Abstract
The bacterial rhizosphere communities of three host plants of the pathogenic fungus Verticillium dahliae, field-grown strawberry (Fragaria ananassa Duch.), oilseed rape (Brassica napus L.), and potato (Solanum tuberosum L.), were analyzed. We aimed to determine the degree to which the rhizosphere effect is plant dependent and whether this effect would be increased by growing the same crops in two consecutive years. Rhizosphere or soil samples were taken five times over the vegetation periods. To allow a cultivation-independent analysis, total community DNA was extracted from the microbial pellet recovered from root or soil samples. 16S rDNA fragments amplified by PCR from soil or rhizosphere bacterium DNA were analyzed by denaturing gradient gel electrophoresis (DGGE). The DGGE fingerprints showed plant-dependent shifts in the relative abundance of bacterial populations in the rhizosphere which became more pronounced in the second year. DGGE patterns of oilseed rape and potato rhizosphere communities were more similar to each other than to the strawberry patterns. In both years seasonal shifts in the abundance and composition of the bacterial rhizosphere populations were observed. Independent of the plant species, the patterns of the first sampling times for both years were characterized by the absence of some of the bands which became dominant at the following sampling times. Bacillus megaterium and Arthrobacter sp. were found as predominant populations in bulk soils. Sequencing of dominant bands excised from the rhizosphere patterns revealed that 6 out of 10 bands resembled gram-positive bacteria. Nocardia populations were identified as strawberry-specific bands.
In the future, biological control of soil-borne fungal or bacterial pathogens will be of increasing importance for a more sustainable agriculture. Furthermore, fungicides such as methyl bromides will be phased out, and thus potential alternatives are needed to control the soil-borne pathogen Verticillium dahliae Kleb. This has prompted the search for reliable antagonists which show a high degree of competitiveness and are active in the rhizospheres of different crops and in different soil types (3, 4, 39). However, to fully exploit the potentials of biological control agents, a better understanding of the structural and functional diversity of microbial populations in the rhizosphere and their succession during plant development is required (49). The rhizosphere, defined as the volume of soil adjacent to and influenced by the plant root (45), is of great importance to plant health and soil fertility. Root exudates stimulate the growth of bacterial and fungal populations in the vicinity of the roots (40). Several studies have indicated that the structural and functional diversity of rhizosphere populations is affected by the plant species due to differences in root exudation and rhizodeposition in different root zones (21, 45). Furthermore, the soil type, growth stage, cropping practices (such as tillage and crop rotation), and other environmental factors (8, 14, 20, 22, 27, 52) seem to influence the composition of the microbial community in the rhizosphere. Rhizosphere microorganisms exert strong effects on plant growth and health by nutrient solubilization, N2 fixation, or the production of plant hormones (19, 36). Increased plant productivity also results from the suppression of deleterious microorganisms by antagonistic bacteria, while soil-borne pathogens can greatly reduce plant growth.
Most studies of the bacterial community structure of rhizospheres indicating a plant-dependent diversity were performed using cultivation-based techniques (12, 23, 25, 28, 30). The major problem of cultivation-based analysis is that only a small proportion of the bacterial populations can be recovered from the rhizosphere and soil by traditional cultivation techniques (1, 46). Cultivation-based limitations can be overcome by analyzing DNA that is directly extracted from rhizosphere and soil samples. Recently the analysis of 16S rDNA fragments that were PCR amplified from community DNA was used to unravel bacterial rhizosphere communities. Molecular fingerprinting techniques, such as denaturing or temperature gradient gel electrophoresis (16, 31), allow analysis of large numbers of samples, which is essential for studying spatial and temporal variations of bacterial rhizosphere populations.
To provide baseline data for biological control of the pathogenic fungus Verticillium dahliae, the bacterial communities of the rhizospheres of three typical Verticillium host plants were studied: strawberry (Fragaria ananassa Duch. [family: Rosaceae]), oilseed rape (Brassica napus L. [family: Brassicaceae]), and potato (Solanum tuberosum L. [family: Solanaceae]). We aimed to find out the degree to which the rhizosphere effect is plant dependent and whether this effect would be increased by planting the same crops in two consecutive years. Rhizosphere or soil samples were taken five times over the vegetation periods. To allow a cultivation-independent analysis, total community DNA was extracted from the microbial pellets recovered from root or soil samples. Fragments of the 16S ribosomal DNA (rDNA) were amplified by PCR with eubacterial primers from soil or rhizosphere DNA, and the PCR products obtained were analyzed by denaturing gradient gel electrophoresis (DGGE). Prominent bands were excised and used for sequence determination in order to obtain further information about the phylogeny of the dominating or plant-specific bacterial populations.
MATERIALS AND METHODS
Field design and sampling.
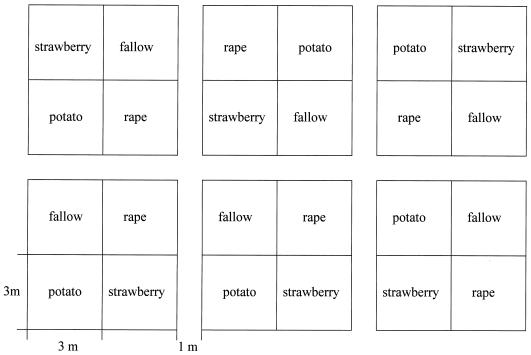
The field test was performed on fields belonging to the Federal Biological Research Centre for Agriculture and Forestry (BBA) in Braunschweig, Germany. The soil type was classified as loamy sand (pH 7) with an organic matter content of 0.9% and a clay content of 12% (data kindly provided by the Institute for Weed Research, BBA). Three different plant species—strawberry (F. ananassa Duch.) cv. Elsanta, oilseed rape (B. napus L.) cv. Licosmos, and potato (S. tuberosum L.) cv. Element—were grown with six replicates per plant type and six unplanted plots (each 3 by 3 m) arranged according to a randomized Latin square (Fig. 1). Potatoes and strawberries were planted, and oilseed rape (surface sterilization done only in the first year) was sown on the same field plots for 2 consecutive years. Samples were taken from each of the plots 1, 2, 3, 4, and 5 months after sowing (labeled 1.1 to 1.5 for the first year and 2.1 to 2.5 for the second). One composite soil sample consisting of five cores (15 cm of topsoil) was taken per fallow plot, and each was mixed by sieving. One composite rhizosphere sample taken per plot consisted of roots of 5 randomly selected strawberry or 5 potato plants or 20 oilseed rape plants, respectively. The roots were shaken vigorously to separate soil not tightly adhering to the roots. Six composite samples of each treatment were obtained per sampling time. A total of 240 composite samples were taken for two consecutive seasons.
