Summary
Background
From consumption of fermented foods and probiotics to emerging applications of faecal microbiota transplantation, the health benefit of manipulating the human microbiota has been exploited for millennia. Despite this history, recent technological advances are unlocking the capacity for targeted microbial manipulation as a novel therapeutic.
Aim
This review summarises the current developments in microbiome‐based medicines and provides insight into the next steps required for therapeutic development.
Methods
Here we review current and emerging approaches and assess the capabilities and weaknesses of these technologies to provide safe and effective clinical interventions. Key literature was identified through Pubmed searches with the following key words, ‘microbiome’, ‘microbiome biomarkers’, ‘probiotics’, ‘prebiotics’, ‘synbiotics’, ‘faecal microbiota transplant’, ‘live biotherapeutics’, ‘microbiome mimetics’ and ‘postbiotics’.
Results
Improved understanding of the human microbiome and recent technological advances provide an opportunity to develop a new generation of therapies. These therapies will range from dietary interventions, prebiotic supplementations, single probiotic bacterial strains, human donor‐derived faecal microbiota transplants, rationally selected combinations of bacterial strains as live biotherapeutics, and the beneficial products or effects produced by bacterial strains, termed microbiome mimetics.
Conclusions
Although methods to identify and refine these therapeutics are continually advancing, the rapid emergence of these new approaches necessitates accepted technological and ethical frameworks for measurement, testing, laboratory practices and clinical translation.
Microbiome‐based medicines include biomarkers, where patients are screened monitored and stratified, and therapeutics, where there are currently nine forms of therapeutics: dietary interventions, prebiotics, probiotics, synbiotics, antibiotics, faecal microbiota transplantation, phage therapy, live biotherapeutics and microbiome mimetics.
1. INTRODUCTION
The gastrointestinal (GI) microbiome is known to play an integral role in overall homeostasis; however, alterations can lead to the development and progression of disease. These complex communities contain between 100 and 1000 bacterial species all of which have the ability to interact with the host in different ways. The concept of altering the GI microbiome to improve health outcomes is now well established in modern medicine. Microbiome‐based medicines can fall into two categories, microbiome‐based biomarkers, and therapeutics (Figure 1). Although some dietary interventions, prebiotics, probiotics, antibiotics and faecal microbiota transplant (FMT) are well‐established therapeutics, recent work has raised the possibility of live biotherapeutics, and phage therapies for managing and treating a large array of diseases 1 , 2 , 3 , 4 , 5 , 6 (Figure 1). With the expansion of diverse microbially targeted therapies, coupled with an increasing availability of cost‐effective gut metagenomic profiling, it is timely to critically evaluate current capabilities and determine fundamental areas on which to focus future research.
FIGURE 1.
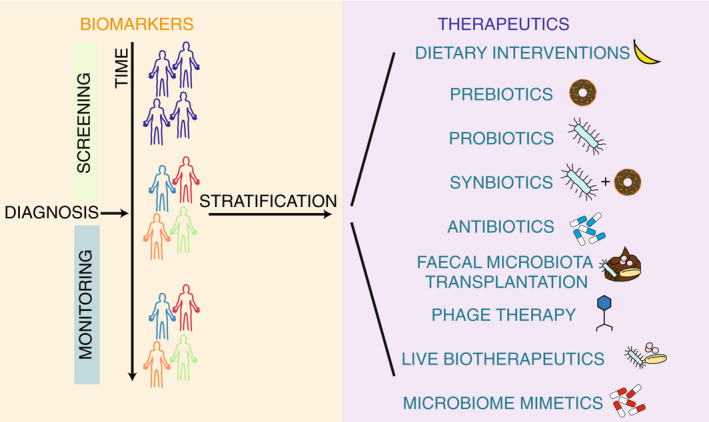
Overview of the different uses of the microbiome for medicine. Microbiota uses include biomarkers (orange box), where patients are screened monitored and stratified, and therapeutics (purple box), where there are currently nine forms of therapeutics: Dietary interventions, prebiotics, probiotics, synbiotics, antibiotics, faecal microbiota transplantation, phage therapy, live biotherapeutics and microbiome mimetics.
2. BIOMARKERS FOR DIAGNOSIS AND TREATMENT
The microbiome, particularly the gut microbiome, shares an expansive interface with the host immune system, rendering it an excellent candidate for biomarker development (Figure 2). Advances in metagenomic sequencing technologies have improved characterisation of microbial communities and provided associations with disease phenotypes. 1 , 7 , 8 This has facilitated the identification of potential microbial disease biomarkers in type 2 diabetes, 1 colorectal cancer, 2 liver cirrhosis 9 and hepatocellular carcinoma. 3 For example, a decrease in the abundance of butyrate producing bacteria is indicative of type 2 diabetes, 1 and an increase in Fusobacterium and Porphyromonas is a biomarker for colorectal cancer. 2 In addition to the identification of biomarkers for diagnosis of disease, microbial biomarkers are being developed to stratify patient cohorts prior to treatment to ensure patients receive the best treatment for them. Furthermore, biomarkers can be used to monitor patients following treatment to ensure treatment efficacy (Figure 2). Currently, the causal relationship between the microbiome and these disease states is unknown and microbial strain level granularity is largely lacking. Given the complexity of these microbiome–disease interactions future biomarkers may require microbial signatures comprised of multiple bacteria or bacterial functions as disease biomarkers. Furthermore, identifying and validating key species or functions may complement or reduce the need for expensive scans and invasive biopsies for patients.
FIGURE 2.
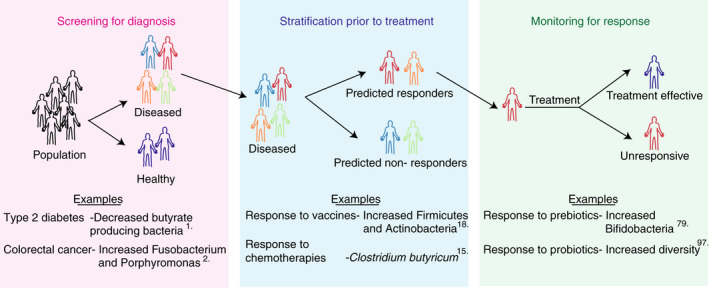
Categorisation of microbiome‐based biomarkers for disease. Microbiome‐based biomarkers can be classed as tools for screening for diagnosis (pink box), stratification prior to treatment (blue box) and monitoring for response to treatment (green box).
Understanding interactions between the microbiome and therapeutic response provides the opportunity for tailored interventions to achieve optimal outcomes or avoid adverse reactions. 10 , 11 In this context, microbiome‐based patient stratification to appropriately target existing therapies to specific patients 12 , 13 , 14 , 15 , 16 and to define responses to vaccines and other therapies, 17 , 18 , 19 , 20 represent two emerging areas for the application of microbiome‐based technologies.
The importance of microbiome diversity on vaccine response is exemplified by studies in paediatric cohorts. These studies suggest greater bacterial diversity in the GI tract correlates with an increased immune response to a variety of vaccines, including the oral rotavirus and polio vaccines, the intramuscular hepatitis B vaccine and the intradermal BCG vaccines. 10 , 18 , 19 Differences in the microbiome have been associated with lower efficacy of these vaccines in lower‐income nations. 17 , 18 , 19 , 20 Specifically, studies have demonstrated that an increase in the abundance of bacteria from the Firmicutes or Actinobacteria phyla is associated with a greater production of antibodies in response to vaccines. 18 Furthermore, patients with a higher relative abundance of Proteobacteria and Bacteroidetes had a lower antibody titre following immunisation. 17 , 18 , 19 , 20 This is consistent with previous studies that link the GI microbiome with immune development, tolerance and priming. 21 In this context, the microbiome has been linked to a decrease in the development of allergies, through anti‐inflammatory effects of microbial metabolites such as long and short‐chain fatty acids (SCFAs). 22 , 23 , 24 Although understanding the proportions of certain bacterial phyla within the microbiome may be the first step in predicting a patient's response to a vaccine, a higher‐resolution taxonomic classification may be important given species and strain‐level functional differences. 25 Determining species and strain‐level variation could allow for determination of causation, leading to personalised microbial therapeutic options with increased efficacy.
Improved patient responses to chemotherapies, radiation and immunotherapies have been associated with a more diverse GI microbiome and key bacterial species.
Reducing microbiome diversity through an antibiotic cocktail, prior to chemotherapy with oxaliplatin or cisplatin for subcutaneous lymphoma in a T‐cell lymphoma–induced mouse model, reduced the efficacy of both treatments. 26 Furthermore, the depletion of microbiome diversity through antibiotic treatment has been observed to decrease patient response to immune checkpoint inhibitors, such as anti‐programmed cell death protein 1 (PD‐1) immune checkpoint inhibitor (anti‐PD‐1) and cytotoxic T‐lymphocyte‐associated protein 4 (CTLA‐4). 27 , 28 Although the relationship between general loss of diversity and adverse outcomes is clear, identification of key microbes as biomarkers requires a more detailed analysis.
Emerging evidence has identified key relationships between Ruminococcaceae/Faecalibacterium strains and Akkermansia muciniphila with some immunotherapies, 23 Clostridium butyricum with chemotherapies 15 and Lactobacillus rhamnosus GG with radiation therapy. 15 , 16 , 29 The association of Ruminococcaceae/Faecalibacterium strains with improved metastatic melanoma patient outcomes was identified following treatment with anti‐PD‐1. 14 Furthermore, when A. muciniphila, is administered to mice through five oral gavages, treatment efficacy of anti‐PD‐1 increased, and was associated with increased microbiota diversity with a specific increase in A. muciniphila. 28 Similarly, administration of C. butyricum, to patients undergoing chemotherapy for lung cancer, 15 and L. rhamnosus GG to patients undergoing radiation therapy, 16 have been shown to decrease diarrhoeal incidents and intestinal mucosal disruption. This correlates with a reduction in adverse event related cessation of treatment. 15 , 16 These results are consistent with murine studies showing administration of L. rhamnosus GG is radioprotective through Toll‐like Receptor 2 and cyclooxygenase‐2 mediated secretion of radioprotective prostaglandin E2, which mitigates intestinal cell damage. 30
As most of the evidence for the use of microbiota as biomarkers has been identified in murine studies, it important to note, that these biomarkers may not be applicable to humans. 31 Indeed, recent studies suggest that only 2.58% of bacterial species are found in both human and mouse GI microbiomes. 31 Therefore, more research is required to ensure functionally equivalent biomarkers are identified in humans. Despite these challenges, in the near future microbiome‐based screening prior to the initiation of some cancer therapies may provide the opportunity to supplement the microbial communities in patients to improve potential responses and outcomes. Similarly, many therapeutic interventions can alter the patient's microbiome composition. This provides the opportunity to develop biomarkers to monitor treatment progression and success. Following treatments for diseases such as irritable bowel syndrome (IBS) and inflammatory bowel diseases (IBDs), the microbiome could be monitored for changes, including increases in diversity or abundance of key species. Thus, biomarkers could be used preceding treatment to determine potential efficacy and post treatment to monitor outcomes.
3. THERAPEUTICS
Unlike prognostic biomarkers, that may be or show a causal relationship, therapeutic intervention requires a causal relationship between the microbes and disease states. There is increasing evidence that changes to microbiome composition, specifically diversity loss and a decrease in bacterial load within the GI system, may be associated with a deleterious effect on the host. 32 This phenomenon is commonly referred to as dysbiosis and is correlated with many GI and metabolic diseases, including diarrhoea, Clostridioides difficile infection (CDI), IBD and type 2 diabetes. 33 , 34 , 35 , 36 , 37 Microbiome‐based therapies include dietary interventions, prebiotics, probiotics, antibiotics, phage therapy, FMT, live biotherapeutics and microbiome mimetics (Figure 3; Table 1), each aiming to modify the microbiome to treat diseases.
FIGURE 3.
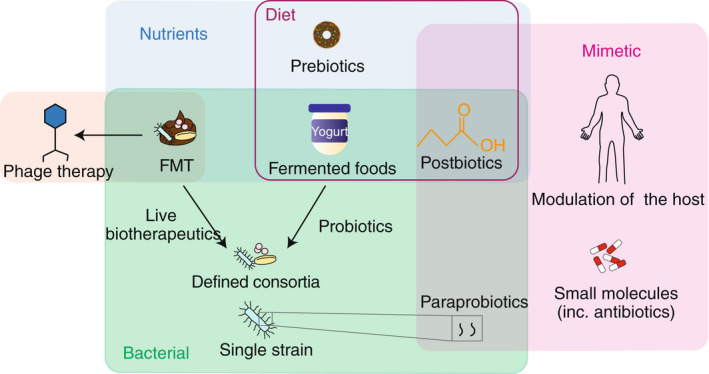
Categorisation of microbiome‐based therapeutics. Microbiome‐based therapeutics can be categorised as nutrients (blue box), bacterial (green box), or microbiome mimetics (pink box). Many therapeutics can be found within the diet (purple box) but are composed of components that are nutrient, bacterial and mimetics.
