Abstract
C−H oxyfunctionalisation remains a distinct challenge for synthetic organic chemists. Oxygenases and peroxygenases (grouped here as “oxygenating biocatalysts”) catalyse the oxidation of a substrate with molecular oxygen or hydrogen peroxide as oxidant. The application of oxygenating biocatalysts in organic synthesis has dramatically increased over the last decade, producing complex compounds with potential uses in the pharmaceutical industry. This review will focus on hydroxyl functionalisation using oxygenating biocatalysts as a tool for drug discovery and development. Established oxygenating biocatalysts, such as cytochrome P450s and flavin‐dependent monooxygenases, have widely been adopted for this purpose, but can suffer from low activity, instability or limited substrate scope. Therefore, emerging oxygenating biocatalysts which offer an alternative will also be covered, as well as considering the ways in which these hydroxylation biotransformations can be applied in drug discovery and development, such as late‐stage functionalisation (LSF) and in biocatalytic cascades.
Keywords: Biocatalysis, Enzymes, Drug discovery, Hydroxylations, Oxyfunctionalisation
C−H bond oxyfunctionalisation is a significant challenge in organic synthetic chemistry. In drug discovery and development, exploiting biocatalytic methods which can introduce hydroxyl groups deftly with high selectivity, under mild conditions, is of paramount interest. This review highlights key classes of oxygenases and peroxygenases as “oxygenating biocatalysts” for C−H hydroxyl functionalisation and discusses their applicability in late‐stage functionalisations and biocatalytic cascades, to access pharmacological relevant compounds and their precursors.
1. Introduction
Within the pharmaceutical industry, the paradigm of drug development is rapidly changing with constant review on how to deliver medicines to patients in a faster, cost‐effective and safe manner. [1] Biocatalysis represents an increasingly utilised tool for the challenges of drug discovery and development.[ 1 , 2 , 3 ] Biocatalysis employs enzymes, used either in their purified form, as a cell lysate or whole cells to convert starting substrates to a desired product(s). [4] The defined three‐dimensional structure of an enzyme enables the chemo‐, regio‐ and stereoselective formation of complex chiral products, a crucial outcome for many pharmaceuticals. [2] This selectivity obviates the need for protecting and deprotecting stages in synthesis, removing wasteful steps which may involve hazardous or toxic reagents. [5] Enzymatic reactions generally work in an aqueous environment, reducing the use of organic solvents and tend to operate under milder temperatures. Therefore, biocatalysis is viewed as a greener approach to chemical synthesis than traditional organic chemistry.[ 6 , 7 , 8 ] Moreover, approaches in protein engineering, such as directed evolution and rational design, have enabled the tailoring of supreme biocatalysts with an expanded repertoire of available reactions.[ 9 , 10 , 11 , 12 ] However, despite these advantages, biocatalysis still has limitations when addressed in industrial settings due to narrow substrate scope, low stability and lack of reusability. [13] Additionally, an evolved biocatalyst, optimised for a specific reaction, cannot generally be used for an alternative substrate or synthetic route. [1] Still, advances in protein engineering have been significant over the past decade, [14] attributed to key innovations in the availability of DNA sequences and gene synthesis. Many reviews have highlighted the benefit of biocatalysis to the pharmaceutical industry.[ 15 , 16 , 17 , 18 ]
Selective (sp3 or sp2 ) C−H oxyfunctionalisation currently represents one of the most challenging reactions in organic chemistry. [19] Biocatalysts have been increasingly utilised to address this organic synthesis challenge, enabling high selectivity that chemical catalysis may fall short.[ 20 , 21 , 22 , 23 ] Hydroxyl groups are ubiquitous functional groups in marketed drugs, directly impacting the pharmacokinetic and physicochemical properties of a compound. [24] Additionally, a hydroxyl group may be used as a synthetic handle to generate lead compounds and/or analogues through chemical modifications, fast‐tracking the drug development process. However, hydroxyl group installation on unreactive C−H sites remains a pivotal challenge in synthetic chemistry. This review will discuss key classes of oxygenases and peroxygenases, covering from 2010, as “oxygenating biocatalysts” for C−H hydroxyl functionalisation. We wish to highlight their potential for performing highly regio‐ and enantioselective biotransformations of drug‐like molecules, or precursors, under mild conditions. The aim of this review is to provide the reader a snapshot of the current hydroxyl functionalisation landscape, covering two classes of established oxygenating biocatalysts, namely cytochrome P450s and flavin‐dependent monooxygenases and providing two classes of emerging oxygenating biocatalysts, iron‐ and α‐ketoglutarate‐dependent oxygenases and unspecific peroxygenases, which offer an alternative to the practical shortcomings of cytochrome P450s. Two avenues for which oxygenating biocatalysts are, and potentially will be, applied in drug discovery and development, particularly in late‐stage functionalisation (LSF) and biocatalytic cascades, will also be covered.
2. Oxygenating Biocatalysts
This section will introduce established and emerging oxygenating biocatalysts for hydroxyl functionalisations. Examples of, where possible, sp 3 and sp 2 C−H hydroxylations are provided to illustrate the breadth of reactions available. Brief descriptions of the catalytic cycles of these enzymes are included for context and to highlight the different ways the oxidant is activated. For detailed discussion of the catalytic mechanisms and enzymatic structures, which would be beyond the scope of the current work, the reader is directed to excellent existing reviews.[ 19 , 25 , 26 ] Notably, there are other enzymes, such as those copper‐dependent, which are also able to perform hydroxyl functionalisation, but for the interest of brevity, and since a detailed review already exists, [27] will not be covered herein.
