Abstract
Background
The COVID-19 is caused by a novel coronavirus SARS-CoV-2, which started from China. It spread rapidly throughout the world and was later declared a pandemic by the WHO. Over the course of time, SARS-CoV-2 has mutated for survival advantages, and this led to multiple variants. Multiple studies on mutations identification in SARS-CoV2 have been published covering extensive sample areas. The purpose of this study was to limit the sample area to the Georgia state in the U.S. and to analyze the genome sequences for mutation profiling across the genome and origin of variants.
Methods
The genome sequences (n = 3,970) were obtained from the NCBI database as of June 12, 2021, with the filter of being complete sequenced genomes, homo-sapiens host, and only from Georgia State of the U.S. NextClade, an online tool was used for the analysis of the sequences using Wuhan-Hu-1/2019 as a reference genome. The algorithm was sequence alignment, translation, mutation calling, phylogenetic placement, clade assignment, and quality control (QC). Thirty-six samples with bad QC were removed from the mutational analysis.
Results
A total 117,743 mutations in the nucleotides were identified (averaging 31.5 mutations per sample). The mutations A23403G, C3037T, C241T, and C14408T were detected in 98% of the samples. Also, a total of 75,517 mutations in the amino acid were identified (averaging 20.2 mutations per sample). The mutations D614G and P314L were identified in >97% samples whereas R203K, G204R, P681H, and N501Y were detected in >50% samples. Analysis also revealed 16 different clades with 20I (49.6%). Clades 20G (24.2%) and 20A (5.5%) being the most abundant, showed that SARS-CoV-2 in the Georgia State originated mainly from Southeast England, other parts of the U.S., and several countries in Western Europe.
Conclusion
Looking at the three most common variants in Georgia State of the U.S., we could determine the primary locations of transmission or origin for the virus, and our analyses indicates that majority of the cases originated from Southeast England (Clade 20I), the U.S. itself (Clade 20G), and from Western Europe (Clade 20C).
Keywords: Coronavirus disease 2019, Severe acute respiratory syndrome coronavirus-2, Genome sequences
1. Introduction
The coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which is a novel coronavirus. This virus was first reported from Wuhan, China, in December 2019, and the disease was later declared a pandemic by the WHO on March 11, 2020 (Cucinotta and Vanelli, 2020). The sequence analysis of SARS-CoV-2 showed that it was close to other members of coronaviruses family, SARS-CoV and Middle East respiratory syndrome-CoV (MERS-CoV) (Zhu et al., 2020), and both were responsible for the large outbreaks within the past two decades (Weissman et al., 2021). The SARS-CoV-2 contains RNA genome, and its genome size is 29.9 kilobase containing 25 genes (Chan et al., 2020).
Mutations or genetic changes arise as a result of viral replications and the RNA viruses tend to have higher rates of mutation than the DNA viruses (Lauring and Hodcroft, 2021). The mutations in viral genomes also occur due to driving force in order to evade the host’s defense mechanism of their immune system (Dos Santos, 2021, Mengist et al., 2021). Mutations lead to changes in the viral infectivity, efficiency of molecular diagnosis, and vaccine efficacy. Studies have shown that the SARS-CoV-2 went through an evolution in which the virus became more transmissible, which is considered quite advantageous for the virus in terms of natural selection (Nagy et al., 2021).
There have been multiple studies on the identification of mutations in SARS-CoV-2 reported in the peer-reviewed literature that covered more extensive sample areas (Badua et al., 2021, Wang et al., 2021, Zhao et al., 2021). The purpose of this study was to limit the sample area to only one state, Georgia. The state of Georgia is one of the most populous South-eastern states of the United States of America and has over 10 million residents with diversity. The main underlying reason for selecting Georgia is that this state is home to the Hartsfield Jackson Airport of Atlanta, the busiest Airport of the world, which serves travelers from all over the globe. Our aim was to analyze the genome sequences from samples submitted to the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) from the state of Georgia in the U.S., and to identify the mutational dynamics of the SARS-CoV-2 as compared to the viral genome, which is believed to be originated from Wuhan, China while also identifying the primary locations of transmission/origin for the virus into the state of Georgia.
2. Methods
2.1. SARS-CoV2 genome sequences
In the present study, we analyzed 3,970 full length SARS-CoV2 genome sequences obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/sars-cov-2/) as of June 12, 2021 (Sample collection date: March 2020 – May 2021). All 3,970 sequences were downloaded with the filter being complete sequenced genomes, homo-sapiens host, as well as only from the Georgia State in the United States as the geographic origins.
2.2. Identification of mutations in the SARS-CoV-2 genome sequences
The SARS-CoV-2 Genome Sequences were downloaded in the FASTA format and online tools NextClade of NextStrain (https://clades.nextstrain.org/) (Rambaut et al., 2020, Aksamentov et al., 2021) were used for the analysis of the genome sequences. NextClade uses SARS-CoV-2 isolate Wuhan-Hu-1/2019 (Accession # MN908947) as a reference genome. The Algorithm was Sequence alignment, Translation, Mutation calling, Detection of PCR primer changes, Phylogenetic placement, Clade assignment and Quality Control. The data summarization was carried out using Microsoft Excel.
