Abstract
The diversity of French fungus-ripened cheeses is due partly to the succession of fungi that colonize the cheese during ripening. Geotrichum candidum appears in the early stages of ripening on soft cheeses such as Camembert and semihard cheeses such as St. Nectaire and Reblochon. Its lipases and proteases promote flavor development, and its aminopeptidases reduce bitterness imparted by low-molecular-weight peptides in cheese. We assessed the genetic diversity of G. candidum strains by using random amplification of polymorphic DNA (RAPD)-PCR correlated with phenotypic tests for carbon assimilation and salt tolerance. Strains were isolated from milk, curd, and cheese collected in seven major cheesemaking regions of France. Sixty-four isolates were characterized. We found high genetic diversity of G. candidum even within the same cheesemaking regions. Strains did not group according to region. All of the strains from the Haute-Savoie were able to assimilate lactate as the sole source of carbon, while lactate assimilation varied among strains from the Auvergne. Strains varied in d-mannitol assimilation, and none used citrate as the sole source of carbon. Yeast-like colony morphology predominated in Reblochon, while all of the strains isolated from St. Nectaire were filamentous. The RAPD-PCR technique readily differentiated Geotrichum fragrans isolated from milk and curd in a St. Nectaire cheesemaking facility. This study reveals an enormous diversity of G. candidum that has been empirically selected through the centuries by the cheesemakers of France.
Over the centuries, cheesemakers have optimized production techniques to select empirically for native strains of microorganisms that produced the best cheeses. Since many traditional cheesemaking methods remain closely guarded family secrets, the diversity of microbial populations that have developed in primitive cheesemaking environments is largely uncharacterized. As cheesemaking has become more industrialized, pasteurized milk and standardized bacterial and fungal inocula have been introduced to ensure consistent product quality. However, the production of raw-milk cheeses is still significant, with 700,000 tons being made annually in Europe, especially in France, Switzerland, Italy, Spain, and Greece (19). The market for artisanal cheeses is growing as consumers seek organic foods with diverse sensory characteristics.
The establishment of the European Economic Community has prompted a new awareness of each country's regional products that reflect the cultural and environmental characteristics of particular locales (3). It has also stimulated a desire to understand and protect the diversity of agricultural products that result from the biochemical activities of bacteria and fungi (6). Efforts have been made to define the biochemical and microbiological characteristics of traditional cheeses unique to specific geographical regions (7, 17, 18, 32, 37).
Geotrichum candidum is a fungus that colonizes nearly all fungal surface-ripened cheeses during the early stages of ripening (4). On some cheeses, like St. Marcellin, it is responsible for the appearance of the cheese, imparting a uniform, white, velvety coat to the surface (26). On soft cheeses, such as Camembert, and semihard cheeses, such as St. Nectaire and Reblochon, the biochemical attributes of G. candidum impact the course of cheese ripening. Lipases and proteases of G. candidum release fatty acids and peptides that can be metabolized by ensuing microbial populations and that contribute to the development of distinctive flavors and other qualities (5, 25, 27, 31). G. candidum reduces bitterness in industrial Camemberts through the activity of its aminopeptidases that hydrolyze low-molecular-weight hydrophobic peptides originating from the degradation of β-casein by Penicillium camemberti (1, 33, 34, 49). It also contributes an aroma similar to that of traditional Norman Camembert (28, 40, 48). G. candidum neutralizes the curd by catabolizing lactic acid produced by lactic acid bacteria and by releasing ammonia during the metabolism of amino acids (20). The latter activity prepares the cheese surface for colonization by acid-sensitive bacteria such as Brevibacterium sp. (12, 43). Metabolites produced by G. candidum can also inhibit Listeria monocytogenes (10, 11).
G. candidum is the anamorph of Galactomyces geotrichum. Both form septate hyphae that disarticulate into arthroconidia and do not form budding yeast cells (47). G. candidum groups phylogenetically with ascomycetous yeast-like fungi along with Dipodascus spp. (51). Within the taxon, considerable morphological variation occurs between strains. Three basic morphologies have been described: strains with yeast-like colonies that produce abundant arthrospores and have generally low proteolytic activity, strains whose colonies are white and resemble filamentous fungi with a predominance of hyphae and high proteolytic activity, and those that fall in between (26). The strain of G. candidum that predominates on a cheese rind helps to determine the texture, cohesiveness, and thickness of the rind. Some less desirable strains create unstable rinds that disintegrate when the young cheeses are turned. Others can lead to irregular growth of ensuing fungal populations or may provide the opportunity for contamination by blue molds or Mucor spp. (9, 23, 26). Some strains of G. candidum, however, inhibit the growth and/or sporulation of Mucor spp. (24). The population density of strains also has an effect on cheese ripening; a less dense covering facilitates gas exchange across the surface of the cheese.
Although G. candidum strains are readily isolated from dairy products, few studies have assessed the genetic diversity of strains that exist in traditional cheesemaking facilities. This issue is important since the industry trend toward standardization of inocula and ripening conditions may lead to the loss of empirically derived biodiversity. Prillinger et al. (38) have used random amplification of polymorphic DNA (RAPD)-PCR to classify isolates of the genus Geotrichum at the species level, but no reports are available on the use of RAPD-PCR for the differentiation of strains of G. candidum or for estimation of the diversity of strains in native cheeses. In this study, we used the RAPD-PCR technique to assess the genetic diversity of G. candidum isolated from a variety of fungus-ripened cheeses in seven regions of France. RAPD-PCR is useful for the assessment of genetic relatedness between closely related species and for the differentiation of strains of a species (29, 35, 45, 50, 52, 53). Phenotypic tests such as morphology, carbon source utilization, and salt tolerance, chosen for their relevance to cheese technology, were also done.
MATERIALS AND METHODS
Collection of G. candidum isolates.
