Abstract
The development of new and effective therapeutics is reliant on the ability to study the underlying mechanisms of potential drug targets in live cells and multicellular systems. A persistent challenge in many drug development programmes is poor selectivity, which can obscure the mechanisms involved and lead to poorly understood modes of action. In efforts to improve our understanding of these complex processes, small molecule inhibitors have been developed in which their OFF/ON therapeutic activity can be toggled using light. Photopharmacology is devoted to using light to modulate drugs. Herein, we highlight the recent progress made towards the development of light‐responsive small molecule inhibitors of selected enzymatic targets. Given the size of this field, literature from 2015 onwards has been reviewed.
Keywords: chemical biology, enzyme, inhibitors, photocaging, photopharmacology, photoswitching
The rapid advancement of the field of photopharmacology has afforded tools to aid the development of light‐responsive entities to probe enzyme function in complex cellular pathways. Here we highlight the recent progress made towards the development of light‐responsive small molecule inhibitors of selected enzymatic targets.
1. Introduction
Binding small molecules that activate or inhibit biological functions is a core concept in drug design. The ideal drug must bind with high selectivity towards the intended target as to avoid interfering with other biological processes. This is one of the many challenges that medicinal chemists face, as small molecule drugs typically suffer from selectivity issues that result in undesired side‐effects. [1] The innovation of small molecules that change structure in response to a stimulus allows one to combat these limitations that are observed with traditional small molecule drugs. As a result of the superior spatiotemporal control that can be achieved with stimuli‐responsive molecules, unwanted off‐target effects can be significantly reduced. This is of paramount importance in drug development where identifying biological targets and their function is crucial. Thus, the ability to precisely control the time and place where a drug is activated overcomes many problems associated with poor drug selectivity.
Photopharmacology is an innovative solution to combat such challenges in which light modulates the therapeutic properties of bioactives. [2] As an input stimulus, light is ideal as it can be applied remotely, the wavelength and intensity of the light can be readily controlled, is highly orthogonal and can be administered with precise spatiotemporal resolution. These features make light an attractive choice of input stimulus for applications in biological settings, and when combined with light‐responsive therapeutics, enables bio‐relevant mechanisms to be investigated while avoiding much of the limitations that arise from drugs with poor selectivity. In photopharmacology, the photoresponsive entities are generally categorised as either photoswitches or photolabile caging groups (often referred to as photocages).
Photoswitchable entities undergo reversible geometric changes upon exposure to defined wavelengths of light. As specific wavelengths of light are required for each direction of photoisomerisation, the pharmacophore can be readily switched between an “active” and “inactive” state (Figure 1). Photoswitchable groups such as azobenzenes, diarylethenes (DTEs) and spiropyrans, can be incorporated into the scaffold of a bioactive so that only one of the isomeric forms acts as an isostere of the active inhibitor that binds the corresponding target.[ 2b , 2c , 3 ]
Figure 1.

Schematic depiction of photoswitching and photocaging approaches utilised in photopharmacology. (top) Reversible light‐mediated activation/deactivation of photoswitchable small molecule inhibitor. (bottom) Irreversible light‐mediated activation of small molecule inhibitors through the use of caging groups.
Photocaging involves modifying the bioactive with additional steric bulk that prevent crucial binding interactions, meaning the compounds are pharmacologically inactive in their caged state. Activation is achieved by irradiation with light, whereby photoinduced bond cleavage results in liberation of the active pharmacophore (Figure 1). Cleavage of a photocaging group from a drug is irreversible so although they cannot be returned to its inactive state via photonic means, contrary to photoswitchable pharmacophores, far greater discrimination between the activity of the two states is typically achieved when using photocaging.
As the field of photopharmacology has gained traction, a growing number of reports have incorporated such light‐responsive therapeutics for targeting subcellular locations such as lipid membranes, [4] ion channels,[ 2b , 3 , 4 ] various receptors[ 2b , 3 , 4 ] and nucleic acids.[ 3 , 5 ] Enzymes are crucial for the regulation of cell processes and function, and are often associated with the dysregulation in disease states. The rapid evolution of the field of photopharmacology has also afforded the tools required to develop valuable light‐responsive entities to probe enzyme function in complex cellular pathways that today are poorly understood. This minireview highlights the recent advances in the development of light‐responsive small molecules that target and control enzymatic activity of popular drug targets including kinase, histone deacetylase (HDAC), dihydrofolate reductase (DHFR) and proteasome enzymes. This article is by no means a comprehensive review of all literature, instead focussing on the more recent advances made to the field since 2015.
2. Light‐Responsive Kinase Inhibitors
Given the critical role that kinases play in a number of cellular functions including apoptosis, cell proliferation and differentiation, kinase inhibition has proven to be a valuable therapeutic approach for the treatment of a number of diseases. [6] As many kinases are ubiquitously expressed, undesired outcomes often arise from their global inhibition. It is therefore no surprise that the development of light‐responsive kinase inhibitors has attracted significant attention over the past decade. Light‐controlled kinase inhibition has been recently reviewed. [7] However, since this publication, the development of light‐responsive kinase inhibitors has continued to evolve, and the more recent caged and photoswitchable inhibitors are described below.
2.1. Photocaged Kinase Inhibitors
The inhibitory properties of the potent bi‐substrate inhibitor 2 of cAMP‐dependent protein kinase (PKAcα) was masked by the inclusion of a nitrodibenzofuran (NDBF)‐derived caging group onto the nitrogen atom of the pyrimidine heterocycle, preventing key hydrogen bond interactions from occurring in the ATP binding site (Figure 2). [8] The binding affinity of the caged inhibitor 1 for PKAcα was 1900 nM K D. Irradiation of the caged inhibitor using a Hg vapour lamp resulted in a significant decrease in affinity for PKAcα by 5‐orders of magnitude (K D <0.1 nM). In a cellular setting, the enzymatic activity of PKAcα is blocked by the formation of a heterotetrametric holoenzyme, whereby the catalytically active subunits of PKAc only become accessible when an increase in concentration of cAMP induces conformational change, or when treated with a potent PKAc inhibitor. [9] The authors demonstrated that, in the absence of light, cell lysates treated with 1 exhibited no effect on the PKA holoenzyme. Only when irradiated with an LED at 398 nm to initiate the release of the active inhibitor 2, was the disruption of the PKA holoenzyme observed.
Figure 2.
UV light induced photoactivation of caged kinase inhibitors.
Photocaged inhibitors of TAM kinases, Tyro3 and Mer, have been achieved by the inclusion of the 4,5‐dimethoxynitrobenzyl (DMNB) caging group onto the nitrogen atom at the 3‐position of the imidazopyridine to prevent key hydrogen bonding interactions within the hinge region of the kinase (Figure 2). [10] Caged compounds, 3 a and 4 a, served as weak inhibitors of Mer, affording IC50 values of 1.97 μM and 3.37 μM, respectively. The residual kinase activity observed for both caged inhibitors suggest that these derivatives may be able to still form key interactions within the active site. Nevertheless, exposure to UV light (λ=350 nm) resulted in the release of the active inhibitors and subsequent increase in inhibitory activity (3 b IC50=0.113 μM; 4 b IC50=0.0146 μM). Inhibitory activity towards Tyro3 was also evaluated. The caged inhibitors, compounds 3 a and 4 a, showed poor inhibitory activity of Tyro3 with IC50 values of 4.39 μM and >100 μM, respectively. Again, inhibitory properties were restored upon photoirradiation (λ=350 nm) to give IC50 values of 1.06 μM and 5.56 μM for compounds 3 b and 4 b, respectively.
The inclusion of the DMNB caging group at the NH moiety of the sulfonamide in the caged PI3K inhibitor 5, resulted in low micromolar IC50 values (PI3Kα IC50=343 nM, Figure 2). [11] Low nanomolar inhibitory was restored following exposure to UV light (λ=365 nm) and subsequent release of the active PI3K inhibitor 6 (PI3Kα IC50=0.58 nM). Similar trends were also observed for other isoforms, in which >1500‐fold difference in inhibitory activity between the caged 5 and uncaged inhibitor 6 was exhibited for PI3Kβ, PIK3γ and PIK3δ. Cancer cell lines were treated with 5, in which exposure to UV light could be used to trigger antiproliferative activity corresponding to PI3K inhibition. The utility of this probe was further demonstrated in a zebrafish model, whereby photocontrol of the anticancer properties of 5 was achieved.
2.2. Photoswitchable Kinase Inhibitors
In efforts to realise reversible photocontrol over the enzymatic activity of p38α MAPK and CK1δ kinases, Schehr et al. [12] pursued the development of the arylazo‐photoswitch 8 (Figure 3). This was achieved by replacing the 2‐methylthioether imidazole unit of the previously reported inhibitor 7, [13] with an arylazo moiety. Exposure to 420 nm light gave the Z‐form (8‐Z ) with a photostationary state (PSS) of 85%, while photoisomerisation to the photo‐enriched E‐form (8‐E ) was initiated with visible light (λ=525 nm) with a PSS of 88% (see Supporting Information, Table S1). Evaluation of the inhibitory properties in a cell‐free assay showed that only a 1.5‐fold difference in kinase activity between the two isomers was obtained for both kinases. With the aid of x‐ray crystallography analysis, it was proposed that the conformational adaptation of the kinase with respect to both photoisomers, as well as the flexible nature of the photoswitch 8 was responsible for the subtle difference in kinase activity.