2.3. Phylogenetic analysis
The phylogenetic tree was visualized by NextStrain Auspice (https://clades.nextstrain.org/tree). The phylogenetic tree generated from NexStrain represented inferred evolutionary relationships among different viral genome samples based on their mutation profile and other sample data.
3. Results
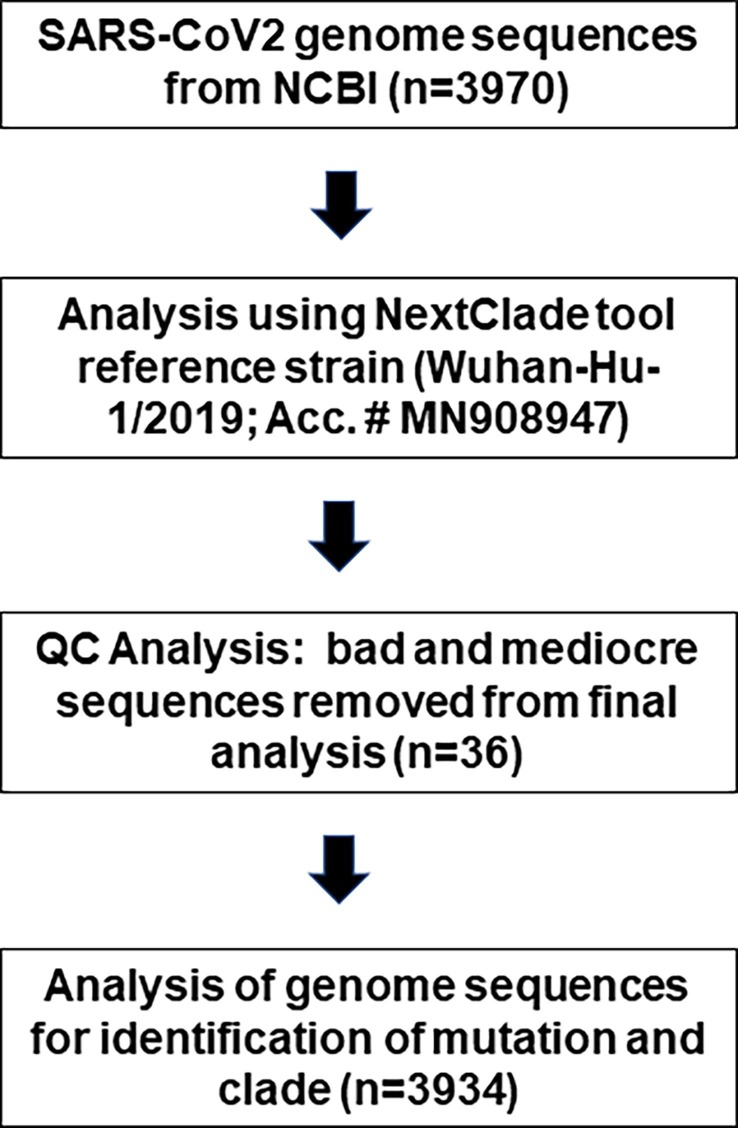
We used the following workflow (Fig. 1 ) for the analysis of SARS-CoV-2 genome sequence. We included all the samples for clade analysis except the removed samples (n = 36) that did not meet the sequencing QC criteria.
Fig. 1.
Workflow for the analysis of SARS-CoV-2 genome sequences that were submitted to the NCBI from Georgia state of the United States (as of June 12, 2021).