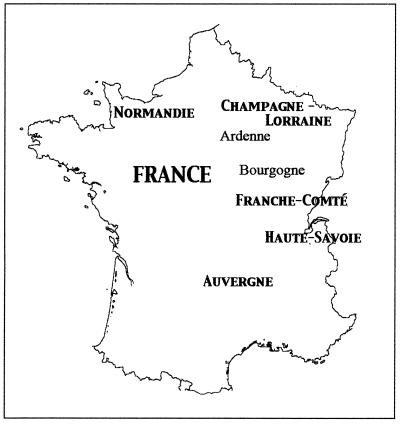
The origins of the isolated strains are shown in Table 1. Samples of milk, curd, and cheese were collected from traditional facilities producing 11 types of soft and semihard cheeses in Normandie, Lorraine, Champagne-Ardenne, Bourgogne, Franche-Comté, Haute Savoie, and Auvergne (Fig. 1). In some cases, samples of cheese from the same batch were collected after 1 week and again at the end of the ripening period. Samples were frozen until processed in the laboratory.
TABLE 1.
Origins of Geotrichum isolates used in this study
| Straina | Sampleb | Cheese | Region |
|---|---|---|---|
| GC1 | F (4) | Chevre | Normandie |
| GC5 | F (6) | Camembert | Normandie |
| GC12 | F (3) | Camembert | Normandie |
| GC13 | F (6) | Camembert | Normandie |
| GC21 | F (7) | Chevre | Normandie |
| GC25 | F (30) | Chevre | Normandie |
| GC28 | F (21) | Chevre | Normandie |
| GC34 | L | Camembert | Normandie |
| GC37 | L | Camembert | Normandie |
| GC39 | F (15) | Chevre | Normandie |
| GC41 | L | Camembert | Normandie |
| GC43 | C | Mont d'Or | Franche-Comté (Haut Doubs) |
| GC44 | C | Reblochon | Haute Savoie |
| GC45 | L | Mont d'Or | Franche-Comté (Haut Doubs) |
| GC46 | C | Reblochon | Haute Savoie |
| GC51 | C | Reblochon | Haute Savoie |
| GC59 | F (7) | Tomme de Savoie | Haute Savoie |
| GC60 | F (35) | Reblochon | Haute Savoie |
| GC63 | F (35) | Reblochon | Haute Savoie |
| GC64 | F (67) | Tomme de Savoie | Haute Savoie |
| GC74 | F (7) | Reblochon | Haute Savoie |
| GC76 | F (8) | Mont d'Or | Franche-Comté (Haut Doubs) |
| GC79 | F (21) | Mont d'Or | Franche-Comté (Haut Doubs) |
| GC82 | C | Epoisses | Bourgogne |
| GC84 | L | Epoisses | Bourgogne |
| GC88 | F (8) | St. Nectaire | Auvergne |
| GC90 | F (21) | Reblochon | Haute Savoie |
| GC94 | F (7) | Reblochon | Haute Savoie |
| GC96 | F (21) | Reblochon | Haute Savoie |
| GC97 | F (7) | Reblochon | Haute Savoie |
| GC100 | F (11) | St. Nectaire | Auvergne |
| GC101 | F (13) | St. Nectaire | Auvergne |
| GC103 | F (11) | St. Nectaire | Auvergne |
| GC105 | L | St. Nectaire | Auvergne |
| GC108 | L | St. Nectaire | Auvergne |
| GC110 | C | St. Nectaire | Auvergne |
| GC120 | C | St. Nectaire | Auvergne |
| GC125 | L | St. Nectaire | Auvergne |
| GC127 | F (21) | Brie de Meaux | Lorraine |
| GC128 | F (21) | Coulommiers | Lorraine |
| GC129 | F (16) | Chaource | Champagne-Ardenne |
| GC139 | F (101) | St. Nectaire | Auvergne |
| GC144 | F (6) | Morbier | Franche-Comté (Haut Doubs) |
| GC146 | C | St. Nectaire | Auvergne |
| GC148 | F (102) | St. Nectaire | Auvergne |
| GC58 | F (7) | St. Nectaire | Auvergne |
| GC159 | F (60) | St. Nectaire | Auvergne |
| GC164 | F (7) | Reblochon | Haute Savoie |
| GC166 | F (7) | Reblochon | Haute Savoie |
| GC172 | F (45) | Bethlehem | Connecticut |
| GC175 | F (10) | Chevre | Switzerland |
| GCU10c | CR | Unknown | Normandie |
| GCU41c | F | St. Nectaire | Auvergne |
| GCU81c | F | Pont l'Eveque | Normandie |
| GCU160c | F | Chevre | Poitou |
| GCU193c | F | Tomme Vaudoise | Switzerland |
| GCU219c | F | Manchego | Spain |
| GCU234c | F | Bleu de Gex | Franche-Comté (Haut Jura) |
| GCU354c | L | Unknown | Normandie |
| GCU442c | L | Unknown | Normandie |
| GCU477c | F | Perladon | Aude |
| GF107 | L | St. Nectaire | Auvergne |
| GF147 | C | St. Nectaire | Auvergne |
| GF152 | L | St. Nectaire | Auvergne |
GC, G. candidum; GF, G. fragrans.
F, cheese, followed by days ripened (in parentheses); L, milk; C, curd; CR, cream.
University of Caen collection.
FIG. 1.
Map of France showing the seven cheesemaking regions from which the isolates of G. candidum characterized in this study were collected. Milk, curd, and cheese samples were collected from traditional facilities making 11 types of soft and semihard cheeses. For details, see Table 1.
To isolate G. candidum, approximately 1 g of milk or cheese rind was added to 5.0 ml of YEG broth (1% yeast extract [bioMérieux, Marcy-l'Etoile, France], 1% glucose) and incubated at 25°C until the formation of a pellicle. YEG agar plates were streaked with the liquid cultures and incubated at 25°C for 48 h. Colonies whose morphology resembled that of G. candidum were restreaked to obtain a pure culture. To confirm the identification of G. candidum, assimilation of d-xylose (+), cellobiose (−), and maltose (−) as sole sources of carbon was tested (26). For long-term storage, sterile skim milk (5.0 ml) was inoculated with purified cultures and incubated at 25°C for 48 h. Samples (0.2 ml) of the culture were lyophilized (Edwards Modulo 4 K Freeze Dryer; RUA instruments, Farmoutiers, France) and stored at 4.0°C until characterized.
