Abstract
Innovative application of surface-enhanced Raman scattering (SERS) for rapid and nondestructive analyses has been gaining increasing attention for food safety and quality. SERS is based on inelastic scattering enhancement from molecules located near nanostructured metallic surfaces and has many advantages, including ultrasensitive detection and simple protocols. Current SERS-based quality analysis contains composition and structural information that can be used to establish an electronic file of the food samples for subsequent reference and traceability. SERS is a promising technique for the detection of chemical, biological, and harmful metal contaminants, as well as for food poisoning, and allergen identification using label-free or label-based methods, based on metals and semiconductors as substrates. Recognition elements, including immunosensors, aptasensors, or molecularly imprinted polymers, can be linked to SERS tags to specifically identify targeted contaminants and perform authenticity analysis. Herein, we highlight recent studies on SERS-based quality and safety analysis for different foods categories spanning the whole food chain, ‘from farm to table’ and processing, genetically modified food, and novel foods. Moreover, SERS detection is a potential tool that ensures food safety in an easy, rapid, reliable, and nondestructive manner during the COVID-19 pandemic.
Keywords: surface-enhanced Raman scattering (SERS), food quality, safety, authenticity, poisoning, contaminant, genetically modified food (GMF), insects food, semiconductor, coronavirus disease 2019 (COVID-19)
1. Introduction
Food quality and safety are prominent components of food science research [1]. In this context, it is essential that reliable systems are developed to detect, eliminate, and control the risks posed by hazardous substances [2]. Surface-enhanced Raman scattering (SERS) is a powerful molecular spectroscopy technique, based on inelastic scattering enhancement from molecules located near the nanostructured metallic surfaces, significantly differing from the gold standard methods, including chromatography and mass spectroscopy [3]. Over the past four decades, the great potential of SERS for the rapid detection of trace chemicals has been demonstrated. Excellent review papers have been published that focus on different perspectives of SERS technology, including its mechanisms, semiconductor SERS substrates, and the applications in food safety and quality, which may be of interest to different readers [4]. Semiconductor materials have more controllable properties than metals, including their band gap, photoluminescence, stability, and degradation resistance [5]. Our research group has also published related reviews and articles that provide details regarding SERS for food safety applications [6].
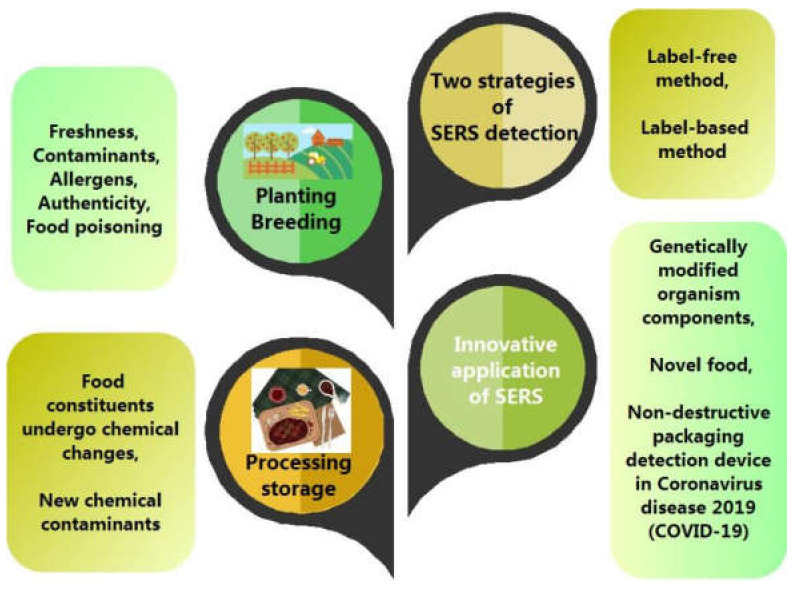
SERS technology is still not completely ready for use as a routine analytical tool to solve real-world food analysis problems, but it is beginning to be applied, especially for conducting ‘Recognition’ to ‘Behavior analysis’ of both food ingredients and pollutants. This can cover the whole food chain, including food planting, breeding, packing, storage, transportation, and processing [7]. Foods can be classified into the following groups: grains; oil-producing plants; vegetables; fruits; nuts; sugars; beverages; edible fungi; flavorings; medicinal plants; and foods of animal origin. The food matrix complexity and different processing technologies produce different concerns regarding the quality and safety of each food type. Herein, we summarize the innovative application of SERS in food quality and safety, based on food classification (Figure 1).
Figure 1.
SERS applications in food quality and safety.
2. SERS Applications in Food Basal Components and Freshness
Food is composed of complex materials and the detection of specific ingredients is an important step. Basal food components include moisture, protein, oil, ash, reducing sugars, and other ingredients. However, the SERS methods are not yet widely available as a tool in food analytical laboratories, especially for the basal components. The analyte response can be increased to achieve an effective SERS signal [8] and the SERS data can be arranged in vector form, belonging to the first-order category. Particularly successful first-order calibrations include partial least-squares (PLS) and PLS regression (PLSR), that permit the quantitation of the selected analytes without knowing the chemical identity of the interfering species [9].
Two strategies to judge freshness include: (1) using the relationship between normally bright matter and Raman signals to judge freshness. In particular, for fruits and vegetables, their respective Raman assignments arise from glucose, carotene, and lycopene and can reflect freshness [10]; (2) monitoring specific marker molecules of corruption and deterioration. For example, ammonia and formaldehyde are spoilage indicators of fish that can be monitored [11]. The SERS methods can detect fructose and pectin in commercial apple juice and pear/apple pulp when combined with the wavelet denoising algorithm and the background subtraction method [12]. Thus, the Raman/SERS spectra may not directly determine the components that influence freshness, but can indirectly monitor the changes in basal components through spectral changes and detecting specific molecules (Table 1) [13]. The SERS spectra also showed that discrimination between products and manufacturers is possible through the fingerprint analysis [14]. For example, when coupled with PCA, the SERS method can be applied for white wine characterization, wherein the main spectral differences arise from adenine, carboxylic acids, and glutathione, with their ratios changing between wine types and producers [15].
Table 1.
Strategies of food freshness and characterization judgment by SERS methods.
| Strategies | Analysis | Algorithms | Ref. |
|---|---|---|---|
| Relationship between normally bright matter and Raman signal | Egg shell | Partial least-squares regression (PLSR) | [8] |
| Oxidation process of nut oils | PLSR and Forest random PLSR (RF-PLSR) | [9] | |
| Citrus fruits | Raman coefficient of freshness (CFresh) | [10] | |
| Red wine | PCA | [14] | |
| White wine | PCA | [15] | |
| Monitoring specific marker molecules of corruption and deterioration | Ammonia and formaldehyde in fish | — | [11] |
| Fructose and pectin in apple juice | Wavelets | [12] | |
| Protein oxidation/denaturation in beef | — | [13] |
—: Not report.