TABLE 1.
The advantages, disadvantages and future direction/ implementations for microbiome‐based therapeutics
| Therapeutic | Advantages | Disadvantages | Future directions/implications |
|---|---|---|---|
| Dietary interventions |
• Safe • Easily manipulated |
• Variable components in each food item • Insufficient dose for therapeutic benefit • Temporary therapeutic response |
Further work required to identify key components of diet that can be altered to allow for a therapeutic response |
| Prebiotics |
• Safe • Components of food • Easily administered |
• Dependent on specific microbe colonisation • Dependent on gut microenvironment • Therapeutic response temporary • Potential adverse responses (e.g. bloating) |
Potential in prevention of paediatric immune diseases (e.g. respiratory disease and allergy). Prebiotics should be examined for their treatment of other conditions |
| Probiotics |
• Relatively safe • Readily available as standardised mix |
• Not targeted to a disease or patient • Dependent on specific microbe colonisation • Dependent on gut microenvironment • Therapeutic response temporary • Viability not requirement of regulator |
Efficacious following antibiotics and in the prevention of NEC. Potential as non‐specific treatments to increase bacterial diversity |
| Synbiotics |
• Relatively safe • Includes all components for efficacy |
• Therapeutic response temporary • Require a specific gut microenvironment • Potential adverse responses (e.g. post antibiotics) |
Efficacious in the treatment of metabolic diseases. Further combinations should be explored for the treatment of other diseases |
| Antibiotics |
• Safe • Cheap • Approved medication • Existing regulatory framework |
• Potential off‐target effects (antibiotic resistance, disruption of colonisation resistance) • Limited to disruption of the microbiota |
Examination for use in targeted microbiome manipulation; however, caution is required to avoid off‐target, adverse effects |
| Phage therapy | • Highly specific |
• Limited to disruption of the microbiota • Targets require specific development • Emerging therapy |
Examination for use in altering microbiome structure due to their highly specific nature |
| FMT |
• Contains all microbes and nutrients • Proven efficacious for Clostridioides difficile treatment |
• Donor variability • Requires rigorous pre‐screening • Efficacy only seen for some conditions • Some administration costly • Inability to standardise composition |
Further work is required to determine causality in FMT treatment. This will allow for FMT to be considered for the treatment of other diseases |
| Live biotherapeutics | • Approved for specific indications |
• Requires maintenance of bacterial viability • Potential adverse long‐term health effects • Difficulty determining causal relationship |
Determination of causality required to allow for development |
| Microbiome mimetics | • Not reliant on current microbiome state | • Limited research to develop mimetics | More research required to identify candidates as mimetics and mechanisms of delivery, including diet should be explored |
Abbreviations: FMT, faecal microbiota transplant; NEC, necrotising enterocolitis.
3.1. Dietary interventions
Diet plays an essential role in health and disease, with intake of dietary fibres from foods, such as whole grains, resistant starch and fruits, being clearly beneficial for the development of a diverse microbiome. 38 , 39 , 40 Alteration of diet has shown to be important for patients with IBS, where it is recommended for some patients to reduce consumption of fermentable oligo‐, di‐, monosaccharides and polyols (FODMAPs). 41 FODMAPs are poorly digested but readily fermented by the microbiome, 42 thus a low FODMAP diet (LFD) provides the opportunity to leverage nutrient availability to shift the microbiome community. This is achieved by decreasing the amount of gas produced by fermentation, therefore decreasing the associated bloating symptoms. 43 , 44 , 45 LFDs have also been linked to a reduction in the amount of Bifidobacteria present in patients with IBS. 43 , 46 As such, a long‐term LFD is not recommended in patients with IBS, given the potential for long‐term health consequences of generating and perpetuating dysbiosis.
Dietary interventions have been extensively explored for the treatment of IBD, with a large variety diets being investigated for uses in either Crohn's disease (CD) or ulcerative colitis (UC). 47 These diets include, exclusive enteral nutrition (EEN), 48 , 49 Crohn's disease exclusion diet (CDED), 50 , 51 , 52 specific carbohydrate diet, 53 LFD, 54 Mediterranean diet (MD), 55 Crohn's Disease TReatment with EATing diet (CD‐TREAT) 56 and partial enteral nutrition. 47 Only two of these diets have proven clinical efficacy in CD treatment and evidence to show their impact on the GI microbiome, EEN 48 , 49 , 57 and CDED. 50 , 51 , 52
Exclusive enteral nutrition involves the replace of all food and beverages with a liquid meal replacement, which can induce remission in 80%–85% of patients with CD. 48 The proposed mechanism of action suggests that inflammatory dietary factors are interacting with the microbiome, host immune system and GI environment. 58 Therefore, removal of these products will decrease inflammation and the associated symptoms of CD. 59 Following the success of EEN, CDED was developed to allow patients to eat whole foods, while restricting the intake of inflammatory dietary factors such as processed foods, gluten, dairy and food additives. Patients on CDED have shown a 70%–75% remission rate, 48 , 50 , 51 , 60 and changes to their microbiome composition have been identified, similar to those seen with EEN. 50 , 51 Both EEN and CDED broadly identified increases in the abundance of bacteria from the Firmicutes phylum, and a decrease in those from the Proteobacteria and Actinobacteria phyla. 48 , 50 , 51 , 60 More specifically, EEN treatment led to an increase in the abundance of bacteria from the Veillonellaceae family 51 and a decrease the amount of Faecalibacterium prausnitzii. 53 Changes at the family or species level have yet to be identified following CDED; however, abundance of bacteria from the Clostridiales class were increased, while those from the Gammaproteobacteria were decreased. 50 , 51 Currently, the causal relationship between these interventions and microbiota changes are unclear; however, it is clear that EEN and CDED are effective diets for the treatment of CD. Furthermore, CDED exemplifies the ability to produce more targeted therapeutics, which may alter microbiome structure and functions.
3.2. Prebiotics
Many dietary fibres act as prebiotics, components in food that are used by the microbiome, confer a health benefit, are easily administered, and support numbers of beneficial bacteria. 42 , 61 Currently, there are five main classes of prebiotics: (1) readily fermentable dietary fibre, 62 (2) phenolics and phytochemicals, 63 (3) human milk oligosaccharides, 64 (4) other oligosaccharides (i.e. fructooligosaccharides [FOS], 65 galactooligosaccharides [GOS] 66 and inulin 67 ) and (5) Conjugated linoleic acid and polyunsaturated fatty acid. 68 , 69 , 70 Although all the aforementioned prebiotics have been used in disease management, the most well studied are FOS and GOS, which have been associated with an increase in Bifidobacteria within the gut. 42 FOS and GOS have shown efficacy in the management of metabolic diseases, such as prediabetes and obesity 66 , 71 ; and GI diseases, including IBS. 72 However, it has been noted that even small doses of prebiotics can cause side effects such as diarrhoea, bloating and flatulence, which may exacerbate some of the conditions being treated. These side‐effects have been attributed to prebiotic‐induced osmotic changes in the GI tract or gasses produced from rapid prebiotic fermentation in the small bowel. 73 Thus, highlighting the patient and disease‐specific efficacy of prebiotics.
Currently, the prebiotics GOS and fructans, are being routinely administered to newborns through infant formula. 74 , 75 These prebiotics increase the abundance of bifidobacteria and lactobacilli within the GI microbiome, 74 and this is correlated with a decreased chance of respiratory infections 75 , 76 and allergic responses. 77 Therefore, GOS and fructans can be used to improve infant outcomes through increasing microbiota diversity; however, many other prebiotics are not as successful.
As observed with other treatments, a subgroup of patients treated with prebiotics will be ‘non‐responders’. One driver of patients being ‘non‐responders’ could be the reliance on the patient being colonised with bacteria that are able to metabolise the treatment. 78 , 79 This observed with the prebiotic fructan, where it is able to increase the amount of bifidobacteria in the GI tract, but this increase is proportional to the amount of bifidobacteria present before treatment. 79 Thus, if a patient only had a small amount of bifidobacteria present before treatment, the increase in abundance would be smaller or non‐existent compared with that of a patient who started with a larger amount. Additionally, other factors such as differences in the host and gut microenvironments can also lead to similar variability in treatment response. In this context, the state of the patients' microbiome and gut before treatment has the capacity to impact the efficacy of prebiotics. Therefore, complementing provision of prebiotics to feed the current microbiota population, with therapies that directly provide beneficial bacteria to the gut may improve patient outcomes.
3.3. Probiotics and synbiotics
The Food and Agriculture Organisation of the United Nations and the World Health Organisation defines probiotics as ‘live microorganisms which when administered in adequate amounts confer a health benefit on the host’. 80 Probiotics have been consumed for at least 10,000 years, 81 where historically they have been components of foods such as yogurt and fermented milk. 82 It was identified that these fermented foods usually contained a mix of lactobacilli, bifidobacteria or other lactic acid producing bacterial strains, which in specific circumstances, can be beneficial. 83 , 84 This discovery has led to the development of modern probiotics that can be administered in controlled doses of purified, live bacteria. 83 , 85 Probiotics function by either colonising or being transiently present at a given body site, where they confer a health benefit. These benefits include increasing colonisation resistance by inhibiting the growth and colonisation of pathogens through competition for nutrients 86 and direct killing by antimicrobials such as bacteriocins. 87 , 88 , 89 Bacteria within the lumen are more likely to interact with probiotics than mucosa‐associated bacteria; therefore, their use as a treatment in diseases associated with mucosal bacteria such as IBD may be limited. 90 Probiotics can also regulate host innate and adaptive immune functions through interactions with epithelial 91 or dendritic cells. 92 , 93 These interactions lead to anti‐inflammatory immune responses from macrophages and T and B lymphocytes. 91 , 92 , 93 Additionally, they can increase mucin production, thereby improving the integrity of the mucosal barrier in the gut, through the production of SCFAs. 94
Generally, most probiotics do not colonise the gut. 95 Therefore, where efficacy is dependent on bacterial presence, sustained or repeated dosing may be required for the benefit to be maintained. 96 This has been observed in elderly, human, subjects where those that consumed probiotics for 13.5 years showed greater changes in their abundance of the beneficial bacterial genera Bifidobacterium than those limited to 3 years of treatment. 96 However, although a probiotic may not colonise the GI system directly, their transient presence appears to allow for colonisation by other beneficial bacteria, such as Lachnoclostridium, Blautia and Clostridium strains. 97 Similarly, in mice where the microbiota has been disrupted by antibiotics, and probiotics were able to restore microbiome diversity to 99.8% of what was observed pre‐treatment, compared with the 80% restoration observed without probiotics. 97 Importantly, the probiotic did not colonise these mice, 97 and it is speculated that these beneficial effects were observed as either the probiotics interacted with the intestinal barrier to increase its integrity, or they interacted with the intestinal epithelium to exert an anti‐inflammatory response. 97 Currently, probiotics are considered food products, in most jurisdictions. As such standardisation of viability or efficacy is limited. To combat this, the American Gastroenterological Association 98 has developed guidelines 99 for the use of probiotics in humans. These do not currently support the use of probiotics for the treatment or prevention of CDI, CD, UC or IBD. 98 , 99 However, preliminary evidence supports the use of probiotics for patients following antibiotic treatment or with pouchitis; however, their use in pouchitis remains controversial. 98 , 99 Additionally, there is strong evidence for probiotic use in preterm babies with low‐birth‐weight to prevent necrotising enterocolitis (NEC). 98 , 99 Conversely, there is moderate evidence against probiotic use for children with acute gastroenteritis. 99 Given the large knowledge gap for the use of probiotics, it is clear that more trials are required to determine if probiotics should be used for each of the above indications.