To characterize the strains, 54 isolates were resuspended from the lyophilized state in YEG broth and streaked onto YEG agar. The provenances of all of the isolates, as well as 10 strains from the collection of the Laboratoire de Microbiologie Alimentaire, Université de Caen Basse-Normandie, Caen, France, and one isolate each from the United States and Switzerland, are listed in Table 1. The 175 isolates collected in France reside in the collection of the Institut National de la Recherche Agronomique, Poligny, France.
Physiological tests.
For carbon source utilization studies, strains from the lyophilized stock were grown on YEG agar for 48 h at 25°C. A loopful of G. candidum was resuspended in 1.0 ml of sterile 0.9% saline. Tubes of yeast nitrogen base broth (Difco, Detroit, Mich.) containing a final concentration of 0.5% d-mannitol, dl-sodium lactate, sodium citrate, or glucose, as well as a control without a carbon source, were then inoculated with 50 μl of the cell suspension in duplicate. The A650 was measured at time zero and 5 days later. Controls were done to determine if residual glucose in unwashed cells of G. candidum grown on YEG interfered with the results; washed and unwashed cells gave the same results.
Since all of the isolates tested did not assimilate citrate under these conditions, they were also tested on Simmons citrate medium (Difco). Testing of a citrate-positive yeast strain in both types of media gave good growth. Since citrate is a chelating agent that can inhibit growth, an experiment was also carried out in which various concentrations of citrate were added to yeast nitrogen base (Difco) medium with other sources of carbon known to be assimilated by G. candidum.
The salt tolerance of the isolates was tested by using solid YEG medium supplemented with 0, 1.0, 1.5, 2.0, 2.5, and 3.0% NaCl. Ten microliters of a suspension of G. candidum was pipetted onto the centers of plates of the medium, and after drying, the diameter of each spot was marked and measured. The increase in the diameter of the spot was calculated after 2, 4, 6, and 8 days of growth at 25°C. Each strain was tested in duplicate.
Morphology.
Isolates were inoculated in duplicate on YEG agar plates and incubated at 25°C in the dark for 3 days. The macroscopic appearance of the colonies was observed to determine the type of morphology: yeast-like, filamentous, or intermediate. In some cases, the colonies were tested for resistance to touch, a technique used to evaluate whether a strain feels greasy, feels powdery, releases water, or peels off the agar when touched. Isolates were grown in the dark since light can inhibit the growth of G. candidum on solid media (23).
DNA extraction.
Cultures were grown in 30 ml of YEG broth in 150-ml Erlenmeyer flasks with agitation at 25°C for 18 h. Each culture was screened microscopically to check for purity and degree of sporulation. DNA was prepared from the cells when arthrospores dominated in the culture at about 18 h. A sample (2 ml) of each culture was pipetted into a microcentrifuge tube and centrifuged for 15 min at 16,000 × g. The supernatant was removed and discarded, and 1.0 ml of sterile filtered, deionized water was added. The tube was vortexed for 10 s and centrifuged for 15 min at 16,000 × g. The washing was repeated twice with 0.5 ml of water, and the pellet was saved.
For DNA isolation, 200 μl of InstaGene Matrix (Bio-Rad, Hercules, Calif.) was added to the pellet of washed cells and the tube was vortexed for 10 s and then incubated in a 56°C water bath for 25 min. The tube was again vortexed for 10 s and then placed in a boiling water bath for 8 min. The tube was vortexed for 10 s and then centrifuged at 16,000 × g for 3 min. The supernatant was transferred to sterile 1.5-ml centrifuge tubes, and 5.0 μl of RNase (Boehringer Mannheim, Meylan, France) was added. After incubation at 37°C for 20 min, the tube was stored at −20°C.
PCR.
To identify discriminatory primers, a total of 55 primers, including Operon Technologies Primer Kits A and T (Operon Technologies, Alameda, Calif.), were tested on 10 isolates. Three primers, OPA-19 (5′ CAAACGTCGG 3′), OPT-12 (5′ GGGTGTGGAG 3′), and OPT-15 (5′ GGATGCCACT 3′), were identified as informative.
The PCR was carried out in 50-μl reaction mixtures that contained the following (final concentrations): 10 mM Tris-HCl buffer (pH 9.0 at 25°C); 200 μM each dATP, dCTP, dGTP, and dTTP (Boehringer Mannheim); 1.5 mM MgCl2; 7.0 to 8.0 μM primer; 2.5 U of Taq DNA polymerase (Qbiogene-Quantum Appligene); and 20 μl of the DNA template solution. DNA was amplified by using a Perkin-Elmer model 9600 thermal cycler (Perkin-Elmer, Roissy, France) with the following temperature profile: 94°C for 5 min; 35 cycles of denaturation at 94°C for 1 min, annealing for 2 min at 34.0°C, and extension for 2 min at 72.0°C; and then holding at 4.0°C. Amplification products were analyzed by horizontal gel electrophoresis in 1.0% SeaKem GTG agarose (FMC Corp., Rockland, Maine). A control containing all of the PCR components minus the DNA template was run on each gel. A 123-bp DNA ladder (Gibco BRL Life Technologies) was included in the middle and outside lanes. Amplicons were detected by ethidium bromide (7.5 μg/ml) staining. The gels were photographed under a UV lamp by using Polaroid 665 negative film. The negatives were scanned, and the images were saved as TIFF files for computerized analysis.
Gel analysis.