FIG. 1.
Experimental field design. See Materials and Methods for details.
Extraction of bacterial cells from soil or roots.
Bacterial cell extraction prior to community DNA extraction was done according to the recommendations given by Bakken and Lindahl (2). Briefly, 3 g of soil or plant roots with firmly adhering soil was resuspended in 9 ml of distilled water and treated in a Stomacher 400 blender (Seward) for 1 min at high speed. After centrifugation at low speed (2 min, 500 × g), the supernatant was collected and the resulting pellet was resuspended in 9 ml of distilled water followed by Stomacher blending and low-speed centrifugation. This step was repeated once. The supernatants of the three centrifugation steps were combined before centrifugation at high speed (10,000 × g) for 30 min to collect the microbial pellet. The resulting pellet was kept at −70°C.
DNA extraction and PCR amplification of 16S rDNA fragments for DGGE analysis.
Total community DNA was extracted from the cell pellet according to the protocol by Smalla and van Elsas (44). The crude DNA was purified by using CsCl and sodium acetate precipitation steps, followed by a purification according to the manufacturer's protocol (GENECLEAN Spin Kit; BIO 101, La Jolla, Calif.).
The 16S rDNA fragments (positions 968 to 1401 [Escherichia coli rDNA sequence]) were amplified by PCR from rhizosphere or soil DNA extracts with the primer pair F984GC and R1378 (17, 18). Amplification was done using the advantage GC-genomic polymerase mixture (25 μl) as described by the manufacturers (Clontech, Palo Alto, Calif.) with 1 M GC-melt and 100 nM of each primer. The template DNA amount was approximately 1 to 5 ng per PCR. Acetamide (50%; 5 μl) was added to the reaction mixture to facilitate the denaturation of double-stranded DNA and to circumvent the formation of secondary structures. After 5 min of denaturation at 94°C and 35 thermal cycles of 1 min at 95°C, 1 min at 53°C, and 2 min at 72°C, PCR was finished by an extension step at 72°C for 10 min. Products were checked by electrophoresis in 1% (wt/vol) agarose gels and ethidium bromide staining (41).
A mixture of the DGGE-PCR products from 11 bacterial species was applied two to three times to each DGGE gel as a marker to check the electrophoresis run and to compare fragment migration between gels as described by Heuer et al. (17). These species were (in the order of the migration distance): Clostridium pasteurianum DSM 525, Erwinia carotovora DSM 30168, Agrobacterium tumefaciens DSM 30205, Pseudomonas fluorescens R2f, Pantoea agglomerans, Nocardia asteroides N3, Rhizobium leguminosarum DSM 30132, Actinomadura viridis DSM 43462, Kineosporia aurantiaca JCM 3230, Nocardiopsis atra ATCC 31511, and Actinoplanes philippiensis JCM 3001.
DGGE.
DGGE analysis was essentially done as described by Heuer et al. (18) with a denaturing gradient of 40 to 58% of the denaturant. Aliquots of PCR samples (4 to 7 μl) were applied on the denaturing gradient gel, and DGGE was performed with 0.5× Tris-acetate-EDTA buffer at 60°C at a constant voltage of 180 V for 4 h. To compare the patterns of all different treatments on one denaturing gradient gel, only PCR products amplified from four replicates per treatment (each representing one composite sample) were loaded on the gel. After silver staining of the gels according to Heuer et al. (18), the gels were air dried and scanned transmissively (pdi 420oe scanner; MWG biotech, Ebersberg, Germany). The GelCompar 4.0 program (Applied Maths, Ghent, Belgium) was used to analyze the bacterial community fingerprints of each denaturing gradient gel as described by Rademaker et al. (37) with a slight modification of some normalization settings. The track resolution was increased to 2,000 pt, curve smoothing was set to 9 pt, and background subtraction was applied using the rolling disk method with an intensity of 8. After normalizing the gel image and background subtraction, the Pearson correlation index (r) for each pair of lanes within a gel was calculated as a measure of similarity between the community fingerprints, and the clustering of patterns was calculated using the unweighted-pair group method using average linkages (UPGMA). According to Rademaker et al. (37), the Pearson product moment correlation coefficient is better suited for identification of fingerprints than band-matching algorithms. The Pearson correlation coefficient is directly applied to the array of densitometric values forming the fingerprint. The coefficient is robust and objective, since whole curves are compared and subjective band scoring is omitted. Moreover, large collections of complex fingerprints can be compared readily using the product moment, since the band-based alternative is more laborious and time consuming. The Pearson correlation coefficient is largely insensitive to relative concentrations of bands between fingerprints, and it is insensitive to differences in the overall intensities of profiles.
For comparison between rhizosphere and soil samples or between rhizosphere samples from different plants, respectively, the similarity of the DGGE profiles within a treatment (natural variability) and between treatments was compared. If the median r values of two treatments (e.g., soil and rhizosphere samples at a certain time) differed more than expected from natural variability (if there is no overlap of interquartile ranges), then a relevant effect was assumed. The approach chosen allowed us to directly compare only lanes (treatments) within one gel and still in one figure to present the comparison for up to 10 different gels.
Cloning and sequencing of DGGE bands.