Foods of an animal origin are often more highly processed than fruits and vegetables, necessitating on-site quality assessments because of their vulnerability to contamination and deterioration. For a qualitative analysis of foods of animal origin, Raman sensors can provide depth-profile information with regards to the purines, proteins, and lipids with repetitive regeneration. Meanwhile, spectral analysis algorithms can simplify the analytical data processing and improve the accuracy and robustness of spectral multivariate analysis [16]. The online monitoring of food freshness and internal ingredients is important, with Raman detection systems well-suited for this application. In addition, the structural information contained in the Raman spectra can be archived into electronic files corresponding to the specific food samples for subsequent reference.
3. SERS Application in “From Farm to Table” Foods
3.1. Chemical Contaminants and Toxin Detection by SERS
Chemical contaminants (e.g., chemical fertilizers, pesticides, veterinary drugs, and hormones) are major food safety issues [17], and many studies have reported comprehensive strategies for chemical contaminant detection [18,19]. Generally, foods can be categorized as liquid, solid, or solid and liquid mixtures, contaminants, or poisons distributed on the surface of these food types and can be detected by SERS [20]. This is usually accomplished using either label-free or label-based methods. The recognition elements of chemical contaminants and poison include probe molecules, modified substrates, and aptamers. Increasing efforts are expected to focus on developing novel SERS substrates and highly sensitive Raman reporters, including semiconductor nanomaterials and related composites (Table 2) [21,22,23,24].
Table 2.
Label-free and label-based strategies for chemical contaminants detection by SERS in food.
| Strategies | Recognition Elements | SERS Substrates | Distribution | Ref. |
|---|---|---|---|---|
| Label-free method | C≡N bond | Au NPs | Orange peels | [18] |
| C–N | Poly(ethylene terephthalate)/indium tin oxide/Ag platform | Apple peels | [25] | |
| N–O bond | Ag NPs | Water | [19] | |
| O–S–O bond | Ag NPs | Wines | [29] | |
| O–S–O bond | Sandwiching the ZnO-paper disc | Wines | [30] | |
| Label-based method | Probe molecules | Ag NPs | Water | [20] |
| Modified substrates | Au nanorod-incorporated melamine foam | Chili sauce, dried chili, and chili powder | [21] | |
| aptamers | PCR sealing membrane | Cabbage | [22] |
The chemical contamination residues in different food categories detected by SERS methods can vary. For the pesticide residues as an example, more reports on beverages, fruits, and vegetables have been published than for other foods [25]. For foods of animal origin, chemical contaminants in the feed and veterinary drug residues and their metabolites may enter the food chain during breeding and raising. In addition to the base drugs, their matrix, transformation products, reaction products, and impurities of toxicological significance can significantly affect food safety [26]. The detection methods for chemical pollutants in foods of animal origin are similar to those used for vegetables and fruits, with the exception of the requirement for efficient lipid separation. Although most foods are solids or solid–liquid mixtures, few studies regarding the detection of contaminations inside solid foods have been reported. This is because an extraction process is necessary prior to analysis to increase the analyte concentration and improve the SERS response.
Beverages and wine are liquids that are very suitable for SERS detection without pretreatment to test food quality or safety. For food quality testing, SERS substances, such as Ag NPs, can react with the components in red or white wine, trapping the color and flavor components and yielding a specific SERS signal for the sensitive detection of certain components [27]. The chemical or biological contaminants in beverages and wine mainly include ethyl carbamate, fusel oil, aldehyde, manganese, aflatoxin, patulin toxin, and N-nitrosodimethylamine, which can be detected using the label-free or label-based SERS methods. In addition to the conventional chemical or biological contaminants, harmful gas residues can be produced during fermentation and beverage production, for example, sulfite residues/SO2 [28]. The SO2 binds to the tertiary amine group on the Ag nanofilm substrate to realize detection by spectral changes, instead of sensing in the solution phase [29]. A gas-diffusion microfluidic paper-based analytical device combined with SERS was also applied for sulfite determination in wine [30], allowing the target analyte to be detected in the gas phase.
3.2. Biological Contaminant Detection by SERS
3.2.1. Mycotoxins Contaminants
In addition to chemical pollution, biological contaminants, including microorganisms and their toxic metabolites, viruses, parasites and their eggs, and vector insects, pose food safety risks. Among these, mold and mycotoxin contamination are the most common [31,32]. Unlike vegetables and fruits, grains and nuts require long-term storage facilities, and are more suitable for storage because of their lower water content. Current SERS detections are usually accomplished by label-free (direct) or label-based (indirect) methods. Although the mycotoxins are usually distributed on the surface of the solid or liquid food, extraction processes are necessary to yield accurate SERS results [33,34]. For label-based mycotoxin detection, the use of recognition elements for biological contaminants, such as antibody/immunoassays, aptamers, molecularly imprinted polymers (MIPs), linear polymer affinity agents (LPAA), and/or specific surface-modified nanomaterials, have been applied (Table 3) [35,36].
Table 3.
Label-free and label-based strategies for mold and its mycotoxins contamination detection by Raman/SERS in food.
| Strategies | Recognition Elements | SERS Substrates | Ref. |
|---|---|---|---|
| Label-free method | Crystal violet (CV) assay | AuNPs | [31] |
| D2O | — | [32] | |
| Label-based method | Antibody/immunoassays | Antigen-modified silica photonic crystal microspheres | [33] |
| Aptamers | Silica photonic crystal microsphere modified AuNPs | [34] | |
| Molecularly imprinted polymers | Surface-Imprinted Gold Nanoparticle | [35] | |
| Linear polymer affinity agents | Poly(N-acryloyl glycinamide) polymers modified AuNPs | [36] |
—: Not report.
3.2.2. Bacterial Contamination Detection by SERS
Biological contaminants are common in foods of animal origin. Label-free and label-based SERS methods can be applied equally here, but with slight differences [37]. Bacillus, a spore-forming bacterium, is commonly detected using a biomarker, such as calcium dipicolinate (CaDPA) or dipicolinic acid (DPA), associated with bacterial spores and can be detected by SERS [38]. The label-free SERS profiles of bacteria offer signatures of molecular structures, cellular compositions, and physiological states, and can be applied to discriminate between bacterial species or to distinguish between live and dead bacteria [39,40]. For the label-based SERS methods to detect bacterial contamination, recognition elements, such as antibodies, aptamers, or small-molecule ligands, must be cross-linked to the SERS tags to improve the specificity for the targeted bacterial pathogen (Table 4) [41,42].
