Summary
Background
In 2021, Delta became the predominant SARS-CoV-2 variant worldwide. While vaccines have effectively prevented COVID-19 hospitalization and death, vaccine breakthrough infections increasingly occurred. The precise role of clinical and genomic determinants in Delta infections is not known, and whether they contributed to increased rates of breakthrough infections compared to unvaccinated controls.
Methods
We studied SARS-CoV-2 variant distribution, dynamics, and adaptive selection over time in relation to vaccine status, phylogenetic relatedness of viruses, full genome mutation profiles, and associated clinical and demographic parameters.
Findings
We show a steep and near-complete replacement of circulating variants with Delta between May and August 2021 in metropolitan New York. We observed an increase of the Delta sublineage AY.25 (14% in vaccinated, 7% in unvaccinated), its spike mutation S112L, and AY.44 (8% in vaccinated, 2% in unvaccinated) with its nsp12 mutation F192V in breakthroughs. Delta infections were associated with younger age and lower hospitalization rates than Alpha. Delta breakthrough infections increased significantly with time since vaccination, and, after adjusting for confounders, they rose at similar rates as in unvaccinated individuals.
Interpretation
We observed a modest adaptation of Delta genomes in breakthrough infections in New York, suggesting an improved genomic framework to support Delta's epidemic growth in times of waning vaccine protection despite limited impact on vaccine escape.
Funding
The study was supported by NYU institutional funds. The NYULH Genome Technology Center is partially supported by the Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center.
Keywords: SARS-CoV-2 variant of concern (VOC) delta, Genomic signatures of vaccine breakthrough, Spike S112L and nsp12 F192V, Selective adaptation, Time since vaccination, Network analysis of clinical variables
Research in context.
Evidence before this study
References for this Original Research article were identified through searches of PubMed, medRxiv, and bioRxiv for SARS-CoV-2 articles published from December 2019 to April 2022, and for general articles in virology and vaccinology without date restriction, by use of the terms “SARS-CoV-2”, “COVID-19”, “vaccination”, “immunization”, “viral evolution”, “epidemiology”, “mutations”, “vaccine escape”, and “breakthrough”. Articles published in English, Spanish, French, and German were included in our search. A few months before the completion of this study, Delta has started to become the predominant SARS-CoV-2 variant in the world causing a new wave of COVID-19 cases that involved both unvaccinated and vaccinated individuals. In 2022, Delta is still circulating globally despite the recent surge of the Omicron variant. A comprehensive view on clinical and genomic determinants of Delta infections relative to vaccine status is lacking.
Added value of this study
Here, we added a comprehensive assessment of clinical and genomic characteristics of SARS-CoV-2 infections in metropolitan New York at a time when Delta nearly completely replaced other circulating variants. Fine-scale variant and mutation analysis included the statistical comparison of full genome base pair and amino acid mutations in unvaccinated and vaccinated individuals. It revealed an increase of the Delta sublineage AY.25 over time, and a slight but significant enrichment of spike mutation S112L as well as nsp12 mutation F192V in breakthrough infections.
Compared with other variants, Delta infections were associated with a specific network of clinical and demographic variables, i.e., they occurred later in 2021 and more likely involved younger individuals, all of which related to lower hospitalization rates. Notably, Delta breakthroughs increased significantly with time since vaccination.
Implications of all the available evidence
After adjusting for confounding variables, we observed that the frequency of Delta infections rose at a similar pace in unvaccinated and vaccinated individuals, which indicates that Delta and its mutations had a limited impact on vaccine escape when compared with other co-circulating variants. The low percentage of vaccine breakthrough infections compared to the total number of vaccinated individuals at NYU and New York City-wide during the time of Delta predominance underline the effectiveness of COVID-19 vaccines against infection with Delta and previous variants. Nevertheless, we found subtle but significant differences in the SARS-CoV-2 mutation profiles in breakthrough infections compared with unvaccinated controls including two enriched and seven decreased mutations, suggesting that selective adaptation was in process that might have played a supportive role. The significant increase of Delta infections relative to time since vaccination highlights Delta's improved potential of epidemic growth in times of waning vaccine protection, backing the implementation of booster vaccinations in the broader population. Emerging SARS-CoV-2 variants will require close monitoring and should include mutation hotspot regions such as the spike N-terminal domain with the S112L mutation as well as nsp12 mutations such as F192V that we found in increased numbers among vaccine breakthroughs.
Alt-text: Unlabelled box
Introduction
The SARS-CoV-2 pandemic has been a showcase for observing viral evolution in real time. New variants emerged in various parts of the globe and caused waves of infection that reached different countries in rapid succession. Variants being monitored (VBM), particularly Alpha, Beta, and Gamma, and variant of concerns (VOC) Delta and, most recently, Omicron emerged and subsequently increased despite worldwide vaccination efforts.1, 2, 3, 4, 5, 6 In the United States, vaccinations started in late December 2020, with three vaccines currently employed: two mRNA-based vaccines, BNT162b2 (Pfizer/BioNTech), now FDA-approved, mRNA-1273 (Moderna), and the adenovirus-based Janssen COVID-19 vaccine, JNJ-78436735.7, 8, 9 All three vaccines utilize the spike sequence of SARS-CoV-2 Wuhan-Hu-1 isolated in January 2020,10,11 and thus their epitopes are not perfectly matched to those of currently circulating variants. Despite this mismatch, the vaccines are highly effective at preventing symptomatic disease, hospitalization, death, and forward transmission.12, 13, 14 However, post-vaccination infections (vaccine breakthroughs) do occur. The epidemiological data on whether these breakthroughs are driven by properties inherent to specific variants are controversial. Two studies earlier in 2021 indicated increased breakthrough rates of the Beta or Gamma variants following two doses of mRNA vaccines.15,16 In contrast, other studies, including our own, found that breakthrough cases had a similar variant distribution as in unvaccinated individuals in the respective cohort at that specific time.17, 18, 19, 20
In 2021, Delta became the predominant variant globally and it remains widespread in 2022. Data from the CDC indicate that coincident with the rise of Delta in the US, vaccine effectiveness against infection decreased from 91% to 66%, which is consistent with waning immunity observed in Israel and Qatar.21, 22, 23 Delta has been associated with higher replication and transmission,24,25 and in vitro studies report slightly decreased neutralizing efficacy of monoclonal antibodies, convalescent sera or sera from vaccinated individuals against Delta, suggesting that some mutations present in Delta may cause partial immune escape.26, 27, 28 Clinical and full SARS-CoV-2 genome data from post-vaccination infections in the era of Delta are still scarce29 but will be key to revealing features critical for vaccine effectiveness/escape and to identify commonalities and differences with the upcoming Omicron variant and other future VOCs. They complement in vitro and in vivo studies on immune escape to identify viral genomic changes and their impact on vaccine escape in a real-world situation, including clinical and demographic aspects in a study population. Here, we determined the SARS-CoV-2 genetic makeup of 132 post-vaccination infections and their associated clinical characteristics in fully vaccinated individuals within NYU Langone Health (NYULH), a large metropolitan New York healthcare system with hospitals across the region. We assessed the probability of Delta to result in breakthrough infection relative to other variants between May and August 2021.
Methods
Study design and sample collection
This study was approved by the NYULH Institutional Review Board, protocol numbers i21-00493 and i21-00561. SARS-CoV-2 infections were studied in vaccinated and unvaccinated individuals from May 1st until August 3rd in the NYULH system. Cases were identified using DataCore, the system's clinical data management and extraction resource. Vaccine breakthroughs were defined by positive real-time (RT)-PCR test for SARS-CoV-2 RNA regardless of Ct at least 14 days after the second dose of BNT162b2 (Pfizer/BioNTech) or mRNA-1273 (Moderna) vaccines, or the single-dose COVID-19 Janssen vaccine. The unvaccinated control group consisted of SARS-CoV-2 positive cases in our healthcare system who had not received a dose of any vaccine at the time of RT-PCR positivity, collected and sequenced in the same period as the breakthrough infections. We sequenced all unvaccinated positive cases with Ct < 38 up to our maximum sequencing capacity of 94 cases per week. Nasopharyngeal swabs were sampled from individuals with exposure to SARS-CoV-2, suspected to have an infection with SARS-CoV-2, or as part of clinical diagnostics or hospital admission.