Probiotics can also be synergistically combined with selected prebiotics and these treatments are called synbiotics. 84 Within synbiotics, there are two sub‐groups: synergistic synbiotics, in which the prebiotic acts by providing nutrients for the probiotic and complementary synbiotics, in which the prebiotic component supports the growth and survival of other bacteria already present within the microbiota, known as autochthonous bacteria. 100 For example, within the synergistic synbiotic mix of FOS with Bifidobacterium longum, B. breve or B. bifidum, 101 , 102 , 103 the bifidobacteria preferentially metabolise FOS, thereby increasing the bacterial numbers. 104 Higher amounts of bifidobacteria within the microbiota have been associated with reduced chances of developing antibiotic‐associated diarrhoea (AAD), 105 CDI, 106 NEC 107 and a reduction of symptoms associated with IBS. 108 Synbiotic combinations also allow for the development of beneficial cross‐feeding networks. 109 This occurs as the synbiotic is metabolised by the probiotic strains and the by‐products can be used to cross‐feed beneficial bacteria. 109 Cross‐feeding has been observed in probiotic strains of Lactobacillus where L. salivarius W57 cannot fully use inulin‐type fructans; however, when co‐cultured with L. paracasei subsp. paracasei W20, an extracellular enzyme from L. paracasei allows for the breakdown of the fructan so that it can be used by the L. salivarius. 110
Synbiotic combinations have shown efficacy in humans for the treatment of non‐alcoholic fatty liver disease using a combination of Lactobacillus, Streptococcus and Bifidobacterium strains and FOS 111 ; IBS using Bacillus coagulans and FOS 112 ; diarrhoea using B. lactis B94 and inulin 113 and type 2 diabetes with L. sporogenes and inulin. 114 Type 2 diabetes is a precursor for many other diseases including polycystic ovary syndrome and cardiovascular disease, therefore synbiotic treatments successfully targeting type 2 diabetes may also be delaying the progression of other metabolic diseases.
As seen with prebiotics, recipients of probiotics and synbiotics include cohorts of ‘non‐responders’, due to factors such as the timing of treatment, disease progression and composition of the microbiome before treatment. 115 Although the bacterial strains found in probiotics and synbiotics can be effective in the management of some diseases, these strains are unable to treat all diseases associated with microbiome composition and can be harmful in some conditions, including following antibiotic treatment. 116 Finally, the transient nature of probiotic treatment may be insufficient in diseases that require a more permanent and substantial restructuring of the bacterial community.
3.4. Antibiotics
Since the discovery of penicillin in 1928 117 and its approval for clinical use in 1945, many antibiotics have been discovered and successfully used to treat bacterial infections. Antibiotics are also commonly administered prophylactically to decrease the risk of post‐operative infections. 118 More recently, the administration of antibiotics prior to delivering microbial therapies has been shown to enhance the efficacy of the treatment in some settings, possibly by opening ecological niches and enabling colonisation. 119 , 120 Although these approaches offer potential, the antibiotic‐associated decrease in bacterial number and diversity may also reduce colonisation resistance 121 or induce other pathologies. 122 Pathogenic strains, including multi‐drug‐resistant C. difficile, 123 Escherichia coli, Enterococcus feacium and Klebsiella pneumoniae 124 can exploit this niche and infect the gut causing AAD. These diseases can be exacerbated by further antibiotic use, which can lead to the development of resistance. In particular, antibiotic‐resistant ‘super‐bugs’ can develop, leading to untreatable infections in patients. 125 Although largely unexplored, microbiota modification through selective administration of antibiotics represents a potential mechanism for targeted community change. However, such interventions will require careful design to minimise off‐target or adverse effects but may form a critical component of future microbiome‐based therapies.
3.5. Phage therapy
Bacteriophages (phages), viruses that exclusively infect bacterial cells, have been used as antibacterial monotherapy for the treatment of bacterial infections for over 100 years. 126 Clinical trials of phage cocktails targeting difficult to treat E. coli, Staphylococcus aureus and Pseudomonas aeruginosa infections 127 and multi‐drug‐resistant bacterial infections including methicillin‐resistant S. aureus 128 and Acinetobacter baumannii 129 have shown substantial promise. Within faecal transplants, phage have been shown to colonise the host following treatment 130 and may be responsible for a proportion of the beneficial effects achieved. It has been postulated that the efficacy of the sterile filtrate of FMT demonstrated in a small cohort of patients with CDI may have been partly attributed to phage. 131 The narrow host range typical of phage 132 provides an opportunity for targeted bacterial depletion and microbiota remodelling as a therapeutic intervention. Coupled with broad‐range antibiotic treatment, these therapies allow for highly controlled microbiome disruption.
3.6. Faecal microbiota transplantation
In contrast to antibiotics and phage therapies, FMT can restore bacterial diversity and health‐associated functions such as colonisation resistance by introducing a faecal‐associated microbiota from a healthy individual. 133 FMT also provides a diversity of bioactive compounds, and other microbes, such as phage. Together these components are a functioning, synbiotic community, which allows for better colonisation within the GI tract. 134
Traditionally, FMT had medicinal use in ancient China and Indigenous Australian culture, 135 however, its potential as a modern medicine was only identified within the last 20–30 years. 136 , 137 , 138 For the treatment of recurrent CDI, FMT has proved highly effective 136 , 139 , 140 with efficacy of 85%–95% reported. 141 , 142 It is thought that the increased diversity and abundance of bacteria that the FMT provides to the GI tract outcompetes C. difficile and prevents reinfection. As FMT has been successful in the treatment of CDI, it is now being examined for efficacy in other GI diseases.
Faecal microbiota transplant has shown promise in the treatment of non‐GI diseases, including insulin resistance, liver disease and autism spectrum disorders, 143 , 144 , 145 and has been used successfully to induce remission in UC. 146 , 147 , 148 Meta‐analysis has shown that multiple forms of FMT administration can induce remission in UC 149 ; however, this is not the case for all diseases, which has led to a call for standardised methods of preparation or administration. 150 Until recently most faecal transplants were prepared in aerobic conditions, leading to the loss of many obligate anaerobic bacteria. 151 , 152 , 153 Studies have now shown that many of the bacteria correlated with a positive treatment outcome, particularly in patients with UC, are obligate anaerobes such as F. prausnitzi, which can be preserved if the treatment is prepared anaerobically. 146 , 154 It is unclear whether anaerobic stool processing confers a clinical benefit relative to aerobic processing; however, there are advantages for microbial drug discovery using anaerobic stool processing in FMT clinical trials. Currently, FMT can be administered to the lower GI tract, by colonoscopy or enema, or the upper GI tract, through gastroscope, or nasogastric, nasojejunal or gastrostomy tube, with each method using varied doses, and frequencies of administration. 155 , 156 , 157 Faecal microbiota transplant delivery via colonoscopy in the lower GI tract has been the most effective (86% success rate, compared with 74% success rate for upper GI tract delivery), 158 particularly in the treatment of CDI. 159 Unfortunately, colonoscopies are relatively invasive procedures and cannot be performed on all patients due to risks such as bowel perforation in groups such as the elderly or critically ill. 160 Administration through the upper GI tract has shown higher rates of adverse events with multiple reports of aspiration pneumonia. 161 To overcome these issues, substantial effort developing encapsulation methods for oral delivery has been undertaken. Specialised acid‐resistant hypromellose capsules have been used, which allow for colonic release of the bacteria and protection from the gastric environment. 162 , 163 However, is it possible that exposure to the GI environment could enhance colonisation efficiency. In addition, lyophilisation of donor stool allows greater stability of the FMT within the capsule. These capsule‐based approaches have shown high rates of clinical success. 164 , 165 Despite this success, a small number of serious complications, including bacteraemia and transient UC flares, have been observed and one death due to infection with an extended spectrum beta‐lactamase (ESBL)‐producing E. coli has been noted with capsule delivered FMT. 166 Therefore, further screening of capsule FMT preparation may be necessary prior to large scale adoption.
Faecal microbiota transplant is remarkably safe. Effective donor and sample screening to prevent transfer of detrimental bacteria including, multi‐drug‐resistant pathogens or other detrimental species is performed. While relatively harmless within the donor, these species may be harmful for an immunocompromised or otherwise susceptible recipient. Introduction of multi‐drug‐resistant bacteria, such as ESBL‐producing E. coli, has resulted in fatal, untreatable sepsis. 167 While screening for pathogens is routine, and should effectively eliminate the risk, identifying patient‐specific detrimental bacteria is substantially more challenging.
Screening samples prior to delivery highlights the substantial donor‐specific variability, which can alter efficacy, allowing for higher rates of treatment success by some donors termed ‘super donors’. 168 This phenomenon was first postulated in a clinical trial testing the efficacy of FMT in inducing UC remission, where 7/9 patients who entered remission received FMT from the same donor. 152 It has been suggested that some donors may be associated with higher efficacy due to having higher bacterial diversity or specific bacteria that are therapeutic for a given disease. 168 It should be noted, studies of 1999 FMTs used to treat CDI have failed to identify evidence of ‘super donors’, 169 highlighting the disease specificity of this relationship. Conditions where ‘super donors’ have been identified, highlight the possibility of donor–patient compatibility and the opportunity for refining microbial therapeutic treatments to contain only the bacterial strains required to induce health benefits. 170
3.7. Live biotherapeutics
To provide more targeted intervention and overcome the risks associated with pathogen or pathobiont transfer, significant research and development effort has been focused on determining bacterial strains that could be used as therapeutics. 171 Termed, live biotherapeutics, these therapies have been defined by the FDA as ‘a biological product that (1) contains live organisms, such as bacteria; (2) is applicable to the prevention, treatment, or cure of a disease or condition of human beings and (3) is not a vaccine’. 172 These are distinct from probiotics as they are microbes that may colonise the gut and have an established clinical benefit for the treatment of a specific disease. 173 Live biotherapeutics may be comprised of a single bacterial species or selected combinations that act synergistically.
To date, very few studies have determined direct causation between bacterial species and disease, 174 due to the complexity of microbiome interactions and limitations of existing experimental models. In the case of CDIs, through both murine and clinical studies in humans, researchers have identified C. scindens as being inversely correlated with the establishment of CDI. 175 It was consequently found that the administration of C. scindens can reduce C. difficile bacterial load in mice 174 , 175 through dehydroxylation of bile acid, which produces a toxic by‐product to C. difficile. 174 Although C. scindens reduced C. difficile levels, colonisation resistance was not restored. 174 However, two studies in mice have identified consortiums of four 175 and six 176 bacterial strains that were able to increase resistance to CDI, with the consortium of six bacterial isolates able to prevent recurrent infection. 176 These studies demonstrate the potential for specifically chosen live biotherapeutics to colonise the gut and provide beneficial health outcomes.
Researchers began developing a more specific treatment for UC using Firmicutes spores derived from ethanol shocked human donor stool, termed SER‐287. 120 This treatment can induce remission in patients and is thought to be superior to FMT as there is less risk of introducing harmful bacteria into patients. 120 It is proposed that SER‐287 is effective because the ratio of metabolites within the gut is observed to change following treatment; however, a direct causation has not been identified. 120 Unfortunately, SER‐287 failed to meet its primary endpoint in a phase 2b clinical trial, and the product is no longer in development with Seres Therapeutics.
As most biotherapeutic development is in its early stages, very little is known about whether long‐term persistent colonisation with these therapies may impact health or chronic conditions. In particular, live biotherapeutics are being developed to treat paediatric conditions, such as paediatric IBD, 177 so it is important to evaluate not only the effect of the bacteria but the dosage, treatment frequency and delivery mode to ensure the safest treatment that will have the least adverse health outcomes later in life. These concerns are one of the reasons that researchers are now also considering microbiome mimetics.
