: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallei M, Luschnig C, Friml J. 2020. Auxin signalling in growth: Schrodinger's cat out of the bag. Current Opinion in Plant Biology 53: 43–49. [DOI] [PubMed] [Google Scholar]
- Gao Y, Dai X, Aoi Y, Takebayashi Y, Yang L, Guo X, Zeng Q, Yu H, Kasahara H, Zhao Y. 2020. Two homologous INDOLE‐3‐ACETAMIDE (IAM) HYDROLASE genes are required for the auxin effects of IAM in Arabidopsis. Journal of Genetics and Genomics 47: 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Flokova K, Pacurar DI, Novak O, Staswick P, Kowalczyk M, Pacurar M, Demailly H, Geiss G et al. 2012. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24: 2515–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes UZ, Murphy AS, Schwechheimer C. 2022. Auxin transporters‐A biochemical view. Cold Spring Harbor Perspectives in Biology 14: a039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanania U, Velcheva M, Sahar N, Perl A. 2004. An improved method for isolating high‐quality DNA from Vitis vinifera nuclei. Plant Molecular Biology Reporter 22: 173–177. [Google Scholar]
- Hayashi K‐I, Arai K, Aoi Y, Tanaka Y, Hira H, Guo R, Hu Y, Ge C, Zhao Y, Kasahara H et al. 2021. The main oxidative inactivation pathway of the plant hormone auxin. Nature Communications 12: 6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R, Shabala S. 2018. Stomata in a saline world. Current Opinion in Plant Biology 46: 87–95. [DOI] [PubMed] [Google Scholar]
- Jung H, Lee DK, Choi YD, Kim JK. 2015. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Science 236: 304–312. [DOI] [PubMed] [Google Scholar]
- Kai K, Horita J, Wakasa K, Miyagawa H. 2007. Three oxidative metabolites of indole‐3‐acetic acid from Arabidopsis thaliana . Phytochemistry 68: 1651–1663. [DOI] [PubMed] [Google Scholar]
- Kakei Y, Yamazaki C, Suzuki M, Nakamura A, Sato A, Ishida Y, Kikuchi R, Higashi S, Kokudo Y, Ishii T et al. 2015. Small‐molecule auxin inhibitors that target YUCCA are powerful tools for studying auxin function. The Plant Journal 84: 827–837. [DOI] [PubMed] [Google Scholar]
- Khan S, Stone JM. 2007. Arabidopsis thaliana GH3.9 influences primary root growth. Planta 226: 21–34. [DOI] [PubMed] [Google Scholar]
- Kirungu JN, Magwanga RO, Lu P, Cai X, Zhou Z, Wang X, Peng R, Wang K, Liu F. 2019. Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genetics 20: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Kamada‐Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi‐Tanaka M, Matsuoka M, Suzuki K et al. 2009. Highly sensitive and high‐throughput analysis of plant hormones using MS‐probe modification and liquid chromatography‐tandem mass spectrometry: an application for hormone profiling in Oryza sativa . Plant and Cell Physiology 50: 1201–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korver RA, Koevoets IT, Testerink C. 2018. Out of shape during stress: a key role for auxin. Trends in Plant Science 23: 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M, Sandberg G. 2001. Quantitative analysis of indole‐3‐acetic acid metabolites in Arabidopsis. Plant Physiology 127: 1845–1853. [PMC free article] [PubMed] [Google Scholar]
- Kubeš M, Yang H, Richter GL, Cheng Y, Młodzińska E, Wang X, Blakeslee JJ, Carraro N, Petrášek J, Zažímalová E et al. 2012. The Arabidopsis concentration‐dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. The Plant Journal 69: 640–654. [DOI] [PubMed] [Google Scholar]
- Lamers J, van der Meer T, Testerink C. 2020. How plants sense and respond to stressful environments. Plant Physiology 182: 1624–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Tellez R, Rampey RA, Matsuda SP, Bartel B. 2002. Characterization of a family of IAA‐amino acid conjugate hydrolases from Arabidopsis. Journal of Biological Chemistry 277: 20446–20452. [DOI] [PubMed] [Google Scholar]
- Leftley N, Banda J, Pandey B, Bennett M, Voss U. 2021. Uncovering how auxin optimizes root systems architecture in response to environmental stresses. Cold Spring Harbor Perspectives in Biology 13: a040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li RJ, Han TT, Cai W, Fu ZW, Lu YT. 2015. Salt stress reduces root meristem size by nitric oxide‐mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiology 168: 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig‐Muller J, Julke S, Bierfreund NM, Decker EL, Reski R. 2009. Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytologist 181: 323–338. [DOI] [PubMed] [Google Scholar]
- Lup SD, Wilson‐Sánchez D, Andreu‐Sánchez S, Micol JL. 2021. Easymap: a user‐friendly software package for rapid mapping‐by‐sequencing of point mutations and large insertions. Frontiers in Plant Science 12. doi: 10.3389/fpls.2021.655286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novák O, Ljung K, Di Paola L et al. 2017. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proceedings of the National Academy of Sciences, USA 114: E7641–E7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo‐Bonmatí E, Casanova‐Sáez R, Šimura J, Ljung K. 2021. Broadening the roles of UDP‐glycosyltransferases in auxin homeostasis and plant development. New Phytologist 232: 642–654. [DOI] [PubMed] [Google Scholar]