3.1. Identification of clade and phylogenetic tree
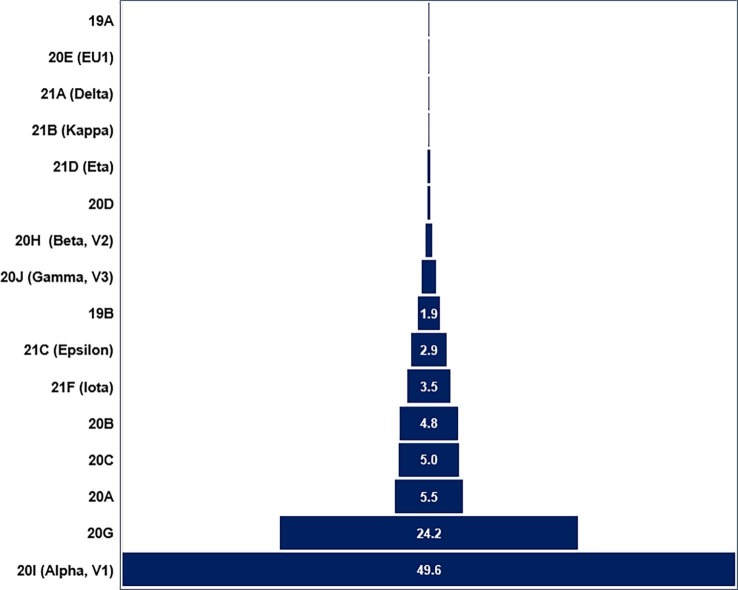
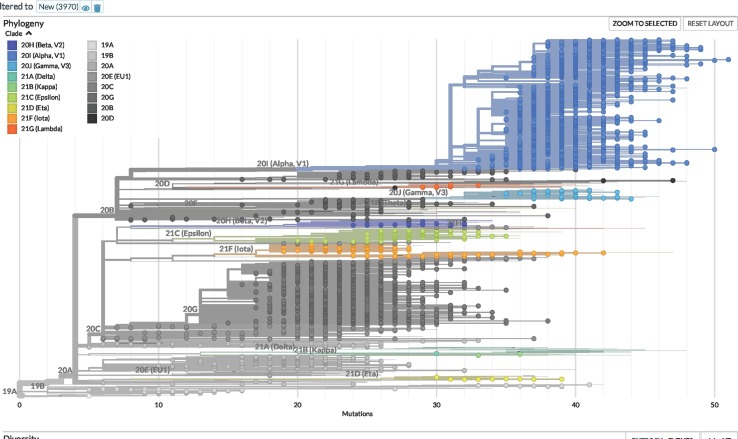
The NextStrain clade analysis revealed 16 different types of clades ranging from an occurrence rate of 0.03% (1 out of 3,970) to 49.6% (1,968 out of 3,970). The most dominant clades were 20I (Alpha, V1; 49.6%) and 20G (24.2%), respectively. Other clades 20A, 20B, and 20C were in the range of 4.8% − 5.5% (Fig. 2 and Table 1 ). The phylogenetic tree generated with the NextStrain analysis tools is shown in the Fig. 3 .
Fig. 2.
Pyramidal presentations of clade identifications and their frequency (%). A total of 16 different clades were identified in the 3,970 SARS-CoV-2 sequences analyzed with an occurrence range of 0.03–49.6%.
Table 1.
List of identified clades its earliest documented samples, and frequency.
| Clade | Number of Clades | Earliest Documented Samples* |
|---|---|---|
| 19A | 1 | China, December-2019 |
| 19B | 76 | China, December-2019 |
| 20A | 219 | Europe (multiple countries), May-2020 |
| 20B | 192 | Multiple countries, November-2020 |
| 20C | 197 | Europe (France and England), July-2020 |
| 20D | 14 | Peru, August-2020 |
| 20E (EU1) | 1 | Spain, June-2020 |
| 20G | 960 | USA, July-2020 |
| 20H (Beta, V2) | 27 | South Africa, May-2020 |
| 20I (Alpha, V1) | 1,968 | United Kingdom, September-2020 |
| 20J (Gamma, V3) | 47 | Brazil, November-2020 |
| 21A (Delta) | 1 | India, October-2020 |
| 21B (Kappa) | 2 | India, October-2020 |
| 21C (Epsilon) | 115 | USA, March-2020 |
| 21D (Eta) | 11 | Multiple countries, December-2020 |
| 21F (Iota) | 139 | USA, November-2020 |
| Total | 3,970 | |
A total of 16 different clades were identified among 3,970 samples, 20I (Alpha, V2) being most prevalent, which originated from the United Kingdom. Other common clades were 20A and 21G, which originated from Europe and the United States of America, respectively.
Fig. 3.
Phylogenetic tree of circulating variants (n = 3,970) of SARS-CoV-2 in the Georgia state in the United States (as of June 12, 2021). The phylogenetic tree was generated using the NextStrain Auspice online tools (https://clades.nextstrain.org/tree).
3.2. Identification of mutation in the genome sequences of SARS-CoV-2
3.2.1. Mutation in the nucleotide sequences
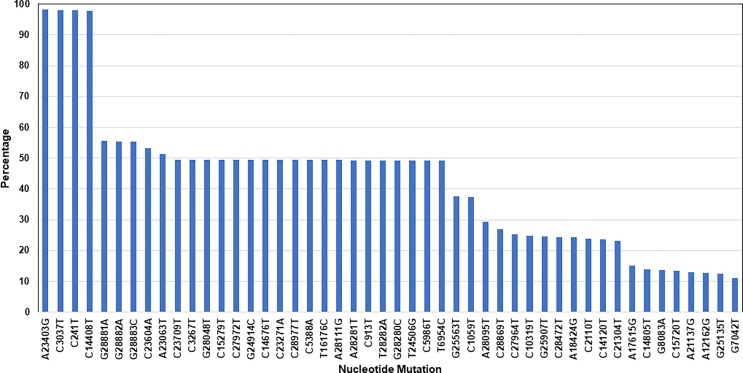
A total of 117,743 mutations in the nucleotides were identified in the 3,734 genome sequences of SARS-CoV-2 (with an average of 31.5 nucleotide mutations per genome sample). There were 6,435 different types of mutations identified in all sequences (Supplementary Data Table S1). The highest mutations rate was 98% detected at locations: A23403G, C3037T, C241T, and C14408T. The second highest mutation rate (50–56%) occurrence was detected at locations: G28881A, G28882A, G28883C, C23604A, A23063T, C23709T, C3267T, G28048T, C15279T, C27972T, G24914C, C14676T, C23271A, C28977T, C5388A, T16176C, A28111G, A28281T, C913T, T28282A, G28280C, T24506G, C5986T, and T6954C (Fig. 4 ).