RNA extraction, library preparation and sequencing
RNA was extracted from nasopharyngeal swab specimens using the MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit. Libraries were prepared using the Swift Normalase Amplicon SARS-CoV-2 Panel (SNAP) and run on the Illumina NovaSeq 6000 system on SP 300 cycle flow cells17 (Supplemental Methods). SARS-CoV-2 sequences that were <23,000 bp or <4000x genome coverage were considered inadequate and not included in the analyses.
Phylogenetic, mutation, and adaptive evolution analyses
Maximum likelihood IQ trees were produced on Mafft-aligned SARS-CoV-2 full genome sequences using 1000 bootstrap replicates.17 Spike amino acid counts and site-specific mutation frequencies were analysed using Fisher exact tests with multiplicity corrections (Benjamini-Hochberg).30 Estimates of the proportion of SARS-CoV-2 sequences with a specific mutation through time were generated using covSpectrum31 with data from GISAID.3 Adaptive evolution in a coding gene was tested using a fast, unconstrained Bayesian approximation for inferring selection (FUBAR), provided by Datamonkey32,33 (Supplemental Methods).
Statistics
The comparison of 132 breakthrough and 283 unvaccinated control samples achieved 96% power in detecting a 15% difference (10% versus 25%) in mutation rates, clinical or demographic variables in a two-tailed chi squared/Fisher Exact test with a type I error of 5% (G*Power v.3.1.9.4).34 We evaluated the variant distribution (Alpha, Delta, Gamma, Iota, and all other variants) in fully vaccinated compared to unvaccinated individuals and addressed confounding variables arising from the use of observational data via matching (chi squared/Fisher Exact tests) and adjustment (logistic regression analyses). We matched the vaccinated breakthrough cases 1:1 to the unvaccinated cases, correcting for the confounding variables: clinical collection date, sex, and age.35 Propensity-score matching was implemented using the nearest neighbour strategy and quality controls of matching included the analysis of propensity score distributions and empirical quantile-quantile (eQQ) plots of distribution balance of co-variates using the R v.4.1.0 MatchIt package. To compare the probability of a variant/mutation for the vaccinated and unvaccinated groups over time, we performed logistic regression analyses on the full data adjusting for sex, age (centred and standardized), and month of test. The relationship between variant distribution and time since vaccination was studied using linear regression analyses in Prism v.8.4.3 using two-sided Pearson tests. We compared clinical and demographic variables between groups using non-parametric Mann-Whitney tests or Kruskal-Wallis tests with Dunn's multiplicity correction in Prism. Correlation analyses were done using two-sided Spearman rank tests in Prism. For all analyses unless otherwise stated, a two-sided Type I error rate of 0.05 was applied. Details are described in Supplemental Methods.
Ethics/Study approval
Ethics approval and informed patient consent were not required on unidentifiable viral sequences. This study was approved by the NYULH Institutional Review Board, protocol numbers i21-00493 and i21-00561.
Role of funders
The funders played no role in the design, conduct, interpretation, or reporting of this study.
Results
Demographic parameters in our cohort of vaccinated and unvaccinated SARS-CoV-2-infected participants
Between May 1st and August 3rd, 2021, a total of n=1613 SARS-CoV-2 infections were recorded in our multicentre healthcare system. The majority of SARS-CoV-2-positive tests (82%) were from unvaccinated individuals (n=1297), whereas 18% were from vaccinated (n=297) among a total of 168,127 fully vaccinated individuals within our system (Table 1). We sequenced all breakthrough cases with available specimens and randomly selected as many specimens from unvaccinated individuals as allowed by our internal weekly processing capacity (up to 96), irrespective of demographic or clinical parameters. We obtained high quality SARS-CoV-2 sequences from 132 vaccinated and 283 unvaccinated individuals. The median age (37 versus 42 years) and sex distribution were similar in both groups. The majority of recorded breakthrough infections (63%) occurred at >120 days after full vaccination with a median of 136 days after vaccination (Table 1).
Table 1.
Demographic and clinical data and SARS-CoV-2 lineages of fully vaccinated vs non-vaccinated COVID-19 cases with full SARS-CoV-2 genomes.
| Fully Vaccinated | Unvaccinated | |
|---|---|---|
| Age | ||
| Median | 37 | 42 |
| Range | 22-90 | <1-96 |
| Age Groups* | ||
| <20 | 0 | 39 (13.8) |
| 20-29 | 31 (23.5) | 45 (15.9) |
| 30-39 | 42 (31.8) | 49 (17.3) |
| 40-49 | 13 (9.8) | 31 (10.9) |
| 50-59 | 19 (14.4) | 45 (15.9) |
| 60-69 | 13 (9.8) | 31 (10.9) |
| 70-79 | 8 (6.1) | 18 (6.4) |
| 80-89 | 5 (3.8) | 21 (7.4) |
| >90 | 1 (0.7) | 4 (1.4) |
| Sex | ||
| Male | 67 | 133 |
| Female | 65 | 155 |
| Vaccine Type | ||
| Pfizer | 115 | Not applicable |
| Moderna | 14 | Not applicable |
| Janssen | 3 | Not applicable |
| Days after vaccination | ||
| Median | 136 | Not applicable |
| Range | 21-221 | |
| <120 | 49 | Not applicable |
| >120 | 83 | Not applicable |
| Hospitalized | 8 (6) | 83 (29) |
| Symptomatic | 72 (54.5) | 207 (84.1)a |
| SARS-CoV-2 variant | ||
| Alpha | 13 (9.8) | 68 (24.0) |
| Iota | 7 (5.3) | 35 (12.4) |
| Gamma | 4 (3.0) | 19 (6.7) |
| Delta | 101 (76.5) | 139 (49.1) |
| Other | 7 (5.3) | 22 (7.8) |
| Total Cases | 132 | 283 |
All figures are absolute numbers except numbers in parenthesis are % from total cases.
information available for 246/283 unvaccinated individuals.
Delta infections were associated with a distinct network of clinical and demographic predictors
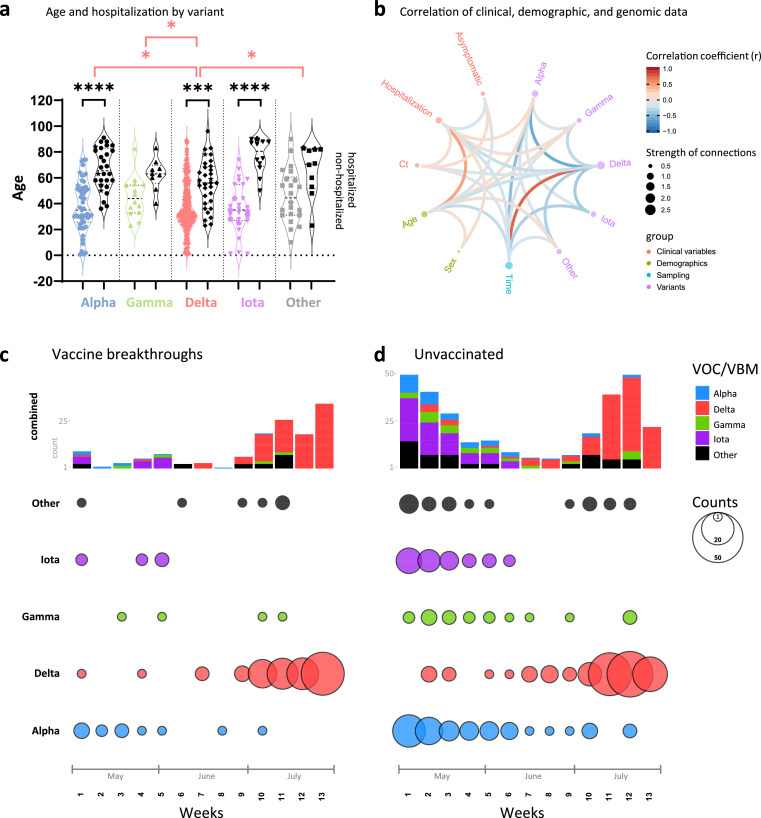
More than 45% of the breakthrough infections in our cohort were asymptomatic (n=60); only 6% (n=8) were hospitalized, all of whom were >50 years (5/8 >70 years) and had co-morbidities. There was one death from metastatic cancer (Table S1, Figures S1, S2). The hospitalization rate was significantly higher in the unvaccinated (29%, n=83, P<0.0001; Fisher Exact test); for 66 of those, COVID-19 was the main reason for hospitalization, 10 required ICU admission, and seven died from COVID-19 (Table S2). To identify differential features of the circulating variants, we compared their clinical and demographic characteristics (Figures 1a, b, S1-S3). Among the SARS-CoV-2-positive cases studied, hospitalizations due to Delta were significantly lower compared with Alpha, Gamma, Iota, and the other variants combined, and significance was maintained against Alpha when assessed separately in vaccinated and unvaccinated individuals (Figure S1; Fisher Exact test). Among Delta infections, we recorded only one COVID-19 related death (0.7%), whereas Alpha had the highest rates with at least four COVID-19-related deaths (5.9%) (Tables S1, S2). Delta affected younger individuals, particularly when compared to Alpha (Kruskal-Wallis test). Hospitalizations were associated with older age, specifically in Alpha, Delta, and Iota infections. There was a trend to lower cycle threshold (Ct) values in Delta infections, which translated into a weak but significant inverse correlation between Delta and Ct values in the overall data set and among breakthrough infections (Figures 1b, S3a; Spearman Rank). The Ct values did not differ in hospitalized versus non-hospitalized. In all, Delta infections exhibited a characteristic clinical profile regarding age (younger), time of infection (later in the year) and hospitalization rate (lower), all of which significantly correlated with each other and might be mutual drivers (Figure 1b).