3.8. Microbiome mimetics
Microbiome mimetics describes any intervention that replicates the interaction between the microbiome and the host, that yields a therapeutically beneficial outcome. This can include bacterial derived products, small molecules, conventional therapeutics or host derived products. The majority of research has focused on postbiotics, which are molecules or components of bacteria that confer a health benefit. 178 Within postbiotics there are two main classes: paraprobiotics and fermented infant formulas (FIFs). Paraprobiotics are non‐viable components of bacteria, including bacterial proteins and polysaccharides. 179 Meanwhile, FIFs are the purified products produced after infant formula is fermented by bacteria. 180 Research into these products is currently aimed at determining which bacterial molecules provide health benefits. This can be achieved through mass spectrometry of bacterial supernatants and targeted purification of these metabolites for use. The most prominent candidate for use as postbiotics are SCFAs. 181 Short‐chain fatty acids are compounds such as butyrate, propionate and acetate, which are produced by bacterial fermentation of prebiotic or dietary fibre, and resistant starches and are the primary energy source of colonocytes. 182 Additionally, SCFAs have beneficial effects on the mucosal immune system, 183 , 184 including anti‐inflammatory effects through blocking inflammatory cytokine production, increasing mucus production, promoting immune tolerance through regulatory T (Treg) cells, and promoting tissue repair. 185 , 186 , 187 While patients with UC have decreased SCFA levels, 188 trials of butyrate enemas have been unsuccessful, likely due to the dysfunction of colonocytes and their reduced ability metabolise butyrate as an energy source in this disease. 189 SCFAs also increase gut mucin production, decreasing the ‘leaky gut’ syndrome in human patients with type 1 diabetes mellitus. 190 Consequently, SCFAs have shown efficacy in delaying the onset of type 1 diabetes mellitus in mice. 191 Therefore, although postbiotics may be used to manage and treat disease without requiring patients to have particular bacterial species present in their microbiome, they may still require their immune system to be primed for an effective response. Interestingly, postbiotics such as SCFAs can also be found in fermented foods such as cheese and yogurt, and beverages such as beer and kombucha. Postbiotics occur at lower doses in these foods; therefore, it is hypothesised that the higher doses used in postbiotics would likely be required for therapeutic efficacy. 192 , 193 , 194 Therefore, future microbiome mimetic treatments may incorporate dietary interventions targeted to replicate the beneficial effects provided by the microbiota.
4. FROM THE LABORATORY TO THE CLINIC
Advances in sequencing technology have revolutionised the study of the microbiome; however, identifying key bacterial species involved in health and disease remains a challenge. Despite the prevalence of microbiome sampling and evidence suggesting that storage time, temperature and storage medium can affect the bacterial strains, 195 there remains no standardised method for sample collection, storage, data analysis or dosage calculation. 196 , 197 , 198
For many microbiome‐based medicines, preclinical safety testing may not be required as the therapeutic may already be approved for use in humans by the FDA or other stringent regulatory authorities. These include commonly consumed foods that may contain prebiotics, probiotics or postbiotics and other medicines that have been repurposed as microbiome mimetics. Importantly, disease‐specific testing of therapeutic efficacy is still required. Preclinical testing and safety profiles of FMT are now well established, although the classification and regulation of the treatment varies from a stringently regulated biological agent in some countries (USA, Canada, Australia), to a medicinal product or treatment with variable regulation (UK, France, Germany, Switzerland), to no regulation (Austria, Denmark, Sweden, Finland). 199 , 200
To improve selection of therapeutic candidates it is necessary to establish acceptable methods for candidate prioritisation and preclinical safety and validation testing. Although animal models can play an important role in preliminary safety testing, limitations of mouse models in replicating human microbiome interactions introduce a unique difficulty in validating microbiome‐based medicines. 201 To address these concerns, gnotobiotic mice with defined microbiome have been used. 202 , 203 , 204 The use of gnotobiotic mice, with a human microbiome for example, have enabled the identification of a T‐cell response integral to the success of FMT, that is associated with increased bacterial abundance in the gut. 205 Many groups have also used human immortalised cell‐culture methods for safety or validation testing, 206 but these methods do not allow replication of the complex physical and mechanistic interactions between the microbiome and host. As a result, cell‐based models, such as organoids and organ‐on‐a‐chip technologies, are emerging as an important component for developing novel microbiome‐based therapeutics.
Organoids are enclosed, three‐dimensional (3D) cultures that mimic the multicellular structure from the corresponding tissue. 207 Methods now exist to establish and maintain patient‐derived GI organoids, which partially recapitulate the environment that the microbiota normally inhabits and provide the opportunity for a ‘personalised’ microbiome cell culture model. 208 , 209 Microinjection technology allows for bacteria to be introduced into the lumen of gut organoids to investigate microbiome–host GI epithelial interactions. 210 , 211 Unfortunately, organoids still contain no stroma or vasculature, limiting the capacity to infer microbiome–host immune interactions beyond those in the epithelium. To overcome this, some studies have used induced pluripotent stem cells to generate organoids containing mesenchymal stem cells, 212 and others have cultured organoids with supporting mesenchymal and/or immune cells. 213 , 214 However, as organoids are enclosed 3D structures, the bacteria within the system are trapped, which leads to the build‐up of detrimental metabolites and other cellular debris that may impact bacterial replication or modify host cell responses. 215 Therefore, when using this technology to investigate novel intervention strategies such as live biotherapeutics, determining bacterial colonisation or transience, host physiology and immune responses following treatment is not possible. These limitations have been addressed with the development of organ‐on‐a‐chip technology.
Organ‐on‐a‐chip is a method that combines microfluidics and cell culture to generate mini human organs on a chip. To date, researchers have modelled brain, 216 lung, 217 heart, 218 skin 219 and the GI tract or gut‐on‐a‐chip systems. 220 Microfluidic channels in these systems enable optimal fluid flow and cyclic mechanical strain on cells to mimic peristalsis. 221 These systems replicate the GI tract as they generate villi‐like structures and exist in two compartments with media perfusion and an oxygen gradient. The upper epithelial layer can be maintained anaerobically, whereas the endothelial layer containing immune cells can be cultured in an aerobic environment. Gut‐on‐a‐chip can include complex cell types such as immortalised cell lines and primary tissues, similar to organoids. 220 Through the use of gut‐on‐a‐chip, microbiome researchers have demonstrated the interactions of bacterial cells, including pathogenic Shigella 222 and E. coli strains, 220 and probiotic Lactobacillus strains, 220 with not only the epithelium, but also immune cells. 220 Other work has examined specific bacterial consortia and the whole microbiome of an individual in this context. 220 , 222 , 223 , 224 , 225 Although some immune cells can be added and the innate immune response to a therapeutic can be examined, gut‐on‐a‐chip cannot replicate complex, adaptive immune responses, which are generated systemically and over longer time periods in response to a stimulus. Further work is required to refine this emerging platform to comprehensively assess host‐microbe responses and test developing medicines. With this collection of current technologies, preclinical testing of microbiome‐based medicines remains dependent on a combination of animal models and cell‐based systems to demonstrate efficacy and safety in the pre‐clinical setting. Following this pre‐clinical testing, each therapeutic will need to be examined under specific clinical settings, to confirm these findings and determine efficacy in humans.
5. CONCLUSION
Microbiome‐based medicines have advanced dramatically over the last decade, from prebiotics and probiotics, to live biotherapeutics and microbiome mimetics. The ability to culture the GI bacteria and new applications of metagenomic sequencing have overcome many of the previous technical hurdles in this area. The primary challenge now faced is identifying clinical diseases amenable to intervention with microbiome‐based medicines and developing appropriate methods to identify, refine and test candidate therapies. Although much progress has been made, further work to optimise methods to identify candidate microbes, develop appropriate preclinical validation models, and progress to personalised targeting of microbiome‐based medicines are the essential next steps.
AUTHORSHIP
Guarantor of article: Samuel Forster.
Author contributions: Emily L Gulliver: Conceptualization (lead); data curation (lead); investigation (lead); project administration (equal); writing – original draft (lead); writing – review and editing (lead). Remy B Young: Data curation (supporting); writing – original draft (supporting); writing – review and editing (supporting). Michelle Chomwerawong: Writing – review and editing (supporting). Gemma L D'Adamo: Writing – review and editing (supporting). Tamblyn Thomason: Writing – review and editing (supporting). James T Widdop: Writing – review and editing (supporting). Emily L Rutten: Writing – review and editing (supporting). Vanessa Rossetto Marcelino: Writing – review and editing (supporting). Robert V Bryant: Writing – review and editing (supporting). Samuel P Costello: Writing – review and editing (supporting). Claire L O'Brien: Writing – review and editing (supporting). Georgina Hold: Writing – review and editing (supporting). Edward M Giles: Writing – review and editing (supporting). Samuel Charles Forster: Conceptualization (equal); writing – review and editing (equal). All authors approved of the final version of this manuscript before publication.
ACKNOWLEDGEMENTS
Declaration of funding interests:This work is supported by an ARC Discovery Project (DP190101504 to SCF) and the Victorian Government's Operational Infrastructure Support Program.
Declaration of personal interests: SPC has received speaker fees from Shire, Ferring, Microbiotica, Pfizer and Janssen and is a shareholder in BiomeBank. RVB has received grant/research support and/or speaker fees [all paid to employer for research support] from AbbVie, Ferring, Janssen, Shire, Takeda and Emerge Health. Open access publishing facilitated by University of Otago, as part of the Wiley ‐ University of Otago agreement via the Council of Australian University Librarians.
Gulliver EL, Young RB, Chonwerawong M, D’Adamo GL, Thomason T, Widdop JT, et al. Review article: the future of microbiome‐based therapeutics. Aliment Pharmacol Ther. 2022;56:192–208. 10.1111/apt.17049
The Handling Editor for this article was Professor Mike Burkitt, and this uncommissioned review was accepted for publication after full peer‐review.
[Correction added on June 04, 2022, after first online publication: Author Claire L. O'Brien affiliation has been updated]
DATA AVAILABILITY STATEMENT
No datasets were generated or analysed as part of this study.