Dominant bands were excised from DGGE gels which contained N,N′-bis-acryl(yl)cystamine instead of N,N′-methylenebisacrylamide (32). Staining was performed with SYBR Green 1 (FMC, Vallensbaek Strand, Denmark), and band excision was done as described by Muyzer et al. (32) with an incubation step of 100 μl of 2-mercaptoethanol for 70 min at 37°C to dissolve the gel. Five microliters of the resulting solution was used in a PCR to reamplify the excised 16S rDNA fragment using the PCR conditions described by Heuer et al. (18), using the same primer pair and temperature program described above, with an additional step of 2 h at 72°C after adding fresh dATP (2 mM) and 2 U of AmpliTaq DNA polymerase (Stoffel fragment; Perkin-Elmer) to support the formation of a 3′ A overhang for improved cloning efficiency (24). Cloning of the PCR products was necessary because DGGE analysis revealed weak bands in addition to the excised bands after reamplification. After confirming the enrichment of the excised band by DGGE, the PCR product was ligated into the pGEM-T vector (Promega, Madison, Wisc.) and transformed into competent cells (E. coli JM109; Promega) as described by the manufacturers. Plasmids with the correct insert, as determined by DGGE, were selected, and inserts were sequenced with the standard primers SP6 and T7 (IIT GmbH; Bielefeld, Germany). Data analysis was done with ARB software (Department of Microbiology, Technical University of Munich, Munich, Germany [http://www.arb-home.de]).
Nucleotide sequence accession numbers.
Nucleotide sequence accession numbers of the partial 16S rDNA sequences determined in this study are given in Table 1.
TABLE 1.
Results of partial sequence analysis and tentative phylogenetic affiliations of bands
| Origin | DGGE band (accession no.a) | Most closely related bacterial sequence(s) | % Identity | Accession no.b | Reference |
|---|---|---|---|---|---|
| Bulk soil | 1b (AJ305337) | Bacillus megaterium IAM1318T | 100 | D16273 | 47 |
| Bulk soil | 2b (AJ305338) | Arthrobacter sp. | 97.7 | AF197029 | 7 |
| Strawberry | 1s (AJ305332) | Frateuria aurantia DSM 40089T | 95.4 | AJ010481 | |
| Strawberry | 2s (AJ305334) | Streptomyces galbus DSM 40089T | 99.0 | X79852 | 51 |
| Strawberry | 3s (AJ305333) | Streptomyces sp. | 97.5 | D63866 | 48 |
| Strawberry | 4s (AJ305331) | Nocardia carnea DSM 43397T | 100 | Z36929 | 6 |
| Strawberry | 5s (AJ305335) | Promicromonospora citrea DSM 43110T | 91.1 | X83808 | 38 |
| Strawberry | 6s (AJ305336) | Promicromonospora citrea DSM 43110T | 91.1 | X83808 | 38 |
| Potato | 1p (AJ305340) | Bacillus megaterium IAM1318T | 100 | D16273 | 47 |
| Potato | 2p (AJ305342) | Unidentified bacterium | 96.2 | AJ229177 | 15 |
| Devosia riboflavina | 95.2 | D49423 | 34 | ||
| Potato | 3p (AJ305341) | Unidentified bacterium | 96.6 | D26248 | |
| Frateuria aurantia | 92.7 | AJO10481 | |||
| Oilseed rape | 1r (AJ305339) | Gordona sp. | 92.4 | AB010909 |
GenBank sequence accession number.
GenBank sequence accession number of most closely related bacterial sequence.
RESULTS
High-molecular-weight DNA was recovered from all rhizosphere and bulk soil samples. DGGE analysis of 16S rDNA fragments from soil or rhizosphere DNA revealed high similarity of the DGGE patterns obtained from each of the six replicates per treatment and time point, suggesting a low degree of variability caused by the sampling, DNA extraction, PCR amplification, and DGGE analysis.
Rhizosphere effect.
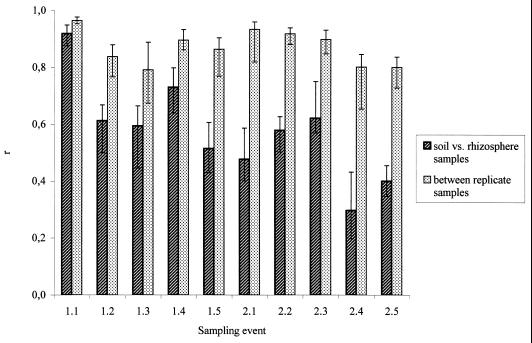
The degree to which bacterial populations are enriched in the rhizospheres of plants compared to the surrounding bulk soil, indicating shifts of relative abundance, was analyzed. For the analysis, 16S rDNA fragments amplified from DNA extracted from the rhizospheres of strawberries, oilseed rape, potatoes, or bulk soil at each sampling time were compared, running them in parallel on one denaturing gradient gel. Since sequencing of prominent bands was done for samples from sampling time 2.3, the DGGE patterns of this sampling time are shown as a representative gel picture in Fig. 2. At all sampling times, the bulk soil patterns consisted of one or two stronger bands and a large number of less intense bands, indicating that in bulk soil samples the 16S rDNA fragments of only one or two populations dominated, while many populations which were less prevalent seemed to be equally abundant. In contrast, the rhizosphere pattern consisted of several strong bands and a lower number of weak bands. Obviously the relative abundance of several bacterial populations was enhanced in the surroundings of the roots, suggesting a rhizosphere effect. To quantify this effect, the Pearson correlation r value for each pair of lanes within a gel was calculated as a measure of similarity between the community fingerprints. For statistical comparison between rhizosphere and soil samples, the similarity of the DGGE profiles within a treatment (Fig. 3) and between treatments (Fig. 3) was compared. Differences between the DGGE patterns of the rhizosphere and bulk soil samples could be detected at all sampling times. Only at sampling time 1.1 did all profiles show a relatively high level of similarity, indicating that at that time the bacterial community in the rhizosphere of the different crops and that of the bulk soil were still rather similar. The patterns from the bulk soil and the rhizosphere became different for all following sampling times. However, compared with the first year (1.1 to 1.5), the similarity between the rhizosphere DGGE patterns and those of the bulk soil clearly decreased in the second year (2.1 to 2.5).