2.1. Established Oxygenating Biocatalysts
2.1.1. Cytochrome P450s
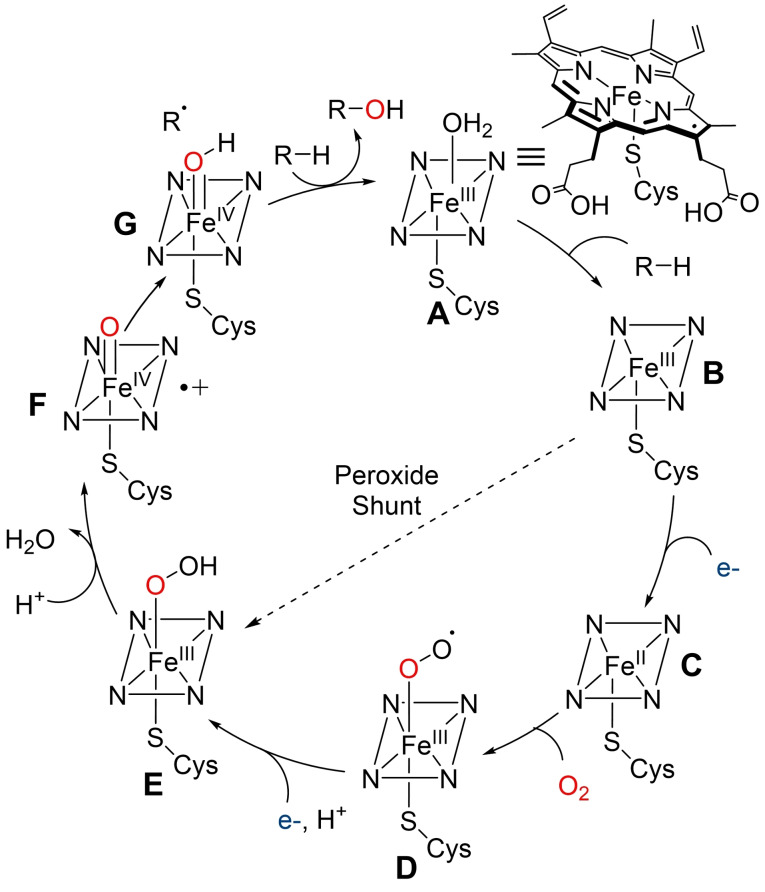
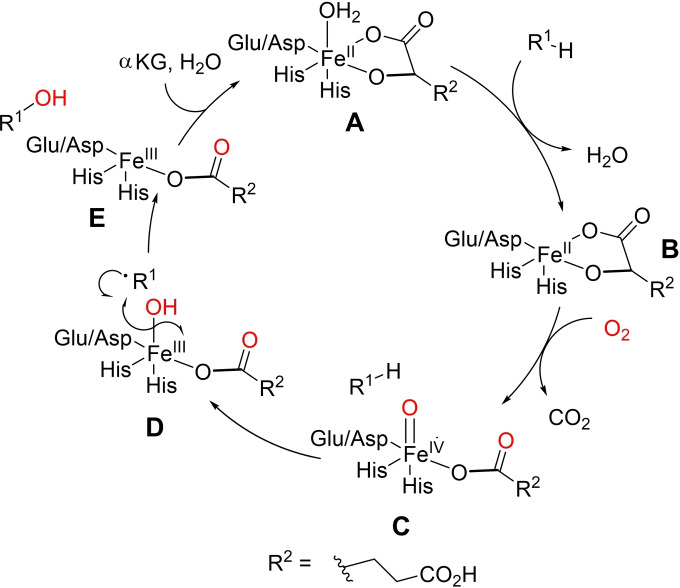
Cytochrome P450s are haem‐dependent monooxygenases found in all kingdoms of life. These enzymes consist of a protoporphyrin IX‐bound iron atom, thiolate‐ligated to a conserved axial cysteine residue. [28] The catalytic cycle is initiated by substrate binding to the Fe(III) resting state A, after loss of a water ligand, to yield intermediate B. Transfer of the first electron from reduced pyridine nucleotide, NAD(P)H, facilitated by auxiliary redox partners, allows reduction of Fe(III) to Fe(II), generating intermediate C. Oxygen binding to C then leads to Fe‐superoxo species D which is further reduced by a second electron transfer, followed by protonation, to produce Fe‐hydroperoxo adduct E. A second protonation event leads to loss of water as E collapses to form a highly electrophilic oxoiron (IV) porphyrin radical cation F (referred to as compound I). F facilitates hydrogen abstraction and subsequent oxygen‐atom transfer to the substrate. Re‐equilibration with water then regenerates the Fe(III)‐resting state.[ 28 , 29 , 30 ] Notably, some cytochrome P450s are able to produce adduct E directly from B, using hydrogen peroxide, through the so‐called peroxide shunt pathway. [22] However, this pathway is limited by low H2O2 tolerance and efficiency which can result in oxidative inactivation (Scheme 1).[ 22 , 31 ]
Scheme 1.
Simplified catalytic cycle for the mechanism of C−H hydroxylation by cytochrome P450s.