REFERENCES
- 1. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome‐wide association study of gut microbiota in type 2 diabetes. Nature. 2012. Oct 4;490(7418):55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 2. Zackular JP, Rogers MAM, MTt R, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014;7(11):1112–21. 10.1158/1940-6207.CAPR-14-0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, et al. Gut microbiome analysis as a tool towards targeted non‐invasive biomarkers for early hepatocellular carcinoma. Gut. 2019. Jun;68(6):1014–23. 10.1136/gutjnl-2017-315084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jackson SE, Chester JD. Personalised cancer medicine. Int J Cancer. 2015. Jul 15;137(2):262–6. 10.1002/ijc.28940 [DOI] [PubMed] [Google Scholar]
- 5. van Vollenhoven RF, L'Ami M, Wolbink G. Personalised medicine in de reumatologie [Personalised medicine in rheumatology]. Tijdschr Psychiatr. 2018;60(3):146–50. [PubMed] [Google Scholar]
- 6. Narod SA. Personalised medicine and population health: breast and ovarian cancer. Hum Genet 2018. Oct;137(10):769–778. 10.1007/s00439-018-1944-6 [DOI] [PubMed] [Google Scholar]
- 7. The Integrative Human Microbiome Project . The integrative human microbiome project. Nature. 2019. May;569(7758):641–8. 10.1038/s41586-019-1238-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leiva‐Gea I, Sanchez‐Alcoholado L, Martin‐Tejedor B, Castellano‐Castillo D, Moreno‐Indias I, Urda‐Cardona A, et al. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case‐control study. Diabetes Care 2018. Nov;41(11):2385–2395. 10.2337/dc18-0253 [DOI] [PubMed] [Google Scholar]
- 9. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014. Sep 4;513(7516):59–64. 10.1038/nature13568 [DOI] [PubMed] [Google Scholar]
- 10. Zimmermann P, Curtis N. The influence of the intestinal microbiome on vaccine responses. Vaccine. 2018. Jul 16;36(30):4433–9. 10.1016/j.vaccine.2018.04.066 [DOI] [PubMed] [Google Scholar]
- 11. Zimmermann P, Messina N, Mohn WW, Finlay BB, Curtis N. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: a systematic review. J Allergy Clin Immunol. 2019;143(2):467–85. [DOI] [PubMed] [Google Scholar]
- 12. Mego M, Chovanec J, Vochyanova‐Andrezalova I, Konkolovsky P, Mikulova M, Reckova M, et al. Prevention of irinotecan induced diarrhea by probiotics: a randomized double blind, placebo controlled pilot study. Complement Ther Med. 2015;23(3):356–62. [DOI] [PubMed] [Google Scholar]
- 13. Lee JR, Muthukumar T, Dadhania D, Taur Y, Jenq RR, Toussaint NC, et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS One. 2015;10(3):e0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti‐PD‐1 immunotherapy in melanoma patients. Science. 2018. Jan 5;359(6371):97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tian Y, Li M, Song W, Jiang R, Li YQ. Effects of probiotics on chemotherapy in patients with lung cancer. Oncol Lett. 2019. Mar;17(3):2836–48. 10.3892/ol.2019.9906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Riehl TE, Alvarado D, Ee X, Zuckerman A, Foster L, Kapoor V, et al. Lactobacillus rhamnosus GG protects the intestinal epithelium from radiation injury through release of lipoteichoic acid, macrophage activation and the migration of mesenchymal stem cells. Gut. 2019. Jun;68(6):1003–13. 10.1136/gutjnl-2018-316226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mullié C, Yazourh A, Thibault H, Odou MF, Singer E, Kalach N, et al. Increased poliovirus‐specific intestinal antibody response coincides with promotion of Bifidobacterium longum‐infantis and Bifidobacterium breve in infants: a randomized, double‐blind, placebo‐controlled trial. Pediatr Res. 2004;56(5):791–5. [DOI] [PubMed] [Google Scholar]
- 18. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, et al. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134(2):e362–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris V, Armah G, Fuentes S, et al. The infant gut microbiome correlates significantly with rotavirus vaccine response in rural Ghana. J Infect Dis. 2016;215:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eloe‐Fadrosh EA, McArthur MA, Seekatz AM, Drabek EF, Rasko DA, Sztein MB, et al. Impact of oral typhoid vaccination on the human gut microbiota and correlations with S. Typhi‐specific immunological responses. PLoS One. 2013;8(4):e62026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017. Feb;20(2):145–55. 10.1038/nn.4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyamoto J, Mizukure T, Park S‐B, Kishino S, Kimura I, Hirano K, et al. A gut microbial metabolite of linoleic acid, 10‐hydroxy‐cis‐12‐octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40‐MEK‐ERK pathway. J Biol Chem. 2015;290(5):2902–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thorburn AN, McKenzie CI, Shen S, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6(1):1–13. [DOI] [PubMed] [Google Scholar]
- 24. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom‐Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159–66. [DOI] [PubMed] [Google Scholar]
- 25. Barnes EM, Carter EL, Lewis JD. Predicting microbiome function across space is confounded by strain‐level differences and functional redundancy across taxa. Front Microbiol. 2020. Feb 7;11:101. 10.3389/fmicb.2020.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013. Nov 22;342(6161):967–70. 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abu‐Sbeih H, Herrera LN, Tang T, Altan M, Chaftari AMP, Okhuysen PC, et al. Impact of antibiotic therapy on the development and response to treatment of immune checkpoint inhibitor‐mediated diarrhea and colitis. J Immunother Cancer. 2019;7(1):242–2. 10.1186/s40425-019-0714-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science. 2018. Jan 5;359(6371):91–7. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 29. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science (New York, NY). 2015;350(6264):1079–84. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ciorba MA, Riehl TE, Rao MS, Moon C, Ee X, Nava GM, et al. Lactobacillus probiotic protects intestinal epithelium from radiation injury in a TLR‐2/cyclo‐oxygenase‐2‐dependent manner. Gut. 2012;61(6):829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beresford‐Jones BS, Forster SC, Stares MD, Notley G, Viciani E, Browne HP, et al. The mouse gastrointestinal bacteria catalogue enables translation between the mouse and human gut microbiotas via functional mapping. Cell Host Microbe. 2022;30(1):124–138.e8. 10.1016/j.chom.2021.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma ZS, Li L, Gotelli NJ. Diversity‐disease relationships and shared species analyses for human microbiome‐associated diseases. ISME J. 2019. Aug;13(8):1911–9. 10.1038/s41396-019-0395-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Palma G, Nadal I, Medina M, et al. Intestinal dysbiosis and reduced immunoglobulin‐coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010. Feb 24;10:63. 10.1186/1471-2180-10-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y. Luminal and mucosal‐associated intestinal microbiota in patients with diarrhea‐predominant irritable bowel syndrome. Gut Pathog. 2010. Dec 9;2(1):19. 10.1186/1757-4749-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cani PD, Bibiloni R, Knauf C, Waget Á, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia‐induced inflammation in high‐fat diet‐induced obesity and diabetes in mice. Diabetes. 2008. Jun;57(6):1470–81. 10.2337/db07-1403 [DOI] [PubMed] [Google Scholar]
- 36. Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol. 2013;51(9):2884–92. 10.1128/jcm.00845-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018. Feb;11(1):1–10. 10.1007/s12328-017-0813-5 [DOI] [PubMed] [Google Scholar]
- 38. Dhingra D, Michael M, Rajput H, Patil RT. Dietary fibre in foods: a review. J Food Science Technol. 2012;49(3):255–66. 10.1007/s13197-011-0365-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol. 1977;34(5):529–33. 10.1128/AEM.34.5.529-533.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roehrig KL. The physiological effects of dietary fiber—a review. Food Hydrocoll. 1988;2(1):1–18. [Google Scholar]
- 41. Gibson PR, Shepherd SJ. Personal view: food for thought‐‐western lifestyle and susceptibility to Crohn's disease. The FODMAP hypothesis. Aliment Pharmacol Ther. 2005. Jun 15;21(12):1399–409. 10.1111/j.1365-2036.2005.02506.x [DOI] [PubMed] [Google Scholar]
- 42. Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995. Jun;125(6):1401–12. 10.1093/jn/125.6.1401 [DOI] [PubMed] [Google Scholar]
- 43. Sloan TJ, Jalanka J, Major GAD, Krishnasamy S, Pritchard S, Abdelrazig S, et al. A low FODMAP diet is associated with changes in the microbiota and reduction in breath hydrogen but not colonic volume in healthy subjects. PLoS One. 2018;13(7):e0201410. 10.1371/journal.pone.0201410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Major G, Pritchard S, Murray K, Hoad C, Marciani L, Gowland P, et al. OC‐065 mechanisms underlying fodmap‐induced symptoms in patients with irritable bowel syndrome: a double‐blind crossover trial using magnetic resonance imaging. Gut. 2015;64(Suppl 1):A33. 10.1136/gutjnl-2015-309861.65 [DOI] [Google Scholar]
- 45. Ong DK, Mitchell SB, Barrett JS, Shepherd SJ, Irving PM, Biesiekierski JR, et al. Manipulation of dietary short chain carbohydrates alters the pattern of gas production and genesis of symptoms in irritable bowel syndrome. J Gastroenterol Hepatol. 2010. Aug;25(8):1366–73. 10.1111/j.1440-1746.2010.06370.x [DOI] [PubMed] [Google Scholar]
- 46. Huaman JW, Mego M, Manichanh C, Cañellas N, Cañueto D, Segurola H, et al. Effects of prebiotics vs a diet low in FODMAPs in patients with functional gut disorders. Gastroenterology. 2018. Oct;155(4):1004–7. 10.1053/j.gastro.2018.06.045 [DOI] [PubMed] [Google Scholar]
- 47. Hart L, Verburgt CM, Wine E, Zachos M, Poppen A, Chavannes M, et al. Nutritional therapies and their influence on the intestinal microbiome in pediatric inflammatory bowel disease. Nutrients. 2021;14(1):4. 10.3390/nu14010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buchanan E, Gaunt WW, Cardigan T, Garrick V, McGrogan P, Russell RK. The use of exclusive enteral nutrition for induction of remission in children with Crohn's disease demonstrates that disease phenotype does not influence clinical remission. Aliment Pharmacol Ther. 2009. Sep 1;30(5):501–7. 10.1111/j.1365-2036.2009.04067.x [DOI] [PubMed] [Google Scholar]
- 49. Logan M, Gkikas K, Svolos V, Nichols B, Milling S, Gaya DR, et al. Analysis of 61 exclusive enteral nutrition formulas used in the management of active Crohn's disease‐new insights into dietary disease triggers. Aliment Pharmacol Ther. 2020. May;51(10):935–47. 10.1111/apt.15695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quince C, Ijaz UZ, Loman N, Eren MA, Saulnier D, Russell J, et al. Extensive modulation of the fecal metagenome in children with Crohn's disease during exclusive enteral nutrition. Am J Gastroenterol. 2015. Dec;110(12):1718–29; quiz 1730. 10.1038/ajg.2015.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaakoush NO, Day AS, Leach ST, Lemberg DA, Nielsen S, Mitchell HM. Effect of exclusive enteral nutrition on the microbiota of children with newly diagnosed Crohn's disease. Clin Transl Gastroenterol. 2015. Jan 15;6(1):e71. 10.1038/ctg.2014.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Day A, Wood J, Melton S, Bryant RV. Exclusive enteral nutrition: an optimal care pathway for use in adult patients with active Crohn's disease. JGH Open. 2020. Apr;4(2):260–6. 10.1002/jgh3.12256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gerasimidis K, Bertz M, Hanske L, Junick J, Biskou O, Aguilera M, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn's disease during enteral nutrition. Inflamm Bowel Dis. 2014. May;20(5):861–71. 10.1097/mib.0000000000000023 [DOI] [PubMed] [Google Scholar]
- 54. Leach ST, Mitchell HM, Eng WR, Zhang L, Day AS. Sustained modulation of intestinal bacteria by exclusive enteral nutrition used to treat children with Crohn's disease. Aliment Pharmacol Ther. 2008. Sep 15;28(6):724–33. 10.1111/j.1365-2036.2008.03796.x [DOI] [PubMed] [Google Scholar]
- 55. Lev‐Tzion R, Ben‐Moshe T, Abitbol G, Ledder O, Peleg S, Millman P, et al. The effect of nutritional therapy on bone mineral density and bone metabolism in pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2021. Jun 1;72(6):877–82. 10.1097/mpg.0000000000003073 [DOI] [PubMed] [Google Scholar]
- 56. Mitrev N, Huang H, Hannah B, Kariyawasam VC. Review of exclusive enteral therapy in adult Crohn's disease. BMJ Open Gastroenterol. 2021. Sep;8(1):1–10. 10.1136/bmjgast-2021-000745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Horwat P, Kopeć S, Garczyk A, et al. Influence of enteral nutrition on gut microbiota composition in patients with Crohn's disease: a systematic review. Nutrients. 2020. Aug 23;12(9):1–11. 10.3390/nu12092551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ashton JJ, Gavin J, Beattie RM. Exclusive enteral nutrition in Crohn's disease: evidence and practicalities. Clin Nutr Feb 2019;38(1):80–89. 10.1016/j.clnu.2018.01.020 [DOI] [PubMed] [Google Scholar]