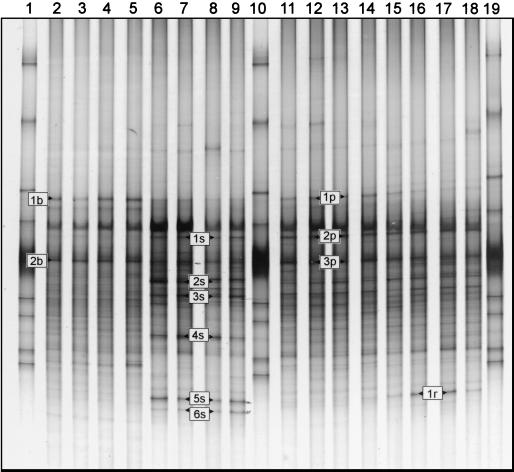
FIG. 2.
Comparison of DGGE patterns of 16S rDNA fragments amplified from bulk soil DNA (lanes 2 to 5) or from rhizosphere DNA of strawberry (lanes 6 to 9), potato (lanes 11 to 14), or oilseed rape (lanes 15 to 18) at sampling time 2.3; lanes 1, 10, and 19, standard.
FIG. 3.
Rhizosphere effect. Median similarity (Pearson correlation coefficient [r]) between soil and rhizosphere communities (left bar) compared with the similarity within a treatment (rhizosphere or soil) (right bar) at different sampling times. If the median r values of two treatments (soil and rhizosphere samples at a certain time) differed more than expected from natural variability (as estimated by quartiles), then a relevant effect was assumed. Sampling event numbers indicate the year (number to left of decimal) and months after planting (number to right of decimal).
Plant-dependent diversity.
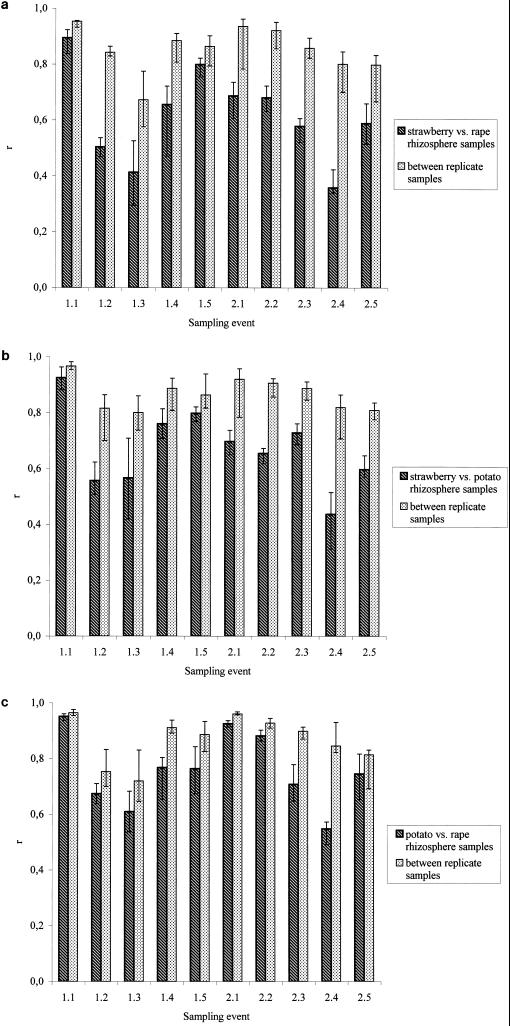
In order to compare the DGGE patterns for strawberry, oilseed rape, and potato at all sampling times in a more quantitative way, the Pearson indices were determined and the similarities of the DGGE profiles within a treatment and between the rhizospheres of two plants were compared (Fig. 4). All DGGE patterns showed a relatively high level of similarity to each other for samples taken 1 month after planting in the first year. The levels of similarity between the profiles of strawberry and oilseed rape were clearly different from each other for all following sampling times except for samples taken at 1.5 (Fig. 4a). When comparing the similarities of DGGE profiles between strawberry and potato rhizosphere, a slightly different picture was found (Fig. 4b). While DGGE patterns for both plants were different at all sampling times in the second year (2.1 to 2.5), the patterns were more similar to each other in the first year. Only at sampling times 1.2 and 1.3 did median r values of the two treatments differ from natural variability more than expected, indicating a relevant effect. Visual inspection of DGGE patterns for all sampling times already indicated that the patterns of oilseed rape and potato rhizosphere communities were more similar to each other than to the strawberry patterns. This could be confirmed (Fig. 4c), since the median r-value differences between the two treatments (potato and oilseed rape) were above variability within each treatment at one sampling time in the first year (1.4) and at two sampling times in the second year (2.3 and 2.4).
FIG. 4.
Plant-dependent diversity. Median similarity (Pearson correlation [r]) between strawberry and oilseed rape (a), strawberry and potato (b), and between potato and oilseed rape (c) rhizosphere communities (left bar) compared with the similarity within a treatment (right bar) at different sampling times. If the median r values of two treatments differed more than expected from natural variability (as estimated by quartiles), then a relevant effect was assumed. See the legend to Fig. 3 for an explanation of sampling event numbers.
Sequence analysis.