The majority of cytochrome P450s in nature exist as multiprotein systems, with reductase partners required for the delivery of electrons from NAD(P)H to the iron centre. The redox partner can consist of flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN) or iron‐sulfur (FeS) centres bound as one protein or separated proteins. [32] However, a few cytochrome P450s are fused to their reductase partners and described as self‐sufficient. [28] The most prominent example of a self‐sufficient cytochrome P450, P450BM3 (CYP1021A1) from Bacillus megaterium has been a focus in biomedical and biotechnological research for its single‐polypeptide nature, leading to an ease of ready application.[ 33 , 34 ] Additionally, P450BM3 can be readily expressed to high levels in Escherichia coli and displays high native activity. [35]
Cytochrome P450s have been utilised for the hydroxyl functionalisation of unactivated sp 3 C−H sites. [19] Urlacher and co‐workers used a minimal enriched P450BM3 library for the selective hydroxylation of inert cyclic and linear alkanes. [36] Isoforms containing F87A/A328V mutations were able to hydroxylate cyclooctane, cyclodecane and cyclododecane. Mutant F87V/A328F catalysed hydroxylation on n‐octane to produce 2‐(R)‐octanol, a precursor for the preparation of pharmaceuticals, in 92 % regioselectivity (46 % ee). [36] Reetz et al. applied directed evolution on P450BM3 to generate mutants capable of catalysing (R)‐selective (>95 % ee) or (S)‐selective (>95 % ee) hydroxylation of cylcohexene‐1‐carboxylic acid methyl ester, with a regioselectivity for the allylic C3‐position of >93 % (Figure 1A). [37] Other cytochrome P450s have also been applied for C(sp 3 )−H hydroxylation. Zhao et al. evolved P450pyr from Sphingomonas sp. HXN‐200 by iterative targeted site‐saturation mutagenesis with the CAST/ISM approach, for the (S)‐ and (R)‐selective hydroxylation of N‐benzyl pyrrolidine at the unactivated C‐3 position. [38] Bell et al. have used cytochrome P450 CYP101B1 from Novosphingobium aromaticivorans to selectively hydroxylate methylene C−H bonds in cycloalkyl rings. [39] The group investigated using ester protecting groups as chemical auxiliaries and directing groups for the biocatalytic hydroxylation, hypothesising these may mimic chemical features of CYP101B1’s known substrates, the norisoprenoids. Hydroxylation of cyclooctyl acetate by CYP101B1 generated trans‐5‐hydroxycyclooctyl in >95 % yield (Figure 1B). [39] Additionally, selective sp 3 C−H hydroxylation may be useful for the functionalisation of fatty acids for pharmaceutical applications. Many cytochrome P450s hydroxylate fatty acid substrates as their natural substrate, producing hydroxy‐fatty acids with potential medicinal uses.[ 40 , 41 ] Pietruszka et al. used P450BM3 mutants to catalyse the allylic hydroxylation of ω‐alkenoic acids and esters, producing therapeutically‐relevant hydroxylated products. [42] The P450BM3 mutant A74G/L188G was found to selectively catalyse the allylic hydroxylation of ethyl 6‐heptenoate with a conversion of 49 %, and alcohol selectivity of 90 % (epoxide as minor product) and 95 % ee (Figure 1C). [42] Recently, Wang et al. have investigated new cytochrome P450 OleT5A from Staphylococcus aureus for its decarboxylation and hydroxylation of fatty acids to generate fatty alkenes and alcohols. [43] The group tested a series of novel fatty acid derivatives to probe the enzyme's activity and found that the minor changes in the substrates’ end group could regulate the hydroxylation or decarboxylation reactions. [43] Chen et al. have employed directed evolution strategies on P450BSβ to provide a quadruple mutant capable of catalysing β‐hydroxylation on a variety of fatty acid substrates, including myristic acid with excellent enantioselectivity (>99 % ee). [44] Zong et al. have conducted mutagenesis studies on the recently discovered self‐sufficient cytochrome P450, BAMF2522, to obtain variants with increased regioselectivity for in‐chain hydroxyl functionalisation (ω‐4 to ω‐9) of medium to long chain fatty acids. [45]
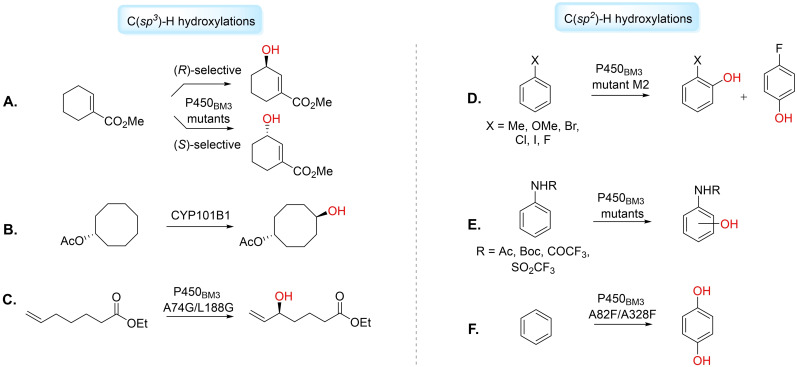
Figure 1.
Illustrative examples of C(sp 3 )−H and C(sp 2 )−H hydroxylations catalysed by cytochrome P450s. A) regioselective allylic hydroxylation of cylcohexene‐1‐carboxylic acid methyl ester. B) CYP101B1‐catalysed hydroxylation of cyclooctyl acetate. C) P450BM3 mutant A74G/L188G catalyses allylic hydroxylation of ethyl 6‐heptenoate. D) Hydroxylations of halogenated benzenes, toluene and anisole using P450BM3 mutant M2. E) P450BM3 mutants catalyse hydroxylation of anilides to mostly 4‐hydroxy derivatives. F) Hydroxylation of benzene to hydroquinone using P450BM3 double mutant A82F/A328F.