The GelCompar Program, version 4.0 (Applied Maths, Kortrijk, Belgium), was used to analyze the gels. The normalization settings used were as follows: a resolution of 500 points, a smoothing factor of 3, and background subtraction by the rolling-disk method with an intensity setting of 12. The Dice similarity coefficient was used for band matching, and the patterns were clustered by the unweighted pair group method using arithmetic averages.
Reproducibility study.
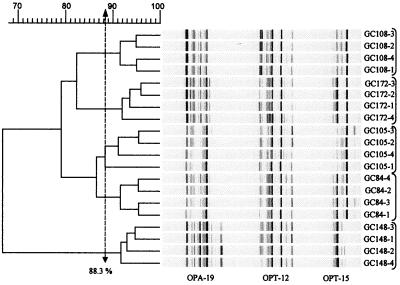
To determine the minimum percent similarity necessary for strain discrimination, a reproducibility study was done on five isolates by using three primers and four iterations of the entire procedure, beginning with culture inoculation. Each isolate was grown in four separate cultures from which DNAs were extracted and amplified by using three different primers in separate reaction tubes for a total of 12 reactions for each strain. The amplification products obtained from the same primer for two replicates of each isolate were run on one gel. Amplicons from the other two replicates were run on another gel to estimate gel effects (Fig. 2).
FIG. 2.
Reproducibility study. RAPD profiles and a similarity dendrogram generated in the study of five strains tested four times by using primers OPA-19, OPT-12, and OPT-15 as described in the text. Photographic negatives of stained gels showing DNA fragments from RAPD-PCR were scanned into TIFF files and compiled and analyzed by GelCompar as described in Materials and Methods. Shown is a combination of six separate gels and 60 separate amplifications that were concatenated and analyzed as a composite gel. The dashed line indicates the 88% level, above which strains are considered the same. The concatenated gels are shown on the right.
RESULTS AND DISCUSSION
Reproducibility of the technique and PCR conditions.
Of the 55 candidate primers tested for discriminatory ability, three (OPA-19, OPT-12, and OPT-15) were identified as the most informative. Five strains (GC84, GC105, GC108, GC148, and GC172) were selected with which to conduct a reproducibility study to estimate the similarity level at which each strain could be reliably differentiated. Banding patterns obtained by using each isolate with each primer were concatenated and analyzed as a composite by the GelCompar program. Figure 2 shows data compiled from six separate gels (duplicate gels for each of three primers). Two replicates (1 and 2) of each strain-primer combination were run on one gel, and the other two replicates (3 and 4) were run on a separate gel. Reproducibility was not gel dependent. For example, the branching order for strain GC108 pairs replicates 3 and 2 as 95% similar and 4 and 1 as 95% similar, with 91% similarity between the pairs of replicates. A similar topology is evident for the other strains in Fig. 2. Isolate GC105 yielded a similarity value of 88.3% for one of the four replicates. Each of the other strains had greater than 91% similarity between replicates. Therefore, we have used 88.0% similarity as the level for strain differentiation.
An annealing temperature of 34°C with 35 cycles gave the best results. The use of a low annealing temperature with many cycles increases the risk of nonspecific amplification (44). For this reason, a negative control without template DNA was included for each set of amplifications. Primer-derived nonspecific amplification products in negative control reactions have been reported to be common when short random primers are used (36).
We found that the age of the culture was important for efficient DNA extraction and amplification. Eighteen-hour cultures in which germinating arthrospores were seen to predominate were optimal for DNA extraction. Older cultures consisting predominantly of vegetative hyphae did not produce strong bands. Although DNA extraction by boiling of cells is rapid, structural and DNA-binding proteins may interfere with PCR amplification. We found that treatment of washed cells with the InstaGene matrix gave stronger and clearer banding patterns, possibly due to the absorption of cell lysis products (42).
Overall diversity of strains.
Figure 3 shows a dendrogram derived by GelCompar analysis of 64 isolates tested with three primers. The banding patterns (not shown) were similar in quality to those shown in Fig. 2. The results of the dl-lactate and d-mannitol assimilation studies are tabulated to the right. Eighty-six percent of the isolates were positive for growth on dl-lactate in yeast nitrogen base. Deacidification of curd through the metabolism of lactate is recognized as a major activity of G. candidum in the early stages of the ripening of mold-ripened cheeses (21). Fresh Brie and Camembert contain about 1% lactate produced by lactose fermentation by lactic acid bacteria (15). The seven isolates from Brie and Camembert in our collection were positive for lactate assimilation.
FIG. 3.
Dendrogram, created by using GelCompar, showing relationships between G. candidum strains. The percent similarity scale indicates the coefficients of similarity between strains calculated from the concatenated gels by the unweighted pair group method using arithmetic averages. G. fragrans strains are indicated. The results for assimilation of dl-lactate and d-mannitol as sole sources of carbon are shown to the right of the strain designations. −, negative; (+), weakly positive; +, positive; ++, strongly positive.
Some strains from St. Nectaire did not grow on lactate. Lactate may be less abundant in St. Nectaire since water added to the curds and whey throughout the cheesemaking process removes much of the lactose from the curd (46). Lactose comprises 70% of the total solids in whey (13).
The carbon source utilization results are generally consistent within clusters of strains that branch above 88%. Below this value, the patterns of carbon utilization vary. These patterns support the conclusion derived from the reproducibility studies that isolates clustering above 88% represent closely related or identical strains of G. candidum. At this threshold value, 48 separate groups can be distinguished.
Biogeography of similar strains.
No correlation was found between the regions from which strains were isolated and clustering patterns. The same or very similar strains were isolated from widely different regions. For example, GC144 and GC146 are from a Morbier in the Haut Doubs and St. Nectaire curd in the Auvergne. The most distantly collected yet similar strains are GC76 and GC172. GC76 is from a Mont d'Or in the Haut Doubs, and GC172 is from a St. Nectaire-type cheese manufactured in Bethlehem, Conn. Therefore, similar or identical strains are ubiquitous throughout France and probably the world.