Figure 3.
Azobenzene‐derived photoswitchable kinase inhibitors and the corresponding parent compound that these are based upon.
Photocontrolled inhibition of the BRAFV600E mutant kinase has been achieved with the development of a photoswitchable derivative of the known inhibitor 9, [14] in which the sulfonamide moiety of 9 was replaced by an azobenzene, affording compound 10 (Figure 3). [15] It was anticipated that the Z‐isomer (10‐Z ) would mimic the bent conformation adopted by the parent compound when bound to the ATP‐binding site of the kinase, thereby serving as the active form. Replacing the amide with an azo group was also considered, however docking studies suggested the thermodynamically stable E‐form would function as the active inhibitor. As E→Z isomerisation can only occur by photonic means, designing the photoswitch so that the metastable Z‐form serves as the active form is favourable, as this approach allows for superior control over the ‘photo‐activation’ of the therapeutic properties. While E→Z photoisomerisation using 365 nm light resulted in a PSS of only 55% of the active Z‐form (10‐Z , Table S1), a 10‐fold difference in inhibitory properties between the E‐form and the photo‐enriched Z‐form was observed (10‐E IC50=1.68 μM; 10‐Z IC50=0.156 μM).
Peifer and co‐workers have developed a photoswitchable derivative of the known VEGFR2 kinase inhibitor, Axitnib, which was achieved by appending a bridged azobenzene moiety (diazocine) to the pharmacophore of the inhibitor (Figure 3). [16] Docking studies showed that the twisted nature of the thermodynamically stable Z‐isomer (11‐Z ) could not be accommodated for in the binding site of the VEGFR2, thereby functioning as a poor inhibitor. Indeed, in the absence of light, 11‐Z exhibited an IC50 value >10,000 nM. Exposure to 405 nm light afforded the photo‐enriched E‐form (11‐E , Table S1) in which over a 47‐fold increase in inhibitory activity was observed (IC50=214 nM).
As a means to regulate CKIα activity and circadian rhythms, Kolarski et al. [17] have developed photoswitchable variants of Longdaysin, a known inhibitor of CKIα. [18] Adopting an azologization approach, the N‐benzyl‐arylamine unit within Longdaysin was replaced with an azobenzene photoswitch (Figure 4, compound 12). [17a] Photoisomerisation of the E→Z isomer was achieved using green light (λ=530 nm), while the thermal back reaction to the E‐isomer (12‐E ) occurred rapidly, with half‐lives ranging from seconds to less than an hour, making this series of azopurines unsuitable for further analysis in circadian rhythm experiments (Table S1). Furthermore, these photoswitches were susceptible to reduction to the corresponding hydrazine when incubated with DTT and GSH.
Figure 4.
Azobenzene‐derived photoswitchable kinase inhibitors and the corresponding parent compound that these are based upon.
The same group also report the extension of the benzylamine moiety of Longdaysin with a photoswitchable azobenzene moiety, affording azopurine 13 (Figure 4). [17b] Upon exposure to 530 nm light, 13‐E was photoisomerised to the corresponding Z‐isomer (13‐Z , Table S1). In contrast to the previous azopurine series 12, 13 exhibited improved thermal stability of the Z‐form in cell culture medium (>50 h). The reverse reaction was achieved when irradiated with 400 nm light. Only a 1.5‐fold difference in inhibitory properties between the two isomers was observed for the CKIα isoform (13‐E IC50=5.3 μM; 13‐Z IC50=8.2 μM). However, the photo‐enriched Z‐form inhibited CKIδ with an IC50 value of 17.3 μM, which is 3.7 times higher than that observed for the E‐isomer (IC50=4.6 μM). The authors also demonstrated that the azobenzene 13 could be used to modulate circadian rhythms in a cellular context in a reversible and light‐controlled manner.
The first example of a photoswitchable DFG‐out kinase inhibitor was recently reported by Xu et al. [19] in 2021 (Figure 4). Utilising the known inhibitor, Ponatinib, as a template, the photoswitchable DFG‐out kinase inhibitor 14 that targets the RET kinase was developed by replacing the acetylene moiety with an azo bridge. Docking studies suggested that the Z‐form (14‐Z ) would not be tolerated in the active site as key interactions with the hinge region cannot be made, while the stable E‐isomer (14‐E ) would serve as the active inhibitor. Irradiation with UV light (λ=365 nm) afforded E→Z isomerisation with a photostationary distribution of 97% for 14‐Z (Table S1). The reverse reaction was achieved when exposed to blue light (λ=460 nm), yielding 64% of 14‐E . Good thermal stability of 14‐Z in aqueous solution was observed (t 1/2=629 h), and the photoswitch proved stable when incubated with DTT/GSH. Inhibitory studies using a RET biochemical assay showed that 14‐E was 17‐fold more potent than the photo‐enriched 14‐Z form, with IC50 values of 3 nM and 50 nM, respectively. Further analysis using a NanoBRET™ TE Intracellular Kinase Assay demonstrated the utility of the photoswitchable RET inhibitor a cellar context, in which IC50 values for the E‐isomer and the photo‐enriched Z‐isomer of 25 nM and 282 nM, respectively, were obtained.
The ROCK inhibitor 15 was originally identified via high throughput screening. [20] Based on the phenylazothiazole scaffold present in 15, a photoswitchable ROCK inhibitor has been developed by Matsuo and co‐workers, in which the amide moiety was replaced with an azo group to give compound 16 (Figure 4). [21] Docking studies showed that the E‐form (16‐E ) of the photoswitch adopted a similar binding pose to that of the parent inhibitor 15, thereby serving as the active form (IC50=19 μM). Photoisomerisation of the E→Z form was achieved when irradiated with 405 nm (Table S1), resulting in a 12‐fold decrease in inhibitory activity (IC50=238 μM). The low micromolar inhibitory properties were restored following exposure to visible light (λ=525 nm), affording an IC50 value of 34 μM. Furthermore, the photoswitchable inhibitor 16 could be used to control the ROCK‐dependent organisation of actin stress fibres in serum starved cells in a light‐dependent manner.
More recently, Reynders et al. [22] detailed the development of the photoswitchable covalent inhibitor 18 for the MAP kinase JNK3 (Figure 4). This was achieved by replacing the diarylamide motif of the known covalent inhibitor 17 [23] with a diazocine photoswitch bearing an electrophilic acrylamide moiety that targets cysteines neighbouring the ATP‐binding site, in which the reactivity of the electrophile could be regulated with light. In the dark, the diazocine photoswitch 18‐Z functions as a poor noncovalent inhibitor of JNK3 (IC50=646 nM). Exposure to 390 nm light yields the metastable E‐form (18‐E ), which can now reach and covalently bind the targeted cysteine residue. Upon doing so, a 30‐fold increase in JNK3 inhibition was observed (IC50=21.4 nM). In the absence of the kinase enzyme, E→Z isomerisation occurs either thermally or when exposed to 565 nm light (Table S1). However, irreversible inhibition of kinase activity was observed once the metastable E‐form bound, suggesting that the pyridinylimidazole unit maintained its presence in the ATP binding site, even after irradiation with visible light (λ=565 nm).
3. Light‐Responsive Histone Deacetylase (HDAC) Inhibitors
Histone deacetylase (HDAC) enzymes are histone modifying enzymes that play a fundamental role in the regulation of gene expression. [24] The dysregulation of HDACs is present in a variety of disease states including cancer, [25] cellular metabolism disorders [26] and inflammation. [27] As such, their inhibition has been of particular interest as potential approaches for the treatment to these diseases. The HDAC family are classified into two major categories: zinc‐dependent (HDACs I, II and IV) and nicotinamide adenine dinucleotide (NAD)‐dependent (class III, sirtuins) HDACs. The photocontrol over the inhibitory activity of small molecules that bind the zinc‐dependent HDACs or sirtuins has been pursued.
3.1. Inhibitors of the Zinc‐dependent HDACs
Optical control over HDAC inhibitors that target zinc‐dependent HDACs has been achieved through the use of photolabile caging groups and the inclusion of photochromic entities into the typical pharmacophore of potent HDAC inhibitors.
3.1.1. Photocaged HDAC Inhibitors
Since the first reports of caged HDAC inhibitors in 2016 [28] only a few examples have been documented in the literature. [29] As depicted in Figure 5, the majority of the reported caged HDAC inhibitors involve the caging of a hydroxamic acid moiety which serves as the zinc binding unit. The introduction of the photolabile caging group onto the hydroxamic acid oxygen prevents the zinc binding group from chelating to the zinc ion that resides within the active site. It is reported that hydroxamic acids readily undergo N−O bond cleavage upon irradiation with UV light, affording the corresponding amide analogue. [30] While the formation of the amide analogue of the HDAC inhibitor upon photoactivation has been observed for the majority of examples depicted in Figure 5, the desired hydroxamic acid was obtained as the major product.