There is a considerable drive for methods of sp 2 C−H hydroxyl functionalisation. Functionalised phenols have applicability as building blocks for high‐value chemicals and drugs. Schwaneberg et al. applied the P450BM3 mutant M2 for ortho‐ and para‐ hydroxylation of halogenated benzenes, toluene and anisole (Figure 1D). [46] The group identified the phenol products were important synthons in the syntheses of FDA‐approved drugs including liothyronine, atomoxetine and vancomycin. [46] Robertson and co‐workers have reported the monohydroxylation of mono‐ and disubstituted N‐trifluoromethanesulfonyl anilides to mostly 4‐hydroxy derivatives using P450BM3 variants (Figure 1E). [47] Watanabe et al. reported P450BM3‐catalysed direct hydroxylation of benzene to phenol by employing amino acid derivatives as decoy molecules, inciting activity of the WT enzyme on the non‐natural substrate. [48] Reetz et al. have demonstrated that a semi‐rational protein engineering strategy could produce P450BM3 mutants capable of the chemo‐ and regioselective dihydroxylation of benzene to hydroquinone, an important intermediate for the preparation of pharmaceutical compounds. [49] A combination of directed evolution and site‐directed mutagenesis led to the creation of double mutant A82F/A328F that catalysed 97 % conversion to hydroquinone (Figure 1F). [49]
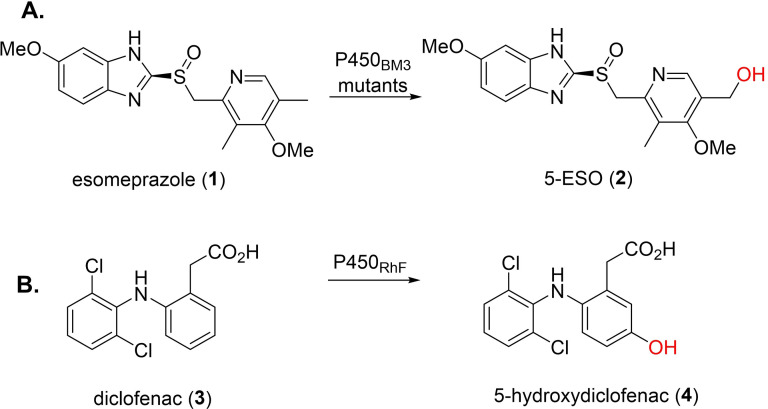
In pharmaceutical research, cytochrome P450s, particularly P450BM3, have been widely used for over a decade in human drug metabolite (HDM) synthesis, [50] for characterisation and toxicity assessment. Human liver cytochrome P450s, largely responsible for drug metabolism, [51] have low applicability due to their multiprotein systems, instability and low activity. In contrast, engineered P450BM3 variants, which exhibit high expression levels and solubility, may be used as a surrogate. [34] Munro et al. demonstrated P450BM3 mutants could oxidise a variety of proton pump inhibitors, including esomeprazole, lansoprazole and rabeprazole. [52] Esomeprazole (1, ESO) was hydroxylated by P450BM3 mutants (A82F, F87V and F87V/A82F) to give 5‐OH ESO (2), the major HDM formed by human cytochrome P450 CYP2C19 (Figure 2A). [52] Commandeur and co‐workers studied P450BM3 for the HDM synthesis of three drugs belonging to the fenamate class of non‐steroidal anti‐inflammatory drugs (NSAIDs). [53] For NSAID mefenamic acid, three monohydroxylated metabolites, 3’‐OH‐methyl‐meclofenamic acid, 5‐OH‐meclofenamic acid and 4’‐OH‐meclofenamic acid, were produced upon screening against the engineered P450BM3 library, which were all HDMs. [53] Furthermore, Wong et al. tested a library of P450BM3 mutants for C−H oxidation of a variety of neutral, cationic and anionic drugs. While not all mutants were active for all the tested drugs, multiple variants showed high activity, with high conversions, to enable full product characterisation. [54] The authors reported mutant RLYF/KSK19 could catalyse the hydroxylation of testosterone to 61 % 2β‐hydroxy‐testosterone, 33 % 15β‐hydroxy‐testosterone, 3 % new compound 2β‐16β‐dihydroxytestesterone and 3 % monohydroxylated testosterone (based on MS). [54] Other cytochrome P450s may also be used to produce HDMs. Flitsch et al. used self‐sufficient P450RhF in a whole cell system for hydroxyl functionalisation of NSAID diclofenac (3) produce human metabolite 5‐hydroxydiclofenac (4) (Figure 2B). [55] Urlacher and co‐workers used a novel cytochrome P450, CYP107L, from Streptomyces platensis DSM 400041 for amodiaquine, ritonavir, amitriptyline and thioridazine metabolising activity. [56] Ritonavir was converted by CYP107L to a monohydroxylated product, in 91 % conversion, with the same retention time as a monohydroxylated product produced by human cytochrome P450s CYP3A4 and CYP3A5. [56] Recently, Winkler et al. used self‐sufficient CYP505X from Aspergillus fumigatus and a quintuple‐mutant, expressed in Pichia pastoris, to hydroxylate ibuprofen, resulting in two major HDMs with hydroxyl functionalisation at the tertiary carbon atom and benzylic position of the lipophilic side chain. [57]
Figure 2.