Prominent bands were excised and sequenced to get further information about the dominating bacterial populations in the different treatments from the DGGE gel of sampling time 2.3 (Fig. 2). We failed to reamplify bands from silver-stained gels, and therefore SYBR Green staining was used. Reamplification products and clones obtained were screened by DGGE analysis of the respective 16S rDNA fragments. Parallel analysis of 16S rDNA fragments amplified from clones or from the community DNA on the same gel allowed us to carefully check whether the cloned 16S rDNA fragments comigrated with the band of the corresponding community pattern. The results of the partial sequence analysis of these bands and their tentative phylogenetic affiliation are shown in Table 1. The majority of bands were excised from the strawberry DGGE patterns, since these patterns showed the strongest shifts in the relative abundance of dominant populations. The sequence of the strong bulk soil band 1b showed 100% similarity to Bacillus megaterium as well as to a dominant band in the rhizosphere pattern of potato (band 1p), which had the same migration behaviour as band 1b. This indicates that B. megaterium seems to be a dominant population not only in the bulk soil but also in the rhizosphere of potato and presumably in the rhizosphere of strawberry and oilseed rape, where bands with identical electrophoretic mobility were also detected. The sequence of the other dominant band in the bulk soil pattern (band 2b) could be assigned to Arthrobacter sp. with a 97.7% similarity, but with a 100% similarity to a clone obtained from the potato rhizosphere analyzed in another project in our laboratory. Although a band with electrophoretic mobility similar to that of band 2b of the bulk soil pattern was excised from the potato rhizosphere DGGE patterns and cloned (band 3p), the sequence obtained showed similarity to that of a γ-proteobacterium (clone sequence from paddy soil [15]). A comparison of the DGGE patterns revealed several bands which were detected in the rhizospheres of all plant species but not in the bulk soil, while a few dominant bands of the rhizosphere patterns were characteristic for one plant species only. As plant-specific bands, which dominated only in the rhizospheres of strawberry plants, sequences related to high-G+C bacteria, such as Nocardia sp. (band 4s) and Streptomyces galbus (band 2s), could be identified. Five out of six dominating bands in the rhizospheres of strawberry plants were assigned to the high-G+C actinomycetes. Band 2p, which showed a migration behavior similar to that of band 1s and dominated in the rhizosphere pattern of potato plants, was related to α-proteobacteria (Devosia riboflavina) at 95.2% similarity and at 98% similarity to clone sequences obtained from the potato rhizosphere in a different project. Two bands (5s and 1r) observed at rather high denaturing concentrations in the DGGE rhizosphere patterns of strawberry and oilseed rape, but not of potatoes, fell in the high-G+C cluster but showed a rather low similarity to bands of Promicromonospora citrea. The sequence of 5s had a 97.2% similarity to the sequence of 1r from oilseed rape. Band 6s, excised from the rhizosphere pattern of strawberry, also showed a high similarity to those sequences, but it showed different migration characteristics.
Seasonal shifts.
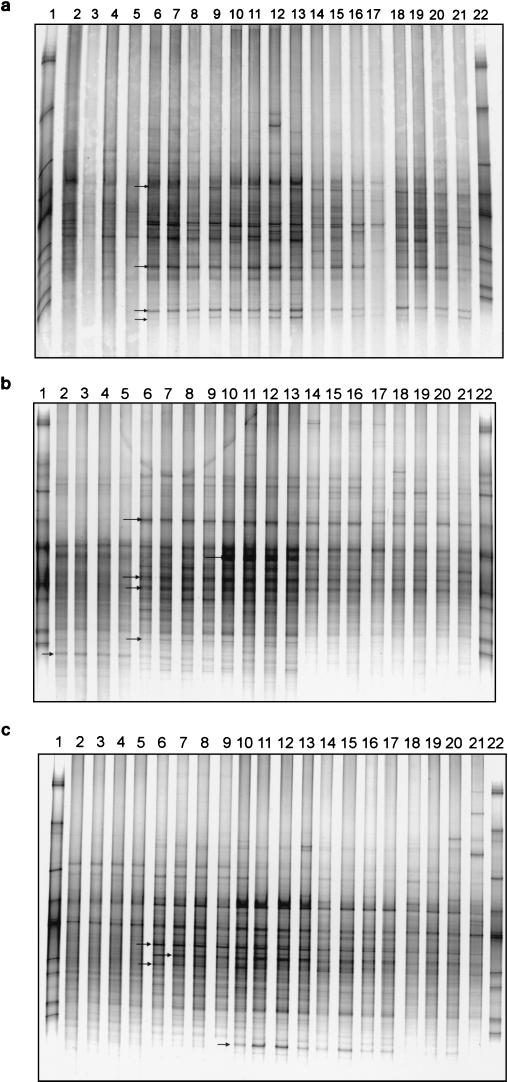
To study the seasonal shifts in the abundance and composition of the bacterial rhizosphere populations, we analyzed the rhizosphere DGGE patterns for all sampling times (1.1 to 1.5 and 2.1 to 2.5) for strawberry, potato, and oilseed rape (Fig. 5). Although many bands were detected at all sampling times, shifts in the bacterial community could be detected. In all treatments, the strongest shift occurred at the beginning of the vegetation period. Independently of the plant species, the patterns of the first sampling time in both years were characterized by the absence of some of those bands which became dominant at the following sampling times. In both years and for all three plants the DGGE patterns of the first sampling time (1.1 and 2.1) formed a separate cluster when UPGMA was used to create a dendrogram describing the similarities between the DGGE patterns. In all treatments, some bands with varying intensities could be detected over time. In the DGGE patterns obtained for strawberry rhizosphere bacteria in the second year (Fig. 5a), the relative abundance of several populations seems to be enhanced at the second sampling time (2.2) compared to the first (2.1). Some very intense bands in the DGGE patterns of 2.2 were much less intense at 2.1, while other bands, such as 4s, 5s, and 6s, were not detected at all at 2.1. In the rhizospheres of strawberry plants, bands 5s and 6s were not observed in the first year. The dominant bands were detected in most of the replicates of sampling times 2.2 to 2.5. Figure 5b shows the seasonal shifts in the rhizospheres of potatoes observed in the second year, and bands showing seasonal shifts in the relative abundance of the bacterial population are indicated. Several bands appeared only from sampling time 2.2 and remained dominant populations in the rhizosphere of potato until the end of the season. The enrichment of bacterial populations was most pronounced at sampling times 2.2 and 2.3. Several bands which according to their melting behavior might belong to high-G+C gram-positive bacteria were detected only at the first three sampling times but not at 2.4 and 2.5. Seasonal shifts in the relative abundances of respective 16S rDNA targets were observed for oilseed rape as well (Fig. 5c). As observed also for strawberry (Fig. 5a) and potato (Fig. 5b), the enrichment of bacterial populations seemed to be most pronounced for sampling times 2.2 and 2.3 when oilseed rape was flowering. Interestingly, band 1r, which shows a high similarity to a sequence obtained from strawberry (band 5s) and a lower similarity to P. citrea, became numerically dominant only at times 2.3 and 2.4.