Table 4.
Label-free and label-based strategies for bacterial contaminations detection by Raman/SERS in food.
| Strategies | Recognition Elements | Algorithms | SERS Substrates | Bacteria Species | Ref. |
|---|---|---|---|---|---|
| Spore | Dipicolinic acid | — | Gold nanoparticle-based substrates | Bacillus anthracis | [38] |
| Label-based method | Complementary DNAs | — | Au nanopillars | Enterococcus faecium and Staphylococcus aureus | [41] |
| Aptamers | — | Dendritic porous silica nanoparticles-Au-MBA-aptamer | Staphylococcus aureus | [42] | |
| Label-free method | — | Principal component analysis (PCA) | Au NPs | Seven foodborne bacteria (Salmonella typhimurium ATCC 50013, Salmonella O7HZ10, Shigella boydii CMCC51514, Shigella sonnei CMCC51529, Shigella dysenteriae CMCC51252, Citrobacter freundii ATCC43864, Enterobacter sakazakii 154) |
[37] |
| — | — | AgNPs | E. coli DSM 1116 | [39] | |
| — | PCA | Ag/Si substrate | C. Jejuni NCTC 11351, C. coli ATCC 33559, C. upsaliensis ATCC 43954, C. lari ATCC BAA-1060, P. aeruginosa ATCC 27853 |
[40] |
—: Not report.
3.3. Harmful Metal Contaminant Detection by SERS
Hg, Pb, Cd, As, Cr, Al, and F are commonly detected harmful elements in food. Their overall quantities and speciation are related to their accumulation capacity and biological toxicity. For SERS detection, these heavy metals are different from other chemical contaminants because they generally do not have chemical bonds that generate Raman scattering. Thus, only inorganic oxyanions and a few oxycations can be directly detected from their characteristic Raman bands. Label-free and label-based methods are suitable for detection, similar to other general chemical contaminants. For the label-free method, semiconductor substrates are prepared using metal oxides, which absorb metal cations [43,44]. For label-based methods, the formation of specific chemical bonds with heavy metal ions or extrinsic SERS probes are required for SERS detection. Generally, label-based methods for metal detection by SERS involves three types of recognition elements: Raman activity dye signal turn-on by the metal; Raman activity dye signal turn-off by the metal; or combination with other analytical techniques (Table 5) [45,46,47]. Therefore, novel SERS substrates should be developed to achieve a better selectivity and replace the requirement for separation.
Table 5.
Label-free and label-based strategies for harmful metal contaminants detection by SERS in food.
| Strategies | Recognition Elements | Harmful Metal Analytes | SERS Substrates | Ref. |
|---|---|---|---|---|
| Label-free method | — | Total As | Cu2O/Ag | [43] |
| — | Trace Cd2+ ions | Flower-like Ag@CuO | [44] | |
| Label-based method | Raman activity dye signal turn-on | Hg2+ | Single-stranded modified-DNA AuNPs | [45] |
| Raman activity dye signal turn-off | Hg2+ | Methimazole-functionalized and cyclodextrin-coated silver nanoparticles | [46] | |
| Combined with other analytical technologies | Cd2+ | Au@Ag core-shell nanoparticles | [47] |
—: Not report.
3.4. Food Allergen Detection by SERS
Food allergies that are triggered by the ingestion of food protein antigens represent an important food safety issue. These reactions are mediated by an immunological mechanism involving specific IgE or cell-mediated mechanisms, with 90% of allergic reactions originating from milk and dairy products, eggs and egg products, fish and shellfish, peanuts, beans and bean products, nuts, and wheat [48,49]. Therefore, it is particularly important to develop sensitive and effective detection methods for these food allergens. This is similar to the biological pollutant detection method described above, and the label-based method (indirect) is an ideal detection strategy. An aptamer-based SERS assay for β-lactoglobulin in milk samples was developed, with a detection limit of 0.07 ng/mL [50]. Peroxidase-mimicking nanozyme-catalyzing signal amplification as part of a portable SERS immunoassay for the food allergy protein α-lactalbumin was developed, with a detection limit of 0.01 ng/mL [51]. The developed label-based methods, such as aptamer-based SERS assays and nanozyme-enhanced SERS immunoassays, demonstrate a broad potential for food allergen supervision (Table 6).
Table 6.
Label-based SERS detection in food allergens.
3.5. Food Poisoning Detection by SERS
Food poisoning is an important food safety problem and encompasses non-infectious polar and subacute diseases that occur after ingestion of food containing biological and chemical toxins. The types of poisoning can be divided into four categories: bacterial, fungal, or chemical food poisoning, toxic animals, and plant-based food poisoning. The first three types are caused by different chemical or biological contaminants and were discussed in previous chapters. Herein, we focus on the phytotoxins and toxic animal-related food poisoning.
The phytotoxins are a class of natural organic compounds with high biological activities and toxicities. Poisonous plants, such as toadstools, cassava, green beans, sprouted potatoes, and fresh day lily, can cause poisoning due to improper cooking. Legumes are the main source of toxin poisoning, with major toxins including plant erythrocyte lectin, trypsin inhibitor, saponin glycoside, and phytic acid. Grains, such as the Solanaceae and Liliaceae plants, can cause poisoning through their secondary metabolites. Toxic animal-based food poisoning is often caused by specific species or tissues, with common examples including the puffer fish, shellfish, animal thyroid, liver, tetrodotoxin, shellfish toxin, and ichthyocholao toxin [52]. Among these chemicals, phytic acid, tetrodotoxin, and saxitoxin have been detected using SERS methods [53,54,55]. These SERS toxin detection methods include both label-free and label-based strategies, similar to chemical contamination detection. SERS has great potential for the identification of trace food poisoning during or after food processing, because of the low toxic analyte concentration and complex food matrix structure.
3.6. Food Authenticity Detection by SERS
The authenticity of food is another important research subject [56]. The adulteration of food products with cheaper materials for economic gain can pose serious health threats to consumers [57]. As vibrational spectroscopic techniques, Raman/SERS have the potential to fulfill the industrial need for food quality and authenticity analyses [56]. Two strategies to judge authenticity include: (1) combination with chemometrics to achieve the identification and quantification of food samples; (2) combination with specific target DNA-modified SERS substrates with a target-responsive Raman dye that can recognize the target (Table 7) [58,59]. The SERS biosensors are regarded as a universal platform for the on-site determination of food quality, authenticity, and safety [60].
Table 7.