Examples of cytochrome P450‐catalysed hydroxylations for HDM synthesis. A) HDM synthesis of 5‐ESO (2) using P450BM3 mutants. B) HDM synthesis of 5‐hydroxydiclofenac (4) using self‐sufficient cytochrome P450RhF.
The full‐integration of cytochrome P450‐based systems for commercial industrial purposes has been limited for a variety of reasons. Many cytochrome P450s are unstable, consist of multiple components, which can be membrane‐bound, and all require expensive cofactors such as NAD(P)H. [58] Additionally, a lack of substrate diversity and selectivity, low activity and limited performance under industrial conditions has made the adoption of cytochrome P450 catalysis in pharmaceutical processes problematic. [59] Whole‐cell systems make stoichiometric amounts of cofactors unnecessary, but are hampered by low substrate solubility in water, product toxicity, [60] (metabolic) by‐product formation or over‐oxidation. Therefore, efforts have been made to find solutions and other systems to whole‐cell approaches. Different enzymatic systems, as well as electrochemical approaches, have been investigated for cofactor regeneration. Mink and co‐workers used wild type P450BM3 permeabilized whole‐cells (whole cells disrupted by a freezing/thawing step) co‐expressed with glucose dehydrogenase (GDH) for NADPH‐cofactor regeneration in E. coli for the hydroxylation of α‐isophorone. [61] After reaction optimisation, the group afforded 4‐hydroxy‐α‐isophorone (4HIP) on 100 L scale with conversions of 80 % and 82 % in two respective batches. 4HIP was produced in high ee (>99 %) and high purity (>98 % HPLC, GC, NMR), delivering a combined 1 kg of hydroxylated product. [61] Song et al. used an electricity‐driven approach for NAD(P)H regeneration in a cytochrome P450 bioelectrocatalytic system (BES) for the 7α‐hydroxylation of dehydroepiandrosterone (DHEA). [62] The group employed cytochrome P450 CYP7B1 from Saccharomyces cerevisiae and a neutral red (NR)‐mediated extracellular electron transfer (EET) pathway to produce 7α‐OH‐DHEA in yields reaching 288.6±7.8 mg L−1. [62] Recently, light has been used to supply redox equivalents for cytochrome P450‐oxidations. In this approach, a photosensitizer, the “light‐harvesting” unit, injects electrons and initiates the cytochrome P450 catalytic cycle when added into the reaction solution, or bound to the cytochrome P450 enzyme.[ 63 , 64 ]
Additionally, protein engineering approaches have been adopted to overcome the need for different auxiliary proteins in cytochrome P450‐systems and/or expand current substrate scope. Work has been reported for the generation of active chimeric cytochrome P450s by fusion of a reductase domain at the genic level of one self‐sufficient cytochrome P450 with another cytochrome P450 domain. [65] Grogan and co‐workers created a cytochrome P450 fusion library using 23 P450 haem domains from Rhodococcus jostii RHA1 expressed with the cytochrome P450 reductase domain of P450RhF, RhFRED. [66] The authors applied this engineered library for screening demethylation and hydroxylation reactions of commercially available drugs. [66] Woodley et al. used the haem domain of CYP153A from Marinobacter aquaeolei fused to the reductase domain of P450BM3 to produce self‐sufficient protein chimera, CYP153A‐CPRBM3. [67] Mutant CYP153A‐CPRBM3 G307A was able to selectively hydroxylate medium and long chain fatty acids to produce terminally‐hydroxylated compounds with pharmaceutical applications. [67] Xu et al. reported the application of artificial self‐sufficient cytochrome P450, using P450SMO reductase domain and a P450cam mutant Y96F/V247L with a linker region (G4S)4. [68] The resultant chimeric cytochrome P450 was capable of hydroxylating (−)‐limonene, α‐pinene and camphor, which were inaccessible reactions for the natural fusion protein P450SMO. [68] Further discoveries of naturally self‐sufficient cytochrome