Figure 5.
Photoactivation of caged HDAC inhibitors, whereby the inclusion of photolabile protecting groups onto the zinc‐binding moiety of known HDAC inhibitors masks inhibitory properties.
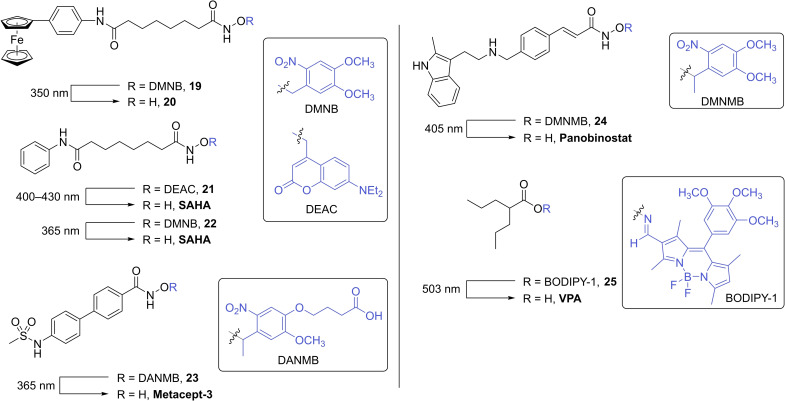
The DMNB caging group was used to mask the inhibitory properties of the known ferrocene‐containing HDAC inhibitor 20 (Figure 5). [28a] The inclusion of the caging group on the zinc binding moiety resulted in poor inhibitory activity towards HDACs 1, 2 and 6, with IC50 values of 3.45 μM, >10 μM and 10 μM, respectively (Table 1). Irradiation of the caged inhibitor 19 with 350 nm resulted in the release of the active inhibitor, 20, in which significant increase in HDAC inhibition was observed (HDAC1 IC50=0.11 μM, HDAC2 IC50=0.23 μM and HDAC6 IC50=0.04 μM).
Table 1.
IC50 (nM) values of photocaged HDAC inhibitors 19 and 24, and uncaged inhibitors 20 and panobinostat.
|
Compound |
HDAC1 |
HDAC2 |
HDAC3 |
HDAC4 |
HDAC5 |
HDAC6 |
HDAC7 |
HDAC8 |
HDAC9 |
HDAC11 |
|---|---|---|---|---|---|---|---|---|---|---|
|
19 [28a] |
3,450 |
>10,000 |
nd |
nd |
nd |
10,000 |
nd |
nd |
nd |
nd |
|
20 [28a] |
130 |
330 |
nd |
nd |
nd |
30 |
nd |
nd |
nd |
nd |
|
19+UV [28a] |
110 |
230 |
nd |
nd |
nd |
40 |
nd |
nd |
nd |
nd |
|
0.31 |
1.51 |
1.69 |
0.42 |
0.84 |
1.98 |
38.6 |
38.9 |
1.65 |
0.32 |
|
|
24 [29c] |
NI |
NI |
16,570 |
29,480 |
NI |
NI |
NI |
NI |
NI |
NI |
Pan=Panobinostat. nd=not determined. NI=no inhibition.
Photocaged derivatives of the FDA approved HDAC inhibitor, SAHA, have also been reported (Figure 5).[ 28b , 29a ] Nakagawa and co‐workers employ the use of a common coumarin caging group (DEAC) in which decaging was achieved when irradiated with blue light (λ=400–430 nm). [28b] In addition to the release of the decaged SAHA inhibitor, analysis of the decaging reaction showed the formation of the corresponding suberanilic acid as a minor product. Inhibitory activity was evaluated using a commercially available luminescence‐based HDAC biochemical assay. The caged inhibitor 21 exhibited no inhibitory activity. However, when irradiated with blue light, HDAC activity was inhibited by 70%. The utility of 21 was further demonstrated in a cell growth inhibition assay using HCT116 cells. In the absence of light, no antiproliferative activity was observed. Cell growth was only inhibited when treated cells were exposed to light.
The zinc binding group of SAHA has also been caged using DMNB. [29a] HDAC activity was evaluated using whole cell lysates from BMDM (bone marrow derived macrophages) as the source of HDACs. When treated with the caged inhibitor 22, no HDAC activity was observed. Irradiation with UV light (λ=365 nm) and the rapid release of the inhibitor resulted in the inhibition of catalytic activity by 40%. The authors also demonstrated that the photoactivation of caged inhibitor 22 in BMDM affect the amount of pro‐inflammatory mediators produced by macrophages with high spatiotemporal precision.
The development of a caged variant of the HDAC inhibitor, metacept‐3, was reported in 2020 (Figure 5). [29b] Adopting an analogous approach to the caged SAHA inhibitors, the 2‐nitrobenzyl derived photolabile protecting group (DANMB) was introduced onto the hydroxamic acid moiety, rendering the inhibitor inactive (compound 23). Authors demonstrated the photocontrol of histone H3 activity in HL‐60 cells. Following the incubation of 23, exposure to UV light (λ=365 nm) and subsequent release of the active form of the HDAC inhibitor afforded an increase in acetylated histone H3 activity with levels comparable to the parent compound. Furthermore, toxicity studies were performed in HUVEC cell lines, in which a pronounced reduction in cell death was observed when treated with the caged inhibitor 23 compared to the active form (metacept‐3).
In 2021, Troelsen et al. [29c] reported the development of a photocaged derivative of known HDAC inhibitor, panobinostat (Figure 5, compound 24). In a cell free biochemical assay, caged inhibitor 24 did not exhibit any inhibitory activity towards HDACs 1–9 and 11 (Table 1). The oesophageal cancer cell line, OE21, was treated with 24, in which no antiproliferative activity was observed. Only the photolysis of 24 in treated cells resulted in apoptotic cell death.
The known HDAC inhibitor, valproic acid (VPA), was rendered inactive with the inclusion of a BODIPY caging group onto the carboxylic acid moiety via an oxime linker (Figure 5, compound 25). [29d] Exposure to green light (λ=503 nm) triggered the cleavage of the N−O bond to afford the active HDAC inhibitor. While no inhibitory data against individual HDAC enzymes is reported, authors demonstrated that the anticancer properties of 25 was light‐dependent, in which cell death was only observed when HeLa cells, incubated with 25, were irradiated with 503 nm light.
3.1.2. Photoswitchable HDAC Inhibitors
A common approach to the development of photochromic HDAC inhibitors has been to merge the azobenzene photoswitch into the molecular scaffold of FDA approved HDAC inhibitors, belinostat, panobinostat and tacedinaline (Figure 6). The first example of photoswitchable HDAC inhibitors were described by Szymanski et al. [32] in 2015. Cell free studies in an HDAC biochemical assay showed that photoswitchable belinostat derivative 26 exhibited an impressive 40‐fold difference in activity between the two isomers against HDAC2. In the absence of light, 26‐E had an IC50 value of 21.65 μM. Exposure to 365 nm triggered E→Z isomerisation to give 92% of 26‐Z (Table S1), which inhibited HDAC2 with an IC50 value of 0.555 μM. Photoresponsive cytotoxicity properties were also achieved in HeLa cells, in which cell death was only observed in the presence of the Z‐form of the photoswitch.
Figure 6.
Photoswitchable HDAC inhibitors utilising the photochromic azobenzene, diarylmaleimide and fulgimide moieties.
In 2021, the same authors reported the development of the photoswitchable HDAC inhibitor 27, in which the azobenzene motif was positioned meta to the zinc‐binding group (Figure 6). [33] Docking studies suggested that the Z‐isomer 27‐Z would adopt a similar binding pose to that of the parent molecule (belinostat), maintaining the key interactions with the active site and therefore serve as the active form. Photoisomerisation of the E‐ to Z‐isomer was achieved by irradiation with 365 nm light (PSS 95%, Table S1). The reverse reaction could be achieved either thermally or upon exposure to 420 nm. Inhibitory studies in a HDAC2 biochemical assay showed a 2‐fold difference in activity between the two photo‐isomers, with IC50 values in the sub‐micromolar range of 0.47 μM and 0.22 μM for the E‐isomer (27‐E ) and photo‐enriched Z‐isomer (27‐Z ), respectively.
In 2016, Reis et al. [34] reported the development of the photoswitchable class I selective HDAC inhibitor 28 (Figure 6). Photoinduced E→Z isomerisation was induced by exposure to 470 nm light (Table S1), in which inhibition of HDACs 1–3 was observed. In contrast to other azobenzene‐derived HDAC inhibitors, the Z‐form of 28 undergoes rapid thermal relaxation back to the E‐form (t 1/2=59.5 μs). Thereby, the active form of the inhibitor only exists when exposed to light.