Strategies of food freshness judgment by SERS methods.
| Strategies | Analysis | Algorithms | Ref. |
|---|---|---|---|
| Combined with chemometrics | Honey | Convolutional neural network | [58] |
| Olive oils | Integral ratio | [60] | |
| Combined with specific target DNA | Spiking little duck meat in lamb roll, pork, beef, mutton, and steak samples | — | [59] |
| Milk | — | [56] |
—: Not report.
4. SERS Applications in Processed Foods
4.1. Contaminants during Processing in Foods of Animal Origin
Notably, contaminants during the processing of foods of animal origin are chemicals generated when food constituents undergo chemical changes, including N-nitroso compounds, polycyclic aromatic hydrocarbons, acrylamide, and heterocyclic amines. There is a certain understanding of the formation mechanism of these compounds, but relevant reports remain sparse [61]. Many outstanding studies have developed SERS detection methods [62], but ultrasensitive, reliable, and facile detection technologies for processing contaminants and trace toxins in foods remain challenging to develop.
4.2. Oil Plants, Fats, and Fried Food
Edible oils can be classified based on their source as vegetable or animal oils, and their improper consumption and storage can endanger human health. Common oil quality and safety problems include rancidity, trans fatty acids produced during heating, and the presence of glucosinolate, erucic acid, and gossypol residues. These can be detected by confirming associated chemical substances. For example, malondialdehyde is a biomarker of lipid peroxidation that is traditionally associated with food rancidity [63]. However, it is worth noting that waste cooking oil from restaurants and street vendors, known as "recycled cooking oil”, contains significant amounts of endogenous pollutants. To this end, capsaicin was developed as a marker as part of a SERS methodology [64]. Moreover, during the processing of starch-rich grains, acrylamide is a major harmful chemical produced by frying and high-temperature baking [65]. Some of these chemical pollutants can be detected by SERS methods, but significant progress needs to be made to achieve widespread applicability.
4.3. Condiments
Soy sauce, vinegar, monosodium glutamate, sauces, and sugar are traditional condiments that can generate harmful substances during their production and fermentation. Chloropropanol is produced by hydrolyzing vegetable protein with hydrochloric acid and can be found in raw soy sauce, old soy sauce, and oyster sauce. However, a SERS detection method for chloropropanol has yet to be reported. Thus, more research is needed to rapidly detect these chemical substances in the condiment supply.
5. Genetically Modified Foods (GMFs) and Novel Foods
5.1. Genetically Modified Foods (GMFs)
The foods derived from genetically modified (GM) organisms are often referred to as GM foods (GMFs). The realization of GM technology in the agricultural system requires due diligence and in-depth analysis of the associated risks and/or benefits to multiple stakeholders on a case-to-case basis before commercialization [66]. For SERS detection, the rapid and simultaneous screening of multiple GM organism components (genes, promoters, codons, and terminators) in rice and soybean has been reported [67,68]. This DNA-conjugated SERS nano-tag detection strategy is similar to the authentication of foods of animal origin. This methodology can be applied in different detection scenarios, using the principle of similarity. Moreover, Raman is capable of extracting sample fingerprints and, in combination with chemometric tools, can discriminate transgenic corn with a predictive accuracy of 87.5% [69]. This also demonstrates the wide applicability of the SERS methodology (Table 8).
Table 8.
Label-free and label-based strategies of SERS detection of genetically modified organisms.
| Strategies | Analytes | Algorithms | Genetically Modified Organism Components | SERS Substrates | Ref. |
|---|---|---|---|---|---|
| Label-free SERS detection | Transgenic corn expressing specific Bacillus thuringiensis and Agrobacterium spp genes | Linear Discriminant Analysis | Chemical composition | — | [69] |
| Label-based SERS detection | Bacillus thuringiensis (Bt) gene-transformed rice | — | Bt gene | Au NPs | [67] |
| Genetically modified organism soybean | — | Promoter, codon, and terminator | Au@Ag | [68] |
—: Not report.
5.2. Novel Foods
‘Novel foods’ are newly developed, innovative food products produced using new technologies and production processes [70]. This includes meat alternatives and replacements produced from plant-based alternatives, as well as insects and fungi [71,72]. Current novel food regulations have adapted quickly to recognize the new food categories and add some to the existing food categories [73]. The qualitative and quantitative analytical data on substances hazardous to human health should also be provided [74]. SERS can provide a useful tool for recording full spectrum information, which is important for distinguishing the contaminants of unknown biological activity, including potential allergens.
6. Application and Major Challenges of SERS Application in Coronavirus Disease 2019
Viruses are common biological contaminants in food, and foodborne viruses use food as carriers to cause human diseases, including those transmitted by the fecal–oral route (such as polio-, rota-, and coronaviruses) and livestock products (e.g., such as avian influenza, prion, foot-and-mouth disease). The proliferation and transmission of viruses have become a threat to biosecurity worldwide, as exemplified by the coronavirus disease 2019 (COVID-19) pandemic. The available evidence shows that the COVID-19 virus does not spread through food, but it can contaminate food. Thus, nondestructive and rapid detection in food packaging, matrix, and processing are particularly important [75]. The SERS materials modified with short-structured oligonucleotides (DNA aptamers) provide excellent specificity for SERS biosensors [76]. If a nondestructive packaging detection device is successfully developed and proven to detect target viruses, it could enable rapid and mass screening of COVID-19 in food samples and be used in future pandemic screening.
7. Conclusions
Emerging Raman spectroscopic techniques, including Raman spectroscopy, Raman mapping, and SERS for rapid and nondestructive analysis, have been developed as effective tools for both qualitative and quantitative analyses in most of the food categories. SERS is based on inelastic scattering enhancement from molecules located near nanostructured metallic surfaces. This mechanism differs from the gold standard methods, chromatography, mass spectroscopy, and other spectroscopy methods, featuring advantages such as ultrasensitive detection, simple protocols, lack of pretreatment, and reduced costs. Semiconductor-metal hybrid substrates have great potential for the highly robust SERS sensing of pesticides with high enhancement ability, good reproducibility, acceptable stability, and reusability. The component and structural information contained in the SERS/Raman spectra can generate an electronic file of the food samples for subsequent reference and food production traceability. Label-free and label-based methods can be used for chemical and biological contaminant detection. The recognition elements including immunosensors, aptasensors, and MIPs can be linked to SERS tags to improve the specificity for target contaminants and allergens in GMF or novel foods. Importantly, the contaminant detection and quality analysis in actual food samples can often differ from the detection in standard solutions, owing to the complexity of the food matrices. Most foods are solid or solid–liquid mixtures, with only a few studies on contamination inside solid foods. Thus, extraction processes are required for the effective detection in both quality and safety analyses. The SERS detection method is a promising tool that can ensure food safety in an easy, rapid, reliable, and nondestructive manner. Thus, despite the persisting limitations for ingredient and freshness determination resulting from technical difficulties and the complexity of the food sample matrix, the continuous development of Raman systems, nanomaterials, and spectral analysis algorithms will facilitate the transfer from laboratory to industry. Industrial application will span the entire food production process and benefit from the on-line application of this technology.