Fig. 4.
Identification of mutation in the nucleotide sequences. A total of 117.743 mutations identified in 3,734 SARS-CoV-2 genome sequences. There were 6,435 different types of mutations identifies with an average of 31.53 mutations per genome sample. The mutations with occurrence rate of >11% are shown here (See Supplementary Data Table S1 for complete list of the identified mutations in the nucleotide sequences.
3.2.2. Mutation in the amino acid sequences
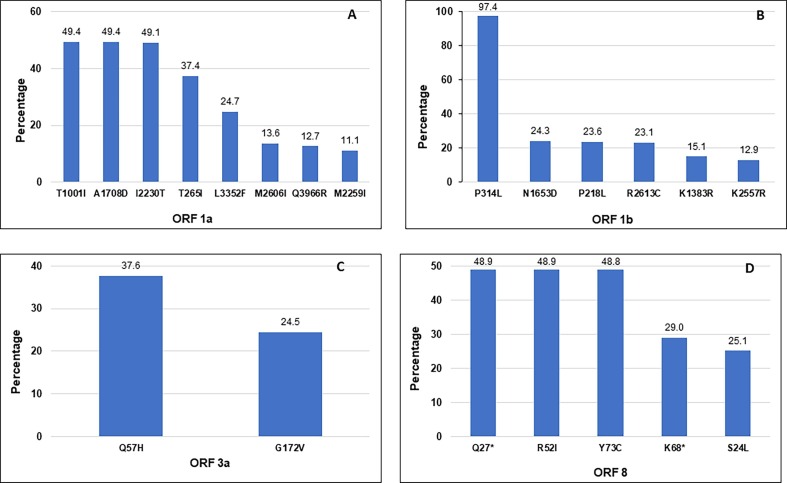
A total of 75,517 mutations in the amino acid were identified in the 3,736 SARS-CoV-2 sequences (with an average of 20.2 amino acid mutations per genome sample). There were 3,633 different types of amino acid mutations identified in all sequences (Supplementary Data Table S2). We further analyzed the presence and distribution of amino acid substitutions in different genes as well as ORFs (Open Reading Frames) of the SARS-CoV-2 genomes (Fig. 5 ). The most common mutations in the ORF1a are presented in Fig. 5 A. The mutations T1001I, A10708D, and I2230T were identified in approximately 50% of the total samples analyzed. The most common mutations in the ORF1b are presented in Fig. 5 B. The mutation P314L was found in 97.4%, whereas mutations N1653D, P218L, and R2613C, were detected in 23–24% of the total samples. Also, there were only two major locations of amnio acid mutations detected in the ORF 3a, Q57H, and G172V with an occurrence rate of 37.6% and 34.5%, respectively (Fig. 5 C). In the ORF 8, Q27* (Stop), R52I, Y73C, K68* (Stop), and S24L were the most frequent mutations (Fig. 5 D). The ORF 8 is the only ORF where mutations Q27* and K68* causes the introduction of premature stop codon (nonsense mutation) with an occurrence rate of 48.9% and 29.0%, respectively, of the total samples were identified.
Fig. 5.
Identification of mutations in the amino acid sequences of four different open reading frames (ORF) of the SARS-CoV-2 sequences analyzed in this study. The mutations with high abundant rate (A) ORF1a, (B) ORF 1b, (C) ORF 3a, and (D) ORF 8 are shown in the figure. The frequency (%) of each mutation is indicated on top of each bar.
The mutation D614G located on the surface glycoprotein (spike protein) was identified in 98.3% of the total samples. Other mutations on the spike protein were P681H, N501Y, T716I, D1118H, A570D, and S982A, with an occurrence rate of 49–53% (Table 2 A). There were six different locations on the nucleocapsid phosphoprotein (N protein), which had an amino acid mutation occurrence rate of higher than 10% of the total samples analyzed (Table 2 B). The topmost mutations were R203K, G204R, S235F, and D3L with the mutation rate between 49 and 55%. The location of mutation in different amino acids in the spike protein is illustrated in Fig. 6 .
Table 2.