- Müller K, Dobrev PI, Pěnčík A, Hošek P, Vondráková Z, Filepová R, Malínská K, Brunoni F, Helusová L, Moravec T et al. 2021. DIOXYGENASE FOR AUXIN OXIDATION 1 catalyzes the oxidation of IAA amino acid conjugates. Plant Physiology 187: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Hayashi K‐I, Suzuki H, Gyohda A, Takaoka C, Sakaguchi Y, Matsumoto S, Kasahara H, Sakai T, Kato J‐I et al. 2014. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. The Plant Journal 77: 352–366. [DOI] [PubMed] [Google Scholar]
- Novak O, Henykova E, Sairanen I, Kowalczyk M, Pospisil T, Ljung K. 2012. Tissue‐specific profiling of the Arabidopsis thaliana auxin metabolome. The Plant Journal 72: 523–536. [DOI] [PubMed] [Google Scholar]
- Ostin A, Kowalyczk M, Bhalerao RP, Sandberg G. 1998. Metabolism of indole‐3‐acetic acid in Arabidopsis. Plant Physiology 118: 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM. 2007. GH3‐mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. Journal of Biological Chemistry 282: 10036–10046. [DOI] [PubMed] [Google Scholar]
- Pěnčík A, Casanova‐Sáez R, Pilarová V, Žukauskaite A, Pinto R, Micol JL, Ljung K, Novák O. 2018. Ultra‐rapid auxin metabolite profiling for high‐throughput mutant screening in Arabidopsis. Journal of Experimental Botany 69: 2569–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencik A, Rolcik J, Novak O, Magnus V, Bartak P, Buchtik R, Salopek‐Sondi B, Strnad M. 2009. Isolation of novel indole‐3‐acetic acid conjugates by immunoaffinity extraction. Talanta 80: 651–655. [DOI] [PubMed] [Google Scholar]
- Pencik A, Simonovik B, Petersson SV, Henykova E, Simon S, Greenham K, Zhang Y, Kowalczyk M, Estelle M, Zazimalova E et al. 2013. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole‐3‐acetic acid catabolite 2‐oxindole‐3‐acetic acid. Plant Cell 25: 3858–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco S, Pěnčík A, Rashed A, Voß U, Casanova‐Sáez R, Bishopp A, Golebiowska A, Bhosale R, Swarup R, Swarup K et al. 2016. Dioxygenase‐encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113: 11016–11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. 2004. A family of auxin‐conjugate hydrolases that contributes to free indole‐3‐acetic acid levels during Arabidopsis germination. Plant Physiology 135: 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M, Li B, Tang M, Katz E, Song L, Ecker JR, Kliebenstein DJ, Estelle M. 2019. Auxin‐sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nature Communications 10: 4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi P, Manna M, Kaur H, Thakur T, Gandass N, Bhatt D, Muthamilarasan M. 2021. Phytohormone signaling and crosstalk in regulating drought stress response in plants. Plant Cell Reports 40: 1305–1329. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al. 2012. Fiji: an open‐source platform for biological‐image analysis. Nature Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real‐time PCR data by the comparative CT method. Nature Protocols 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shani E, Salehin M, Zhang Y, Sanchez SE, Doherty C, Wang R, Mangado CC, Song L, Tal I, Pisanty O et al. 2017. Plant stress tolerance requires auxin‐sensitive Aux/IAA transcriptional repressors. Current Biology 27: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Chen L, Ye T, Liu X, Ding K, Chan Z. 2014. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiology and Biochemistry 82: 209–217. [DOI] [PubMed] [Google Scholar]
- Simura J, Antoniadi I, Siroka J, Tarkowska D, Strnad M, Ljung K, Novak O. 2018. Plant hormonomics: multiple phytohormone profiling by targeted metabolomics. Plant Physiology 177: 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolko A, Bauer N, Pavlovic I, Pencik A, Novak O, Salopek‐Sondi B. 2021. Altered root growth, auxin metabolism and distribution in Arabidopsis thaliana exposed to salt and osmotic stress. International Journal of Molecular Sciences 22: 7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole‐3‐acetic acid. Plant Cell 17: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML. 2002. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole‐3‐acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H. 2009. Biochemical analyses of indole‐3‐acetaldoxime‐dependent auxin biosynthesis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106: 5430–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Li C. 2014. Cross talk of signaling pathways between ABA and other phytohormones. In: Zhang D‐P, ed. Abscisic acid: metabolism, transport and signaling. Dordrecht, the Netherlands: Springer, 243–253. [Google Scholar]