Figure 1.
Relationship of clinical, demographic, and genomic data. a, Violin plot summarizing the age distribution of the combined data set of 132 vaccinated and 283 unvaccinated SARS-CoV-2-positive individuals by variant. The pairs of coloured and black violins show non-hospitalized versus hospitalized cases per variant. Horizontal lines indicate the median and interquartile ranges of values. Breakthrough cases are shown as stars. Statistical comparisons of age were made using Kruskal-Wallis tests between variants (red brackets) and, for each variant, between non-hospitalized and hospitalized cases (black brackets). All statistically significant results are shown: * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. b, Correlation analysis of clinical, demographic, and genomic data of SARS-CoV-2 infected individuals (as in A). Red and blue edges represent positive and negative correlations between connected variables, respectively, according to the scale of r values to the right. Only significant correlations (P<0.05, Spearman rank test) are displayed. Nodes are color-coded based on the grouping of variables. Node size corresponds to the strength of correlations. Ct: Cycle threshold in RT-PCR; sex: male sex; time: date of sampling. c, d, Distribution of variants (top) and absolute variant counts (bottom), coloured and separated into to the most frequently detected variants of concern (VOC) and being monitored (VBM) and all other variants combined (Other). Vaccine breakthrough infections (c) are shown side-by-side with unvaccinated controls (d) on a weekly basis starting May 1st 2021 (all full weeks shown).
Delta rapidly replaced Alpha and Iota variants in both unvaccinated and vaccine breakthrough cases recorded at NYULH
At the beginning of our study period (May 2021), several variants circulated in the New York metropolitan area, mainly Alpha, Iota and Gamma (Figure 1c, d). In June, we observed a nadir of case numbers (<10 per week), followed by a rise of Delta infections starting end of June that caused a dramatic increase in total cases. This rapid shift in variants towards Delta was detected in both vaccinated and unvaccinated individuals. By the end of our study, we saw a near complete replacement of the locally-originating Iota and the imported Alpha and Gamma variants by Delta.
Delta infections including breakthroughs exhibit a characteristic subvariant makeup
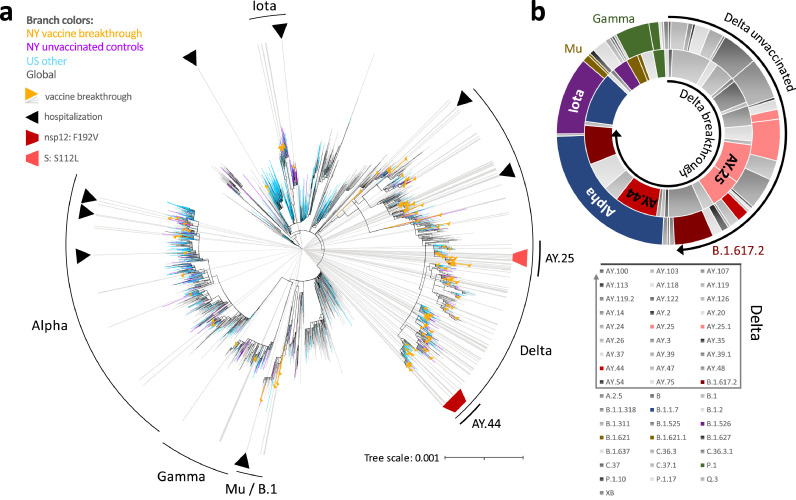
To determine the diversity and relatedness of SARS-CoV-2 genomes, we performed phylogenetic analyses including our 132 breakthrough and 283 unvaccinated cases, using US and global reference sequences (Figures 2a, S4 and Table S3). Our SARS-CoV-2 sequences covered a broad set of variants with predominance of Delta and its numerous subvariants, Alpha, Gamma, Iota, Mu, and additional B.1 lineages (Figure 2b). Delta sequences appeared more densely packed and clustered, raising the possibility of transmission chains (Figures 2A, S4). Two of these clusters consisted of the Delta subvariants AY.25 and AY.44 that included sequences with spike:S112L and nsp12:F192V mutations, respectively. We found no evidence of transmission clusters composed solely of breakthrough infections. Alpha (10% versus 24%) and Iota (3% versus 11%) were overall less frequently found among breakthrough infections compared to unvaccinated controls (odds ratios and 95% confidence intervals (95%CI) of 0.35 (0.19-0.66) and 0.25 (0.09-0.69), respectively), whereas Delta counts were more frequent over the study period (77% versus 49%)(odds ratio and 95%CI of 2.12 (1.34-3.36))(Figure 2b). The spike:S112L-positive AY.25 sub-cluster contained SARS-CoV-2 sequences from one unvaccinated and five vaccinated cases, with four cases occurring in New York County. The AY.44 cluster included eleven vaccinated and five unvaccinated cases, all of which carried the nsp12:F192V mutation (Figures 3, S4, S5).
Figure 2.
Phylogenetic analysis and variant distribution of SARS-CoV-2 vaccine breakthrough and unvaccinated control sequences. a, Maximum likelihood (IQ) tree of 3511 SARS-CoV-2 full genome sequences (base pairs 202-29,666 according to Wuhan-Hu-1 as reference), including 132 vaccine breakthrough (orange) and 283 unvaccinated control SARS-CoV-2 sequences from the NYU Langone Health cohort (greater NYC area) (purple) together with 920 other US (non-NYU; cyan) and 2176 global (non-US; black) reference sequences. The substitution scale of the tree, generated with 1000 bootstrap replicates and Wuhan/WH01/2019-12-26 as root, is indicated at the bottom right. Vaccine breakthrough sequences are highlighted by orange triangles (as branch symbols) and grey rays radiating from the root to the outer rim of the tree. Hospitalizations among vaccine breakthrough infections are indicated by black triangles. The variants responsible for most vaccine breakthrough infections are labelled. The Delta plus spike:S112L (AY.25) and nsp12:F192V (AY.44) sub-lineages are labelled and highlighted with light and carmine red trapezoid symbols, respectively. b, Double-donut plot to compare the variant distribution of breakthrough (inner ring) and unvaccinated control sequences (outer ring). The most abundant variants and Delta subvariants (highlighted by black arrows) are shown in colour and labelled in the plot (outer ring only). All detected variants (Pango lineages) and their colour code in the plot are shown below.