In efforts to probe the role of bacterial HDAC homologues in infectious disease, Fuchter and co‐workers reported the development of photoswitchable HDAC inhibitor 29 (Figure 6). [35] This was achieved by merging the photochromic arylazopyrazole group in the pharmacophore of belinostat and panobinostat. Irradiation of the thermally stable E‐form (29‐E ) with 365 nm light afforded the Z‐form (29‐Z ) in high PSS (97%, Table S1). Isomerisation back to the E‐form was achieved upon irradiation with 532 nm light (PSS 93%). It was envisaged that the bent/twisted nature of the Z‐isomer would not be accommodated in the narrow active site of the enzyme, thereby serving as the inactive form of the inhibitor. However, in the absence of light, E‐form (29‐E ) exhibited an IC50 value of 0.77 μM for the bacterial HDAC enzyme PA1409, while a modest 6‐fold increase in inhibitory activity was observed for the photo‐enriched Z‐form (29‐Z ), with an IC50 value of 0.13 μM. This trend is consistent with that observed for the azobenzene derived HDAC inhibitors reported by Szymanski and Mazitschek.[ 32 , 34 ]
In efforts to overcome the main drawbacks of using azobenzene derivatives, thermal reversibility and incomplete photoconversion due to spectral overlap of both photoisomers, König and co‐workers have functionalised DTE and fulgimide photoswitches with aromatic hydroxamic acids that should bind to the zinc ion in the active site of HDAC enzymes (Figure 6, compounds 30–33). [36] Unfortunately, reversible photoswitching of 30 could only be realised in DMSO, as the photoinduced ring closing reaction did not proceed in aqueous solution. Due to the high dependency of the photoinduced ring closing reaction of diarylmaleimide derivatives on the polarity of its environment, photoisomerisation of 31‐o to 31‐c in either DMSO or aqueous solution was not observed. [37] In contrast to the DTE derivatives, the open form of the fulgimide photoswitches 32 and 33 could be isomerised to the corresponding closed form in aqueous solution upon exposure to 365 nm light, while irradiation with 530 nm afforded the open form (Table S2). While low micromolar affinity was observed against several HDACs, little‐to‐no difference in HDAC activity between the open and closed forms of the fulgimides 32 and 33 was observed.
3.2. Photoswitchable Sirtuin Inhibitors
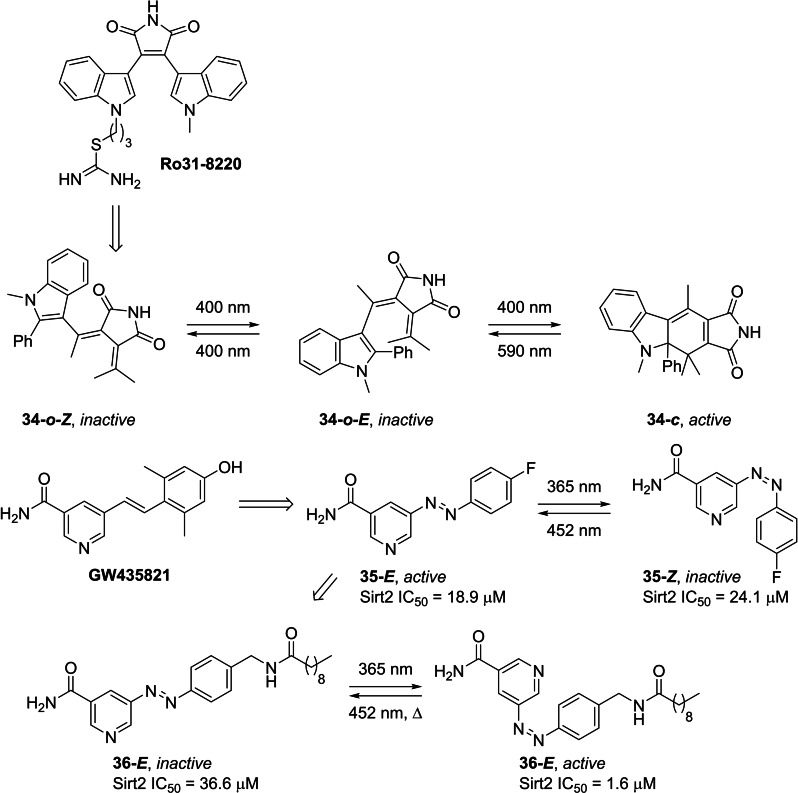
In efforts to further our understanding of the role that sirtuins play in health and disease, a small number of photoswitchable sirtuin inhibitors have been reported. [38] Simeth et al. [38a] replaced the diarylmaleimide moiety of known sirtuin inhibitor, Ro31–8220, with an indolyl fulgimide photoswitch, affording the light‐responsive sirtuin inhibitor 34 (Figure 7). The open form of the fulgimide photoswitch adopts two configurations, the 34‐o ‐ Z and 34‐o ‐ E isomers, in which irradiation with 400 nm results in the photoisomerisation between these two forms. Under these conditions, photoisomerisation to the corresponding closed‐form 34‐c only occurs from the E‐isomer (34‐o ‐ E ), affording 32% of 34‐c at the PSS (Table S2). Exposure to 590 nm initiates the ring‐opening reaction. Despite only realising a modest amount of the closed form at the PSS, the closed form exhibited superior inhibitory properties towards Sirt2 (IC50=29.6 μM). Whilst an IC50 value for the open form at the thermodynamic equilibrium could not be obtained, the maximum enzymatic inhibition observed for 34‐o at 500 μM was <50%. The difference in inhibitory activity was attributed to the orientation of the phenyl moiety. Upon simulating the structures of both isomers (34‐o and 34‐c ), it was observed that the phenyl moiety as the 2‐position of the indole in the open form is parallel to the succinimide motif, thus preventing the succinimide from binding to the active site.
Figure 7.
Photoswitchable sirtuin inhibitors derived from known sirtuin inhibitors Ro31–8220 and GW435821.
In 2019, Link and co‐workers replaced the stilbene moiety of known, moderately active, Sirt2 inhibitor GW435821, [39] with an azobenzene photoswitch (Figure 7). [38b] The authors anticipated that the bent nature of the Z‐isomer (35‐Z ) would not be accommodated for in the active site, thereby serving as the inactive form, while the E‐isomer (35‐E ) would mimic the inhibitory properties of the parent compound. Irradiation with 365 nm afforded E→Z photoisomerisation with a PSS consisting of 84% of the Z‐form (Table S1). 35‐Z exhibited good thermal stability, with a half‐life of 300 hours. Exposure to visible light (λ=452 nm) initiated the reverse photoisomerisation. Evaluation of the inhibitory properties of the two photoisomers against Sirt2 only showed a modest 1.3‐fold difference in enzymatic activity, with IC50 values of 18.9 μM and 24.1 μM for the E‐form (35‐E ) and photo‐enriched Z‐form (35‐Z ), respectively.
In efforts to increase the difference in inhibitory activity between the two isomers, a follow‐up study by the same group considered the inclusion of sterically demanding groups in place of the p‐fluoro substituent on the azobenzene (Figure 7). [38c] Molecular docking studies showed that the Z‐form (36‐Z ) binds more favourably to the active site than the E‐form (36‐E ). Indeed, this was also observed in a fluorescence‐based ZMAL activity assay. In the absence of light, 36‐E inhibited Sirt2 with an IC50 value of 36.6 μM. Exposure to UV light (λ=365 nm) initiated E→Z photoisomerisation (Table S1), in which a 23‐fold increase in inhibitory activity was observed (36‐Z IC50=1.6 μM).
4. Light‐Responsive Dihydrofolate Reductase (DHFR) Inhibitors
Dihydrofolate reductase (DHFR) catalyses the reduction of dihydrofolate to tetrahydrofolate, an essential cofactor for the biosynthesis of proteins and nucleic acids. [40] The inhibition of DHFR results in the depletion of tetrahydrofolate, in turn inducing cell death. As such, DHFR has gained attention as a therapeutic target for antibacterial [41] and anticancer [42] agents. The optical control over DHFR inhibition has posed as an effective means to reduce the occurrence of drug resistance. Recent examples of photoswitchable inhibitors of bacterial and human DHFR are discussed below (Figure 8). [43]
Figure 8.
Photoswitchable inhibitors of DHFR derived from known DHFR inhibitors trimethoprim and methotrexate.