- Takehara S, Sakuraba S, Mikami B, Yoshida H, Yoshimura H, Itoh A, Endo M, Watanabe N, Nagae T, Matsuoka M et al. 2020. A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin. Nature Communications 11: 2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J, Domingo C, Talon M. 2006. The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371: 279–290. [DOI] [PubMed] [Google Scholar]
- Thalmann M, Pazmino D, Seung D, Horrer D, Nigro A, Meier T, Kolling K, Pfeifhofer HW, Zeeman SC, Santelia D. 2016. Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 28: 1860–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin MN, Hossain MA, Burritt DJ. 2016. Salinity and drought stress: similarities and differences in oxidative responses and cellular redox regulation. In: Ahmad P, ed. Water stress and crop plants: a sustainable approach. Singapore, Singapore: John Wiley & Sons, 86–101. [Google Scholar]
- Verslues PE, Agarwal M, Katiyar‐Agarwal S, Zhu J, Zhu JK. 2006. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. The Plant Journal 45: 523–539. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li K, Li X. 2009. Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana . Journal of Plant Physiology 166: 1637–1645. [DOI] [PubMed] [Google Scholar]
- Ware A, Walker CH, Simura J, Gonzalez‐Suarez P, Ljung K, Bishopp A, Wilson ZA, Bennett T. 2020. Auxin export from proximal fruits drives arrest in temporally competent inflorescences. Nature Plants 6: 699–707. [DOI] [PubMed] [Google Scholar]
- Westfall CS, Sherp AM, Zubieta C, Alvarez S, Schraft E, Marcellin R, Ramirez L, Jez JM. 2016. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proceedings of the National Academy of Sciences, USA 113: 13917–13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson‐Sánchez D, Rubio‐Díaz S, Munóz‐Viana R, Pérez‐Pérez JM, Jover‐Gil S, Ponce MR, Micol JL. 2014. Leaf phenomics: a systematic reverse genetic screen for Arabidopsis leaf mutants. The Plant Journal 79: 878–891. [DOI] [PubMed] [Google Scholar]
- Yu Z, Duan X, Luo L, Dai S, Ding Z, Xia G. 2020. How plant hormones mediate salt stress responses. Trends in Plant Science 25: 1117–1130. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lin JE, Harris C, Campos Mastrotti Pereira F, Wu F, Blakeslee JJ, Peer WA. 2016. DAO1 catalyzes temporal and tissue‐specific oxidative inactivation of auxin in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 113: 11010–11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li J, Zhang W, Yan S, Wang R, Zhao J, Li Y, Qi Z, Sun Z, Zhu Z. 2012. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. The Plant Journal 72: 805–816. [DOI] [PubMed] [Google Scholar]
- Zhang SW, Li CH, Cao J, Zhang YC, Zhang SQ, Xia YF, Sun DY, Sun Y. 2009. Altered architecture and enhanced drought tolerance in rice via the down‐regulation of indole‐3‐acetic acid by TLD1/OsGH3.13 activation. Plant Physiology 151: 1889–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li Y, Hassan MJ, Li Z, Peng Y. 2020. Indole‐3‐acetic acid improves drought tolerance of white clover via activating auxin, abscisic acid and jasmonic acid related genes and inhibiting senescence genes. BMC Plant Biology 20: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhang H, Song C, Zhu J‐K, Shabala S. 2020. Mechanisms of plant responses and adaptation to soil salinity. Innovation 1. doi: 10.1016/j.xinn.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. 2018. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annual Review of Plant Biology 69: 417–435. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang Y, Liu XI, Zhang X, Liu S, Yu X, Ren Y, Zheng X, Zhou K, Jiang L et al. 2013. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Developmental Cell 27: 113–122. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novak O, Chen W, Ljung K, Noel JP, Chory J. 2016. Local auxin metabolism regulates environment‐induced hypocotyl elongation. Nature Plants 2: 16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The gh3oct mutant is an octuple knockout.
Fig. S2 The gh3oct mutant harbours two extra T‐DNA insertions at intergenic regions.
Fig. S3 The gh3oct is a photoperiod‐independent early‐flowering mutant.
Fig. S4 Root growth in the gh3oct mutant is hypersensitive to IAA.
Fig. S5 Transcriptional response of Arabidopsis thaliana group II GH3 genes to salinity according to published datasets.
Fig. S6 Root growth in the gh3oct mutant is tolerant to different osmolytes.
Fig. S7 Concentrations of IAA and IAA metabolites in shoots and roots from Arabidopsis thaliana group II gh3 single mutants.
Fig. S8 Tolerance to salinity and water deficit of the Arabidopsis thaliana gh3 single mutants.
Fig. S9 Concentrations of IAA and stress‐related phytohormones in response to salinity in the gh3oct mutant.
Fig. S10 Col‐0 and gh3oct plants lose water at the same rate.
Table S1 Insertion lines combined to obtain the gh3oct mutant.
Table S2 Primer sets used in this work.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The sequencing data generated in this study are openly available at the Short Read Archive (SRA) with the code SRX8771538.