- 59. Yu Y, Chen KC, Chen J. Exclusive enteral nutrition versus corticosteroids for treatment of pediatric Crohn's disease: a meta‐analysis. World J Pediatr. 2019. Feb;15(1):26–36. 10.1007/s12519-018-0204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee D, Baldassano RN, Otley AR, Albenberg L, Griffiths AM, Compher C, et al. Comparative effectiveness of nutritional and biological therapy in north American children with active Crohn's disease. Inflamm Bowel Dis. 2015. Aug;21(8):1786–93. 10.1097/mib.0000000000000426 [DOI] [PubMed] [Google Scholar]
- 61. Slavin J. Fiber and prebiotics: mechanisms and health benefits. Nutrients. 2013. Apr 22;5(4):1417–35. 10.3390/nu5041417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Delcour JA, Aman P, Courtin CM, Hamaker BR, Verbeke K. Prebiotics, fermentable dietary fiber, and health claims. Adv Nutr. 2016. Jan;7(1):1–4. 10.3945/an.115.010546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Duenas M, Munoz‐Gonzalez I, Cueva C, et al. A survey of modulation of gut microbiota by dietary polyphenols. Biomed Res Int. 2015;2015:850902. 10.1155/2015/850902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Oozeer R, van Limpt K, Ludwig T, Ben Amor K, Martin R, Wind RD, et al. Intestinal microbiology in early life: specific prebiotics can have similar functionalities as human‐milk oligosaccharides. Am J Clin Nutr. 2013. Aug;98(2):561s–71s. 10.3945/ajcn.112.038893 [DOI] [PubMed] [Google Scholar]
- 65. Hidaka H, Eida T, Takizawa T, Tokunaga T, Tashiro Y. Effects of fructooligosaccharides on intestinal flora and human health. Bifidobacteria Microflora. 1986;5(1):37–50. 10.12938/bifidus1982.5.1_37 [DOI] [Google Scholar]
- 66. Vulevic J, Juric A, Tzortzis G, Gibson GR. A mixture of trans‐galactooligosaccharides reduces markers of metabolic syndrome and modulates the fecal microbiota and immune function of overweight adults. J Nutr. 2013. Mar;143(3):324–31. 10.3945/jn.112.166132 [DOI] [PubMed] [Google Scholar]
- 67. Costabile A, Kolida S, Klinder A, Gietl E, Bäuerlein M, Frohberg C, et al. A double‐blind, placebo‐controlled, cross‐over study to establish the bifidogenic effect of a very‐long‐chain inulin extracted from globe artichoke (Cynara scolymus) in healthy human subjects. Br J Nutr. 2010. Oct;104(7):1007–17. 10.1017/s0007114510001571 [DOI] [PubMed] [Google Scholar]
- 68. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: the international scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017. Aug;14(8):491–502. 10.1038/nrgastro.2017.75 [DOI] [PubMed] [Google Scholar]
- 69. Sanlier N, Gokcen BB, Sezgin AC. Health benefits of fermented foods. Crit Rev Food Sci Nutr. 2019;59(3):506–27. 10.1080/10408398.2017.1383355 [DOI] [PubMed] [Google Scholar]
- 70. Costantini L, Molinari R, Farinon B, Merendino N. Impact of omega‐3 fatty acids on the gut microbiota. Int J Mol Sci. 2017. Dec 7;18(12):1–18. 10.3390/ijms18122645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Genta S, Cabrera W, Habib N, Pons J, Carillo IM, Grau A, et al. Yacon syrup: beneficial effects on obesity and insulin resistance in humans. Clin Nutr. 2009. Apr;28(2):182–7. 10.1016/j.clnu.2009.01.013 [DOI] [PubMed] [Google Scholar]
- 72. Silk DB, Davis A, Vulevic J, Tzortzis G, Gibson GR. Clinical trial: the effects of a trans‐galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharmacol Ther. 2009. Mar 1;29(5):508–18. 10.1111/j.1365-2036.2008.03911.x [DOI] [PubMed] [Google Scholar]
- 73. Svensson U, Håkansson J. Safety of food and beverages: safety of probiotics and prebiotics. Cambridge, Massachusetts, United States: Academic Press; 2014. [Google Scholar]
- 74. Ivakhnenko OS, Nyankovskyy SL. Effect of the specific infant formula mixture of oligosaccharides on local immunity and development of allergic and infectious disease in young children: randomized study. Pediatr Pol. 2013. Sep 1;88(5):398–404. 10.1016/j.pepo.2013.07.002 [DOI] [Google Scholar]
- 75. Shahramian I, Kalvandi G, Javaherizadeh H, et al. The effects of prebiotic supplementation on weight gain, diarrhoea, constipation, fever and respiratory tract infections in the first year of life. J Paediatr Child Health. 2018;54(8):875–80. [DOI] [PubMed] [Google Scholar]
- 76. Arslanoglu S, Moro GE, Boehm G. Early supplementation of prebiotic oligosaccharides protects formula‐fed infants against infections during the first 6 months of life. J Nutr. 2007;137(11):2420–4. [DOI] [PubMed] [Google Scholar]
- 77. Arslanoglu S, Moro GE, Schmitt J, Tandoi L, Rizzardi S, Boehm G. Early dietary intervention with a mixture of prebiotic oligosaccharides reduces the incidence of allergic manifestations and infections during the first two years of life. J Nutr. 2008;138(6):1091–5. [DOI] [PubMed] [Google Scholar]
- 78. Disha T, Haque MM, Manoj G, et al. A prospective randomized, double‐blind, placebo‐controlled, dose‐response relationship study to investigate efficacy of fructo‐oligosaccharides (FOS) on human gut microflora. Sci Rep. 2019;9(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Healey G, Murphy R, Butts C, Brough L, Whelan K, Coad J. Habitual dietary fibre intake influences gut microbiota response to an inulin‐type fructan prebiotic: a randomised, double‐blind, placebo‐controlled, cross‐over, human intervention study. Br J Nutr. 2018. Jan;119(2):176–89. 10.1017/s0007114517003440 [DOI] [PubMed] [Google Scholar]
- 80. Cordoba A. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Prevention. 2001;5(1):1–10. [Google Scholar]
- 81. Ozen M, Dinleyici EC. The history of probiotics: the untold story. Benef Microbes. 2015;6(2):159–65. 10.3920/bm2014.0103 [DOI] [PubMed] [Google Scholar]
- 82. Gasbarrini G, Bonvicini F, Gramenzi A. Probiotics history. J Clin Gastroenterol. 2016;50:S116–9. [DOI] [PubMed] [Google Scholar]
- 83. Markowiak P, Slizewska K. Effects of probiotics, prebiotics, and Synbiotics on human health. Nutrients. 2017. Sep 15;9(9):1–30. 10.3390/nu9091021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schrezenmeir J, de Vrese M. Probiotics, prebiotics, and synbiotics‐approaching a definition. Am J Clin Nutr 2001. Feb;73(2 Suppl):361s‐364s. 10.1093/ajcn/73.2.361s [DOI] [PubMed] [Google Scholar]
- 85. Nakazawa Y, Hosono A. editors. Fermented milk in the orient. Functions of fermented milk: challenges for the health sciences. 1. Barking: Elsevier; 1992. p. 1–518. [Google Scholar]
- 86. Freter R, Brickner H, Botney M, Cleven D, Aranki A. Mechanisms that control bacterial populations in continuous‐flow culture models of mouse large intestinal flora. Infect Immun. 1983. Feb;39(2):676–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Klaenhammer TR. Bacteriocins of lactic acid bacteria. Biochimie. 1988. Mar;70(3):337–49. 10.1016/0300-9084(88)90206-4 [DOI] [PubMed] [Google Scholar]
- 88. Gillor O, Giladi I, Riley MA. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol. 2009. Aug 12;9:165. 10.1186/1471-2180-9-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hillman JD, Dzuback AL, Andrews SW. Colonization of the human oral cavity by a Streptococcus mutans mutant producing increased bacteriocin. J Dent Res. 1987. Jun;66(6):1092–4. 10.1177/00220345870660060101 [DOI] [PubMed] [Google Scholar]
- 90. Jonkers D, Stockbrügger R. Probiotics and inflammatory bowel disease. J R Soc Med. 2003;96(4):167–71. [PMC free article] [PubMed] [Google Scholar]
- 91. Yan F, Polk DB. Probiotic bacterium prevents cytokine‐induced apoptosis in intestinal epithelial cells. J Biol Chem. 2002. Dec 27;277(52):50959–65. 10.1074/jbc.M207050200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Veckman V, Miettinen M, Pirhonen J, Sirén J, Matikainen S, Julkunen I. Streptococcus pyogenes and Lactobacillus rhamnosus differentially induce maturation and production of Th1‐type cytokines and chemokines in human monocyte‐derived dendritic cells. J Leukoc Biol. 2004. May;75(5):764–71. 10.1189/jlb.1003461 [DOI] [PubMed] [Google Scholar]
- 93. Smits HH, Engering A, van der Kleij D, de Jong EC, Schipper K, van Capel T, et al. Selective probiotic bacteria induce IL‐10‐producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell‐specific intercellular adhesion molecule 3‐grabbing nonintegrin. J Allergy Clin Immunol. 2005. Jun;115(6):1260–7. 10.1016/j.jaci.2005.03.036 [DOI] [PubMed] [Google Scholar]
- 94. Burger‐van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, et al. The regulation of intestinal mucin MUC2 expression by short‐chain fatty acids: implications for epithelial protection. Biochem J. 2009. May 13;420(2):211–9. 10.1042/bj20082222 [DOI] [PubMed] [Google Scholar]
- 95. Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019. May;25(5):716–29. 10.1038/s41591-019-0439-x [DOI] [PubMed] [Google Scholar]
- 96. Gao R, Zhang X, Huang L, Shen R, Qin H. Gut microbiota alteration after long‐term consumption of probiotics in the elderly. Probiotics Antimicrob Proteins. 2019. Jun;11(2):655–66. 10.1007/s12602-018-9403-1 [DOI] [PubMed] [Google Scholar]
- 97. Grazul H, Kanda LL, Gondek D. Impact of probiotic supplements on microbiome diversity following antibiotic treatment of mice. Gut Microbes. 2016;7(2):101–14. 10.1080/19490976.2016.1138197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Preidis GA, Weizman AV, Kashyap PC, Morgan RL. AGA technical review on the role of probiotics in the management of gastrointestinal disorders. Gastroenterology. 2020. Aug;159(2):708–738.e4. 10.1053/j.gastro.2020.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Su GL, Ko CW, Bercik P, Falck‐Ytter Y, Sultan S, Weizman AV, et al. AGA clinical practice guidelines on the role of probiotics in the Management of Gastrointestinal Disorders. Gastroenterology. 2020. Aug;159(2):697–705. 10.1053/j.gastro.2020.05.059 [DOI] [PubMed] [Google Scholar]
- 100. Swanson KS, Gibson GR, Hutkins R, Reimer RA, Reid G, Verbeke K, et al. The international scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nature reviews . Gastroenterol Hepatol. 2020. Aug 21;17:687–701. 10.1038/s41575-020-0344-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Crittenden R, Playne M. In: Lee YK, Salminen S, editors. Handbook of probiotics and prebiotics. New Jersey: John Wiley & Sons; 2009. p. 535–84. [Google Scholar]
- 102. Olveira G, González‐Molero I. Actualización de probióticos, prebióticos y simbióticos en nutrición clínica. Endocrinol Nutr. 2016. Nov 1;63(9):482–94. 10.1016/j.endonu.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 103. Saez‐Lara MJ, Robles‐Sanchez C, Ruiz‐Ojeda FJ, Plaza‐Diaz J, Gil A. Effects of probiotics and synbiotics on obesity, insulin resistance syndrome, type 2 diabetes and non‐alcoholic fatty liver disease: a review of human clinical trials. Int J Mol Sci. 2016. Jun 13;17(6):1–27. 10.3390/ijms17060928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, et al. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl Environ Microbiol. 2005;71(10):6150–8. 10.1128/AEM.71.10.6150-6158.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Corrêa NB, Péret Filho LA, Penna FJ, Lima FM, Nicoli JR. A randomized formula controlled trial of Bifidobacterium lactis and Streptococcus thermophilus for prevention of antibiotic‐associated diarrhea in infants. J Clin Gastroenterol. 2005. May‐Jun;39(5):385–9. 10.1097/01.mcg.0000159217.47419.5b [DOI] [PubMed] [Google Scholar]
- 106. Plummer S, Weaver MA, Harris JC, Dee P, Hunter J. Clostridium difficile pilot study: effects of probiotic supplementation on the incidence of C. difficile diarrhoea. Int Microbiol. 2004. Mar;7(1):59–62. [PubMed] [Google Scholar]
- 107. Lin HC, Hsu CH, Chen HL, Chung MY, Hsu JF, Lien RI, et al. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: a multicenter, randomized, controlled trial. Pediatrics. 2008. Oct;122(4):693–700. 10.1542/peds.2007-3007 [DOI] [PubMed] [Google Scholar]
- 108. Kajander K, Hatakka K, Poussa T, Färkkilä M, Korpela R. A probiotic mixture alleviates symptoms in irritable bowel syndrome patients: a controlled 6‐month intervention. Aliment Pharmacol Ther. 2005. Sep 1;22(5):387–94. 10.1111/j.1365-2036.2005.02579.x [DOI] [PubMed] [Google Scholar]
- 109. MJAv H, RMH M. Emergence of microbial diversity due to cross‐feeding interactions in a spatial model of gut microbial metabolism. BMC Syst Biol. 2017. May 16;11(1):56. 10.1186/s12918-017-0430-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. MCL B, Lammerts van Bueren A, Dijkhuizen L. Cross‐feeding among probiotic bacterial strains on prebiotic inulin involves the extracellular Inulinase of lactobacillus paracasei strain W20. Appl Environ Microbiol. 2018;84(21):e01539‐18. 10.1128/aem.01539-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Eslamparast T, Poustchi H, Zamani F, Sharafkhah M, Malekzadeh R, Hekmatdoost A. Synbiotic supplementation in nonalcoholic fatty liver disease: a randomized, double‐blind, placebo‐controlled pilot study. Am J Clin Nutr. 2014;99(3):535–42. [DOI] [PubMed] [Google Scholar]