P450s, such as the new class VII cytochrome P450s, will see the continued growth of chimeric technologies with greater incidences of fusion with heterogenous cytochrome P450 domains for pharmaceutical applications. [69] In addition, cytochrome P450 “fingerprinting” methods, developed by the Fasan group, map out the P450BM3 enzyme active site, in order to predict the regioselectivity towards terpene substrates. This method has been used to select focused libraries of P450BM3 variants with potentially higher regio‐ and stereoselectivity towards target substrates, based on fingerprint probes of pre‐tested substrates of similar structures. [70] Moreover, Houk et al., in collaboration with the Tang and Sherman/Montgomery groups, have adopted computational analysis using both DFT calculations and subsequent MD simulations to elucidate cytochrome P450 mechanisms and design new enzyme mutants. [71]
2.1.2. Flavin‐dependent Monooxygenases
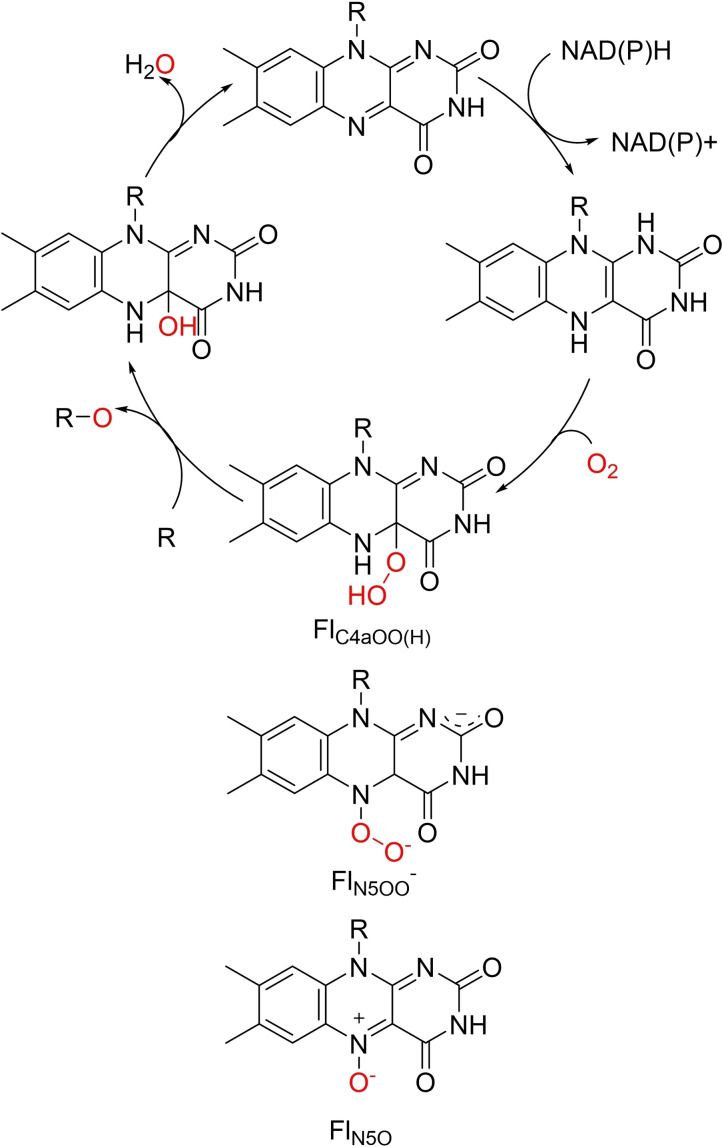
Flavin‐dependent monooxygenases (FMOs) are an extremely diverse class of enzyme which catalyse substrate oxidation utilising molecular oxygen in a NAD(P)H‐mediated catalytic cycle. FMOs can be categorised into eight groups (A–H) based on a combination of structural and biochemical features, and additionally each group belongs to one of three classifications depending on the method of flavin cofactor reduction.[ 72 , 73 , 74 ] Unlike the metal‐based cytochrome P450s, FMOs activate oxygen by flavin cofactors such as FMN or FAD. The catalytic cycle is initiated when NAD(P)H reduces the flavin group, which then binds molecular oxygen to produce the active oxygenating species, C4a‐(hydro)peroxoflavin (FlC4aOO(H)). After elimination of water, the oxidised flavin group is reformed (Scheme 2).[ 22 , 75 ] While FlC4aOO(H) has been widely accepted, and verified, as the key oxygen transferring agent in FMO catalysis, recent reports have discovered other oxygen transfer species flavin‐N5‐oxide (FlN5O) and flavin‐N5‐peroxide (FlN5OO) in FMO‐catalysed reactions.[ 26 , 76 , 77 , 78 , 79 ] These species will not be discussed further as they have been excellently described elsewhere. [76]
Scheme 2.
Simplified catalytic cycle of flavin‐dependent monooxygenases. Flavin prosthetic groups FMN, R=PO3 2− or FAD R=ADP are reduced by NAD(P)H followed by molecular oxygen binding. The activating oxygenating species FlC4aOO(H) facilitates O‐transfer. Other reported oxygenating species FlN5OO and FlN5O are drawn underneath for structural comparison.
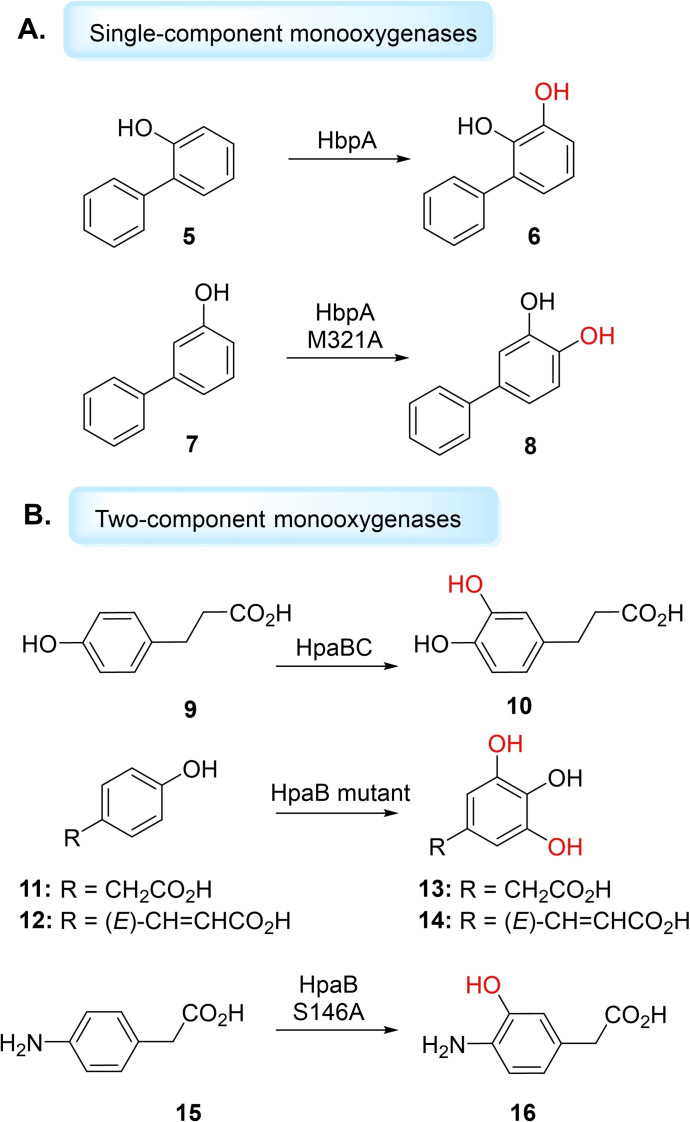
FMOs that catalyse aromatic C−H‐ hydroxyl functionalisations can be divided into two types: single‐component monooxygenases (from group A), where the flavin is tightly bound as a prosthetic group, and two‐component monooxygenases (from group D), in which reduced flavin acts as a substrate, and is delivered by a flavin reductase partner.[ 26 , 80 , 81 ] An extensively studied single‐component aromatic hydroxylase is 4‐hydroxy‐benzoate 3‐hydroxylase (PHBH) from Pseudomonas fluorescens, which catalyses hydroxylation of 4‐hydroxybenzoate (pOHB) to generate 3,4‐dihydroxybenzoate (3,4‐DOHB). [82] However, PHBH acts only on phenolic acids, demonstrating narrow substrate scope. [83] Another representative single‐component monooxygenase is 3‐hydroxybenzoate‐6‐hydroxylase (3HB6H), which catalyses the para‐hydroxylation of 3‐hydroxybenzoate to 2,5‐dihydroxybenzoate. [84] Yu et al. have reported MabA, a 3‐aminobenzoate 6‐hydroxylase, that catalyses the para‐hydroxylation of 3‐aminobenzoate to yield 2‐hydroxy‐5‐aminobenzoate (mesalazine), a drug used to treat inflammatory bowel disease. [85] Single‐component monooxygenase hydroxybiphenyl monooxygenase (HbpA), which catalyses hydroxylation of 2‐hydroxybiphenyl to 3‐phenylcatechol, has been shown to catalyse a broad substrate range to form 3‐aryl, 3‐alkyl and 3‐halo catechol products, important pharmaceutical synthons.