In efforts to achieve photo‐control of antibacterial activity, Wegener et al. [43a] developed photochromic inhibitors of bacterial DHFR. Utilising the well‐established antifolate, trimethoprim, as a valuable starting point, a series of photoswitchable inhibitors were realised by appending an azobenzene photoswitch to the methoxyphenyl moiety of trimethoprim. In the absence of light, 37‐E inhibited E. coli CS1562 bacterial growth with a MIC50 value of 20 μM. Upon exposure to UV light (λ=365 nm) diaminopyridine 37‐E was photoisomerised to the corresponding active form (Table S1), 37‐Z , in which a 2‐fold increase in antibacterial activity was observed (MIC50=10 μM). The inclusion of fluoro groups in all positions ortho to the azo moiety (compound 38), resulted in visible light mediated photoisomerisation. More specifically, E→Z isomerisation was possible when irradiated with 527 nm, while the reverse reaction occurred upon exposure to 400 nm (Table S1). Compared to 37, a more pronounced difference in antibacterial was observed, in which a MIC50 value of 5 μM was obtained following irradiation with green light. Authors also demonstrated that substitution of fluoro atoms for chloro groups (Figure 8, compound 39) resulted in a significant red‐shift in the wavelength used for reversible photoisomerisation. [43a] Irradiation with 652 nm resulted in E→Z photoisomerisation, affording a PSS of 87% of 39‐Z (Table S1). An 8‐fold difference in antibacterial activity was observed against E. coli, in which MIC50 values of >80 μM and 10 μM were obtained for 39‐E and the photo‐enriched 39‐Z form, respectively. In 2021, the same group studied the mechanism of resistance development in E. coli against the photoswitchable DHFR inhibitor 39. [44] While in vitro studies showed that 39 exhibits DHFR inhibitory activity analogous to that of the parent compound, transcriptome analysis indicated that the acquired resistance for the photochromic and classical DHFR inhibitors occurred via different mechanisms. In a follow‐up study, Kobauri et al. [43b] reported the IC50 values for the photoswitchable DHFR inhibitors 37–39 (Figure 8). Despite previously reporting a significant difference in antibacterial activity between the two photoisomers, little‐to‐no difference in inhibitory activity was observed. In efforts to increase the bioactivity difference between the E‐ and Z‐isomers, the authors undertook a detailed SAR study, whereby 40 was identified as the lead compound. In the absence of light, 40‐E inhibited DHFR with an IC50 value of 52 nM. Irradiation with 365 nm light initiated E→Z isomerisation (Table S1), affording a 2‐fold increase in inhibitory activity, with an IC50 value of 27 nM for 40‐Z at the PSS. A 2‐fold difference in antibacterial activity was also observed, in which MIC50 values of 40 μM and 20 μM were obtained before and after irradiation of 40 with UV light, respectively.
Both Gorostiza [43c] and Mizukami [43d] independently reported the development of a photochromic derivative of the known DHFR inhibitor, methotrexate. This was achieved by replacing the benzylamine motif of methotrexate with an azobenzene group. Molecular docking studies suggested that the Z‐form of 41 mimicked the conformation adopted by the parent compound when bound to DHFR, thereby serving as the active form of the inhibitor.[ 43b , 43c ] In cell free assays, Gorostiza and co‐workers demonstrated that 21% of human DHFR activity was inhibited when treated with 41‐E . [43c] However, irradiation with 375 nm induced E→Z isomerisation (Table S1), in which 70% of DHFR catalytic activity was inhibited. The authors also demonstrated the utility of 41 in zebrafish embryos, whereby DHFR inhibition and its effect on folate metabolism could be controlled with light.
Mizukami and co‐workers employed 41 as a means to obtain dynamic control over E. coli DHFR activity. [43d] Light induced E→Z isomerisation was achieved upon irradiation with 394 nm, while the reverse Z→E isomerisation was induced with green light (λ=560 nm, Table S1). A competitive binding assay of 41 and E. coli DHFR showed a 10‐fold difference in inhibitory properties between the two isomers, with IC50 values of 45 nM and 3.4 nM for 41‐E and photo‐enriched 41‐Z , respectively.
More recently, Kunsági‐Máté and co‐workers have utilised 41 as a molecular tool to gain further insight into the antioxidant properties of the parent compound, methotrexate. [45] The reactive oxygen species (ROS) of both photoisomers of 41 and methotrexate were evaluated using ESR spectroscopy, in which the rate of ROS production was significantly slower in the presence of 41‐E , compared to that of the photo‐enrich 41‐Z or methotrexate.
5. Light‐Responsive Protease Inhibitors
Due to the broad function that proteases exhibit in cellular processes, such as protein degradation, cell signalling and apoptosis, inhibition of dysregulated proteases is highly sort after as a means to treat a number of diseases. [46] The light‐dependent inhibition of proteases has been motivated by the need to further elucidate their role in fundamental cellular processes and disease.
In efforts to achieve rapid and selective inhibition of the cysteine protease, cathepsin B (CTSB), Toupin et al. [47] developed a photocaged variant of the known CTSB inhibitor, 43 (Figure 9). [48] A BODIPY‐derived caging group was introduced onto the carboxylic acid moiety of 43, preventing the key interactions between the inhibitor and the CTSB active site and in turn, rendering compound 42 a poor CTSB inhibitor (K i=320 μM). Exposure to blue light (λ=460–470 nm) initiated the decaging reaction and the release of the active inhibitor 43, which exhibited a K i value of 0.51 μM. The photo‐controlled inhibition of CTSB was further demonstrated in the MDA‐MB‐231 breast cancer cell line, in which cell toxicities were observed following treatment with 42 and exposure to light. Interestingly, the generation of singlet oxygen was also observed during the light induced decaging reaction, working in synergy with the inhibitory properties of the active inhibitor 43 to induce necrosis.
Figure 9.
Photocaged inhibitors of cathepsin B and caspase‐3, as well as a DTE‐derived photoswitchable serine protease inhibitor.
Spatiotemporal control of apoptosis has been achieved through the development of caged caspase inhibitor 44 (Figure 9). [49] A nitroindoline caging group was introduced onto the carboxylate motif of the aspartate unit of the peptidyl inhibitor 45 via an amide linker, preventing binding to the primary recognition pocket of caspase. In a cell free biochemical assay, caged 44 inhibited caspase‐3 with an IC50 value of 89 nM exhibiting significantly lower inhibitory activity when compared to the uncaged inhibitor, 45 (IC50=4.2 nM). Additional studies in Jurkat cells were performed, whereby, in the absence of light, inhibition of anti‐FAS‐induced apoptosis was not observed when treated with 44. Only after exposure to UV light and release of the active form of the caspase inhibitor was 90% inhibition of apoptosis observed.
The photochromic DTE moiety has also been utilised as a means to achieve photoswitchable inhibition of serine proteases (Figure 9). [50] Known bicyclic peptidic inhibitor, SFTI‐1, of the serine protease Bos taurus typsin 1 (T1) consists of two loops stabilised by a disulfide bridge, whereby the reactive loop binds the serine protease. The DTE‐derived peptidic inhibitor 46 was realised by merging the photoswitching unit into the structural loop of SFTI‐1, which does not interact with the active site of the enzyme. In doing so, the authors anticipated that the open form would serve as the active form, whilst the closed form would render the inhibitor inactive as photoisomerisation to the closed form would induce conformational changes to the bicyclic peptidic inhibitor that would be unfavourable for binding. The open form of 46 exhibited a K i value of 5.2 nM. Exposure to UV light (λ=365 nm) resulted in the photoinduced ring closing reaction to give 46‐C with a K i value of 125 nM.
Proteasome is a multi‐component enzyme responsible for the degradation of proteins that regulate the cell cycle. As proteasome inhibition induces apoptosis, the development of proteasome inhibitors as anticancer agents has attracted considerable attention. [51] A small number of light‐responsive proteasome inhibitors have recently dotted the literature in efforts to gain optical control over apoptosis in cancer cells. [52]
In the pursuit to obtain remote control over cell cycle arrest and apoptosis, Zanin and co‐workers developed a photocaged variant of the well‐known proteasome inhibitor, MG132. [52a] More specifically, the reactive aldehyde of MG132 that covalently binds to the proteasome was rendered inactive by converting the aldehyde to a mixed acetal of a DMNB caging group (Figure 10). Indeed, 47 served as a poor proteasome inhibitor. HeLa cells treated with 47 and maintained in the dark did not exhibit any signs of perturbed cell viability or proliferation. Only when treated cells were exposed to 405 nm and the active inhibitor released, was apoptosis induced.
Figure 10.
Photocaged and photoswitchable inhibitors of proteasome as light‐responsive anticancer agents.
Blanco et al. [52b] have reported the development of the photoswitchable variant of bortezomib, a known anticancer agent that inhibits proteasome (Figure 10). Irradiation with UV light (λ=365 nm) gave the photo‐enriched Z‐isomer (48‐Z ). Inhibitory activity against the β5 subunit of proteasome was evaluated, in which a 5‐fold difference in activity was observed (48‐E IC50=11 nM, 48‐Z IC50=54 nM). Cytotoxicity studies were performed in the HCT‐116 cancer cell line. In contrast to that observed in the cell free assays, the photo‐enriched Z‐form was more active that the E‐form, inducing cell death with an LD50 value of 212 nM, compared to 1.03 μM observed for the E‐form.
6. Other Enzymatic Targets
A handful of examples have demonstrated light controlled inhibition against other enzymatic targets of interest. We highlight these reports as they demonstrate the versatility and universality of photopharmacology (Figure 11).