GC34, isolated from the milk of a Camembert-making facility in Normandie, and GC128, from a 21-day-old Coulommiers made in Lorraine, also exhibit more than 90% similarity. However, both dairies use a commercial inoculum (Arôme Norman) at certain times of the year and although the starter was not added when our samples were obtained, this strain may have colonized the cheesemaking environments.
Seven of the 10 strains tested from the University of Caen collection (GCU prefix in Fig. 3) grouped closely together despite their diverse geographic origins (Table 1). The remaining three strains from this collection (GCU41, GCU219, and GCU354) were very different and constitute separate strains in Fig. 3.
Diversity within a cheesemaking facility.
Some strain groups were found within the same cheesemaking facility at different times during ripening. For example, the four closely related strains at the top of Fig. 3, GC63, GC59, GC64, and GC60, were isolated from two 35-day-old Reblochon cheeses and two 7-day-old Tomme de Savoie cheeses produced and ripened in the same facility in Annecy, Haute Savoie. The isolates have very similar yeast-like morphologies and are positive for lactate utilization and strongly positive for d-mannitol utilization. Isolate GC60, which is the only one of the four that is strongly positive for lactate assimilation, has 87% similarity to the other three isolates. The latter are 94% similar to one another and are probably the same strain. A different strain (GC74) was also isolated from a 7-day-old Reblochon cheese in the same facility. This strain has a filamentous morphology.
A similarly diverse set of five strains was isolated from a farm that was located 19 km from Annecy and that also produced Reblochon. Strain GC44 from the curd had the same morphology as four isolates from Annecy but its RAPD profile is only 81% similar and it does not grow on d-mannitol. Strains GC97, GC94, and GC90 are similar to each other but show less than 30% similarity to the rest of the collection. GC97 and GC94 were isolated from two different 7-day-old cheeses, and GC90 and GC96 came from 21-day-old cheeses. GC96 constitutes a separate strain. Altogether, this farm yielded at least four different strains.
Similar diversity was found among strains isolated from individual facilities making Camembert (GC5, GC12, GC13, and GC41) and goat cheese (chevre) (GC21, GC25, GC28, and GC39) in Normandie, Mont d'Or in Haut Doubs (GC43, GC45, GC76 and GC79), and St. Nectaire in the Auvergne (GC103, GC107, GC108, GC139, GC147, and GC152). Strains GF107, GC147, and GC152 have been identified as a separate species of Geotrichum, G. fragrans (see below).
Some pairs of isolates from milk and curd of the same facilities showed 90% or greater similarity and had similar morphology and carbon assimilation profiles. For example, GC84 and GC82 from Epoisses and GC45 and GC43 from Mont d'Or were isolated from the milk and curd, respectively, of the same facility. This observation suggests that a strain can be followed during the cheesemaking process.
Although isolates did not group according to cheese type, all 12 strains from Reblochon from three different facilities were positive for lactate assimilation (Fig. 3). The cheesemaking operations used yogurt as the source of their starter cultures. The lactate content of yogurt may select G. candidum capable of using lactate as a source of carbon. The whitish orange rind characteristic of Reblochon is due to the presence of Brevibacterium linens and G. candidum, the only fungus found on this cheese. Bärtschi et al. (2) found that from the time the curd is pressed and drained to day 8 of ripening, the population of G. candidum increased 1,000-fold. In metabolizing lactate, the fungus lowers the acidity in the environment of the rind, which allows subsequent colonization by the acid-sensitive bacterium B. linens (30).
Regional diversity.
Figure 4 shows the RAPD and carbon assimilation profiles, as well as the salt tolerances, of G. candidum and G. fragrans isolates from seven St. Nectaire cheesemaking facilities. St. Nectaire has been made since the 17th century in the Departments of Cantal and Puy de Dome, volcanic mountainous regions in the Auvergne (Fig. 1). Of the 37 French cheeses legally protected with the Appellation d'Origine Contrôlée designation, St. Nectaire's production is the fifth largest nationally. About 13,873 tonnes was made in 1998, 44% of which was farmstead cheese made with raw milk (http://www.beulet.fr/aoc.htm). These cheeses thus provide an excellent source of regional diversity.
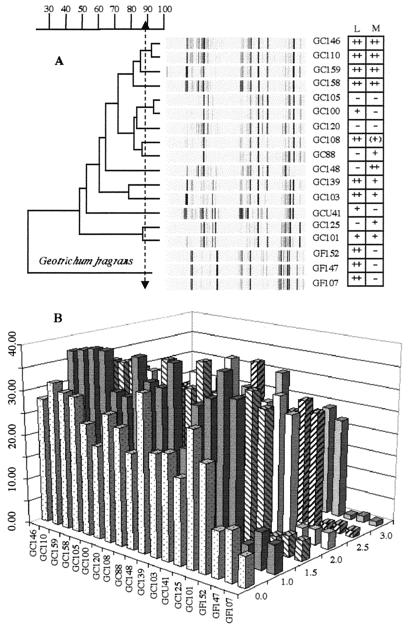
FIG. 4.
Collection of G. candidum and G. fragrans strains from the Auvergne. (A) RAPD profiles and dendrogram, obtained by using primers OPA-19, OPT-12, and OPT-15, showing relationships between strains. According to the results of the reproducibility study (Fig. 2), isolates showing less than 88.0% similarity are probably different strains. Assimilation of dl-lactate (L) and d-mannitol (M) as sole sources of carbon is indicated to the right of the strain designations. −, negative; (+), weakly positive; +, positive; ++, strongly positive. (B) NaCl tolerances of the same strains after 8 days of growth in YEG medium containing different concentrations of NaCl (z axis). The y axis indicates the colony diameter (in millimeters) from day 0 to day 8.