FIG. 5.
Seasonal shifts of rhizosphere bacterial communities. DGGE analysis of 16S rDNA fragments amplified from rhizosphere DNA taken at different sampling times in the second year. Analyses of strawberry (a), potato (b), and oilseed rape (c) are shown. Lanes 1 and 22, standard; lanes 2 to 5, sampling time 2.1; lanes 6 to 9, sampling time 2.2; lanes 10 to 13, sampling time 2.3; lanes 14 to 17, sampling time 2.4; lanes 18 to 21, sampling time 2.5. Arrows indicate bands showing seasonal shifts in the relative abundance of the bacterial population.
DISCUSSION
DGGE fingerprints of PCR-amplified 16S rDNA genes were used to study dominant bacterial populations in the rhizospheres of the three V. dahliae Kleb. host plants, strawberry, potato, and oilseed rape, over two growing seasons. In contrast to other recently published papers, in which DGGE fingerprints have been used to analyze bacterial rhizosphere communities (9, 10, 35, 53), the rhizosphere samples investigated in this study originated from plants grown under field conditions in six replicated plots per treatment. We detected a rhizosphere effect, namely an increased relative abundance of some populations in the vicinity of the roots for all three plants. Several bands observed in rhizosphere DGGE patterns were not detected in patterns of soil from unplanted plots or were detected only as weak bands. The rhizosphere effect became more pronounced in the second year. In contrast, Duineveld et al. (9) observed no differences or only minor differences between bulk soil and the rhizospheres of chrysanthemum plants grown in pots in a growth room. No differences between bulk and rhizosphere patterns were found for barley grown in pots in a growth chamber (35). Due to the cell extraction technique applied in our study, the DNA obtained originated from rhizosphere and rhizoplane bacteria. Although several bands occurred in the DGGE patterns of all plants, a plant-dependent diversity of the bacterial patterns could be shown for most of the sampling times. The differences between the DGGE patterns of the three different plants became more pronounced in the second year, in particular when the similarities of strawberry rhizosphere patterns were compared with potato or oilseed rape. The finding that the patterns of oilseed rape and potato rhizosphere communities were more similar to each other than to the strawberry patterns might be explained by the fact that oilseed rape and potato are annual plants while strawberry is a perennial plant. Our data provide further evidence for the assumption that different plant species select different bacterial communities in the proximity of their roots and that these plant-specific enrichments can be increased by repeated cultivation of the plant species in the same field. Recently, Schwieger and Tebbe (42) reported differences in the single-strand conformation polymorphism fingerprints of 16S rDNA fragments amplified from rhizosphere DNA of Chenopodium album and Medicago sativa grown under field conditions. We observed seasonal shifts of a similar trend in the rhizospheres of strawberry, oilseed rape, and potato plants for 2 years. The most pronounced differences were noticed between the patterns of the first and second samplings of both years. The patterns of rhizosphere samples taken when the plants were flowering (1.2 and 1.3; 2.2 and 2.3) showed the strongest enrichment of some bacterial populations. Lottmann et al. (26) also observed the appearance of additional dominant bands in the DGGE rhizosphere patterns of potatoes at the time of flowering. The variability between replicates was highest for samples taken at the end of both growing seasons. The seasonal shifts observed in our study were less dramatic than those reported for bacterial communities in the rhizosphere of maize grown in tropical soil (13). In contrast, experiments performed in controlled pot experiments showed no changes in relation to the age of the barley plants (35), and only minor shifts in the rhizosphere of chrysanthemum (9) were found. However, both studies followed potential temporal shifts in the composition of bacterial rhizosphere communities for a much shorter period of time (up to 36 days after sowing and up to 10 weeks after planting, respectively) under growth chamber conditions, one factor which might have contributed to the contrasting results. Semenov et al. (43) found a moderate rhizosphere effect in one experiment with soil rich in fresh plant debris and a very pronounced rhizosphere effect in a second experiment with soil low in organic matter content using cultivation techniques (by enumeration of copiotrophic and oligotrophic bacteria).
Sequencing of DGGE bands revealed an astonishingly high proportion of dominant populations in the rhizosphere belonging to high-G+C gram-positive bacteria. Thus, five out of six sequences obtained from dominant bands in the rhizospheres of strawberry plants belonged to different high-G+C gram-positive bacteria, such as Nocardia and Streptomyces. One dominant band which was only detected in the rhizosphere of strawberry showed a 100% similarity to those of four different Nocardia species in the database. Interestingly, the sequences of the two dominant bands from bulk soil and four bands from the rhizosphere shared a similarity of more than 97% with those of cultured isolates. The seasonal shifts that were followed for strawberry, potato, and oilseed rape indicated that the abundance of high-G+C populations was different during the developmental stages of these plants. However, conclusions regarding their activity can hardly be drawn, since our analysis was based on 16S rDNA fragments amplified from DNA.
There is increasing evidence that gram-positive bacteria might be more dominant in the rhizosphere than previously supposed. Several recently published papers support this notion. Bacillus species were found as dominant populations in the rhizospheres of chrysanthemum (9), of barley (35), and of grass (11). McCaig et al. (29) reported that in a clone library obtained from grass rhizospheres, Actinomycetes spp. were the second-most-abundant group after the most frequently found α-proteobacteria. Arthrobacter spp. were also found as dominant populations in the molecular fingerprints of 16S rDNA fragments amplified from the rhizosphere DNA of maize grown in tropical soil (13), from the rhizosphere DNA of M. sativa and C. album (42), and from the rhizosphere DNA of chrysanthemum (9). One dominant DGGE band obtained at all locations of the barley rhizospheres grown in controlled pot experiments performed by Yang and Crowley (53) was identified as Microbacterium. However, in none of the recently published cultivation-independent studies of the bacterial diversity of the rhizosphere was such a high proportion of dominant populations detected belonging to a diverse range of high-G+C gram-positive bacteria.