Author Contributions
Conceptualization, M.-L.X.; writing—original draft preparation, M.-L.X. and Y.G.; writing—review and editing, X.-X.H. and B.Z.; funding acquisition, B.Z. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Education Department of Jilin Province (No. JJKH20211127KJ), the National Natural Science Foundation of the People’s Republic of China (No. 22011540378), the Development Program of the Science and Technology of Jilin Province (No. 20190701003GH), the Interdisciplinary Integration and Innovation Project of JLU (No. JLUXKJC2021QZ11), the China Postdoctoral Science Foundation (No. 2018M631876), the Open Research Fund of National Research Center of Engineering and Technology of Tea Quality and Safety (No. 2017NTQS0201), China Agriculture Research System of MOF and MARA (No. CARS-04). The APC was funded by 22011540378.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Xu M.-L., Gao Y., Wang X., Han X.X., Zhao B. Comprehensive strategy for sample preparation for the analysis of food contaminants and residues by GC–MS/MS: A review of recent research trends. Foods. 2021;10:2473. doi: 10.3390/foods10102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu M.-L., Liu J.-B., Lu J. Determination and control of pesticide residues in beverages: A review of extraction techniques, chromatography, and rapid detection methods. Appl. Spectrosc. Rev. 2014;49:97–120. [Google Scholar]
- 3.Zhu J., Jiang M., Ma H., Zhang H., Cheng W., Li J., Cai L., Han X.X., Zhao B. Redox-state-mediated regulation of cytochromec release in apoptosis revealed by surface-enhanced Raman scattering on nickel substrates. Angew. Chem. Int. Ed. 2019;58:16499–16503. doi: 10.1002/anie.201909638. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y., Ma H., Han X.X., Zhao B. Metal–Semiconductor heterostructures for surface-enhanced Raman scattering: Synergistic contribution of plasmons and charge transfer. Mater. Horiz. 2021;8:370–382. doi: 10.1039/D0MH01356K. [DOI] [PubMed] [Google Scholar]
- 5.Han X.X., Ji W., Zhao B., Ozaki Y. Semiconductor-enhanced Raman scattering: Active nanomaterials and applications. Nanoscale. 2017;9:4847–4861. doi: 10.1039/c6nr08693d. [DOI] [PubMed] [Google Scholar]
- 6.Xu M.-L., Gao Y., Han X.X., Zhao B. Detection of pesticide residues in food using surface-enhanced Raman spectroscopy: A review. J. Agric. Food Chem. 2017;65:6719–6726. doi: 10.1021/acs.jafc.7b02504. [DOI] [PubMed] [Google Scholar]
- 7.Xu M.L., Gao Y., Jin J., Xiong J.F., Han X.X., Zhao B. Role of 2−13C isotopic glyphosate adsorption on silver nanoparticles based on ninhydrin reaction: A study based on surface-enhanced Raman spectroscopy. Nanomaterials. 2020;10:2359. doi: 10.3390/nano10122539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., Ren X., Yu H., Cheng Y., Guo Y., Yao W., Xie Y. Non-destructive and online egg freshness assessment from the egg shell based on Raman spectroscopy. Food Control. 2020;118:107426–107437. doi: 10.1016/j.foodcont.2020.107426. [DOI] [Google Scholar]
- 9.Wang C., Sun Y., Zhou Y., Cui Y., Yao W., Yu H., Guo Y., Xie Y. Dynamic monitoring oxidation process of nut oils through Raman technology combined with PLSR and RF-PLSR model. LWT. 2021;146:111290–111298. [Google Scholar]
- 10.Nekvapil F., Brezestean I., Barchewitz D., Glamuzina B., Chiş V., Cintă Pinzaru S. Citrus fruits freshness assessment using Raman spectroscopy. Food Chem. 2018;242:560–567. doi: 10.1016/j.foodchem.2017.09.105. [DOI] [PubMed] [Google Scholar]
- 11.Naghdi S., Rezaei M., Bahramifar N., Kuswandi B. Preparation and characterization of intelligent color-changing nanosensor based on bromophenol blue and GONH2 nanosheet for freshness evaluation of minced Caspian sprat (Clupeonella cultriventris caspia) stored at 4 °C. Chem. Pap. 2022;76:3133–3146. [Google Scholar]
- 12.Camerlingo C., Portaccio M., Tatè R., Lepore M., Delfino I. Fructose and pectin detection in fruit-based food products by surface-enhanced Raman spectroscopy. Sensors. 2017;17:839. doi: 10.3390/s17040839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q., Xie Y., Yu H., Guo Y., Cheng Y., Yao W. Application of Raman spectroscopy in a correlation study between protein oxidation/denaturation and conformational changes in beef after repeated freeze–thaw. Int. J. Food Sci. Technol. 2022;57:719–727. doi: 10.1111/ijfs.15394. [DOI] [Google Scholar]
- 14.Qu Y., Tian Y., Chen Y., He L. Chemical profiling of red wines using surface-enhanced Raman spectroscopy (SERS) Anal. Methods. 2020;12:1324–1332. doi: 10.1039/D0AY00099J. [DOI] [Google Scholar]
- 15.Zanuttin F., Gurian E., Ignat I., Fornasaro S., Calabretti A., Bigot G., Bonifacio A. Characterization of white wines from north-eastern Italy with surface-enhanced Raman spectroscopy. Talanta. 2019;203:99–105. doi: 10.1016/j.talanta.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 16.Wang K., Li Z., Li J., Lin H. Raman spectroscopic techniques for nondestructive analysis of agri-foods: A state-of-the-art review. Trends Food Sci. Technol. 2021;118:490–504. doi: 10.1016/j.tifs.2021.10.010. [DOI] [Google Scholar]
- 17.Wu J., Zhang L., Bu X., Li P., Zhao B., Tian Y. Determination of the illegal adulteration of natural healthcare products with chemical drugs using surface-enhanced Raman scattering. Analyst. 2018;143:5202–5209. doi: 10.1039/C8AN01286E. [DOI] [PubMed] [Google Scholar]
- 18.Yu H., Xu L., Yang F., Xie Y., Guo Y., Cheng Y., Yao W. Rapid surface-enhanced Raman spectroscopy detection of chlorothalonil in standard solution and orange peels with pretreatment of ultraviolet irradiation. Bull. Environ. Contam. Toxicol. 2021;107:221–227. doi: 10.1007/s00128-021-03258-9. [DOI] [PubMed] [Google Scholar]