The isolates characterized here were taken from raw-milk farmstead facilities in the Puy de Dome. These operations all had their own milk sources and ripening facilities. The diversity within this region is striking. The RAPD profiles and carbon assimilation patterns of the four isolates shown at the top of the dendrogram form a distinct cluster (Fig. 4A). Isolates GC146 and GC110 seem to be the same strain, with 93% similarity in their RAPD profiles. These isolates originated in the curds of two different facilities located approximately 3.5 km apart. The other two isolates (GC158 and GC159) were from 7- and 60-day-old cheese samples made from the same curd from which GC146 was isolated.
GC105 and GC100 also appear to have the same RAPD profile. They originated in milk and cheese from two facilities within 2.6 km of one another. GC125 and GC101 cluster together, with 86% similarity between them, but as a pair are less than 50% similar to the other strains. GC101 and GC148 were isolated from cheese samples made with milk from which GC125 originated. They differ from each other in carbon assimilation and morphology. All of the isolates from St. Nectaire, except GCU41, from the University of Caen collection, had a filamentous type of morphology.
The concentration of NaCl in cheese can vary from 0.7 to 8.0% (wt/wt). Salting affects flavor and moisture content and inhibits the growth of undesirable microorganisms (14). G. candidum is sensitive to salt (22, 26). Growth is inhibited at 1 to 2% NaCl, and no growth is observed at 5%. Soft cheeses like Camembert and Brie have 1.5 to 2.0% (wt/wt) NaCl (13). In contrast, blue cheeses, such as Roquefort, contain about 4.0% (wt/wt) salt that helps select for Penicillium sp. strains that can tolerate 5.0% NaCl (13). The salt content of the cheese and the salt tolerance of fungi help determine the pattern of microbial succession on a rind. Figure 4B shows the salt tolerance of the 18 isolates from St. Nectaire. Growth in 1% NaCl was better than growth with no salt, and the colony diameters generally dropped by about 30 to 50% as NaCl increased from 1 to 3%. These results are consistent with the fact that traditional St. Nectaire generally contains 1 to 1.3% (wt/wt) NaCl (23, 39).
G. fragrans
In Figures 3 and 4A and B, GF107, GF147, and GF152 stand out from the rest of the strains. GF107 and GF147 were isolated from one St. Nectaire facility (Table 1), and GF152 was isolated from the same farm 3 months later. The characteristics of the isolates are consistent with those of G. fragrans. The colonies were tough, embedded, and difficult to streak, and an unmistakable fruity fragrance was detected when petri dishes were opened. Microscopically, arthrospores and branched septate hyphae were present but the hyphae were narrower than the hyphae of G. candidum. An annelation was noted at the point of disarticulation of the hyphae. The isolates were also negative for the assimilation of d-xylose, cellobiose, and maltose and positive for lactate assimilation. These are all characteristics typical of G. fragrans (8). The low similarity between G. candidum and G. fragrans (Fig. 3) agrees with previous studies showing that 20% genome similarity is sufficient for differentiation of species (38). G. fragrans strains are very sensitive to NaCl (Fig. 4B), which may explain why the strain was isolated from milk and curd but not from cheese that was salted on the day of manufacture.
The industrial production of St. Nectaire using pasteurized milk began in 1964. At that time, it was feared that the industrial product, considered by some to lack the character and flavor of the traditional cheese, would supplant the farmstead St. Nectaire (41). However, the production of artisanal cheeses in the European community increased 65% from 1984 to 1996 (16), which may be due to the fact that consumers prefer the diverse sensory characteristics of artisanal products to the uniformity made possible by pasteurization (19). One study found that G. candidum is rarely found on industrial St. Nectaire, while it was routinely isolated from farmstead St. Nectaire made with raw milk throughout all stages of ripening (9). Our study found 14 strains of G. candidum from seven farmstead St. Nectaire facilities in the Puy de Dome region alone. Only two pairs of strains were potential duplicates.
We have found a high degree of biodiversity in G. candidum strains from seven cheesemaking regions of France. The results of carbon assimilation and salt tolerance tests suggest that cheesemaking techniques play a role in strain selection. Native strains of G. candidum could provide a rich source of cheese-ripening enzymes that develop aroma and texture. Since much of the flavor development associated with cheese ripening is due to microbial activity, it seems likely that the diversity of G. candidum strains that we have found may contribute to the acknowledged diversity of flavor found in French cheeses. As traditional cheesemaking techniques are threatened or have been abandoned, the collection, characterization, and preservation of native strains of cheese-ripening microorganisms are critical.
ACKNOWLEDGMENTS
This research was funded by the Fulbright Foundation; the Institut National de la Recherche Agronomique; the Research Foundation of the University of Connecticut, the American Institute of Wine and Food, Paris; and Pomposello Productions, New York, N.Y.
We thank Sydney Ruth Palmer for assistance, André Dasen and Franck Dufrene for technical advice and assistance, and Françoise Berthier for generously sharing her expertise. The support of the Abbey of Regina Laudis and the cheesemakers of France who contributed samples is gratefully acknowledged.