In this study DGGE analysis of 16S rDNA fragments amplified from community DNA was used for the molecular analysis of a large number of rhizosphere and bulk soil samples taken over two growing seasons. The DGGE profiles mainly reflect the evenness of populations in an environmental sample, and in this study they indicated that a reduced evenness was found in the rhizosphere compared to soil. Interpretation of DGGE patterns needs to be done cautiously as discussed in several reviews (16, 33). Amplified 16S rDNA fragments of different but phylogenetically related species might have the same electrophoretic mobility because they share the identical or similar sequence in the stretch analyzed, as found for the Nocardia sequence from the strawberry-specific band in this study or for Arthrobacter clones (13) and isolates (5). But phylogenetically nonrelated species also by coincidence might have a similar melting behavior, as observed in this study for bands 1s and 2p as well as 2b and 3p. However, since only one clone per band was sequenced, we cannot exclude the possibility that both sequences 1s and 2p and sequences 2b and 3p are present in the same band. Thus, a rather large diversity of populations sharing the identical 16S rDNA sequence might be hidden behind a DGGE band. Despite several pitfalls of PCR-based rRNA analysis (50), profiling of bacterial rhizosphere and bulk communities by denaturing gradient gels proved to be a powerful method allowing a cultivation-independent analysis of large numbers of rhizosphere and bulk soil samples. Currently we are applying group-specific primers (13, 17) to amplify 16S rDNA fragments of different phylogenetic groups, which should enable a better level of resolution.
ACKNOWLEDGMENTS
This study was funded by a DFG grant to K.S. and G.B. The DGGE approach used had already been established in BMBF research project 0311295.
We are grateful to S. Kropf, Leipzig University, for discussing statistical data treatment with us.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakken L R, Lindahl V. Recovery of bacterial cells from soil. In: Trevors J T, van Elsas J D, editors. Nucleic acids in the environment. Berlin, Germany: Springer-Verlag; 1995. pp. 9–27. [Google Scholar]
- 3.Berg G, Kurze S, Buchner A, Wellington E M H, Smalla K. Successful strategy for the selection of new strawberry associated rhizobacteria antagonistic to Verticillium wilt. Can J Microbiol. 2000;46:1128–1137. doi: 10.1139/w00-101. [DOI] [PubMed] [Google Scholar]
- 4.Berg, G., A. Fritze, N. Roskot, and K. Smalla. Evaluation of potential biocontrol rhizobacteria from different host plants of Verticillium dahliae Kleb. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 5.Brim H, Heuer H, Krögerrecklenfort E, Mergeay M, Smalla K. Characterization of the bacterial community of a zinc-polluted soil. Can J Microbiol. 1999;45:326–338. doi: 10.1139/w99-012. [DOI] [PubMed] [Google Scholar]
- 6.Chun J, Goodfellow M. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol. 1995;45:240–245. doi: 10.1099/00207713-45-2-240. [DOI] [PubMed] [Google Scholar]
- 7.Crocker F H, Fredrickson J K, White D C, Ringelberg D B, Balkwill D L. Phylogenetic and physiological diversity of Arthrobacter strains isolated from unconsolidated subsurface sediments. Microbiology. 2000;146:1295–1310. doi: 10.1099/00221287-146-6-1295. [DOI] [PubMed] [Google Scholar]
- 8.De Leij F A A M, Whipps J M, Lynch J M. The use of colony development for the characterization of bacterial communities in soil and on roots. Microb Ecol. 1994;27:81–97. doi: 10.1007/BF00170116. [DOI] [PubMed] [Google Scholar]
- 9.Duineveld B M, Kowalchuk G A, Keijzer A, van Elsas J D, van Veen J A. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl Environ Microbiol. 2001;67:172–178. doi: 10.1128/AEM.67.1.172-178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duineveld B M, Rosado A S, van Elsas J D, van Veen J A. Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl Environ Microbiol. 1998;64:4950–4957. doi: 10.1128/aem.64.12.4950-4957.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felske A, Wolterink A, van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Germida J J, Siciliano S D, Renato de Freitas J, Seib A M. Diversity of root-associated bacteria associated with field-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.) FEMS Microbiol Ecol. 1998;26:43–50. [Google Scholar]
- 13.Gomes N C M, Heuer H, Schönfeld J, Costa R, Hagler-Mendonca L, Smalla K. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil. 2001;232:167–180. [Google Scholar]
- 14.Grayston S J, Wang S, Campbell C D, Edwards A C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem. 1998;30:369–378. [Google Scholar]
- 15.Hengstmann U, Chin K J, Janssen P H, Liesack W. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl Environ Microbiol. 1999;65:5050–5058. doi: 10.1128/aem.65.11.5050-5058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heuer H, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities. In: Van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker; 1997. pp. 353–373. [Google Scholar]
- 17.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heuer H, Hartung K, Wieland G, Kramer I, Smalla K. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl Environ Microbiol. 1999;65:1045–1049. doi: 10.1128/aem.65.3.1045-1049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Höflich G, Wiehe W, Kühn G. Plant growth stimulation by inoculation with symbiotic and associative rhizosphere microorganisms. Experientia. 1994;50:897–905. [Google Scholar]
- 20.Horwath W R, Elliott L F, Lynch J M. Influence of soil quality on the function of inhibitory rhizobacteria. Lett Appl Microbiol. 1998;26:87–92. [Google Scholar]
- 21.Jaeger C H, III, Lindow S E, Miller W, Clark E, Firestone M K. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol. 1999;65:2685–2690. doi: 10.1128/aem.65.6.2685-2690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latour X, Philippot L, Corberand T, Lemanceau P. The establishment of an introduced community of fluorescent pseudomonads in the soil and in the rhizosphere is affected by the soil type. FEMS Microbiol Ecol. 1996;30:163–170. doi: 10.1111/j.1574-6941.1999.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 23.Lemanceau P, Corberand T, Gardan L, Latour X, Laguerre G, Boeufgras J M, Alabouvette C. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl Environ Microbiol. 1995;61:1004–1012. doi: 10.1128/aem.61.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q-B, Guy C L. Prolonged final extension time increases cloning efficiency of PCR products. BioTechniques. 1996;21:192–196. doi: 10.2144/96212bm04. [DOI] [PubMed] [Google Scholar]