- 19.Gao Y., Xu M.-L., Xiong J. Raman and SERS spectra of thiamethoxam and the Ag3–thiamethoxam complex: An experimental and theoretical investigation. J. Environ. Sci. Health Part B. 2019;54:665–675. doi: 10.1080/03601234.2019.1631099. [DOI] [PubMed] [Google Scholar]
- 20.Xu M.-L., Gao Y., Li Y., Li X., Zhang H., Han X.X., Zhao B., Su L. Indirect glyphosate detection based on ninhydrin reaction and surface-enhanced Raman scattering spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018;197:78–82. doi: 10.1016/j.saa.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y., Li W., Zhao L., Li F., Xie Y., Yao W., Liu W., Lin Z. Simultaneous SERS detection of illegal food additives rhodamine B and basic orange II based on Au nanorod-incorporated melamine foam. Food Chem. 2021;357:129741–129749. doi: 10.1016/j.foodchem.2021.129741. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J., Wang D., Yang H., Wang F. Specific detection of acetamiprid with aptamer based on flexible and adhesive SERS membrane. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022;270:120801–120807. doi: 10.1016/j.saa.2021.120801. [DOI] [PubMed] [Google Scholar]
- 23.Sun C., Chen T., Ruan W., Jung Y.M., Cong Q., Zhao B. A simple strategy to improve the sensitivity of probe molecules on SERS substrates. Talanta. 2019;195:221–228. doi: 10.1016/j.talanta.2018.11.040. [DOI] [PubMed] [Google Scholar]
- 24.Jin J., Song W., Zhang N., Li L., Liu H., Yang B., Zhao B. Highly efficient core–shell Ag@carbon dot modified TiO2 nanofibers for photocatalytic degradation of organic pollutants and their SERS monitoring. RSC Adv. 2020;10:26639–26645. doi: 10.1039/D0RA00168F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowicka A.B., Czaplicka M., Kowalska A.A., Szymborski T., Kamińska A. Flexible PET/ITO/Ag SERS platform for label-free detection of pesticides. Biosensors. 2019;9:111. doi: 10.3390/bios9030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Y., Zhao M., Hu Q., Cheng Y., Guo Y., Qian H., Yao W. Selective detection of chloramphenicol in milk based on a molecularly imprinted polymer–surface-enhanced Raman spectroscopic nanosensor. J. Raman Spectrosc. 2017;48:204–210. doi: 10.1002/jrs.5034. [DOI] [Google Scholar]
- 27.Pinzaru S.C., Magdas D.A. Ag nanoparticles meet wines: SERS for wine analysis. Food Anal. Methods. 2018;11:892–900. doi: 10.1007/s12161-017-1056-2. [DOI] [Google Scholar]
- 28.Kong D., Zhu W., Li M. A facile and sensitive SERS-based platform for sulfite residues/SO2 detection in food. Microchem. J. 2021;165:106174–106184. doi: 10.1016/j.microc.2021.106174. [DOI] [Google Scholar]
- 29.Mandrile L., Cagnasso I., Berta L., Giovannozzi A.M., Petrozziello M., Pellegrino F., Asproudi A., Durbiano F., Rossi A.M. Direct quantification of sulfur dioxide in wine by surface enhanced Raman spectroscopy. Food Chem. 2020;326:127009–127016. doi: 10.1016/j.foodchem.2020.127009. [DOI] [PubMed] [Google Scholar]
- 30.Chen M., Yang H., Rong L., Chen X. A gas-diffusion microfluidic paper-based analytical device (μPAD) coupled with portable surface-enhanced Raman scattering (SERS): Facile determination of sulphite in wines. Anal. 2016;141:5511–5519. doi: 10.1039/C6AN00788K. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y., Yu H., Cheng Y., Guo Y., Yao W., Xie Y. Non-destructive monitoring of staphylococcus aureus biofilm by surface-enhanced Raman scattering spectroscopy. Food Anal. Methods. 2020;13:1710–1716. [Google Scholar]
- 32.Feng J., Yao W., Guo Y., Cheng Y., Qian H., Xie Y. Incorporation of heavy water for rapid detection of Salmonella typhimurium by Raman microspectroscopy. Food Anal. Methods. 2018;11:3551–3557. doi: 10.1007/s12161-018-1328-5. [DOI] [Google Scholar]
- 33.Sun J., Li W., Zhu X., Jiao S., Chang Y., Wang S., Dai S., Xu R., Dou M., Li Q., et al. A novel multiplex mycotoxin surface-enhanced Raman spectroscopy immunoassay using functional gold nanotags on a silica photonic crystal microsphere biochip. J. Agric. Food Chem. 2021;69:11494–11501. doi: 10.1021/acs.jafc.1c03469. [DOI] [PubMed] [Google Scholar]
- 34.Song L., Li J., Li H., Chang Y., Dai S., Xu R., Dou M., Li Q., Lv G., Zheng T. Highly sensitive SERS detection for Aflatoxin B1 and Ochratoxin A based on aptamer-functionalized photonic crystal microsphere array. Sens. Actuators B Chem. 2022;364:131778–131788. doi: 10.1016/j.snb.2022.131778. [DOI] [Google Scholar]
- 35.Wu L., Yan H., Li G., Xu X., Zhu L., Chen X., Wang J. Surface-imprinted gold nanoparticle-based surface-enhanced Raman scattering for sensitive and specific detection of patulin in food samples. Food Anal. Methods. 2019;12:1648–1657. doi: 10.1007/s12161-019-01498-4. [DOI] [Google Scholar]
- 36.Szlag V.M., Rodriguez R.S., Jung S., Bourgeois M.R., Bryson S., Purchel A., Schatz G.C., Haynes C.L., Reineke T.M. Optimizing linear polymer affinity agent properties for surface-enhanced Raman scattering detection of Aflatoxin B1. Mol. Syst. Des. Eng. 2019;4:1019–1031. [Google Scholar]
- 37.Xie Y., Xu L., Wang Y., Shao J., Wang L., Wang H., Qian H., Yao W. Label-free detection of the foodborne pathogens of enterobacteriaceae by surface-enhanced Raman spectroscopy. Anal. Methods. 2013;5:946–952. doi: 10.1039/C2AY26107C. [DOI] [Google Scholar]
- 38.Cheng H.-W., Huan S.-Y., Yu R.-Q. Nanoparticle-based substrates for surface-enhanced Raman scattering detection of bacterial spores. Analyst. 2012;137:3601–3608. doi: 10.1039/c2an35448a. [DOI] [PubMed] [Google Scholar]