Identification of mutations in the amino acid sequences of (A) Surface glycoprotein (Spike or S-protein), and (B) Nucleocapsid phosphoprotein (N-protein). A total of 75,517 mutations were identified in the 3,734 SARS-CoV-2 sequences. There were 3,633 different types of amino acid mutations identified and the mutations with high abundant rate are shown (complete list of mutations identified in the amino acids is given in Supplementary Data Table S2).
| Mutation (Amino Acid) | Mutation (Nucleotide) | Occurance Rate (%) | Mutation Type |
|---|---|---|---|
| D614G | A23403G | 98.30 | Non-Synonymous |
| P681H | C23604A | 53.15 | Non-Synonymous |
| N501Y | A23063T | 51.30 | Non-Synonymous |
| T716I | C23709T | 49.47 | Non-Synonymous |
| D1118H | G24914C | 49.42 | Non-Synonymous |
| A570D | C23271A | 49.39 | Non-Synonymous |
| S982A | T24506G | 49.26 | Non-Synonymous |
| K1191N | G25135T | 12.51 | Non-Synonymous |
| E484K | G23012A | 6.18 | Non-Synonymous |
| L5F | C21575T | 6.08 | Non-Synonymous |
| Mutation (Amino Acid) | Mutation (Nucleotide) | Occurrence Rate (%) | Mutation Type |
|---|---|---|---|
| R203K | G28881A | 55.5 | Non-Synonymous |
| G204R | G28883C | 55.2 | Non-Synonymous |
| S235F | C28977T | 49.4 | Non-Synonymous |
| D3L | A28281T | 49.3 | Non-Synonymous |
| P199L | C28869T | 27.0 | Non-Synonymous |
| P67S | C28472T | 24.4 | Non-Synonymous |
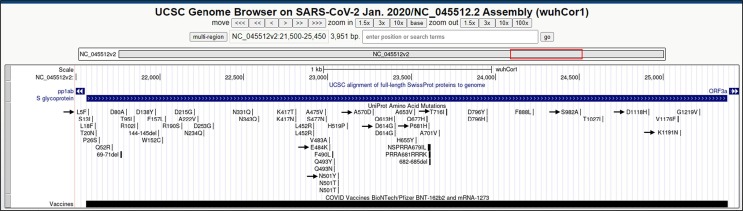
Fig. 6.
Mutations in Surface glycoprotein gene: Illustration of location of the identified amino acid mutations in Surface glycoprotein (Spike protein) gene. The identified mutations are indicated with black arrows.
4. Discussion
In the present study, we sought to analyze 3,970 SARS-CoV2 genome sequence samples submitted to the NCBI from Georgia State of the United States. The analyses revealed several interesting mutations in the nucleotide and amino acid sequences. The virus found in Wuhan (China) was designated as clade 19A. Within just a few weeks later, clade 19B emerged and it was hypothesized that mutations in the spike protein occurred for survival advantages. Herein, we identified the spike protein amino acid mutation, D614G, in >98% of all the samples (Table 2 A), which caused by a nucleotide mutation, A23403G, in the Wuhan original strain (Korber et al., 2020). It has recently been demonstrated that the virus with the D614G mutation transmitted faster than the wild type and increases in infectivity and fatality (Korber et al., 2020, Trucchi et al., 2021). This variant was dominant during the early COVID-19 pandemic and was highly prevalent in Southeast England (Arora et al., 2021). Three other mutations, viz., C241T (5′ UTR region), C3037T (F106F, synonymous mutation), and C14408T, were almost always accompanied by D614G (A2303G) (Korber et al., 2020) (Fig. 4). The C14408T mutation in the ORF 1b (Fig. 5 B) resulted an amino acid change, P314L, near the potential docking sites of viral RNA-dependent RNA polymerase (RdRp) (Mishra et al., 2021). The strains with D614G and P314L mutations were dominant variant in Europe and to some extent in the U.S. during the early COVID-19 pandemic (Eskier et al., 2020).(Lubinski et al., 2022) reported that P681H mutation in the S-protein may slightly increase the S1/S2 cleavage while not showing high impact viral entry or cell–cell spread. However, another mutation in the S-protein, N501Y, improved viral affinity for cellular receptor ACE2 and provided a major adaptive spike mutation in variants from Brazil and South Africa (Tian et al., 2021, Liu et al., 2022). The mutations N501Y, S477N, N439K, D364Y, and E484K were responsible for higher transmissibility of the virus compared with original strain (Thakur et al., 2022). Similarly, mutations P681H and D1118H in the S-protein, increases the infectivity and transmissibility of the virus (Farkas et al., 2021, Li et al., 2021). We did not identify any unique mutations in the samples from Georgia state, but the analyses of the data revealed three of the top ten mutations (K1191N, E484K, and L5F) in the S-protein of SARS-CoV-2 in the samples from Georgia state are different as compared to the samples from entirety of the United States of America ( Table 3 ). Interestingly, the mutation K1191N is not listed in the top 10 mutations of the S-protein in the samples from any other country (e.g., UK, China, India, Japan, and South Africa, etc.) listed in Table 3. Interestingly, the mutation K1191N in not listed in the top 10 mutations in the S-protein in the samples from any other country (e.g., UK, China, India, Japan, South Africa etc.) listed in the Table 3. It is reported that the mutations K1191N and L5F are in sites recognized by Human Leukocyte Antigen (HLA) (Timmers et al., 2021). The mutation P681H (C23604A), detected in 53% samples and located in the spike protein, was reported in the variants from Hawaii (USA), England, and Nigeria (Maison et al., 2021).