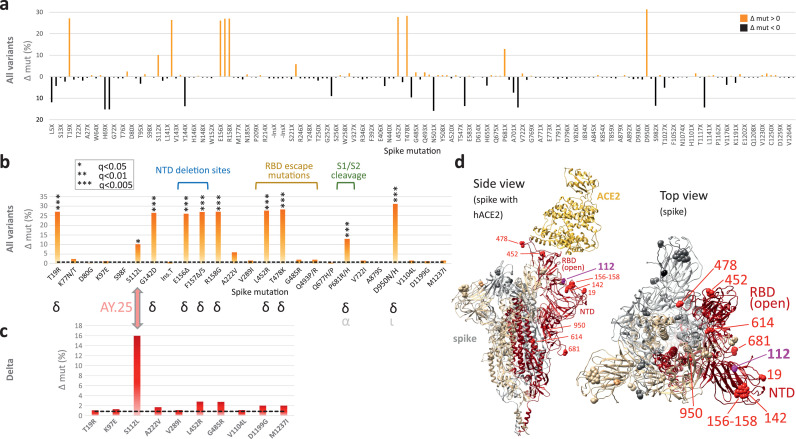
Figure 3.
Site-specific spike mutation analysis in SARS-CoV-2 vaccine breakthrough sequences compared to unvaccinated controls. a, Site-specific amino acid mutation (mut) frequencies in spike in 132 vaccine breakthrough sequences compared to 283 unvaccinated controls from the same cohort. The Wuhan-Hu-1 sequence served as reference. The mirror plot displays differences of mutation frequencies per spike residue between vaccinated and unvaccinated groups, shown along the x-axis (n=168); orange (facing up) and black bars (facing down) refer to elevated mutation frequencies in vaccinated or unvaccinated individuals, respectively. b, Enriched spike mutations in vaccine breakthrough sequences compared to unvaccinated controls. Unique occurrences of mutations in breakthrough cases were disregarded. The dashed black line indicates the average mutation frequency across all spike residues in the unvaccinated control data set (n=283) compared to Wuhan-Hu-1. Significantly enriched mutations in Fisher exact tests are indicated by asterisks (* P<0.05, *** P<0.005) and the variants in which these mutations were found are shown below (black: main source, grey: secondary source). Mutations in the spike N-terminal domain (NTD), receptor binding domain (RBD), and near the S1/S2 interface associated with neutralization escape and/or affecting important biological functions are labeled. c, The same analysis as in (b) but focusing on Delta sequences exclusively. 101 Delta vaccine breakthrough sequences were compared to 139 Delta unvaccinated controls. The dashed black line indicates the average mutation frequency across all spike residues in the Delta unvaccinated control data set compared to Wuhan-Hu-1 as reference (n=139). d, Structural analysis of mutation sites on a spike trimer bound to human ACE2 (hACE2) (pdb S_ACE2). Each protomer is coloured differently. The hACE2-bound protomer with the RBD in the “up” position is shown in red. Statistically enriched mutation sites in vaccine breakthroughs (according to b) are shown as spheres, labelled in one protomer in red (Delta) or pink (AY.25).
Potential sites of adaptive evolution of the Delta variant under vaccine pressure
To screen for genomic signatures of vaccine breakthrough and possible adaptive selection, we differentially assessed all base pair mutations over the full genome, all amino acid mutations across spike, and compared synonymous (dS) and non-synonymous (dN) substitution rates (Figures 3, 4, S6-S8, Tables S4-S9). The numbers of full genome and spike mutations were comparable in viruses obtained from vaccinated and unvaccinated individuals. Breakthrough sequences had on average 35.8 base pair mutations over the full genome (range: 24-44) and 9.7 amino acid mutations per spike (range: 3-14), compared to 35.6 base pair mutations/full genome (range: 19-56) and 9.6 amino acid mutations/spike (range: 3-15) in the unvaccinated. In spike, we found a total of 168 amino acid changes compared to Wuhan-Hu-1: 115 sites were more frequently mutated in unvaccinated controls, compared to 52 sites in breakthroughs (Figure 3a). To identify spike amino acid mutations that are enriched in breakthrough sequences, we performed a two-pronged approach: I) to investigate the enrichment of mutations across variants, we statistically analysed sequences from all variants combined with a post-hoc variant assessment to identify potential bias introduced by unequal variant distribution between groups (Figure 3b). II) We performed a Delta-specific mutation analysis to identify mutations enriched among Delta sequences and to confirm the results using all variants (Figure 3c). Among the 52 mutations preferentially found in breakthroughs, 10 significantly differed between breakthrough and unvaccinated cases (Figure 3b, Table S4; Fisher exact test). The post-hoc analysis revealed that nine of these 10 sites were Delta-defining mutations, whilst one, S112L, is not Delta-defining but contributes to the array of Delta subvariant AY.25 mutations. The comparative spike mutation analysis exclusive for the subset of Delta sequences confirmed S112L as the only substantially enriched mutation among Delta breakthroughs (P<0.05, Figure S5 and Table S5). The S112L mutation locates at the surface-exposed top part of NTD, one of the major antigenic regions of the spike protein (Figure 3d).36
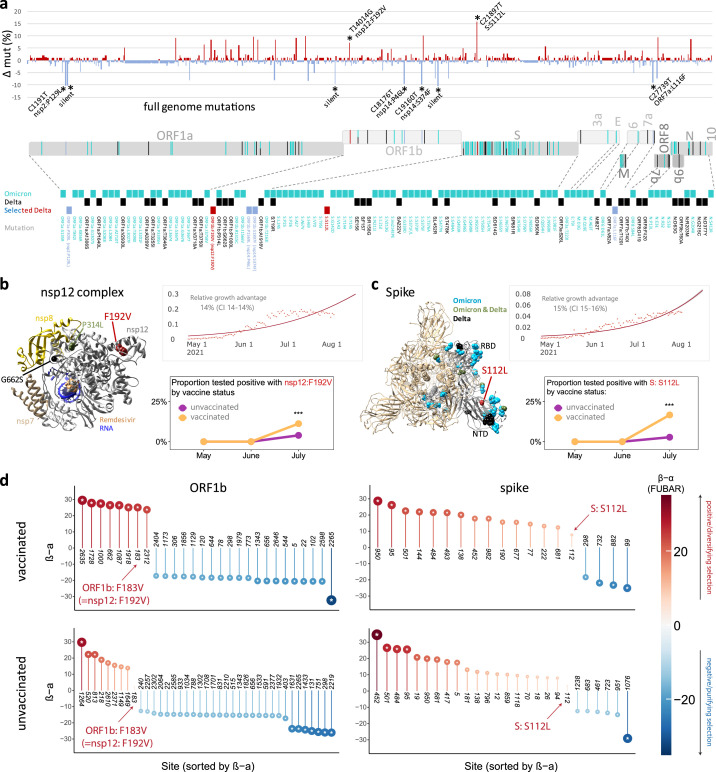
Figure 4.