[ 86 , 87 ] Buehler et al. have applied HbpA for the hydroxylation of 2‐hydroxybiphenyl (5) in a continuous segmented flow tube‐in‐tube reactor (Figure 3) to generate 6. [88] The authors applied formate dehydrogenase (FDH) for concomitant NADH regeneration, enabling production of 1 g of 3‐phenylcatechol at a high space time yield of 14.5 g L−1h−1. [88] Fishman et al. produced the HbpA variant, M321A, through saturation mutagenesis studies, which displayed altered regioselectivity and activity. [89] M321A catalysed hydroxylation of non‐natural substrate 3‐hydroxybiphenyl (7) to produce a new antioxidant, 3,4‐hydroxybiphenyl (8) (Figure 3A). [89]
Figure 3.
Aromatic hydroxylations catalysed by FMO aromatic hydroxylases. A) Single component FMO HbpA and HbpA mutant with hydroxybiphenyl substrates. B) Two‐component FMO hydroxyl functionalisation using HPAH systems, HpaBC (monooxygenase and oxidoreductase components) and HpaB (FMO monooxygenase).
Unlike single‐component monooxygenases, which have only been reported to use FAD as a cofactor, two‐component monooxygenases can either use reduced FAD or FMN. [80] 4‐hydroxyphenylacetate 3‐hydroxylases (HPAHs) are extensively‐studied two‐component monooxygenases which catalyse the hydroxylation of 4‐hydroxyphenylacetate (4‐HPA) to yield 3,4‐dihydroxyphenylacetate (3,4‐DHPA). [90] Kino et al. studied HpaBC (HPAH monooxygenase (HpaB) and oxidoreductase (HpaC) components) from Pseudomonas aeruginosa, expressed in E. coli, for the synthesis of hydroxycinnamic acids, harbouring antioxidant activity. [91] HpaBC displayed high activity toward 3‐(4‐hydroxyphenyl)propanoic acid (9), to produce 3‐(3,4‐dihydroxyphenyl)propanoic acid (10), a compound with high medicinal potential with antiproliferative effect against human cancer cell lines. HpaBC was also the first bacterial oxygenating biocatalyst found to hydroxylate caffeic acid, ferulic acid and coniferaldehyde. [91] HpaBC has also been studied for its ortho‐hydroxylation of ‐tyrosine to produce ‐3,4‐dihydroxyphenylalanine ( ‐DOPA), a prescription drug for Parkinson's disease. [92] Chaiyen and co‐workers have shown that rational engineering of HPAH monooxygenase HpaB, from Acinetobacter baumanii, could catalyse the double hydroxylation of 4‐HPA (11) and p‐coumaric acid (12) to produce 2‐(3,4,5‐trihydroxyphenyl)acetic acid (13) (3,4,5‐THPA) and 3,4,5‐hydroxycinnamic acid (3,4,5‐THCA) (14), respectively. [93] The Chaiyen group have also engineered HpaB to expand the reactivity of the FMO to beyond its natural substrates. By a single mutation, the group yielded the S146A variant which accepted an aniline, instead of the native phenolic substate. S146A hydroxyllated 4‐aminophenylacetic acid (4‐APA) (15) to form 3‐hydroxy‐4‐aminphenylacetic acid (3‐OH‐4‐APA) (16) in ∼100 % yield at pH 6 (Figure 3B). [94] The group have also shown that mutations at R263 residue, which is speculated for the specificity towards substrates containing negatively charged carboxylic acid moieties, could enable hydroxylation of 4‐hydroxyphenylethylamine derivatives. [95] Mutant R263D was found to hydroxylate tyramine to dopamine (57 % yield) and native‐substrate 4‐HPA to 3,4‐DHPA (86 % yield). Double mutant R263D/Y398D could also hydroxylate octopamine to form norepinephrine, albeit lacking in stereoselectivity and activity (∼10 %), and represents a good starting‐point for engineering HPAH systems for the synthesis of catecholamine drugs. [95] Furuya et al. have demonstrated that a genome‐mining approach in the HPAH family can produce new enzymes capable of the synthesis of 3’‐ and 6’‐hydroxyequols from biologically‐active equol. [96] Deng and co‐workers have functionally and structurally characterised HpaB from E. coli and found it can hydroxylate a range of phenolic substrates including tyrosol, hydroxymandelic acid, coumaric acid, hydroxybenzoic acid and