Table 3.
Comparison of top 10 mutations in amino acid of S-protein of SARS-CoV-2. Three mutations (indicated in Bold) are unique in the samples from Georgia state as compared to the samples from the entirety of the United State of America. The mutation K1191N in not listed in the top 10 mutations from samples from any country listed in the table. All data in this table are taken from ADDIN EN.CITE (Negi et al., 2022).
| Country |
Mutation frequency in Amino Acid of S-Protein (in decreasing order) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Georgia State (USA)* | D614G | P681H | N501Y | T716I | D1118H | A570D | S982A | K1191N | E484K | L5F |
| USA | D614G | P681H | N501Y | T716I | A570D | D1118H | S982A | L452R | W152C | S13I |
| UK | D614G | P681H | N501Y | T716I | A570D | S982A | D1118H | A222V | L18F | L5F |
| France | D614G | N501Y | T716I | P681H | A570D | S982A | D1118H | S477N | A222V | E484K |
| Germany | D614G | N501Y | P681H | S982A | T716I | D1118H | A570D | A222V | L18F | S98F |
| Spain | D614G | P681H | T716I | N501Y | A570D | S982A | D1118H | A222V | D138Y | L18F |
| Italy | D614G | N501Y | P681H | T716I | D1118H | S982A | A570D | A222V | P272L | A262S |
| China | D614G | S12F | H49V | M153T | S50L | A688N | D1084E | Q498H | V1228I | F32S |
| India | D614G | P681R | E484Q | L452R | N440K | G142D | Q1071H | E154K | N501Y | Q677H |
| South Korea | D614G | N501Y | P681H | T716I | D1118H | A570D | S982A | L452R | S13I | W152C |
| Japan | D614G | M153T | Q675H | E484K | W152L | G769V | P681H | L54F | Q677H | G184S |
| Brazil | D614G | V1176F | E484K | N501Y | L18F | H655Y | P26S | D138Y | T20N | T1027I |
| South Africa | D614G | A701V | E484K | D80A | K417N | N501Y | D215G | L18F | R246I | A688V |
| All | D614G | P681H | N501Y | T716I | D1118H | A570D | S982A | A222V | L18F | S477N |
* Data from current study.
Our analyses revealed 55% samples with mutations R203K (G28881A) and G204R (G28883C) on the SARS-CoV-2 nucleocapsid (N) protein (Table 2 B), which had a similar frequency (58%) reported from Oceania continent (Wu et al., 2021). It is reported that co-occurring of R203K/G204R mutations were associated with the appearance of the high-transmissibility variant 20I (Alpha, V1 or B.1.1.7) (Wu et al., 2021). However, the R203K/G204R mutations were reported in 25% cases from North America, 22% in Asia and Africa, and 87–89% from Europe and Latin America (Wu et al., 2021). The mutations R203K/G204R, Q57H, and G251V in ORF3a and S194L in N-protein caused the changes in protein structure and affected the binding affinity of intra-viral protein interactions and increases viral replication by increasing nucleocapsid phosphorylation which confers resistance to inhibition of the GSK-3 kinase (Wu et al., 2021, Johnson et al., 2022). Our data indicates that most dominant clade was 20I (Alpha, V1 or B.1.1.7) (Fig. 2) and it was first discovered in mid-December of 2020. It originated from Southeast England and was subsequently reported from numerous other countries around the world, including the U.S. The 20I variant contains 23 mutations with 17 amino acid changes (Abdool Karim and de Oliveira, 2021). The mutations T265I (C1059T; ORF1a) and Q57H (Q25563G; ORF3a) most of the time occurred together and these two mutations were first detected in Singapore in February of 2020 (Wang et al., 2021). This pair of mutations were identified in 37% of the samples in this study. (Fig. 5 A and 5D).
The ORF8 is involved in viral evolution (alter the expression of surface MHC-Ι expression) and pathogenesis (Flower et al., 2021, Zhang et al., 2021). Three hotspot mutations identified in ORF8, Q27* (Stop) (C27972T), R52I (G28048T) and Y73C (A28111G), were also associated with the 20I variant. (Zekri et al., 2021). The G172V mutation of the ORF3a possibly contributes to the stabilization of the β-barrel by increasing the hydrophobic interactions while decreasing local flexibility (Bianchi et al., 2021). The mutations of ORG8 e.g., Q27*, R52I, Y73C, and K68*, increase the infectivity and transmissibility of the virus (Farkas et al., 2021, Li et al., 2021).
Our data shows that the second most populous clade was 20G with an occurrence rate of 24% (Fig. 2). This clade was originated from the United States itself and evolved from clade 20C (derived from 20A bearing ORF3a, Q57H and ORF1a, T265I). The clade 20A (basal pandemic lineage bearing D614G) was the third most common with an occurrence rate of 5.5%. This clade was a very dominant variant in the Western Europe right after the 20E variant.