Full genome mutation analysis, relative growth, and adaptive evolution of SARS-CoV-2 Delta vaccine breakthrough sequences and associated mutations compared to Delta unvaccinated controls.
a, Site-specific base pair mutation frequencies in full genomes (bp 202-29666) of 101 Delta vaccine breakthrough sequences compared to 139 Delta unvaccinated controls from the same cohort. The Wuhan-Hu-1 sequence served as reference. The mirror plot displays differences of mutation frequencies per site between vaccinated and unvaccinated groups, shown along the x-axis (n=791); red (facing up) and blue bars (facing down) refer to elevated mutation rates in vaccinated or unvaccinated individuals, respectively. Significantly enriched mutations in Fisher exact tests are indicated by asterisks (* P<0.05) and are labelled. SARS-CoV-2 coding genomic regions are shown below the plot. Non-synonymous mutations in Omicron (cyan; including B.1.1.529, BA.1, and BA.2 mutations), Delta (black), or Delta breakthrough-enriched mutations (red and blue) are shown by colored ticks. The mutation sites/names are indicated below. b, c, Structural analysis of Delta breakthrough-enriched mutations in comparison to Omicron- and Delta-defining mutations (briefly labelled as Omicron, Delta, or Omicron & Delta, the latter common in both variants). Structures are shown for the nsp12 complex (b) with bound nsp7, nsp8, template-primer RNA, and remdesivir triphosphate (pdb: 7bv2) and for spike (c) in the activated state with one RBD in the up position (pdb: S_ACE2; mutations only shown in the grey, activated protomer). Upper right: The estimated daily proportion of SARS-CoV-2 sequences with indicated mutation through time in the USA is shown as light red dots. The dark red line is the logistic fit. The provided relative growth estimate with confidence interval (CI) reflects the advantage compared to co-circulating strains if variants spread pre-dominantly by local transmission across demographic groups. Lower right: Probability of the detection of a mutation by month in vaccinated (n=132) and unvaccinated (n=283) individuals, adjusted for month of test, sex, and age of participants. *** P<0.001 in a chi-squared test. d, Adaptive evolution analysis of individual sites of a coding gene using a fast, unconstrained Bayesian approximation for inferring selection (FUBAR, Datamonkey), done for vaccinated (n=132) and unvaccinated (n=283) cases. Sites of interest (labelled) are studied in comparison to all sites with significant positive (red, facing up) and negative (blue, facing down) selection per gene. Posterior probabilities >0.9 are considered significant and are indicated by an asterisk inside the circles.
α: mean posterior synonymous mutation rate at a site; β: mean posterior non-synonymous mutation rate at a site; 3a: ORF3a; 7a: ORF7a; 7b: ORF7b; 9b: ORF9b; 10: ORF10; bp: base pairs; E: envelope; mut: mutation; nsp: non-structural protein; N: nucleocapsid; NTD: N-terminal domain; ORF: open reading frame; RBD: receptor-binding domain; S: spike.
To identify sites of potential adaptive evolution in Delta beyond spike, we extended our statistical analysis to the full genome (Figure 4a-d). Among 240 Delta sequences (101 breakthroughs and 139 unvaccinated controls), we observed 791 mutation sites, of which 448 were more frequently mutated in controls, 328 more frequent in breakthroughs, and 15 equally mutated in both groups. In total, we found nine sites with significantly different mutation rates between groups (Figure 4a, Table S6; Fisher exact test). Among the seven mutations that were more frequent in unvaccinated individuals, there were four non-synonymous mutations, i.e., nsp2 P129L, nsp14 P46L, nsp14 S374F, and ORF7a L116F, and three synonymous mutations, i.e., c1267t, t12946c, and a20262g. Two mutations were enriched in breakthrough sequences compared to unvaccinated controls, which, besides spike mutation S112L included mutation ORF1b:F183V that corresponds to F192V in nsp12, the RNA-dependent RNA polymerase (RdRp) gene (Figure 4b,c). In addition, seven mutations were less frequent in breakthrough sequences, which included four non-synonymous mutations in nsp2, nsp14 (2x), and ORF7a, and three synonymous mutations in nsp2, nsp9, and nsp15. A genomic region-specific mutation analysis further revealed that most coding regions had higher numbers of mutations in unvaccinated control sequences, whereas ORF3a (56%) and ORF8 (67%) had higher numbers of sites with enriched mutations in breakthrough sequences, though at low mutation rates (Table S7).
Mutations in Omicron and mutations enriched in Delta breakthroughs involve common genomic hot and cold spot regions
Omicron, first detected in October/November 2021 in Nigeria and Botswana, has caused a massive SARS-CoV-2 outbreak in South Africa starting November 2021, and eventually reached the US, Europe and many countries worldwide with rising numbers as of December 2021. While a zoonotic emergence of Omicron cannot be ruled out, Omicron supposedly evolved in an immunocompromised host and acquired a broad set of mutations associated with immune escape.4, 5, 6,37 Thus, both Omicron and Delta breakthrough viruses were presumably selected against immune selection pressure, which prompted us to perform a comparative analysis of their full-genome mutational landscapes including regions with high (hotspots) or low (coldspots) evolutionary mutation rates.38 Omicron is characterized by more than 30 mutations in spike, complemented by smaller mutation hotspot regions in ORF1ab, M, and N genes (Figure 4a). One of the two mutations that we found positively associated with Delta breakthrough (spike:S112L) is situated in the middle of the spike NTD domain, an Omicron hotspot region that carries abundant mutations and deletions (Figure 4a-c). The second breakthrough-enriched mutation in Delta (nsp12:F192V) is located in the functionally relevant nsp12 domain, coding for the RdRp polymerase, which also entails the Omicron- and/or Delta-defining mutations P314L and G662S (Figure 4a-c). In contrast, the seven sites that we identified as being inversely correlated with Delta breakthrough fell into mutation coldspots harbouring a low density of Omicron mutations. The inverse correlations suggest that these Delta mutations had no benefit in conferring breakthrough, or it is possible these mutations were deleterious for other Delta mutations enabling escape from immune pressure.
Increase of Delta subvariants AY.25 and AY.44, and evidence of diversifying selection involving AY.44’s nsp12:F192V mutation
Spike:S112L and nsp12:F192V (=ORF1b:F183V) are the clade-defining mutations for the Delta variants AY.25 and AY.44, respectively, which were most frequently detected in the USA (Figure 4b,c, S7). Both mutations have been strictly associated with Delta, thus they were hardly found before Delta and they subsequently disappeared with the decline and replacement of Delta by Omicron (Figure S7a,b). In our study period from May to August 2021, there was a significant growth advantage US-wide for viruses carrying either of these mutations (14–15%) compared to all other co-circulating strains, under the premise that variants spread predominantly by local transmission across demographic groups (Figure 4b,c). Furthermore, viruses with either spike:S112L or nsp12:F192V mutations were significantly overrepresented among breakthrough cases compared to unvaccinated controls by July 2021 (Figure 4b,c, S7c; covariate-adjusted logistic regression analysis). Bayesian analyses for adaptive selection based on dN-dS rates revealed a number of sites in ORF1b and spike with evidence (posterior probability >0.9) of positive or negative adaptive selection (Figure 4b,c, S8, Table S8, S9). These sites included Delta-defining sites, such as ORF1b:662, 1000, and 1918, and spike:19, 222, 452, 681, and 950, but also sites that have been associated with enhanced ACE2 binding or immune escape, such as spike:144, 501, and 484. Notably, the ORF1b:183 (=nsp12:192) site, which we found more frequently mutated in vaccinated compared to unvaccinated NYULH study participants (Figure 4a), was also significantly associated with positive, diversifying selection among the vaccinated participants, but not in the unvaccinated individuals, supporting our statistical counting-based approach from above (Figure 4d, S8, Table S8). Spike:S112L did not yield posterior probabilities of >0.9, but still had a tendency towards positive selection that was more pronounced among vaccinated compared to unvaccinated participants (Figure 4d, Table S9).