- 112. Rogha M, Esfahani MZ, Zargarzadeh AH. The efficacy of a synbiotic containing bacillus Coagulans in treatment of irritable bowel syndrome: a randomized placebo‐controlled trial. Gastroenterol Hepatol. 2014;7(3):156. [PMC free article] [PubMed] [Google Scholar]
- 113. Islek A, Sayar E, Yilmaz A, Baysan BO, Mutlu D, Artan R. The role of Bifidobacterium lactis B94 plus inulin in the treatment of acute infectious diarrhea in children. Turk J Gastroenterol. 2014;25(6):628–33. [DOI] [PubMed] [Google Scholar]
- 114. Asemi Z, Khorrami‐Rad A, Alizadeh S‐A, Shakeri H, Esmaillzadeh A. Effects of synbiotic food consumption on metabolic status of diabetic patients: a double‐blind randomized cross‐over controlled clinical trial. Clin Nutr. 2014;33(2):198–203. [DOI] [PubMed] [Google Scholar]
- 115. Reid G, Gaudier E, Guarner F, et al. Responders and non‐responders to probiotic interventions: how can we improve the odds? Gut Microbes. 2010. May‐Jun;1(3):200–4. 10.4161/gmic.1.3.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Suez J, Zmora N, Zilberman‐Schapira G, et al. Post‐antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018. Sep 6;174(6):1406–1423.e16. 10.1016/j.cell.2018.08.047 [DOI] [PubMed] [Google Scholar]
- 117. Fleming A. Classics in infectious diseases: on the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae by Alexander Fleming, reprinted from the British Journal of experimental pathology 10:226‐236, 1929. Rev Infect Dis. 1980. Jan‐Feb;2(1):129–39. [PubMed] [Google Scholar]
- 118. Toor AA, Farooka MW, Ayyaz M, Sarwar H, Malik AA, Shabbir F. Pre‐operative antibiotic use reduces surgical site infection. J Pak Med Assoc. 2015. Jul;65(7):733–6. [PubMed] [Google Scholar]
- 119. Ji SK, Yan H, Jiang T, et al. Preparing the gut with antibiotics enhances gut microbiota reprogramming efficiency by promoting Xenomicrobiota colonization. Front Microbiol. 2017;8:1208–8. 10.3389/fmicb.2017.01208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Henn MR, O'Brien EJ, Diao L, et al. A phase 1b safety study of SER‐287, a spore‐based microbiome therapeutic, for active mild to moderate ulcerative colitis. Gastroenterology. 2021;160(1):115–127.e30. 10.1053/j.gastro.2020.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Vollaard EJ, Clasener HA, van Saene HK, Muller NF. Effect on colonization resistance: an important criterion in selecting antibiotics. DICP. 1990. Jan;24(1):60–6. 10.1177/106002809002400113 [DOI] [PubMed] [Google Scholar]
- 122. Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host–microbiota mutualism. Nat Rev Microbiol. 2011. Apr 1;9(4):233–43. 10.1038/nrmicro2536 [DOI] [PubMed] [Google Scholar]
- 123. Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. Antibiotic‐associated pseudomembranous colitis due to toxin‐producing clostridia. N Engl J Med. 1978. Mar 9;298(10):531–4. 10.1056/nejm197803092981003 [DOI] [PubMed] [Google Scholar]
- 124. Larcombe S, Hutton ML, Riley TV, Abud HE, Lyras D. Diverse bacterial species contribute to antibiotic‐associated diarrhoea and gastrointestinal damage. J Infect. 2018. Nov;77(5):417–26. 10.1016/j.jinf.2018.06.006 [DOI] [PubMed] [Google Scholar]
- 125. Sahra S, Jahangir A, De Chavez V. Antimicrobial stewardship: a review for internal medicine physicians. Cureus. 2021;13(4):e14385. 10.7759/cureus.14385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. d'Herelle F. Sur une épizootie de nature bactérienne sévissant sur les sauterelles au Mexique. CR Acad Sci. 1911;152:1413–5. [Google Scholar]
- 127. Rhoads D, Wolcott R, Kuskowski M, Wolcott B, Ward L, Sulakvelidze A. Bacteriophage therapy of venous leg ulcers in humans: results of a phase I safety trial. J Wound Care. 2009;18(6):237–43. [DOI] [PubMed] [Google Scholar]
- 128. MacNeal W, Frisbee F. One hundred patients with Staphylococcus septicemia receiving bacteriophage service. Am J Med Sci. 1936;191(2):179–95. [Google Scholar]
- 129. La Vergne S, Hamilton T, Biswas B, Kumaraswamy M, Schooley RT, Wooten D. Phage therapy for a multidrug‐resistant Acinetobacter baumannii craniectomy site infection. Open Forum Infect Dis. 2018;5(4):ofy064. 10.1093/ofid/ofy064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Draper LA, Ryan FJ, Smith MK, et al. Long‐term colonisation with donor bacteriophages following successful faecal microbial transplantation. Microbiome. 2018. Dec 10;6(1):220. 10.1186/s40168-018-0598-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ott SJ, Waetzig GH, Rehman A, et al. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology. 2017. Mar;152(4):799–811.e7. 10.1053/j.gastro.2016.11.010 [DOI] [PubMed] [Google Scholar]
- 132. Koskella B, Meaden S. Understanding bacteriophage specificity in natural microbial communities. Viruses. 2013;5(3):806–23. 10.3390/v5030806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. York A. FMT in the clinic. Nat Rev Microbiol. 2019. Mar;17(3):127. 10.1038/s41579-019-0157-x [DOI] [PubMed] [Google Scholar]
- 134. Goldenberg SD, Batra R, Beales I, Digby‐Bell JL, Irving PM, Kellingray L, et al. Comparison of different strategies for providing fecal microbiota transplantation to treat patients with recurrent Clostridium difficile infection in two English hospitals: a review. Infect Dis Ther. 2018. Mar;7(1):71–86. 10.1007/s40121-018-0189-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Stripling J, Rodriguez M. Current evidence in delivery and therapeutic uses of fecal microbiota transplantation in human diseases‐Clostridium difficile disease and beyond. Am J Med Sci. 2018. Nov;356(5):424–32. 10.1016/j.amjms.2018.08.010 [DOI] [PubMed] [Google Scholar]
- 136. Tvede M, Rask‐Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet. 1989. May 27;1(8648):1156–60. 10.1016/s0140-6736(89)92749-9 [DOI] [PubMed] [Google Scholar]
- 137. Borody TJ, George L, Andrews P, et al. Bowel‐flora alteration: a potential cure for inflammatory bowel disease and irritable bowel syndrome? Med J Aust. 1989. May 15;150(10):604. 10.5694/j.1326-5377.1989.tb136704.x [DOI] [PubMed] [Google Scholar]
- 138. Bennet JD, Brinkman M. Treatment of ulcerative colitis by implantation of normal colonic flora. Lancet. 1989. Jan 21;1(8630):164. 10.1016/s0140-6736(89)91183-5 [DOI] [PubMed] [Google Scholar]
- 139. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile . N Engl J Med. 2013;368(5):407–15. 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- 140. Petrof EO, Gloor GB, Vanner SJ, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013. Jan 9;1(1):3. 10.1186/2049-2618-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Greenberg SA, Youngster I, Cohen NA, Livovsky DM, Strahilevitz J, Israeli E, et al. Five years of fecal microbiota transplantation ‐ an update of the Israeli experience. World J Gastroenterol. 2018;24(47):5403–14. 10.3748/wjg.v24.i47.5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. SMD B, Lee MM, Eriksen MK, et al. Faecal microbiota transplantation for recurrent Clostridioides difficile infection: an updated systematic review and meta‐analysis. EClinicalMedicine. 2020. Dec;29–30:100642. 10.1016/j.eclinm.2020.100642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–916.e7. [DOI] [PubMed] [Google Scholar]
- 144. Kang DW, Adams JB, Gregory AC, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open‐label study. Microbiome. 2017. Jan 23;5(1):10. 10.1186/s40168-016-0225-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Zhou D, Pan Q, Shen F, et al. Total fecal microbiota transplantation alleviates high‐fat diet‐induced steatohepatitis in mice via beneficial regulation of gut microbiota. Sci Rep. 2017. May 8;7(1):1529. 10.1038/s41598-017-01751-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Costello SP, Hughes PA, Waters O, et al. Effect of fecal microbiota transplantation on 8‐week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321(2):156–64. 10.1001/jama.2018.20046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Fuentes S, Rossen NG, van der Spek MJ, Hartman JHA, Huuskonen L, Korpela K, et al. Microbial shifts and signatures of long‐term remission in ulcerative colitis after faecal microbiota transplantation. ISME J. 2017;11(8):1877–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Hui W, Li T, Liu W, Zhou C, Gao F. Fecal microbiota transplantation for treatment of recurrent C. difficile infection: an updated randomized controlled trial meta‐analysis. PLoS One. 2019;14(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 149. Costello SP, Soo W, Bryant RV, Jairath V, Hart AL, Andrews JM. Systematic review with meta‐analysis: faecal microbiota transplantation for the induction of remission for active ulcerative colitis. Aliment Pharmacol Ther. 2017. Aug 1;46(3):213–24. 10.1111/apt.14173 [DOI] [PubMed] [Google Scholar]
- 150. Wang S, Xu M, Wang W, Cao X, Piao M, Khan S, et al. Systematic review: adverse events of fecal microbiota transplantation. PLoS One. 2016;11(8):e0161174. 10.1371/journal.pone.0161174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Rossen NG, Fuentes S, van der Spek MJ, et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology. 2015. Jul;149(1):110–118.e4. 10.1053/j.gastro.2015.03.045 [DOI] [PubMed] [Google Scholar]
- 152. Moayyedi P, Surette MG, Kim PT, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015. Jul;149(1):102–109.e6. 10.1053/j.gastro.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 153. Paramsothy S, Kamm MA, Kaakoush NO, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo‐controlled trial. Lancet. 2017. Mar 25;389(10075):1218–28. 10.1016/s0140-6736(17)30182-4 [DOI] [PubMed] [Google Scholar]
- 154. Papanicolas LE, Choo JM, Wang Y, et al. Bacterial viability in faecal transplants: which bacteria survive? EBioMedicine. 2019. Mar;41:509–16. 10.1016/j.ebiom.2019.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, Park T, et al. Long‐term follow‐up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012. Jul;107(7):1079–87. 10.1038/ajg.2012.60 [DOI] [PubMed] [Google Scholar]
- 156. Juul FE, Garborg K, Bretthauer M, et al. Fecal microbiota transplantation for primary Clostridium difficile infection. N Engl J Med. 2018. Jun 28;378(26):2535–6. 10.1056/NEJMc1803103 [DOI] [PubMed] [Google Scholar]
- 157. Kelly CR, de Leon L, Jasutkar N. Fecal microbiota transplantation for relapsing Clostridium difficile infection in 26 patients: methodology and results. J Clin Gastroenterol. 2012. Feb;46(2):145–9. 10.1097/MCG.0b013e318234570b [DOI] [PubMed] [Google Scholar]
- 158. Bhutiani N, Schucht JE, Miller KR, McClave SA. Technical aspects of fecal microbial transplantation (FMT). Curr Gastroenterol Rep. 2018. Jun 9, 30;20(7):1–6. 10.1007/s11894-018-0636-7 [DOI] [PubMed] [Google Scholar]
- 159. Ianiro G, Maida M, Burisch J, et al. Efficacy of different faecal microbiota transplantation protocols for Clostridium difficile infection: a systematic review and meta‐analysis. United Eur Gastroenterol J. 2018. Oct 1;6(8):1232–44. 10.1177/2050640618780762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Makkar R, Bo S. Colonoscopic perforation in inflammatory bowel disease. Gastroenterol Hepatol. 2013;9(9):573–83. [PMC free article] [PubMed] [Google Scholar]
- 161. Marcella C, Cui B, Kelly CR, Ianiro G, Cammarota G, Zhang F. Systematic review: the global incidence of faecal microbiota transplantation‐related adverse events from 2000 to 2020. Aliment Pharmacol Ther. 2021. Jan;53(1):33–42. 10.1111/apt.16148 [DOI] [PubMed] [Google Scholar]
- 162. Youngster I, Mahabamunuge J, Systrom HK, et al. Oral, frozen fecal microbiota transplant (FMT) capsules for recurrent Clostridium difficile infection. BMC Med. 2016;14(1):134–4. 10.1186/s12916-016-0680-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Chehri M, Christensen AH, Halkjær SI, Günther S, Petersen AM, Helms M. Case series of successful treatment with fecal microbiota transplant (FMT) oral capsules mixed from multiple donors even in patients previously treated with FMT enemas for recurrent Clostridium difficile infection. Medicine. 2018;97(31):e11706. 10.1097/MD.0000000000011706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kao D, Roach B, Silva M, et al. Effect of oral capsule‐ vs colonoscopy‐delivered fecal microbiota transplantation on recurrent Clostridium difficile infection: a randomized clinical trial. JAMA. 2017. Nov 28;318(20):1985–93. 10.1001/jama.2017.17077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Rao K, Young VB, Malani PN. Capsules for fecal microbiota transplantation in recurrent Clostridium difficile infection: the new way forward or a tough pill to swallow? JAMA. 2017;318(20):1979–80. 10.1001/jama.2017.17969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. De Filipp Z, Bloom PP, Torres Soto M, et al. Drug‐resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med. 2019. Nov 21;381(21):2043–50. 10.1056/NEJMoa1910437 [DOI] [PubMed] [Google Scholar]
- 167. FDA . Important safety alert regarding use of fecal microbiota for transplantation and risk of serious adverse reactions due to transmission of multi‐drug resistant organisms. Fda.gov. 2019.