REFERENCES
- 1.Auberger B, Lenoir J, Bergère J. Caractérisation partielle des exopeptidases d'une souche de Geotrichum candidum. Sci Aliments. 1997;17:655–670. [Google Scholar]
- 2.Bärtschi C, Berthier J, Valla G. Inventaire et évolution des flores fongiques de surface du reblochon de Savoie. Lait. 1994;74:105–114. [Google Scholar]
- 3.Bérard L, Marchenay P. Lieux, temps et preuves: la construction sociale des produits de terroir. Terrain Carnets Patrimoine Ethnol. 1995;24:153–164. [Google Scholar]
- 4.Berger C, Khan J, Molimard P, Martin N, Spinnler H. Production of sulfur flavors by ten strains of Geotrichum candidum. Appl Environ Microbiol. 1999;65:5510–5514. doi: 10.1128/aem.65.12.5510-5514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertolini M C, Schrag J D, Cygler M, Ziomek E, Thomas D Y, Vernet T. Expression and characterization of Geotrichum candidum lipase I gene, comparison of specificity profile with lipase II. Eur J Biochem. 1995;228:863–869. [PubMed] [Google Scholar]
- 6.Bertozzi L, Panari G. Cheeses with Appellation d'Origine Contrôlée (AOC): factors that affect quality. Int Dairy J. 1993;3:297–312. [Google Scholar]
- 7.Corroler D, Mangin I, Desmasures N, Gueguen M. An ecological study of lactococci isolated from raw milk in the Camembert cheese Registered Designation of Origin area. Appl Environ Microbiol. 1998;64:4729–4735. doi: 10.1128/aem.64.12.4729-4735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Hoog G S, Smith M T, Guého E. A revision of the genus Geotrichum and its teleomorphs. Stud Mycol. 1986;29:1–131. [Google Scholar]
- 9.Delespaul G, Guéguen M, Lenoir J. La flore fongique superficielle des fromages de St-Nectaire et de Tomme de Savoie. Rev Lait Frse. 1973;325:715–729. [Google Scholar]
- 10.Dieuleveux V, Lemarinier S, Guéguen M. Antimicrobial spectrum and target site of D-3-phenyllactic acid. Int J Food Microbiol. 1998;40:177–183. doi: 10.1016/s0168-1605(98)00031-2. [DOI] [PubMed] [Google Scholar]
- 11.Dieuleveux V, Van Der Pyl D, Chataud J, Guéguen M. Purification and characterization of anti-Listeria compounds produced by Geotrichum candidum. Appl Environ Microbiol. 1998;64:800–803. doi: 10.1128/aem.64.2.800-803.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eliskases-Lechner F, Ginzinger W. The yeast flora of surface-ripened cheeses. Milchwissenschaft. 1995;50:458–462. [Google Scholar]
- 13.Fox P F, Guinee T P, Cogan T M, McSweeney P L H. Fundamentals of cheese science. Gaithersburg, Md: Aspen Publishers, Inc.; 2000. [Google Scholar]
- 14.Fox P F, McSweeney P L H. Chemistry and biochemistry of cheese and fermented milks. In: Fox PF, editor. Dairy chemistry and biochemistry. London, England: Blackie Academic and Professional; 1998. pp. 379–434. [Google Scholar]
- 15.Fox P F, Wallace J M. Formation of flavor compounds in cheese. In: Neidleman S L, Lostein A I, editors. Advances in applied microbiology. New York, N.Y: Academic Press, Inc.; 1997. pp. 17–85. [DOI] [PubMed] [Google Scholar]
- 16.Freitas C, Malcata F X. Microbiology and biochemistry of cheeses with Appelation d'Origine Protégée and manufactured in the Iberian Peninsula from ovine and caprine milks. J Dairy Sci. 2000;83:584–602. doi: 10.3168/jds.s0022-0302(00)74918-6. [DOI] [PubMed] [Google Scholar]
- 17.Fresno J M, Tornadijo M E, Carballo J, GonzalezPrieto J, Bernardo A. Characterization and biochemical changes during the ripening of a Spanish craft goat's milk cheese (Armada variety) Food Chem. 1996;55:225–230. [Google Scholar]
- 18.Gobbetti M, Lowney S, Smacchi E, Battistotti B, Damiani P, Fox P F. Microbiology and biochemistry of Taleggio cheese during ripening. Int Dairy J. 1997;7:509–517. [Google Scholar]
- 19.Grappin R, Beuvier E. Possible implications of milk pasteurization on the manufacture and sensory quality of ripened cheese. Int Dairy J. 1997;7:751–761. [Google Scholar]
- 20.Greenberg R S, Ledford R A. Deamination of glutamic and aspartic acids by Geotrichum candidum. J Dairy Sci. 1979;62:368–372. doi: 10.3168/jds.s0022-0302(79)83253-1. [DOI] [PubMed] [Google Scholar]
- 21.Gripon J C. Flavour and texture in soft cheese. In: Law B A, editor. Microbiology and biochemistry of cheese and fermented milk. London, England: Blackie Academic & Professional; 1997. pp. 193–206. [Google Scholar]
- 22.Guéguen M. Contribution à la connaissance de Geotrichum candidum et notamment de sa variabilité, conséquences pour l'industrie fromagère. Ph.D. dissertation. Caen, France: Université de Caen; 1984. [Google Scholar]
- 23.Guéguen M, Delespaul G, Lenoir J. La flore fongigue superficielle des fromages de St-Nectaire et de Tomme de Savoie. Rev Lait Frse. 1974;325:795–816. [Google Scholar]
- 24.Guéguen M, Jacquet J, Allain-Garnier M, Pineau A, Kogbo W. Sur les interactions microbiennes de Geotrichum candidum Link. Microbiol Alim Nutr. 1984;2:139–152. [Google Scholar]
- 25.Guéguen M, Lenoir J. Aptitude de l'espèce Geotrichum candidumà la production d'enzymes protéolytiques. Lait. 1975;55:145–162. [Google Scholar]
- 26.Guéguen M, Schmidt J L. Les levures et Geotrichum candidum. In: Hermier J, Lenoir J, Weber F, editors. Les groupes microbiens d'intérêt laitier. Paris, France: CEPIL; 1992. pp. 165–219. [Google Scholar]