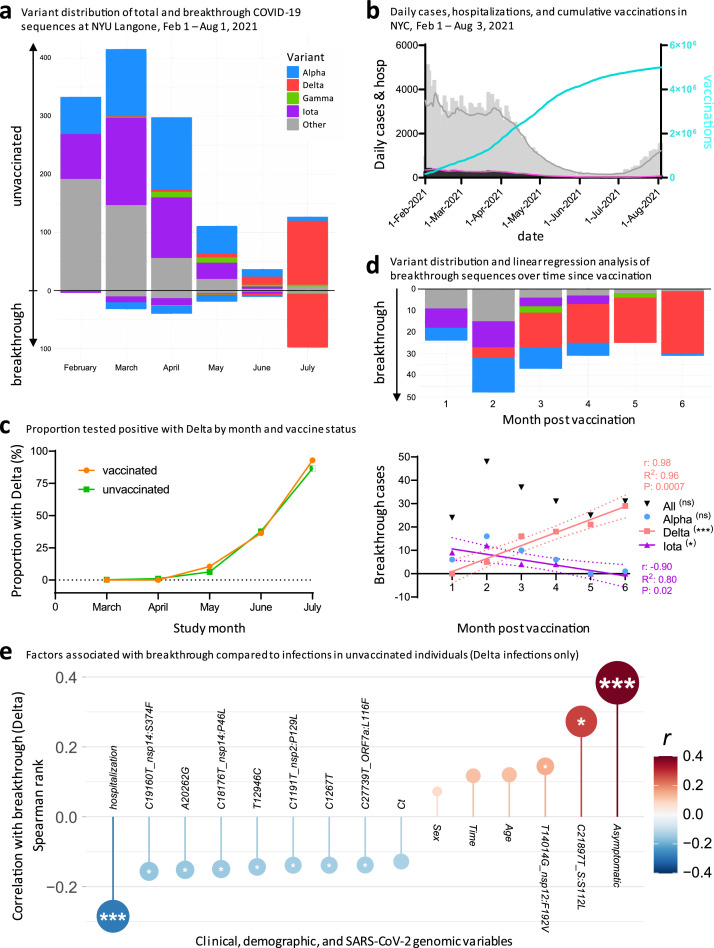
Delta cases increased at comparable rates in vaccinated and unvaccinated individuals and Delta breakthrough is associated with a distinct set of clinical and genomic factors
We expanded our current analysis of recorded breakthrough infections in our cohort, by adding data from our previous study17 that covered February to April 2021, for a total of 208 breakthrough and 1329 control sequences (Figure 5). The pattern of total COVID-19 case numbers in New York State over time is comparable to our SARS-CoV-2 sequences, supporting our attempt to sequence a representative set of viruses (Figure 5a, b). While increasing vaccination rates were associated with an overall decline in COVID-19 cases and SARS-CoV-2 sequences obtained, emerging Delta infections partially reverted this trend (Figure 5b). Iota breakthrough significantly decreased and Alpha breakthrough exhibited no significant change over time since vaccination, whereas Delta breakthrough infections rose significantly in near linear fashion within the first six months post full vaccination, both in absolute and relative counts (Figure 5d, S9-S11).
Figure 5.
Increasing rates of SARS-CoV-2 breakthrough infections with Delta over six months post vaccination, and clinical and genomic factors associated with Delta breakthrough. a, Variant distribution of SARS-CoV-2 sequences from unvaccinated (facing up) and vaccinated individuals (facing down), obtained at NYU Langone Health between February and July 2021. b, Daily COVID-19 cases (grey bars) and 7-day averages (grey line), daily COVID-19 deaths (black bars) and 7-day averages (pink line), and cumulative vaccination numbers (turquoise line) in New York City between February 1st and August 3rd. Source of data: NYC Open Data and NYC Health, Citywide Immunization Registry (CIR). c, Probability of positive test with Delta by month in vaccinated and unvaccinated individuals, adjusted for month of test, sex, and age of participants. d, Variant distribution among all NYULH breakthrough sequences, displayed according to months post full vaccination (starting at day 14 after the last dose for full vaccination). The chart below shows a linear regression analysis of breakthrough infections per variant against time post vaccination. Significant results are highlighted by asterisks, labelled with the correlation coefficient (r), goodness of fit (R2), and P value (t-test), and the fitted line with 95% confidence intervals shown. * P<0.05, ** P<0.01, *** P<0.005. e, Correlation of clinical, demographic, and SARS-CoV-2 genomic factors with breakthrough by comparing Delta infections in vaccinated (n=101) and unvaccinated individuals (n=139). Spearman rank correlation is displayed on the y-axis and colour-coded. Multiple comparison-corrected P values (q, Benjamini-Hochberg) are indicated by asterisks within the circles (* q<0.05, ** q<0.01, *** q<0.005). Ct: Cycle threshold in RT-PCR; sex: male sex; time: date of sampling.
To assess whether Delta infections are statistically enriched among breakthrough infections compared to other variants, we performed covariate-adjusted logistic regression analyses and, as sensitivity analysis, matched the 208 vaccinated breakthrough patients 1:1 to the 1329 unvaccinated patients, correcting for the confounding variables clinical collection date, sex, and age (Table S10). While unmatched analyses suggested elevated numbers of Delta infections in vaccinated compared to unvaccinated individuals (Table S11, upper part), the propensity score-matched analysis revealed no significant difference between both groups, as confirmed by the covariate-adjusted logistic regression analysis (Figure 5c, Tables S11,S12). Recorded Delta cases increased significantly compared to all other variants starting in May 2021 (Table S13). Notably, the rise of Delta occurred at comparable rates in vaccinated and in unvaccinated individuals (Figure 5c), indicating no major advantage of Delta per se in conferring breakthrough after adjustment for confounding variables.
In summary, the study of Delta infections revealed that, compared to unvaccinated controls, breakthrough infections were associated with asymptomatic disease, lower rates of hospitalization, spike mutation S112L, nsp12 mutation F192V, and the absence of seven mutations across different regions of the SARS-CoV-2 full genome (Figure 5e).
Discussion
Since emergency-use authorization of the first COVID-19 vaccine in December 2020, the spotlight has been on viral sequences from vaccinated individuals to closely monitor the potential emergence of vaccine escape mutations. While the first half of 2021 was characterized by a wealth of variants, some more prevalent than others regionally or globally, the Delta variant rapidly replaced all other variants in the second half of 2021 and has remained prevalent worldwide in 2022.39 Our cohort in metropolitan New York is a prime example of Delta's takeover in 2021, both in unvaccinated and vaccinated individuals with a near linear increase of Delta breakthrough infections according to elapsed time post vaccination (Figures 1, 5). Breakthrough numbers in our cohort approach the absolute numbers in the unvaccinated, which is in line with other reports.40,41 The reasons for this change are multifactorial, including epidemiological and virological factors. First, the probability of a positive case being a breakthrough versus an unvaccinated case rose over time due to increased vaccination rates in our area and elsewhere (Figure 5).42 Second, Delta was detected in increased rates in younger hosts, who tend to be vaccinated at lower rates than older individuals, or were ineligible for vaccination at the time of our study. The younger population is also associated with lower hospitalization rates (Figures 1, S1).43 Third, Delta carries the P681R substitution in the spike furin cleavage site, which triggers enhanced S1/S2 cleavage and might explain Delta's increased replication rate in vitro.24,44,45 A substitution at the same site (P681H) is found in Alpha, which evolved independently of Delta and dominated the global infection landscape between January and June 2021.46, 47, 48, 49, 50 P681H/R mutations were both detected more frequently among breakthrough infections in our previous study of breakthrough infections in New York when Alpha and Iota variants were dominant regionally,17 and in this study during the current Delta wave (Figure 3). Although Delta shows compromised sensitivity to some RBD and NTD neutralizing antibodies with up to 8-fold reduced sensitivity in vitro to vaccine-induced antibodies compared to D614G viruses (including infectious virus assays),24,26,51 neutralization escape is substantially lower in magnitude as compared to Beta, Gamma, Mu, and Omicron.52,53 Efficient spike cleavage (P681H/R) and replicative competence appear to be central checkpoints for SARS-CoV-2’s epidemiological success, with Delta being improved compared to Alpha.24,44,54, 55, 56
In line with data from other laboratories, the pre-Delta breakthrough infections that we studied in early 2021 displayed a variant distribution similar to infections in unvaccinated individuals,17, 18, 19, 20 though with signs of a starting sieve effect of neutralizing antibody escape mutations, e.g., E484K.15, 16, 17,57 The variant pattern in both breakthrough and unvaccinated cases dramatically changed towards the second half of 2021 with a clear dominance of Delta-associated mutations (Figure 3). In the second half of 2021, breakthrough appeared to be primarily shaped by virological factors that increased transmissibility, facilitated by immunological permeability in times of waning vaccine efficacy.58 Delta's global rise in the second half of 2021 coincided when large parts of high-risk populations were past six months post full vaccination. The lift of US mask recommendations in May 2021, right before the ignition of the Delta wave, may have also added to its spread. Mask mandates were reinstated halfway through the Delta wave, i.e., on July 28th 2021, at NYU Langone Health, whereas New York State only issued a recommendation to wear masks in early August.59
Delta remains a primary source for the evolution of next generation variants, evidenced by the rapid diversification into more than 190 Delta subvariants and the emergence of Delta–Omicron recombinants.60,61 We observed an uptick of the subvariant AY.25 with S112L mutation in spike NTD as well as AY.44 with F192V mutation in nsp12, which, while still at low numbers, preferentially spread among the vaccinated individuals in our cohort. In our phylogenetic analysis, we observed multiple Delta clusters that suggest efficient spread of distinct sublineages in the population. As shown for AY.25 and AY.44, they involve unvaccinated and vaccinated individuals, and their presence in clusters is suggestive of transmission chains.