- 168. Wilson BC, Vatanen T, Cutfield WS, O'Sullivan JM. The super‐donor phenomenon in fecal microbiota transplantation. Front Cell Infect Microbiol. 2019;9:2. 10.3389/fcimb.2019.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Osman M, Stoltzner Z, O'Brien K, et al. Donor efficacy in fecal microbiota transplantation for recurrent Clostridium difficile: evidence from a 1,999‐patient cohort. Open Forum Infect Dis. 2016;3(suppl_1):1–1. 10.1093/ofid/ofw194.48 [DOI] [Google Scholar]
- 170. Giles EM, D'Adamo GL, Forster SC. The future of faecal transplants. Nat Rev Microbiol. 2019. Dec 1;17(12):719. 10.1038/s41579-019-0271-9 [DOI] [PubMed] [Google Scholar]
- 171. Tarantino V, Savaia V, D'Agostino R, et al. Bacteriotherapy in children with recurrent upper respiratory tract infections. Eur Rev Med Pharmacol Sci. 2019. Mar;23(1 Suppl):39–43. 10.26355/eurrev_201903_17347 [DOI] [PubMed] [Google Scholar]
- 172. FDA . Early clinical trials with live biotherapeutic products: chemistry, manufacturing and control information. 2016.
- 173. Olle B. Medicines from microbiota. Nat Biotechnol. 2013;31(4):309–15. [DOI] [PubMed] [Google Scholar]
- 174. Studer N, Desharnais L, Beutler M, et al. Functional intestinal bile acid 7alpha‐Dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front Cell Infect Microbiol. 2016;6:191. 10.3389/fcimb.2016.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile . Nature. 2015;517(7533):205–8. 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8(10):e1002995. 10.1371/journal.ppat.1002995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Papamichael K, Afif W, Drobne D, et al. Therapeutic drug monitoring of biologics in inflammatory bowel disease: unmet needs and future perspectives. Lancet Gastroenterol Hepatol. 2022. Feb;7(2):171–85. 10.1016/s2468-1253(21)00223-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Wegh CAM, Geerlings SY, Knol J, Roeselers G, Belzer C. Postbiotics and their potential applications in early life nutrition and beyond. Int J Mol Sci. 2019. Sep 20;20(19):1–42. 10.3390/ijms20194673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Taverniti V, Guglielmetti S. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: proposal of paraprobiotic concept). Genes Nutr. 2011;6(3):261–74. 10.1007/s12263-011-0218-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Aguilar‐Toalá JE, Garcia‐Varela R, Garcia HS, et al. Postbiotics: an evolving term within the functional foods field. Trends Food Sci Technol. 2018. May 1;75:105–14. 10.1016/j.tifs.2018.03.009 [DOI] [Google Scholar]
- 181. Gill PA, van Zelm MC, Muir JG, Gibson PR. Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Ther. 2018. Jul;48(1):15–34. 10.1111/apt.14689 [DOI] [PubMed] [Google Scholar]
- 182. Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Gd B, Lange K, Havinga R, et al. Gut‐derived short‐chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am J Physiol Gastrointest Liver Physiol. 2013;305(12):G900–10. 10.1152/ajpgi.00265.2013 [DOI] [PubMed] [Google Scholar]
- 184. Koh A, De Vadder F, Kovatcheva‐Datchary P, Bäckhed F. From dietary fiber to host physiology: short‐chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–45. 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 185. Nastasi C, Candela M, Bonefeld CM, et al. The effect of short‐chain fatty acids on human monocyte‐derived dendritic cells. Sci Rep. 2015. Nov 6;5:16148. 10.1038/srep16148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014. Feb 11;111(6):2247–52. 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Macia L, Tan J, Vieira AT, et al. Metabolite‐sensing receptors GPR43 and GPR109A facilitate dietary fibre‐induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015. Apr 1;6:6734. 10.1038/ncomms7734 [DOI] [PubMed] [Google Scholar]
- 188. Roediger WE. The colonic epithelium in ulcerative colitis: an energy‐deficiency disease? Lancet. 1980. Oct 4;2(8197):712–5. 10.1016/s0140-6736(80)91934-0 [DOI] [PubMed] [Google Scholar]
- 189. Roediger WE, Nance S. Metabolic induction of experimental ulcerative colitis by inhibition of fatty acid oxidation. Br J Exp Pathol. 1986;67(6):773–82. [PMC free article] [PubMed] [Google Scholar]
- 190. Brown CT, Davis‐Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6(10):e25792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Li N, Hatch M, Wasserfall CH, Douglas‐Escobar M, Atkinson MA, Schatz DA, et al. Butyrate and type 1 diabetes mellitus: can we fix the intestinal leak? J Pediatr Gastroenterol Nutr. 2010;51(4):414–7. [DOI] [PubMed] [Google Scholar]
- 192. Wolfe BE, Dutton RJ. Fermented foods as experimentally tractable microbial ecosystems. Cell. 2015;161(1):49–55. [DOI] [PubMed] [Google Scholar]
- 193. Sieuwerts S, de Bok FA, Hugenholtz J, van Hylckama Vlieg JE. Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl Environ Microbiol. 2008. Aug;74(16):4997–5007. 10.1128/aem.00113-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Montel MC, Buchin S, Mallet A, Delbes‐Paus C, Vuitton DA, Desmasures N, et al. Traditional cheeses: rich and diverse microbiota with associated benefits. Int J Food Microbiol. 2014. May 2;177:136–54. 10.1016/j.ijfoodmicro.2014.02.019 [DOI] [PubMed] [Google Scholar]
- 195. Choo JM, Leong LEX, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015;5:16350. 10.1038/srep16350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Vandeputte D, Tito RY, Vanleeuwen R, Falony G, Raes J. Practical considerations for large‐scale gut microbiome studies. FEMS Microbiol Rev. 2017;41(Supplement_1):S154–67. 10.1093/femsre/fux027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Macpherson AJ, McCoy KD. Standardised animal models of host microbial mutualism. Mucosal Immunol. 2015. May 1;8(3):476–86. 10.1038/mi.2014.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. A framework for human microbiome research. Nature. 2012. Jun 13;486(7402):215–21. 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Merrick B, Allen L, Masirah M, Zain N, Forbes B, Shawcross DL, et al. Regulation, risk and safety of Faecal microbiota transplant. Infect Prev Pract. 2020;2(3):100069. 10.1016/j.infpip.2020.100069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. FitzGerald MJ, Spek EJ. Microbiome therapeutics and patent protection. Nat Biotechnol. 2020. Jul 1;38(7):806–10. 10.1038/s41587-020-0579-z [DOI] [PubMed] [Google Scholar]
- 201. Nguyen TLA, Vieira‐Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8(1):1–16. 10.1242/dmm.017400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Reyniers JA. The pure‐culture concept and gnotobiotics. Ann N Y Acad Sci. 1959;78(1):3–16. [Google Scholar]
- 203. Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci. 2011;108(15):6252–7. 10.1073/pnas.1102938108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Faith JJ, Rey FE, O'Donnell D, Karlsson M, McNulty NP, Kallstrom G, et al. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010. Sep;4(9):1094–8. 10.1038/ismej.2010.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Britton GJ, Contijoch EJ, Spindler MP, Aggarwala V, Dogan B, Bongers G, et al. Defined microbiota transplant restores Th17/RORyt regulatory T cell balance in mice colonized with inflammatory bowel disease microbiotas. Proc Natl Acad Sci. 2020;117(35):21536–45. 10.1073/pnas.1922189117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Pearce SC, Coia HG, Karl JP, Pantoja‐Feliciano IG, Zachos NC, Racicot K. Intestinal in vitro and ex vivo models to study host‐microbiome interactions and acute stressors. Front Physiol. 2018;9:1584–4. 10.3389/fphys.2018.01584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt‐villus structures in vitro without a mesenchymal niche. Nature. 2009. May 14;459(7244):262–5. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 208. Sato T, Stange DE, Ferrante M, Vries RGJ, van Es JH, van den Brink S, et al. Long‐term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011. Nov;141(5):1762–72. 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- 209. Jung P, Sato T, Merlos‐Suarez A, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011. Sep 4;17(10):1225–7. 10.1038/nm.2470 [DOI] [PubMed] [Google Scholar]
- 210. Hill DR, Huang S, Tsai YH, Spence JR, Young VB. Real‐time Measurement of epithelial barrier permeability in human intestinal organoids. J Vis Exp. 2017. Dec 18;(1: Dec 18;(130):1–25. 10.3791/56960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Dutta D, Heo I, O'Connor R. Studying cryptosporidium infection in 3D tissue‐derived human organoid culture systems by microinjection. J Vis Exp. 2019. Sep 14;(1: Sep 14;(151):1–10. 10.3791/59610 [DOI] [PubMed] [Google Scholar]
- 212. Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011. Feb 3;470(7332):105–9. 10.1038/nature09691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018. Dec 13;175(7):1972–1988.e16. 10.1016/j.cell.2018.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of tumor‐reactive T cells by co‐culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018. Sep 6;174(6):1586–1598.e12. 10.1016/j.cell.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Aguilar C, Alves da Silva M, Saraiva M, Neyazi M, Olsson IAS, Bartfeld S. Organoids as host models for infection biology – a review of methods. Exp Mol Med. 2021. Oct 1;53(10):1471–82. 10.1038/s12276-021-00629-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Huang Y, Williams JC, Johnson SM. Brain slice on a chip: opportunities and challenges of applying microfluidic technology to intact tissues. Lab Chip. 2012. Jun 21;12(12):2103–17. 10.1039/c2lc21142d [DOI] [PubMed] [Google Scholar]
- 217. Nalayanda DD, Puleo C, Fulton WB, Sharpe LM, Wang TH, Abdullah F. An open‐access microfluidic model for lung‐specific functional studies at an air‐liquid interface. Biomed Microdevices. 2009. Oct;11(5):1081–9. 10.1007/s10544-009-9325-5 [DOI] [PubMed] [Google Scholar]
- 218. Cheng W, Klauke N, Sedgwick H, Smith GL, Cooper JM. Metabolic monitoring of the electrically stimulated single heart cell within a microfluidic platform. Lab Chip 2006. Nov;6(11):1424–31. 10.1039/b608202e [DOI] [PubMed] [Google Scholar]
- 219. Wufuer M, Lee G, Hur W, Jeon B, Kim BJ, Choi TH, et al. Skin‐on‐a‐chip model simulating inflammation, edema and drug‐based treatment. Sci Rep. 2016;6:37471–1. 10.1038/srep37471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut‐on‐a‐chip. Proc Natl Acad Sci. 2016;113(1):E7–15. 10.1073/pnas.1522193112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Allen JW, Bhatia SN. Formation of steady‐state oxygen gradients in vitro: application to liver zonation. Biotechnol Bioeng. 2003;82(3):253–62. [DOI] [PubMed] [Google Scholar]
- 222. Grassart A, Malarde V, Gobaa S, et al. Bioengineered human organ‐on‐Chip reveals intestinal microenvironment and mechanical forces impacting shigella infection. Cell Host Microbe. 2019. Sep 11;26(3):435–444.e4. 10.1016/j.chom.2019.08.007 [DOI] [PubMed] [Google Scholar]
- 223. Bein A, Shin W, Jalili‐Firoozinezhad S, Park MH, Sontheimer‐Phelps A, Tovaglieri A, et al. Microfluidic organ‐on‐a‐Chip models of human intestine. Cell Mol Gastroenterol Hepatol. 2018. Jan 1;5(4):659–68. 10.1016/j.jcmgh.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Tovaglieri A, Sontheimer‐Phelps A, Geirnaert A, et al. Species‐specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome. 2019. Mar 20;7(1):43. 10.1186/s40168-019-0650-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Jalili‐Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, et al. A complex human gut microbiome cultured in an anaerobic intestine‐on‐a‐chip. Nat Biomed Eng. 2019. Jul;3(7):520–31. 10.1038/s41551-019-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed as part of this study.