- 27.Holmquist M. Insights into the molecular basis for fatty acyl specificities of lipases from Geotrichum candidum and Candida rugosa. Chem Phys Lipids. 1998;93:57–65. doi: 10.1016/s0009-3084(98)00029-2. [DOI] [PubMed] [Google Scholar]
- 28.Jollivet N, Chataud J, Vayssier Y, Bensoussan M, Belin J. Production of volatile compounds in model milk and cheese media by eight strains of Geotrichum candidum Link. J Dairy Res. 1994;61:241–248. [Google Scholar]
- 29.Karp A, Edwards K J, Bruford M, Funk S, Vosman B, Morgante M, Seberg O, Kremer A, Boursof P, Arctander P, Tautz D, Hewitt G. Molecular technologies for biodiversity evaluation: opportunities and challenges. Nat Biotechnol. 1997;15:625–628. doi: 10.1038/nbt0797-625. [DOI] [PubMed] [Google Scholar]
- 30.Lecocq J, Gueguen M. Effects of pH and sodium chloride on the interactions between Geotrichum Candidum and Brevibacterium linens. J Dairy Sci. 1994;77:2890–2899. [Google Scholar]
- 31.Litthauer D, Louw C H, Du Toit P. Geotrichum candidum P-5 produces an intracellular serine protease resembling chymotrypsin. Int J Biochem Cell Biol. 1996;28:1123–1130. doi: 10.1016/1357-2725(96)00065-9. [DOI] [PubMed] [Google Scholar]
- 32.Medina M I R, Tornadijo M E, Carballo J, Sarmiento R M. Microbiological study of Leon raw cow-milk cheese, a Spanish craft variety. J Food Prot. 1995;58:998–1006. doi: 10.4315/0362-028X-58.9.998. [DOI] [PubMed] [Google Scholar]
- 33.Molimard P, Bouvier I, Issanchou S, Lesschaeve I, Vassal L, Spinnler H E. Cooperation between Penicillium camemberti and Geotrichum candidum: effect on taste and flavour qualities of Camembert type cheese. In: Etievant P, Schreier P, editors. Bioflavour 95. Colloques de l'INRA (FRA) no. 75. 75:167–172. 1995. pp. 167–172. [Google Scholar]
- 34.Molimard P, Lesschaeve I, Bouvier I, Vassal L, Schlich P, Issanchou S, Spinnler H E. Amertume et fractions azotées de fromages à pâte molle de type camembert: rôle de l'association de Penicillium camemberti avec Geotrichum candidum. Lait. 1994;74:361–374. [Google Scholar]
- 35.Paffetti D, Barberio C, Casalone E, Cavalieri D, Fani R, Fia G, Mori E, Polsinelli M. DNA fingerprinting by random amplified polymorphic DNA and restriction length polymorphism is useful for yeast typing. Res Microbiol. 1995;146:587–594. doi: 10.1016/0923-2508(96)80565-1. [DOI] [PubMed] [Google Scholar]
- 36.Pan Y-B, Burner D M, Ehrlich K C, Grisham M P, Wei Q. Analysis of primer-derived, nonspecific amplification products in RAPD-PCR. BioTechniques. 1997;22:1071–1077. doi: 10.2144/97226bm13. [DOI] [PubMed] [Google Scholar]
- 37.Prieto B, Urdiales R, Franco I, Tornadijo M E, Fresno J M, Carballo J. Biochemical changes in Picon Bejes-Tresviso cheese, a Spanish blue-veined variety, during ripening. Food Chem. 1999;67:415–421. [Google Scholar]
- 38.Prillinger H, Molnar O, Eliskases-Lechner F, Lopandic K. Phenotypic and genotypic identification of yeasts from cheeses. Antonie van Leeuwenhoek. 1999;75:267–283. doi: 10.1023/a:1001889917533. [DOI] [PubMed] [Google Scholar]
- 39.Ratomahenina R, Chabalier C, Galzy P, Dieu B. Study of Chrysosporum sulfureum, the mould responsible for “fleur jaune”on Saint-Nectaire cheese. Milchwissenschaft. 1995;50:266–267. [Google Scholar]
- 40.Ribadeau-Dumas B. Maîtrise de l'affinage des fromages de type Camembert. Lait. 1984;64:448–468. [Google Scholar]
- 41.Ricard D. CERAMAC, Université Blaise Pascal, Clermont-Ferrand, France. 1994. Les Montagnes Fromagères en France. [Google Scholar]
- 42.Richner S M, Meirung J, Kirby R. A study of the genetic diversity of Mycobacterium tuberculosis isolated from patients in the eastern province of South Africa using random amplified polymorphic DNA profiling. Electrophoresis. 1997;18:1570–1576. doi: 10.1002/elps.1150180915. [DOI] [PubMed] [Google Scholar]
- 43.Rossi J, Gobbetti M, Buzzini P, Corsetti A, Smacchi E, De Angelis M. Yeasts in dairy. Ann Microbiol Enzimol. 1997;47:169–183. [Google Scholar]
- 44.Roux K H. Optimization and troubleshooting in PCR. PCR Methods Appl. 1995;4:5185–5194. doi: 10.1101/gr.4.5.s185. [DOI] [PubMed] [Google Scholar]
- 45.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 46.Scott R. Cheesemaking practice. New York, N.Y: Elsevier Applied Science; 1986. [Google Scholar]
- 47.Smith M T, Poot G A. Diapodascus capitatus, Dipodascus spicifer and Geotrichum clavatum: genomic characterization. Antonie Van Leeuwenhoek. 1998;74:229–235. doi: 10.1023/a:1001762024710. [DOI] [PubMed] [Google Scholar]
- 48.Spinnler H E, Molimard P, Lesschaeve I, Vassal L, Issanchou S. Impact of associations of microorganisms on the sensory properties of fermented foods. In: Etievant P, Schreier P, editors. Bioflavour 95. Colloques de l'INRA (FRA) no. 75. 75:173–175. 1995. pp. 173–175. [Google Scholar]
- 49.Trieu-Cuot P, Gripon J C. A study of proteolysis during Camembert cheese ripening using isoelectric focusing and two-dimensional electrophoresis. J Dairy Res. 1982;49:501–510. [Google Scholar]
- 50.Tyler K D, Wang G, Tyler S D, Johnson W M. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueda-Nishimura K, Mikata K. Two distinct 18S rRNA secondary structures in Dipodascus (Hemiascomycetes) Microbiology. 2000;146:1045–1051. doi: 10.1099/00221287-146-5-1045. [DOI] [PubMed] [Google Scholar]
- 52.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]