- 39.Zhou H., Yang D., Ivleva N.P., Mircescu N.E., Schubert S., Niessner R., Wieser A., Haisch C. Label-free in situ discrimination of live and dead bacteria by surface-enhanced Raman scattering. Anal. Chem. 2015;87:6553–6561. doi: 10.1021/acs.analchem.5b01271. [DOI] [PubMed] [Google Scholar]
- 40.Witkowska E., Niciński K., Korsak D., Dominiak B., Waluk J., Kamińska A. Nanoplasmonic sensor for foodborne pathogens detection. Towards development of ISO-SERS methodology for taxonomic affiliation of Campylobacter spp. J. Biophotonics. 2020;13:e201960227. doi: 10.1002/jbio.201960227. [DOI] [PubMed] [Google Scholar]
- 41.Lee S.H., Lee W.-C., Koh E.H., Ansah I.B., Yang J.-Y., Mun C., Lee S., Kim D.-H., Jung H.S., Park S.-G. Organometallic hotspot engineering for ultrasensitive EC-SERS detection of pathogenic bacteria-derived DNAs. Biosens. Bioelectron. 2022;210:114325–114332. doi: 10.1016/j.bios.2022.114325. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Du X., Wang X., Yan T., Yuan M., Yang Y., Jurado-Sánchez B., Escarpa A., Xu L.-P. Patterned liquid-infused nanocoating integrating a sensitive bacterial sensing ability to an antibacterial surface. ACS Appl. Mater. Interfaces. 2022;14:23129–23138. doi: 10.1021/acsami.1c24821. [DOI] [PubMed] [Google Scholar]
- 43.Barimah A.O., Guo Z., Agyekum A.A., Guo C., Chen P., El-Seedi H.R., Zou X., Chen Q. Sensitive label-free Cu2O/Ag fused chemometrics SERS sensor for rapid detection of total arsenic in tea. Food Control. 2021;130:108341–108351. doi: 10.1016/j.foodcont.2021.108341. [DOI] [Google Scholar]
- 44.Cheng M., Li C., Li W., Liu Y. Trace Cd2+ ions detection on the flower-like Ag@CuO substrate. Nanomaterials. 2020;10:1664. doi: 10.3390/nano10091664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun B., Jiang X., Wang H., Song B., Zhu Y., Wang H., Su Y., He Y. Surface-enhancement Raman scattering sensing strategy for discriminating trace mercuric ion (II) from real water samples in sensitive, specific, recyclable, and reproducible manners. Anal. Chem. 2015;87:1250–1256. doi: 10.1021/ac503939d. [DOI] [PubMed] [Google Scholar]
- 46.Ma P., Liang F., Yang Q., Wang D., Sun Y., Wang X., Gao D., Song D. Highly sensitive SERS probe for mercury(II) using cyclodextrin-nanoprotected silver particles functionalized with methimazole. Microchim. Acta. 2014;181:975–981. doi: 10.1007/s00604-014-1196-7. [DOI] [Google Scholar]
- 47.Fu Q., Liu H.L., Wu Z., Liu A., Yao C., Li X., Xiao W., Yu S., Luo Z., Tang Y. Rough surface Au@Ag core–shell nanoparticles to fabricating high sensitivity SERS immunochromatographic sensors. J. Nanobiotech. 2015;13:81–90. doi: 10.1186/s12951-015-0142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xi J., Yu Q. The development of lateral flow immunoassay strip tests based on surface enhanced Raman spectroscopy coupled with gold nanoparticles for the rapid detection of soybean allergen β-conglycinin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020;241:118640–118648. doi: 10.1016/j.saa.2020.118640. [DOI] [PubMed] [Google Scholar]
- 49.Styles M.J., Rodriguez R.S., Szlag V.M., Bryson S., Gao Z., Jung S., Reineke T.M., Haynes C.L. Optimization of film over nanosphere substrate fabrication for SERS sensing of the allergen soybean agglutinin. J. Raman Spectrosc. 2021;52:482–490. doi: 10.1002/jrs.6019. [DOI] [Google Scholar]
- 50.Duan N., Yao T., Li C., Wang Z., Wu S. Surface-enhanced Raman spectroscopy relying on bimetallic Au–Ag nanourchins for the detection of the food allergen β-lactoglobulin. Talanta. 2022;245:123445–123453. doi: 10.1016/j.talanta.2022.123445. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X., Li G., Liu J., Su Z. Bio-inspired nanoenzyme synthesis and its application in a portable immunoassay for food allergy proteins. J. Agric. Food Chem. 2021;69:14751–14760. doi: 10.1021/acs.jafc.1c04309. [DOI] [PubMed] [Google Scholar]
- 52.Neng J., Zhang Q., Sun P. Application of surface-enhanced Raman spectroscopy in fast detection of toxic and harmful substances in food. Biosens. Bioelectron. 2020;167:112480. doi: 10.1016/j.bios.2020.112480. [DOI] [PubMed] [Google Scholar]
- 53.Shen S., Guo X.-y., Song P., Pan Y.-C., Wang H.-q., Wen Y., Yang H.-F. Phytic acid adsorption on the copper surface: Observation of electrochemistry and Raman spectroscopy. Appl. Surf. Sci. 2013;276:167–173. doi: 10.1016/j.apsusc.2013.03.061. [DOI] [Google Scholar]
- 54.Yin L., Fan M., She Q., You R., Lu Y., Lu D., Li M. Facilely self-assembled and dual-molecule calibration aptasensor based on SERS for ultra-sensitive detection of tetrodotoxin in pufferfish. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022;279:121275. doi: 10.1016/j.saa.2022.121275. [DOI] [PubMed] [Google Scholar]
- 55.Cao C., Li P., Liao H., Wang J., Tang X., Yang L. Cys-functionalized AuNP substrates for improved sensing of the marine toxin STX by dynamic surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 2020;412:4609–4617. doi: 10.1007/s00216-020-02710-9. [DOI] [PubMed] [Google Scholar]
- 56.Pan R., Liu J., Wang P., Wu D., Chen J., Wu Y., Li G. Ultrasensitive CRISPR/Cas12a-driven SERS biosensor for on-site nucleic acid detection and its application to milk authenticity testing. J. Agric. Food Chem. 2022;70:4484–4491. doi: 10.1021/acs.jafc.1c08262. [DOI] [PubMed] [Google Scholar]
- 57.Lohumi S., Lee S., Lee H., Cho B.-K. A review of vibrational spectroscopic techniques for the detection of food authenticity and adulteration. Trends Food Sci. Technol. 2015;46:85–98. [Google Scholar]