- 25.Liljeroth E, Burgers S L G E, van Veen J A. Changes in bacterial populations along roots of wheat seedings. Biol Fertil Soils. 1991;10:276–280. [Google Scholar]
- 26.Lottmann J, Heuer H, de Vries J, Mahn A, Düring K, Wackernagel W, Smalla K, Berg G. Establishment of introduced antagonistic bacteria in the rhizosphere of transgenic potatoes and their effect on the bacterial community. FEMS Microbiol Ecol. 2000;33:41–49. doi: 10.1111/j.1574-6941.2000.tb00725.x. [DOI] [PubMed] [Google Scholar]
- 27.Lupwayi N Z, Rice W A, Clayton G W. Soil microbial diversity and community structure under wheat as influenced by tillage and crop rotation. Soil Biol Biochem. 1998;30:1733–1741. [Google Scholar]
- 28.Mahaffee W F, Kloepper J W. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field-grown cucumber (Cucumis sativus L.) Microb Ecol. 1997;34:210–223. doi: 10.1007/s002489900050. [DOI] [PubMed] [Google Scholar]
- 29.McCaig A E, Glover L A, Prosser J I. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl Environ Microbiol. 1999;65:1721–1730. doi: 10.1128/aem.65.4.1721-1730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller H J, Henken G, van Veen J A. Variation and composition of bacterial populations in the rhizospheres of maize, wheat, and grass cultivars. Can J Microbiol. 1989;35:656–660. [Google Scholar]
- 31.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, 3.4.4. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 33.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa Y, Sakane T, Yokota A. Transfer of “Pseudomonas riboflavina” (Foster 1944), a gram-negative, motile rod with long-chain 3-hydroxy fatty acids, to Devosia riboflavina gen. nov., sp. nov., nom. rev. Int J Syst Bacteriol. 1996;46:16–22. doi: 10.1099/00207713-46-1-16. [DOI] [PubMed] [Google Scholar]
- 35.Normander B, Prosser J I. Bacterial origin and community composition in the barley phytosphere as a function of habitat and presowing conditions. Appl Environ Microbiol. 2000;66:4372–4377. doi: 10.1128/aem.66.10.4372-4377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patten C L, Glick B R. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol. 1996;42:207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- 37.Rademaker J L W, Louws F J, Rossbach U, Vinuesa P, de Bruijn F J. Computer-assisted pattern analysis of molecular fingerprints and database construction, 7.1.3. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. [Google Scholar]
- 38.Rainey F A, Weiss N, Stackebrandt E. Phylogenetic analysis of the genera Cellulomonas, Promicromonospora, and Jonesia and proposal to exclude the genus Jonesia from the family Cellulomonadaceae. Int J Syst Bacteriol. 1995;45:649–652. doi: 10.1099/00207713-45-4-649. [DOI] [PubMed] [Google Scholar]
- 39.Ross I L, Alami Y, Harvey P R, Achouak W, Ryder M H. Genetic diversity and biological control activity of novel species of closely related pseudomonads isolated from the wheat fields in South Australia. Appl Environ Microbiol. 2000;66:1609–1616. doi: 10.1128/aem.66.4.1609-1616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rovira A D. Interactions between plant roots and soil micro-organisms. Annu Rev Microbiol. 1965;19:241–266. doi: 10.1146/annurev.mi.19.100165.001325. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Schwieger F, Tebbe C C. Effect of field inoculation with Sinorhizobium meliloti L33 on the composition of bacterial communities in rhizospheres of a target plant (Medicago sativa) and a non-target plant (Chenopodium album)—linking of 16S rRNA gene-based single-strand conformation polymorphism community profiles to the diversity of cultivated bacteria. Appl Environ Microbiol. 2000;66:3556–3565. doi: 10.1128/aem.66.8.3556-3565.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semenov A M, van Bruggen A H C, Zelenev V V. Moving waves of bacterial populations and total organic carbon along roots of wheat. Microbiol Ecol. 1999;37:116–128. doi: 10.1007/s002489900136. [DOI] [PubMed] [Google Scholar]
- 44.Smalla K, van Elsas J D. Application of the PCR for detection of antibiotic resistance genes in environmental samples. In: Trevors J T, van Elsas J D, editors. Nucleic acids in the environment. Berlin, Germany: Springer-Verlag; 1995. pp. 241–256. [Google Scholar]
- 45.Sørensen J. The rhizosphere as a habitat for soil microorganisms. In: Van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc; 1997. pp. 21–45. [Google Scholar]
- 46.Staley J T, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki T, Yamasato K. Phylogeny of spore-forming lactic acid bacteria based on 16S rRNA gene sequences. FEMS Microbiol Lett. 1994;115:13–17. doi: 10.1111/j.1574-6968.1994.tb06607.x. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi T, Sawada H, Tanaka F, Matsuda I. Phylogenetic analysis of Streptomyces spp. causing potato scab based on 16S rRNA sequences. Int J Syst Bacteriol. 1996;46:476–479. doi: 10.1099/00207713-46-2-476. [DOI] [PubMed] [Google Scholar]
- 49.Van Veen J A, van Overbeek L S, van Elsas J D. Fate and activity of microorganisms introduced into soil. Microbiol Mol Biol Rev. 1997;61:121–135. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang G C, Wang Y. Rapid differentiation of bacterial species with multiple probes of different lengths in a single slot blot hybridization. Appl Environ Microbiol. 1995;61:4269–4273. doi: 10.1128/aem.61.12.4269-4273.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westover K M, Kennedy A C, Kelley S E. Patterns of rhizosphere microbial community structure associated with co-occurring plant species. J Ecol. 1997;85:863–873. [Google Scholar]
- 53.Yang C H, Crowley D E. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl Environ Microbiol. 2000;66:345–351. doi: 10.1128/aem.66.1.345-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]