Figure 11.
Other examples of light‐responsive enzyme inhibitors highlighted in Section 6.
The inhibitory activity of the known cyclo‐oxygenase‐2 (COX‐2) inhibitor, Celecoxib, has been masked through the inclusion of a two‐photon caging group (compound 49). [53] Two‐photon excitation at 800 nm initiated the cleavage of the caging group and the release of the active inhibitor in an in vitro assay.
The photo‐regulation of an allosteric bienzyme complex, ImGP−S, has been achieved through the use of the DTE‐derived competitive inhibitor 50. [54] Catalytic activity and allostery was readily modulated upon switching between the open and closed form.
The photoswitchable allosteric covalent inhibitor 51 of small GTPases has been realised by merging an azobenzene motif with a covalent inhibitor. [55] The photocontrol of GTP affinity to K‐Ras was demonstrated in a nucleotide exchange assay.
The GTPase, dynamin, plays a critical role in most endocytic pathways. [56] The development of the photoswitchable inhibitor 52 has afforded optical control over dynamin activity, in which dynamin‐dependent endocytosis was regulated upon ON/OFF exposure to UV light. [57]
In efforts to achieve optical control of acid sphingomyelinase (ASM) inhibition, photolabile caging groups were introduced onto the phosphate groups of a known ASM inhibitor, rendering the inhibitor inactive (compound 53). [58] The inclusion of biologically labile butyryl groups on the hydroxy moieties were also needed to enhance cellular uptake. Light‐responsive inhibition of ASM in live cells was observed following irradiation with UV light.
Recently Adibekian and co‐workers reported photonic control over the activity of carboxylesterases CES1 and CES2, a class of a serine hydrolase that is involved in ester metabolisation. [59] Inspired from serine hydrolase inhibitors based on triazole urea scaffolds, the analogous arylazopyrazole urea and sulfone derived inhibitors (compound 54) were found to undergo efficient E to Z switching while being shown to modulate the metabolism of the immunosuppressant mycophenolate mofetil.
A report of azobenzene‐based inhibitors for tryptophan synthase, a target enzyme of interest in the development of new antibiotics, has shown reversible switching of 55 can be used to modulate enzyme activity. [60] It is hoped that such photocontrollable pharmacophores will assist in the development of smart antibiotic agents and combating the pressing issue of antibiotic resistance.
An azobenzene‐based bistacrine acetylcholinesterase inhibitor (compound 56) has been reported by Decker and co‐workers. [61] The highly active (single‐digit nanomolar) azobenzene inhibitors showed 8‐fold higher inhibition in the Z enriched form. Such inhibitors are of interest for elucidating mechanisms involved in neurodegenerative diseases.
A fascinating example of an azobenzene‐based probe has been reported for targeting the mammalian cryptochrome protein CRY1, which is involved in the regulation of circadian clock. [62] The authors report impressive isoform selectivity, along with many desirable properties of the tetra‐o‐fluoroazobenzene switch 57. These probes allow great pharmacological control and opens possibilities for a deeper understanding of CRY1 and its role in many diseased states.
7. Summary and Outlook
Light is an ideal non‐invasive external stimulus that can be applied with high spaciotemporal resolution. The field of photopharmacology continues to evolve at a remarkable pace. Over the past 5 years, a large number of light‐responsive tools have been developed to combat issues with regards to off‐target binding and the development of drug resistance, whilst also serving as a valuable means to probe the role of various enzymes in health and disease, which today are poorly understood.
The strategic inclusion of photolabile caging groups onto small molecule inhibitors to mask their innate inhibitory properties, allows for the light‐induced activation at pre‐defined time points. Unlike photoswitching, decaging of photolabile protecting groups is a point of no return where activity can only be switched ON. Photocaging groups are generally capable of achieving much greater differences between their ‘active’ and ‘inactive’ states. The synthetic simplicity of installation of photocaging groups is another appealing quality as relatively minor synthetic work can be performed on known inhibitors. These advantages make photocaged inhibitors an excellent approach for photoactivatable molecular tools.
Embedding photoswitchable entities into the core of small molecule inhibitors allows for reversible OFF/ON optical control over enzymatic inhibition. In this context, azobenzenes have historically been a popular choice, while other photoswitches, namely fulgimides and DTE‐derivatives have gained popularity in the more recent literature. The power of dynamic control over photoswitchable compounds comes with additional challenges, primarily those associated with the inability to quantitatively convert between the two states, achieving pronounced differences in activity of the two isomers and unwanted thermally induced isomerisation.
In order to achieve distinct OFF/ON switching of inhibitory activity, the difference in the affinity for the target between the active and inactive form must be significant (>100‐fold). Whilst this is largely dependent on the judicious molecular design, ensuring significant structural changes when switching between the two isomers. Similarly, the ratio of the two photo‐isomers at the photostationary state is also a contributing factor. In reality, a mixture of the active and inactive forms is present at the photostationary state as spectral overlap of both isomers precludes quantitative photoisomerisation.
In addition to optimising the photophysical properties of the photoswitch, the thermal and chemical stability of the photoswitch in biologically relevant media must also be considered. Ideally, both isomeric forms are stable and do not interconvert, until irradiation initiates the desired isomerisation. It is also known that thiol‐mediated reduction of azobenzenes to the corresponding hydrazine can occur in biological assays.[ 12 , 63 ] This is particularly evident in the development of azobenzene‐derived kinase inhibitors, as dithiothreitol (DTT) and glutathione (GSH) are typically employed for protein stabilisation.
Traditionally, UV light has been required as the input stimulus in the majority of the examples in photopharmacology. Biological samples are prone to damage from UV radiation and the poor tissue penetration of UV light prevents the full potential of these molecular tools from being reached. [64] The emergence of new or modified photoactive moieties that are responsive towards longer wavelengths of light is crucial in the progression of photopharmacology, and accordingly the research community has responded with numerous reports focussed on the development of photoresponsive systems that can be manipulated with visible to near‐infrared light, or adopt other methods in order to avoid UV light such as two‐photon adsorption. [65]
To date, many examples have demonstrated the feasibility of photoresponsive molecules for modulating biological processes with high spatiotemporal resolution. As this fascinating field of research matures there will no doubt be further advancements and applications in multicellular systems. This unparalleled level of control over when and where small molecular bioactives exhibit their therapeutic properties affords unique opportunities to perform detailed studies on the mode of action of drugs and further enrich our understanding of such complex diseased states.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Dóra Laczi completed her undergraduate studies in medicinal chemistry at Loughborough University in 2020. In 2021, she obtained her MRes from Imperial College London in the group of Professor Matthew J Fuchter. Currently, she is pursuing her PhD in organic synthesis at Auckland University of Technology under the supervision of Dr Cassandra Fleming, looking at the development of light‐responsive drug delivery systems. Her research interests include photopharmacology and the development of photocages as drug delivery systems.

Biographical Information
Mark Johnstone obtained his PhD from Deakin University, Australia. In 2015, he joined the group of Professor Guido Clever at University of Göttingen and in 2016, commenced an Alexander von Humboldt postdoctoral fellowship with the same group at the Technical University of Dortmund. In 2018, he moved to Chalmers Univesity of Technology as a postdoctoral researcher in the group of Associate Professor Henrik Sundén, before joining Auckland University of Technology as a Lecturer in Chemistry. His main research interests are related to the design and application of supramolecular systems, primarily coordination cages and stimuli‐responsive supramolecular gels.

Biographical Information
Cassandra Fleming obtained her PhD in organic chemisty in 2015 from Deakin University, Australia. Following her doctorate, she joined the group of Professor Joakim Andréasson at Chalmers University of Technology. She spent a further two years in Sweden as a Marie Curie postdoctoral fellow in the group of Professor Morten Grøtli at the University of Gothenburg. In 2020, Cassandra moved to Auckland University of Technology as a Lecturer in Chemistry. Her research interests include the development of light‐responsive molecular tools for the study of disease progression in a cellular setting.

Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
DL and CLF acknowledges funding from the Royal Society of New Zealand Marsden Fast‐Start grant. CLF also acknowledges funding from the Health Research Council of New Zealand. Open access publishing facilitated by Auckland University of Technology, as part of the Wiley ‐ Auckland University of Technology agreement via the Council of Australian University Librarians.
D. Laczi, M. D. Johnstone, C. L. Fleming, Chem. Asian J. 2022, 17, e202200200.
References
- 1.
- 1a. Muller P. Y., Milton M. N., Nat. Rev. Drug Discovery 2012, 11, 751–761; [DOI] [PubMed] [Google Scholar]
- 1b. Edwards I. R., Aronson J. K., Lancet 2000, 356, 1255–1259. [DOI] [PubMed] [Google Scholar]
- 2.