High vaccine efficacy was reported against Delta infections, particularly at the beginning of the Delta wave; however, there is growing evidence that vaccine efficacy against Delta decreases over time.23,25,62, 63, 64 A central question remains whether reduced vaccine efficiency is due to Delta per se or whether it is a matter of waning immunity and dependent on elapsed time since full vaccination. Current knowledge indicates that it may be both. Statistical analyses of retrospective cohort studies indicated that waning immunity with time, coinciding with a period of easing societal public health restrictions, played a larger role than Delta's vaccine escape.63,64 However, an observational study on SARS-Cov-2-infected index cases and contacts indicated that vaccination reduced transmission more effectively in Alpha compared to Delta.25 Our data suggest that selective adaptation processes were in process that eventually led to the accumulation of mutations (spike S112L, nsp12 F192V) or new (sub)variants (AY.25, AY.44) under vaccine immune pressure (Figures 4 and 5e).
Of note, when we compared the selectively enriched Delta breakthrough mutations with the ones present in Omicron, a variant which has presumably overcome immune selective pressure during its formation,4, 5, 6,37 we found certain commonalities. Delta breakthrough-enriched mutations and Omicron mutations involve the NTD of spike and nsp12, whereas sites that inversely correlated with Delta breakthrough were found in cold spot regions in terms of Omicron mutations. The latter suggests that mutations in these cold spot regions have no benefit in conferring breakthrough, whereas the wild-type residues (Wuhan-Hu-1) might be superior in supporting breakthrough, immune escape, or transmission/replication. Although none of the breakthrough-associated Delta mutations fell onto Omicron's clade-defining mutations, both share comparable hot and cold spot regions, indicating common features of selective adaptation with fine specificities differing by variant, particularly for Delta and Omicron, which originate from different branches of the phylogenetic tree. Notably, the adaptive evolution analysis based on dN-dS rates confirmed the full-genome statistical mutation analysis in that nsp12:F192V is significantly overrepresented among vaccinated compared to unvaccinated participants. The fact that we did not observe significant adaptive selection at spike:112 using dN-dS rates was presumably based on the overall lower number of viruses with this mutation in our cohort; however, we still obtained higher dN-dS rates in vaccinated compared to unvaccinated individuals. It remains elusive whether spike:S112L, located at the apical part of spike NTD, nsp12:F192V, located distant from nsp12’s active site, or the absence of mutations in the cold spot regions played an active role in breakthrough, immune evasion, enhanced transmission, or acted as bystander mutations. Overall, the modest adaptation of Delta genomes in breakthrough infections in New York, including two mutations that were significantly enriched and seven mutations that were less frequent in sequences from vaccinated compared to unvaccinated individuals, suggests an improved genomic framework that assisted Delta's epidemiologic success in individuals with waning immunity.
As with all observational genomic surveillance studies, limitations and confounding factors exist. These include demographic, temporal, and behavioural factors, diagnostic testing practices, sequencing sensitivity thresholds, and sampling, e.g., undersampling of mild or asymptomatic cases. Some but not all of these caveats can be adjusted or minimized by unbiased sample collection, consistent testing procedures and guidelines, and matched data analyses. We matched by age, sex, and month of collection, but we had no available data on body mass index (BMI), ethnicity or comorbidities, thus we cannot exclude confounding effects by these demographic/clinical variables. The study was limited to the available number of SARS-CoV-2 sequences from our healthcare institution, i.e., sequences from 132 vaccinated and 283 unvaccinated individuals. Our adjusted model indicates that the probability of Delta infection has increased at similar rates in vaccinated and unvaccinated individuals. However, Delta breakthrough cases rose continuously with extended time post vaccination (Figure 5), which justified the use of booster doses even before Omicron emerged.65,66 The monitoring of new variants including Delta's ongoing evolution of subvariants and their early signs of adaptive evolution will be critical to adequately address upcoming SARS-CoV-2 outbreaks and the increasing numbers of vaccine breakthrough infections.
Contributors
Ralf Duerr, MD, PhD designed the study, analysed data, created figures, and wrote the manuscript; Dacia Dimartino, PhD and Paul Zappile made libraries and sequenced the samples; Christian Marier analysed genomic data; Samuel Levine and Fritz Francois, MD, generated total numbers of vaccinations for the entire healthcare system; Guiqing Wang, MD, PhD collected samples and performed clinical SARS-CoV-2 detection; Eduardo Iturrate, MD, and Brian Elbel, PhD monitored epidemiological data and generated lists of breakthrough infections; Meike Dittmann, PhD wrote the manuscript; Jennifer Lighter, MD reviewed clinical information; Andrea Troxel, ScD and Keith S. Goldfeld, DrPH performed statistical analyses; and Adriana Heguy, PhD designed the study, generated genomic data and wrote the manuscript. All authors directly accessed and verified the underlying data, reviewed, and edited the manuscript, and read and approved the final version of the manuscript.
Data sharing statement
All deidentified viral sequences and metadata are publicly available in SRA (BioProject PRJNA769411) and GISAID (Tables S1–S3).
Declaration of interests
The authors have no conflict of interest to declare.
Acknowledgements
We thank NYU Langone Health DataCore for support extracting the data for this study from clinical databases and the clinical laboratory technicians for assistance in testing, saving and retrieving specimens, especially Joanna Fung. We also thank Dr. Joan Cangiarella for her continuous support of genomic surveillance for SARS-CoV-2 at NYULH, including providing institutional funding for this study. We are grateful to the submitting laboratories who deposited data in GISAID, in particular to those whose sequences we used to create the phylogenetic tree (Table S3). The NYULH Genome Technology Center is partially supported by the Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104141.
Contributor Information
Ralf Duerr, Email: ralf.duerr@nyulangone.org, Ralf.Duerr@nyumc.org.
Adriana Heguy, Email: adriana.heguy@nyulangone.org.