- 58.Wu X., Xu B., Ma R., Niu Y., Gao S., Liu H., Zhang Y. Identification and quantification of adulterated honey by Raman spectroscopy combined with convolutional neural network and chemometrics. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022;274:121133. doi: 10.1016/j.saa.2022.121133. [DOI] [PubMed] [Google Scholar]
- 59.Liu J., Chen J., Wu D., Huang M., Chen J., Pan R., Wu Y., Li G. CRISPR-/Cas12a-mediated liposome-amplified strategy for the surface-enhanced Raman scattering and naked-eye detection of nucleic acid and application to food authenticity screening. Anal. Chem. 2021;93:10167–10174. doi: 10.1021/acs.analchem.1c01163. [DOI] [PubMed] [Google Scholar]
- 60.Chavez-Angel E., Puertas B., Kreuzer M., Soliva Fortuny R., Ng R.C., Castro-Alvarez A., Sotomayor Torres C.M. Spectroscopic and thermal characterization of extra virgin olive oil adulterated with edible oils. Foods. 2022;11:1304. doi: 10.3390/foods11091304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M., Yu H., Cheng Y., Guo Y., Yao W., Xie Y. Simultaneous and rapid determination of polycyclic aromatic hydrocarbons by facile and green synthesis of silver nanoparticles as effective SERS substrate. Ecotoxicol. Environ. Saf. 2020;200:110780. doi: 10.1016/j.ecoenv.2020.110780. [DOI] [PubMed] [Google Scholar]
- 62.Liu S., Huo Y., Deng S., Li G., Li S., Huang L., Ren S., Gao Z. A facile dual-mode aptasensor based on AuNPs@MIL-101 nanohybrids for ultrasensitive fluorescence and surface-enhanced Raman spectroscopy detection of tetrodotoxin. Biosens. Bioelectron. 2022;201:113891. doi: 10.1016/j.bios.2021.113891. [DOI] [PubMed] [Google Scholar]
- 63.Zhang D., Haputhanthri R., Ansar S.M., Vangala K., De Silva H.I., Sygula A., Saebo S., Pittman C.U. Ultrasensitive detection of mahylondialdede with surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 2010;398:3193–3201. doi: 10.1007/s00216-010-4225-3. [DOI] [PubMed] [Google Scholar]
- 64.Liu S.-H., Lin X.-M., Yang Z.-L., Wen B.-Y., Zhang F.-L., Zhang Y.-J., Li J.-F. Label-free SERS strategy for rapid detection of capsaicin for identification of waste oils. Talanta. 2022;245:123488. doi: 10.1016/j.talanta.2022.123488. [DOI] [PubMed] [Google Scholar]
- 65.Wu L., Zhang W., Liu C., Foda M.F., Zhu Y. Strawberry-like SiO2/Ag nanocomposites immersed filter paper as SERS substrate for acrylamide detection. Food Chem. 2020;328:127106. doi: 10.1016/j.foodchem.2020.127106. [DOI] [PubMed] [Google Scholar]
- 66.Sendhil R., Nyika J., Yadav S., Mackolil J., Workie E., Ragupathy R., Ramasundaram P. Genetically modified foods: Bibliometric analysis on consumer perception and preference. GM Crops Food. 2022;13:65–85. doi: 10.1080/21645698.2022.2038525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen K., Han H., Luo Z., Wang Y., Wang X. A practicable detection system for genetically modified rice by SERS-barcoded nanosensors. Biosens. Bioelectron. 2012;34:118–124. doi: 10.1016/j.bios.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 68.Yao L., Xu J., Cheng J., Yao B., Zheng L., Liu G., Chen W. Simultaneous and accurate screening of multiple genetically modified organism (GMO) components in food on the same test line of SERS-integrated lateral flow strip. Food Chem. 2022;366:130595. doi: 10.1016/j.foodchem.2021.130595. [DOI] [PubMed] [Google Scholar]
- 69.Dib S.R., Silva T.V., Neto J.A.G., Guimarães L.J.M., Ferreira E.J., Ferreira E.C. Raman spectroscopy for discriminating transgenic corns. Vib. Spectrosc. 2021;112:103183–103188. doi: 10.1016/j.vibspec.2020.103183. [DOI] [Google Scholar]
- 70.Tuorila H., Hartmann C. Consumer responses to novel and unfamiliar foods. Curr. Opin. Food Sci. 2020;33:1–8. [Google Scholar]
- 71.Parodi A., Leip A., De Boer I.J.M., Slegers P.M., Ziegler F., Temme E.H.M., Herrero M., Tuomisto H., Valin H., Van Middelaar C.E., et al. The potential of future foods for sustainable and healthy diets. Nat. Sustain. 2018;1:782–789. doi: 10.1038/s41893-018-0189-7. [DOI] [Google Scholar]
- 72.Gao Y., Zhao Y.-J., Xu M.-L., Shi S.-S. Soybean hawkmoth (Clanis bilineata tsingtauica) as food ingredients: A review. CyTA—J. Food. 2021;19:341–348. doi: 10.1080/19476337.2021.1903082. [DOI] [Google Scholar]
- 73.Gao Y., Wang D., Xu M.-L., Shi S.-S., Xiong J.-F. Toxicological characteristics of edible insects in China: A historical review. Food Chem. Toxicol. 2018;119:237–251. doi: 10.1016/j.fct.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 74.Ververis E., Ackerl R., Azzollini D., Colombo P.A., de Sesmaisons A., Dumas C., Fernandez-Dumont A., Ferreira da Costa L., Germini A., Goumperis T., et al. Novel foods in the European Union: Scientific requirements and challenges of the risk assessment process by the European food safety authority. Food Res. Int. 2020;137:109515. doi: 10.1016/j.foodres.2020.109515. [DOI] [PubMed] [Google Scholar]
- 75.Abid S.A., Ahmed Muneer A., Al-Kadmy I.M.S., Sattar A.A., Beshbishy A.M., Batiha G.E.-S., Hetta H.F. Biosensors as a future diagnostic approach for COVID-19. Life Sci. 2021;273:119117. doi: 10.1016/j.lfs.2021.119117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zavyalova E., Ambartsumyan O., Zhdanov G., Gribanyov D., Gushchin V., Tkachuk A., Rudakova E., Nikiforova M., Kuznetsova N., Popova L., et al. SERS-based aptasensor for rapid quantitative detection of SARS-CoV-2. Nanomaterials. 2021;11:1394. doi: 10.3390/nano11061394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.