- 2a. Welleman I. M., Hoorens M. W. H., Feringa B. L., Boersma H. H., Szymański W., Chem. Sci. 2020, 11, 11672–11691; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b. Hüll K., Morstein J., Trauner D., Chem. Rev. 2018, 118, 10710–10747; [DOI] [PubMed] [Google Scholar]
- 2c. Lerch M. M., Hansen M. J., van Dam G. M., Szymanski W., Feringa B. L., Angew. Chem. Int. Ed. 2016, 55, 10978–10999; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 11140–11163; [Google Scholar]
- 2d. Velema W. A., Szymanski W., Feringa B. L., J. Am. Chem. Soc. 2014, 136, 2178–2191. [DOI] [PubMed] [Google Scholar]
- 3. Szymański W., Beierle J. M., Kistemaker H. A. V., Velema W. A., Feringa B. L., Chem. Rev. 2013, 113, 6114–6178. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Broichhagen J., Frank J. A., Trauner D., Acc. Chem. Res. 2015, 48, 1947–1960; [DOI] [PubMed] [Google Scholar]
- 4b. Mayer G., Heckel A., Angew. Chem. Int. Ed. 2006, 45, 4900–4921; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2006, 118, 5020–5042. [Google Scholar]
- 5.
- 5a. Lubbe A. S., Szymanski W., Feringa B. L., Chem. Soc. Rev. 2017, 46, 1052–1079; [DOI] [PubMed] [Google Scholar]
- 5b. Liu Q., Deiters A., Acc. Chem. Res. 2014, 47, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ferguson F. M., Gray N. S., Nat. Rev. Drug Discovery 2018, 17, 353–377. [DOI] [PubMed] [Google Scholar]
- 7. Fleming C. L., Grøtli M., Andréasson J., ChemPhotoChem 2019, 3, 318–326. [Google Scholar]
- 8. Sõrmus T., Lavogina D., Enkvist E., Uri A., Viht K., Chem. Commun. 2019, 55, 11147–11150. [DOI] [PubMed] [Google Scholar]
- 9. Ivan T., Enkvist E., Viira B., Manoharan G. B., Raidaru G., Pflug A., Alam K. A., Zaccolo M., Engh R. A., Uri A., Bioconjugate Chem. 2016, 27, 1900–1910. [DOI] [PubMed] [Google Scholar]
- 10. Le Bescont J., Mouawad L., Boddaert T., Bombard S., Piguel S., ChemPhotoChem 2021, 5, 989–994. [Google Scholar]
- 11. Zhang K., Ji M., Lin S., Peng S., Zhang Z., Zhang M., Zhang J., Zhang Y., Wu D., Tian H., Chen X., Xu H., J. Med. Chem. 2021, 64, 7331–7340. [DOI] [PubMed] [Google Scholar]
- 12. Schehr M., Ianes C., Weisner J., Heintze L., Müller M. P., Pichlo C., Charl J., Brunstein E., Ewert J., Lehr M., Baumann U., Rauh D., Knippschild U., Peifer C., Herges R., Photochem. Photobiol. Sci. 2019, 18, 1398–1407. [DOI] [PubMed] [Google Scholar]
- 13. Halekotte J., Witt L., Ianes C., Krüger M., Bührmann M., Rauh D., Pichlo C., Brunstein E., Luxenburger A., Baumann U., Knippschild U., Bischof J., Peifer C., Molecules 2017, 22, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wenglowsky S., Ahrendt K. A., Buckmelter A. J., Feng B., Gloor S. L., Gradl S., Grina J., Hansen J. D., Laird E. R., Lunghofer P., Mathieu S., Moreno D., Newhouse B., Ren L., Risom T., Rudolph J., Seo J., Sturgis H. L., Voegtli W. C., Wen Z., Bioorg. Med. Chem. Lett. 2011, 21, 5533–5537. [DOI] [PubMed] [Google Scholar]
- 15. Hoorens M. W. H., Ourailidou M. E., Rodat T., van der Wouden P. E., Kobauri P., Kriegs M., Peifer C., Feringa B. L., Dekker F. J., Szymanski W., Eur. J. Med. Chem. 2019, 179, 133–146. [DOI] [PubMed] [Google Scholar]
- 16. Heintze L., Schmidt D., Rodat T., Witt L., Ewert J., Kriegs M., Herges R., Peifer C., Int. J. Mol. Sci. 2020, 21, 8961–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.
- 17a. Kolarski D., Sugiyama A., Rodat T., Schulte A., Peifer C., Itami K., Hirota T., Feringa B. L., Szymanski W., Org. Biomol. Chem. 2021, 19, 2312–2321; [DOI] [PubMed] [Google Scholar]
- 17b. Kolarski D., Miró-Vinyals C., Sugiyama A., Srivastava A., Ono D., Nagai Y., Iida M., Itami K., Tama F., Szymanski W., Hirota T., Feringa B. L., Nat. Commun. 2021, 12, 3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hirota T., Lee J. W., Lewis W. G., Zhang E. E., Breton G., Liu X., Garcia M., Peters E. C., Etchegaray J.-P., Traver D., PLoS Biol. 2010, 8, e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu Y., Gao C., Håversen L., Lundbäck T., Andréasson J., Grøtli M., Chem. Commun. 2021, 57, 10043–10046. [DOI] [PubMed] [Google Scholar]
- 20. Green J., Cao J., Bandarage U. K., Gao H., Court J., Marhefka C., Jacobs M., Taslimi P., Newsome D., Nakayama T., Shah S., Rodems S., J. Med. Chem. 2015, 58, 5028–5037. [DOI] [PubMed] [Google Scholar]
- 21. Matsuo K., Thayyil S., Kawaguchi M., Nakagawa H., Tamaoki N., Chem. Commun. 2021, 57, 12500–12503. [DOI] [PubMed] [Google Scholar]
- 22. Reynders M., Chaikuad A., Berger B.-T., Bauer K., Koch P., Laufer S., Knapp S., Trauner D., Angew. Chem. Int. Ed. 2021, 60, 20178–20183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muth F., El-Gokha A., Ansideri F., Eitel M., Döring E., Sievers-Engler A., Lange A., Boeckler F. M., Lämmerhofer M., Koch P., Laufer S. A., J. Med. Chem. 2017, 60, 594–607. [DOI] [PubMed] [Google Scholar]
- 24. Xu W. S., Parmigiani R. B., Marks P. A., Oncogene 2007, 26, 5541–5552. [DOI] [PubMed] [Google Scholar]
- 25. Zhao C., Dong H., Xu Q., Zhang Y., Expert Opin. Ther. Pat. 2020, 30, 263–274. [DOI] [PubMed] [Google Scholar]
- 26. Nijhawan P., Behl T., Khullar G., Pal G., Kandhwal M., Goyal A., Obesity Medicine 2020, 18, 100212. [Google Scholar]
- 27. Zwinderman M. R. H., de Weerd S., Dekker F. J., Epigenomes 2019, 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.
- 28a. Leonidova A., Mari C., Aebersold C., Gasser G., Organometallics 2016, 35, 851–854; [Google Scholar]
- 28b. Ieda N., Yamada S., Kawaguchi M., Miyata N., Nakagawa H., Bioorg. Med. Chem. 2016, 24, 2789–2793. [DOI] [PubMed] [Google Scholar]
- 29.
- 29a. Parasar B., Chang P. V., Chem. Sci. 2017, 8, 1450–1453; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29b. Sama G. R., Liu H., Mountford S., Thompson P., Robinson A., Dear A. E., Bioorg. Med. Chem. Lett. 2020, 30, 127291; [DOI] [PubMed] [Google Scholar]
- 29c. Troelsen K. S., Calder E. D. D., Skwarska A., Sneddon D., Hammond E. M., Conway S. J., ChemMedChem 2021, 16, 3691–3700; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29d. Sambath K., Zhao T., Wan Z., Zhang Y., Chem. Commun. 2019, 55, 14162–14165. [DOI] [PubMed] [Google Scholar]
- 30. Grither W. R., Korang J., Sauer J. P., Sherman M. P., Vanegas P. L., Zhang M., McCulla R. D., J. Photochem. Photobiol. A 2012, 227, 1–10. [Google Scholar]
- 31. Arts J., King P., Mariën A., Floren W., Beliën A., Janssen L., Pilatte I., Roux B., Decrane L., Gilissen R., Hickson I., Vreys V., Cox E., Bol K., Talloen W., Goris I., Andries L., Du Jardin M., Janicot M., Page M., van Emelen K., Angibaud P., Clin. Cancer Res. 2009, 15, 6841–6851. [DOI] [PubMed] [Google Scholar]
- 32. Szymanski W., Ourailidou M. E., Velema W. A., Dekker F. J., Feringa B. L., Chem. Eur. J. 2015, 21, 16517–16524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kobauri P., Szymanski W., Cao F., Thallmair S., Marrink S. J., Witte M. D., Dekker F. J., Feringa B. L., Chem. Commun. 2021, 57, 4126–4129. [DOI] [PubMed] [Google Scholar]
- 34. Reis S. A., Ghosh B., Hendricks J. A., Szantai-Kis D. M., Törk L., Ross K. N., Lamb J., Read-Button W., Zheng B., Wang H., Salthouse C., Haggarty S. J., Mazitschek R., Nat. Chem. Biol. 2016, 12, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weston C. E., Krämer A., Colin F., Yildiz Ö., Baud M. G. J., Meyer-Almes F.-J., Fuchter M. J., ACS Infect. Dis. 2017, 3, 152–161. [DOI] [PubMed] [Google Scholar]
- 36. Wutz D., Gluhacevic D., Chakrabarti A., Schmidtkunz K., Robaa D., Erdmann F., Romier C., Sippl W., Jung M., König B., Org. Biomol. Chem. 2017, 15, 4882–4896. [DOI] [PubMed] [Google Scholar]
- 37.