We also identified 20C and 20B clades being present in the samples analyzed. The clade 20C primarily originated from 10 different countries across Europe. However, clade 20B was reported to be mainly from Northern European countries such as Norway, Denmark, and Sweden. Some reports showed that clade 20B had also arised from Africa. Our analyses also revealed the presence of clade 21F (Epsilon), 21C (Iota), 20 J (Gamma), 20H (Beta), and 21D (Eta) but the occurrence rate of these clades was relatively low (0.3–5.0%) in our samples (Fig. 2).
Thus, looking at the three most common variants in Georgia state of the United States, we could determine the primary locations of transmission or origin for the virus. We conclude that the majority of cases originated from Southeast England (Clade 20I), the U.S. itself (Clade 20G), and from Western Europe (Clade 20C). We did not identify any region-specific mutation but identified three of the top ten mutations (K1191N, E484K, and L5F) in the S-protein of SARS-CoV-2 in the samples from Georgia state are different as compared to the samples from entirety of the United States of America.
Authors contributions
WA, RB conceived and designed the study and have diligently contributed to its preparation, including the literature search, concept organization, data collection & interpretation, and writings. SA also collaborated with valuable inputs in research guidance, data interpretation, and manuscript writing. All authors approved the final draft for publication.
Funding
RB is supported by National Institutes of Health (NIH) grant (NIH/NIMHD # 1S21MD012472).
Credit author statement
Waqas Ahmad, Riyaz Basha conceived and designed the study and have diligently contributed to its preparation, including the literature search, concept organization, data collection & interpretation, and writings. Sarfraz Ahmad also collaborated with valuable inputs in research guidance, data interpretation, and manuscript writing. All authors approved the final draft for publication.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Edited by Dr. X. Carette
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gene.2022.146774.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications. N. Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksamentov, I., Roemer, C., Hodcroft, E.B. and Neher, R.A., 2021. Nextclade: clade assignment, mutation calling and quality control for viral genomes.
- Arora P., Pohlmann S., Hoffmann M. Mutation D614G increases SARS-CoV-2 transmission. Signal. Transduct. Target. Ther. 2021;6:101. doi: 10.1038/s41392-021-00502-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badua C., Baldo K.A.T., Medina P.M.B. Genomic and proteomic mutation landscapes of SARS-CoV-2. J. Med. Virol. 2021;93:1702–1721. doi: 10.1002/jmv.26548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Borsetti A., Ciccozzi M., Pascarella S. SARS-Cov-2 ORF3a: Mutability and function. Int. J. Biol. Macromol. 2021;170:820–826. doi: 10.1016/j.ijbiomac.2020.12.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos W.G. Impact of virus genetic variability and host immunity for the success of COVID-19 vaccines. Biomed. Pharmacother. 2021;136 doi: 10.1016/j.biopha.2021.111272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskier D., Suner A., Karakulah G., Oktay Y. Mutation density changes in SARS-CoV-2 are related to the pandemic stage but to a lesser extent in the dominant strain with mutations in spike and RdRp. PeerJ. 2020;8 doi: 10.7717/peerj.9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas, C., Mella, A., Turgeon, M. and Haigh, J.J., 2021. A Novel SARS-CoV-2 Viral Sequence Bioinformatic Pipeline Has Found Genetic Evidence That the Viral 3′ Untranslated Region (UTR) Is Evolving and Generating Increased Viral Diversity. Front. Microbiol. 12. [DOI] [PMC free article] [PubMed]
- Flower, T.G., Buffalo, C.Z., Hooy, R.M., Allaire, M., Ren, X. and Hurley, J.H., 2021. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci U S A 118. [DOI] [PMC free article] [PubMed]
- Johnson B.A., Zhou Y., Lokugamage K.G., Vu M.N., Bopp N., Crocquet-Valdes P.A., Kalveram B., Schindewolf C., Liu Y., Scharton D., Plante J.A., Xie X., Aguilar P., Weaver S.C., Shi P.Y., Walker D.H., Routh A.L., Plante K.S., Menachery V.D. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.-G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182(812–827) doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Hodcroft E.B. Genetic Variants of SARS-CoV-2-What Do They Mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang L., Chen S., Ji W., Li C., Ren L. Recent progress on the mutations of SARS-CoV-2 spike protein and suggestions for prevention and controlling of the pandemic. Infect. Genet. Evol. 2021;93 doi: 10.1016/j.meegid.2021.104971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., Widen S.G., Menachery V.D., Shi P.Y., Weaver S.C. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2022;602:294–299. doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski B., Fernandes M.H.V., Frazier L., Tang T., Daniel S., Diel D.G., Jaimes J.A., Whittaker G.R. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience. 2022;25 doi: 10.1016/j.isci.2021.103589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison D.P., Ching L.L., Shikuma C.M., Nerurkar V.R. Genetic Characteristics and Phylogeny of 969-bp S Gene Sequence of SARS-CoV-2 from Hawai'i Reveals the Worldwide Emerging P681H Mutation. Hawaii. J. Health. Soc. Welf. 2021;80:52–61. [PMC free article] [PubMed] [Google Scholar]