Appendix. Supplementary materials
References
- 1.World Health Organization (WHO) 2021. WHO Coronavirus (COVID-19) Dashboard.https://covid19.who.int/ Accessed 3 October 2021. [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) 2021. SARS-CoV-2 Variant Classifications and Definitions.https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html Accessed 3 October 2021. [Google Scholar]
- 3.GISAID . 2021. GISAID Database.https://www.gisaid.org/ Accessed 25 September 2021. [Google Scholar]
- 4.Pulliam JRC, van Schalkwyk C, Govender N, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376(6593) doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602(7898):654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bansal K, Kumar S. Mutational cascade of SARS-CoV-2 leading to evolution and emergence of omicron variant. Virus Res. 2022;315 doi: 10.1016/j.virusres.2022.198765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadoff J, Gray G, Vandebosch A, et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against covid-19. N Engl J Med. 2021;38(23):2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagan N, Barda N, Kepten E, et al. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N Engl J Med. 2021;384(15):1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas EJ, Angulo FJ, McLaughlin JM, et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. 2021;397(10287):1819–1829. doi: 10.1016/S0140-6736(21)00947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris RJ, Hall JA, Zaidi A, Andrews NJ, Dunbar JK, Dabrera G. Effect of vaccination on household transmission of SARS-CoV-2 in England. N Engl J Med. 2021;385(8):759–760. doi: 10.1056/NEJMc2107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kustin T, Harel N, Finkel U, et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat Med. 2021;27(8):1379–1384. doi: 10.1038/s41591-021-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McEwen AE, Cohen S, Bryson-Cahn C, et al. Variants of concern are overrepresented among postvaccination breakthrough infections of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Washington State. Clin Infect Dis. 2021;74(6):1089–1092. doi: 10.1093/cid/ciab581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duerr R, Dimartino D, Marier C, et al. Dominance of Alpha and Iota variants in SARS-CoV-2 vaccine breakthrough infections in New York City. J Clin Invest. 2021;131(18) doi: 10.1172/JCI152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollett SD, Richard SA, Fries AC, et al. The SARS-CoV-2 mRNA vaccine breakthrough infection phenotype includes significant symptoms, live virus shedding, and viral genetic diversity. Clin Infect Dis. 2021;74(5):897–900. doi: 10.1093/cid/ciab543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CDC Covid- Vaccine Breakthrough Case Investigations Team COVID-19 vaccine breakthrough infections reported to CDC - United States, January 1-April 30, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(21):792–793. doi: 10.15585/mmwr.mm7021e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergwerk M, Gonen T, Lustig Y, et al. Covid-19 breakthrough infections in vaccinated health care workers. N Engl J Med. 2021;385(16):1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chemaitelly H, Tang P, Hasan MR, et al. Waning of BNT162b2 vaccine protection against SARS-CoV-2 infection in Qatar. N Engl J Med. 2021;385(24):e83. doi: 10.1056/NEJMoa2114114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin EG, Lustig Y, Cohen C, et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N Engl J Med. 2021;385(24):e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowlkes A, Gaglani M, Groover K, et al. Effectiveness of COVID-19 vaccines in preventing SARS-CoV-2 infection among frontline workers before and during B.1.617.2 (Delta) variant predominance - eight U.S. locations, December 2020-August 2021. MMWR Morb Mortal Wkly Rep. 2021;70(34):1167–1169. doi: 10.15585/mmwr.mm7034e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mlcochova P, Kemp S, Dhar MS, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eyre DW, Taylor D, Purver M, et al. Effect of Covid-19 vaccination on transmission of alpha and delta variants. N Engl J Med. 2022;386(8):744–756. doi: 10.1056/NEJMoa2116597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Ginn HM, Dejnirattisai W, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184(16):4220–4236. doi: 10.1016/j.cell.2021.06.020. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas C, Vogels CBF, Yildirim I, et al. Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity. Nature. 2021;600(7889):523–529. doi: 10.1038/s41586-021-04085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statistic Soc Ser B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 31.Chen C, Nadeau SA, Topolsky I, et al. Quantification of the spread of SARS-CoV-2 variant B.1.1.7 in Switzerland. Epidemics. 2021;37 doi: 10.1016/j.epidem.2021.100480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosakovsky Pond SL, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 33.Murrell B, Moola S, Mabona A, et al. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol. 2013;30(5):1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39(2):175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 35.D'Agostino RB., Jr. Propensity score methods for bias reduction in the comparison of a treatment to a non-randomized control group. Stat Med. 1998;17(19):2265–2281. doi: 10.1002/(sici)1097-0258(19981015)17:19<2265::aid-sim918>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 36.Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt F, Muecksch F, Weisblum Y, et al. Plasma neutralization of the SARS-CoV-2 omicron variant. N Engl J Med. 2022;386(6):599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nextstrain team. Genomic epidemiology of novel coronavirus - Global subsampling. 2021. https://nextstrain.org/ncov/global. Accessed 3 October 2021.
- 40.Brown CM, Vostok J, Johnson H, et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings - barnstable county, Massachusetts, July 2021. MMWR Morb Mortal Wkly Rep. 2021;70(31):1059–1062. doi: 10.15585/mmwr.mm7031e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keehner J, Horton LE, Binkin NJ, et al. Resurgence of SARS-CoV-2 infection in a highly vaccinated health system workforce. N Engl J Med. 2021;385(14):1330–1332. doi: 10.1056/NEJMc2112981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi A, Velasquez J, Welch W. Coronavirus in New York City - Tracking the spread of the pandemic. 2021. https://projects.thecity.nyc/2020_03_covid-19-tracker/. Accessed 6 October 2021.
- 43.Lam-Hine T, McCurdy SA, Santora L, et al. Outbreak associated with SARS-CoV-2 B.1.617.2 (Delta) variant in an elementary school - Marin County, California, May-June 2021. MMWR Morb Mortal Wkly Rep. 2021;70(35):1214–1219. doi: 10.15585/mmwr.mm7035e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lubinski B, Frazier LE, VTP M, et al. Spike protein cleavage-activation mediated by the SARS-CoV-2 P681R mutation: a case-study from its first appearance in variant of interest (VOI) A.23.1 identified in Uganda. In: bioRxiv. 2022. Preprint; 2021.06.30.450632.
- 45.Peacock TP, Sheppard CM, Brown JC, et al. The SARS-CoV-2 variants associated with infections in India, B.1.617, show enhanced spike cleavage by furin, bioRxiv. 2021:2021.05.28.446163. Preprint.
- 46.Martin DP, Weaver S, Tegally H, et al. The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell. 2021;184(20):5189–5200.e7. doi: 10.1016/j.cell.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collier DA, De Marco A, Ferreira IATM, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021 doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- 48.Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu H, Chen Q, Yang G, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng B, Kemp SA, Papa G, et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the variant of concern lineage B.1.1.7. Cell Rep. 2021, 35(13):109292. [DOI] [PMC free article] [PubMed]
- 51.Chia PY, Ong SWX, Chiew CJ, et al. Virological and serological kinetics of SARS-CoV-2 Delta variant vaccine breakthrough infections: a multicentre cohort study. Clin Microbiol Infect. 2022;28(4):612.e1–612.e7. doi: 10.1016/j.cmi.2021.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyakawa K, Jeremiah SS, Kato H, Ryo A. Neutralizing efficacy of vaccines against the SARS-CoV-2 Mu variant. medRxiv. 2021: 2021.09.23.21264014. Preprint.
- 53.Uriu K, Kimura I, Shirakawa K, et al. Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N Engl J Med. 2021;385(25):2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Liu J, Johnson BA, et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv. 2021: 2021.08.12.456173. Preprint. [DOI] [PMC free article] [PubMed]
- 55.Zhang J, Xiao T, Cai Y, et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021;374(6573):1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saito A, Irie T, Suzuki R, et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 delta P681R mutation. Nature. 2022;602(7896):300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feder KA, Patel A, Vepachedu VR, et al. Association of E484K spike protein mutation with SARS-CoV-2 infection in vaccinated persons—Maryland, January - May 2021. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas SJ, Moreira ED, Kitchin N, et al. Safety and Efficacy of the BNT162b2 mRNA covid-19 vaccine through 6 months. N Engl J Med. 2021 doi: 10.1056/NEJMoa2110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matthews K, Hill M. Masks encouraged in New York, but no city or state mandate. 2021. https://apnews.com/article/lifestyle-business-health-coronavirus-pandemic-97863fa563eadaad177e0ccba8000cb4. Accessed 15 September 2021.
- 60.Pango Network. New AY lineages. 2021. https://www.pango.network/new-ay-lineages/. Accessed 5 April 2022.
- 61.Duerr R, Dimartino D, Marier C, et al. Delta-Omicron recombinant SARS-CoV-2 in a transplant patient treated with Sotrovimab. bioRxiv. 2022:2022.04.06.487325. Preprint.
- 62.Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenberg ES, Holtgrave DR, Dorabawila V, et al. New COVID-19 cases and hospitalizations among adults, by vaccination status - New York, May 3-July 25, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(34):1150–1155. doi: 10.15585/mmwr.mm7034e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tartof SY, Slezak JM, Fischer H, et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet. 2021, 398(10309):1407–1416. [DOI] [PMC free article] [PubMed]
- 65.Bar-On YM, Goldberg Y, Mandel M, et al. Protection of BNT162b2 vaccine booster against covid-19 in Israel. N Engl J Med. 2021;385(15):1393–1400. doi: 10.1056/NEJMoa2114255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.World Health Organization (WHO). Interim statement on booster doses for COVID-19 vaccination. 2021. https://www.who.int/news/item/04-10-2021-interim-statement-on-booster-doses-for-covid-19-vaccination. Accessed 5 October 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.