- 37a. Irie M., Sayo K., J. Phys. Chem. 1992, 96, 7671–7674; [Google Scholar]
- 37b. Fleming C., Remón P., Li S., Simeth N. A., König B., Grøtli M., Andréasson J., Dyes Pigm. 2017, 137, 410–420. [Google Scholar]
- 38.
- 38a. Simeth N. A., Altmann L.-M., Wössner N., Bauer E., Jung M., König B., J. Org. Chem. 2018, 83, 7919–7927; [DOI] [PubMed] [Google Scholar]
- 38b. Grathwol C. W., Wössner N., Swyter S., Smith A. C., Tapavicza E., Hofstetter R. K., Bodtke A., Jung M., Link A., Beilstein J. Org. Chem. 2019, 15, 2170–2183; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38c. Grathwol C. W., Wössner N., Behnisch-Cornwell S., Schulig L., Zhang L., Einsle O., Jung M., Link A., ChemMedChem 2020, 15, 1480–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDonald O., Lackey K., Davis-Ward R., Wood E., Samano V., Maloney P., Deanda F., Hunter R., Bioorg. Med. Chem. Lett. 2006, 16, 5378–5383. [DOI] [PubMed] [Google Scholar]
- 40. Kompis I. M., Islam K., Then R. L., Chem. Rev. 2005, 105, 593–620. [DOI] [PubMed] [Google Scholar]
- 41. Zhou W., Scocchera E. W., Wright D. L., Anderson A. C., MedChemComm 2013, 4, 908–915. [Google Scholar]
- 42. Gonen N., Assaraf Y. G., Drug Resist. Updates 2012, 15, 183–210. [DOI] [PubMed] [Google Scholar]
- 43.
- 43a. Wegener M., Hansen M. J., Driessen A. J. M., Szymanski W., Feringa B. L., J. Am. Chem. Soc. 2017, 139, 17979–17986; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43b. Kobauri P., Galenkamp N. S., Schulte A. M., de Vries J., Simeth N. A., Maglia G., Thallmair S., Kolarski D., Szymanski W., Feringa B. L., J. Med. Chem. 2022, 65, 4798–4817; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43c. Matera C., Gomila A. M. J., Camarero N., Libergoli M., Soler C., Gorostiza P., J. Am. Chem. Soc. 2018, 140, 15764–15773; [DOI] [PubMed] [Google Scholar]
- 43d. Mashita T., Kowada T., Takahashi H., Matsui T., Mizukami S., ChemBioChem 2019, 20, 1382–1386. [DOI] [PubMed] [Google Scholar]
- 44. Lauxen A. I., Kobauri P., Wegener M., Hansen M. J., Galenkamp N. S., Maglia G., Szymanski W., Feringa B. L., Kuipers O. P., Pharmaceuticals 2021, 14, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Preisz Z., Hartvig N., Bognár B., Kálai T., Kunsági-Máté S., Pharmaceuticals 2021, 14, 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.
- 46a. Drag M., Salvesen G. S., Nat. Rev. Drug Discovery 2010, 9, 690–701; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46b. Turk B., Nat. Rev. Drug Discovery 2006, 5, 785–799. [DOI] [PubMed] [Google Scholar]
- 47. Toupin N. P., Arora K., Shrestha P., Peterson J. A., Fischer L. J., Rajagurubandara E., Podgorski I., Winter A. H., Kodanko J. J., ACS Chem. Biol. 2019, 14, 2833–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murata M., Miyashita S., Yokoo C., Tamai M., Hanada K., Hatayama K., Towatari T., Nikawa T., Katunuma N., FEBS Lett. 1991, 280, 307–310. [DOI] [PubMed] [Google Scholar]
- 49. Chakrabarty S., Verhelst S. H. L., Cell Chem. Biol. 2020, 27, 1434–1440. [DOI] [PubMed] [Google Scholar]
- 50. Babii O., Afonin S., Diel C., Huhn M., Dommermuth J., Schober T., Koniev S., Hrebonkin A., Nesterov-Mueller A., Komarov I. V., Ulrich A. S., Angew. Chem. Int. Ed. 2021, 60, 21789–21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ping Dou Q., Zonder J. A., Curr. Cancer Drug Targets 2014, 14, 517–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.
- 52a. Uhl E., Wolff F., Mangal S., Dube H., Zanin E., Angew. Chem. Int. Ed. 2021, 60, 1187–1196; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 1207–1216; [Google Scholar]
- 52b. Blanco B., Palasis K. A., Adwal A., Callen D. F., Abell A. D., Bioorg. Med. Chem. 2017, 25, 5050–5054. [DOI] [PubMed] [Google Scholar]
- 53. Anstaett P., Pierroz V., Ferrari S., Gasser G., Photochem. Photobiol. Sci. 2015, 14, 1821–1825. [DOI] [PubMed] [Google Scholar]
- 54. Kneuttinger A. C., Winter M., Simeth N. A., Heyn K., Merkl R., König B., Sterner R., ChemBioChem 2018, 19, 1750–1757. [DOI] [PubMed] [Google Scholar]
- 55. Ge Z., Yang Z., Liang J., Dong D., Zhu M., ChemBioChem 2019, 20, 2916–2920. [DOI] [PubMed] [Google Scholar]
- 56.
- 56a. Doherty G. J., McMahon H. T., Annu. Rev. Biochem. 2009, 78, 857–902; [DOI] [PubMed] [Google Scholar]
- 56b. Ferguson S. M., De Camilli P., Nat. Rev. Mol. Cell Biol. 2012, 13, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Camarero N., Trapero A., Pérez-Jiménez A., Macia E., Gomila-Juaneda A., Martín-Quirós A., Nevola L., Llobet A., Llebaria A., Hernando J., Giralt E., Gorostiza P., Chem. Sci. 2020, 11, 8981–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Prause K., Naseri G., Schumacher F., Kappe C., Kleuser B., Arenz C., Chem. Commun. 2020, 56, 14885–14888. [DOI] [PubMed] [Google Scholar]
- 59. Dwyer B. G., Wang C., Abegg D., Racioppo B., Qiu N., Zhao Z., Pechalrieu D., Shuster A., Hoch D. G., Adibekian A., Angew. Chem. Int. Ed. 2021, 60, 3071–3079; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 3108–3116. [Google Scholar]
- 60. Simeth N. A., Kinateder T., Rajendran C., Nazet J., Merkl R., Sterner R., König B., Kneuttinger A. C., Chem. Eur. J. 2021, 27, 2439–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Scheiner M., Sink A., Spatz P., Endres E., Decker M., ChemPhotoChem 2021, 5, 149–159. [Google Scholar]
- 62. Kolarski D., Miller S., Oshima T., Nagai Y., Aoki Y., Kobauri P., Srivastava A., Sugiyama A., Amaike K., Sato A., Tama F., Szymanski W., Feringa B. L., Itami K., Hirota T., J. Am. Chem. Soc. 2021, 143, 2078–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boulègue C., Löweneck M., Renner C., Moroder L., ChemBioChem 2007, 8, 591–594. [DOI] [PubMed] [Google Scholar]
- 64.
- 64a. Hoorens M. W. H., Szymanski W., Trends Biochem. Sci. 2018, 43, 567–575; [DOI] [PubMed] [Google Scholar]
- 64b. Banerjee G., Gupta N., Kapoor A., Raman G., Cancer Lett. 2005, 223, 275–284; [DOI] [PubMed] [Google Scholar]
- 64c. Kamarajan P., Chao C. C. K., Biosci. Rep. 2000, 20, 99–108; [DOI] [PubMed] [Google Scholar]
- 64d. Brash D. E., Rudolph J. A., Simon J. A., Lin A., McKenna G. J., Baden H. P., Halperin A. J., Pontén J., Proc. Nat. Acad. Sci. 1991, 88, 10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.
- 65a. Weinstain R., Slanina T., Kand D., Klán P., Chem. Rev. 2020, 120, 13135–13272; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65b. Samanta S., Beharry A. A., Sadovski O., McCormick T. M., Babalhavaeji A., Tropepe V., Woolley G. A., J. Am. Chem. Soc. 2013, 135, 9777–9784; [DOI] [PubMed] [Google Scholar]
- 65c. Bléger D., Schwarz J., Brouwer A. M., Hecht S., J. Am. Chem. Soc. 2012, 134, 20597–20600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information