- Mengist H.M., Kombe Kombe A.J., Mekonnen D., Abebaw A., Getachew M., Jin T. Mutations of SARS-CoV-2 spike protein: Implications on immune evasion and vaccine-induced immunity. Semin. Immunol. 2021;55 doi: 10.1016/j.smim.2021.101533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra D., Suri G.S., Kaur G., Tiwari M. Comparative insight into the genomic landscape of SARS-CoV-2 and identification of mutations associated with the origin of infection and diversity. J. Med. Virol. 2021;93:2406–2419. doi: 10.1002/jmv.26744. [DOI] [PubMed] [Google Scholar]
- Nagy A., Pongor S., Gyorffy B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int. J. Antimicrob. Agents. 2021;57 doi: 10.1016/j.ijantimicag.2020.106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S.S., Schein C.H., Braun W. Regional and temporal coordinated mutation patterns in SARS-CoV-2 spike protein revealed by a clustering and network analysis. Sci. Rep. 2022;12:1128. doi: 10.1038/s41598-022-04950-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, S., Sasi, S., Pillai, S.G., Nag, A., Shukla, D., Singhal, R., Phalke, S. and Velu, G.S.K., 2022. SARS-CoV-2 Mutations and Their Impact on Diagnostics, Therapeutics and Vaccines. Front. Med. 9. [DOI] [PMC free article] [PubMed]
- Tian F., Tong B., Sun L., Shi S., Zheng B., Wang Z., Dong X., Zheng P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife. 2021;10 doi: 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers L., Peixoto J.V., Ducati R.G., Bachega J.F.R., de Mattos Pereira L., Caceres R.A., Majolo F., da Silva G.L., Anton D.B., Dellagostin O.A., Henriques J.A.P., Xavier L.L., Goettert M.I., Laufer S. SARS-CoV-2 mutations in Brazil: from genomics to putative clinical conditions. Sci. Rep. 2021;11:11998. doi: 10.1038/s41598-021-91585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucchi E., Gratton P., Mafessoni F., Motta S., Cicconardi F., Mancia F., Bertorelle G., D'Annessa I., Di Marino D. Population Dynamics and Structural Effects at Short and Long Range Support the Hypothesis of the Selective Advantage of the G614 SARS-CoV-2 Spike Variant. Mol. Biol. Evol. 2021;38:1966–1979. doi: 10.1093/molbev/msaa337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chen J., Gao K., Hozumi Y., Yin C., Wei G.W. Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Commun. Biol. 2021;4:228. doi: 10.1038/s42003-021-01754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., Mansouri K., Gobeil S., McDanal C., Pardi N., Hengartner N., Lin P.J.C., Tam Y., Shaw P.A., Lewis M.G., Boesler C., Sahin U., Acharya P., Haynes B.F., Korber B., Montefiori D.C. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell. Host. Microbe. 2021;29(23–31) doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Xing N., Meng K., Fu B., Xue W., Dong P., Tang W., Xiao Y., Liu G., Luo H., Zhu W., Lin X., Meng G., Zhu Z. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell. Host. Microbe. 2021;29(1788–1801) doi: 10.1016/j.chom.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri A.N., Bahnasy A.A., Hafez M.M., Hassan Z.K., Ahmed O.S., Soliman H.K., El-Sisi E.R., Dine M., Solimane M.S., Latife L.S.A., Seadawy M.G., Elsafty A.S., Abouelhoda M. Characterization of the SARS-CoV-2 genomes in Egypt in first and second waves of infection. Sci. Rep. 2021;11:21632. doi: 10.1038/s41598-021-99014-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Chen, Y., Li, Y., Huang, F., Luo, B., Yuan, Y., Xia, B., Ma, X., Yang, T., Yu, F., Liu, J., Liu, B., Song, Z., Chen, J., Yan, S., Wu, L., Pan, T., Zhang, X., Li, R., Huang, W., He, X., Xiao, F., Zhang, J. and Zhang, H., 2021. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota. Proc. Natl. Acad. Sci. USA 118. [DOI] [PMC free article] [PubMed]
- Zhao, L.P., Lybrand, T.P., Gilbert, P.B., Hawn, T.R., Schiffer, J.T., Stamatatos, L., Payne, T.H., Carpp, L.N., Geraghty, D.E. and Jerome, K.R., 2021. Tracking SARS-CoV-2 Spike Protein Mutations in the United States (January 2020-March 2021) Using a Statistical Learning Strategy. Viruses 14. [DOI] [PMC free article] [PubMed]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G.F., Tan, W., China Novel Coronavirus, I. and Research, T., 2020. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl. J. Med. 382, 727-733. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.