phenol. [97] Recently, Wessjohann et al. have rationally engineered HpaB from E. coli to catalyse hydroxylation on bulky aromatic substrates including ferulic acid, naringenin and alkaloid mimetic 2‐hydroxycarbaozole. [98] The group exchanged selected residues in HpaB from E. coli with those found in homolog HpaB from P. aeuginosa, leading to mutants that performed highly regiospecific aromatic hydroxyl functionalisations with no byproducts. [98] This work has attested the viability to synthesise pharmaceutically‐relevant hydroxylated aromatic compounds via single‐component or two‐component FMOs.
The occurrence of FMO‐catalysed sp 3 C−H hydroxylation has been rarer. The first reported FMO capable of hydroxylating non‐activated alkanes was LadA from Geobacillus thermodenitrificans NG80‐2, isolated in 2007. [99] LadA catalyses the selective, but slow, terminal hydroxylation on n‐alkanes (C15 to C36) with the utility of FMN cofactor. While reports of LadA application have been low, work by Dong et al. demonstrated that random‐ and site‐directed mutagenesis studies could enhance LadA activity towards hexadecane substrate. [100]
In summary, FMOs are attractive, well‐established, oxygenating biocatalyst for hydroxyl functionalisations. At present, there is still a limited substrate scope, with mainly phenolic compound substrates, for single‐ and two‐component FMO aromatic hydroxylases and only one FMO capable of alkane hydroxylation has thus‐far been reported. Additionally, like cytochrome P450s, many FMOs depend on expensive NAD(P)H cofactors for electron transfer. Similarly to work described for cytochrome P450s, dehydrogenase systems have been employed for cofactor regeneration, or using FMOs as whole‐cell systems.[ 101 , 102 ] Promising alternatives have used nicotinamide analogues for direct reduction of flavins.[ 103 , 104 ] To this end, cheap nicotinamide coenzyme biomimetics (NCBs) can be used in stoichiometric amounts and have application in both single‐ and two‐component monooxygenase reactions. [103]
2.2. Emerging Oxygenating Biocatalysts
2.2.1. Iron‐ and α‐Ketoglutarate‐dependent Oxygenases
Iron‐ and α‐ketoglutarate‐dependent oxygenases (Fe/αKGs) have proven to be a robust, scalable enzyme class that may be evolved to expand substrate scope for pharmaceutical intermediates. [105] These oxygenases contain a conserved ligation of two His residues and a carboxylate residue, either Glu or Asp (the His1‐X‐Asp/Glu‐Xn‐His2 motif). [106] Catalysis initiates with the primary substrate binding to a αKG‐bound Fe(II)‐species (A), displacing a water ligand. Molecular oxygen is activated by binding to the resulting Fe(II)‐species (B), followed by expulsion of CO2, to form an Fe(IV)‐oxo species (C, also known as the ferryl intermediate) bound to succinate. The ferryl intermediate mediates hydrogen‐abstraction of the primary substrate to facilitate hydroxylation, via D. Subsequent product dissociation and H2O/αKG ligand‐rebinding regenerates the resting state of the enzyme (E) (Scheme 3).[ 105 , 106 , 107 , 108 ]
Scheme 3.
Simplified catalytic cycle of Fe/αKG hydroxylations. FeII‐binding amino acid residues His1‐X‐Asp/Glu‐Xn‐His2 motif displayed. Molecular oxygen is activated when bound to the FeII‐species to form reactive FeIV‐oxo intermediate. Subsequent hydrogen abstraction is followed by combination of the substrate radical with the ferric hydroxyl group to complete the O‐transfer. [108]
Therefore, the cycle utilises the oxidative decarboxylation of αKG, forming CO2 and succinate as coproducts. [25] Unlike cytochrome P450s, Fe/αKGs do not depend on expensive cofactors NAD(P)H nor require a reductase partner to facilitate electron transfer.[ 105 , 108 ] Thus, their cost‐effective synthetic utility, coupled with an innate activity toward amino acid substrates, is highly attractive for C−O bond formation with pharmaceutical applications.[ 109 , 110 ]
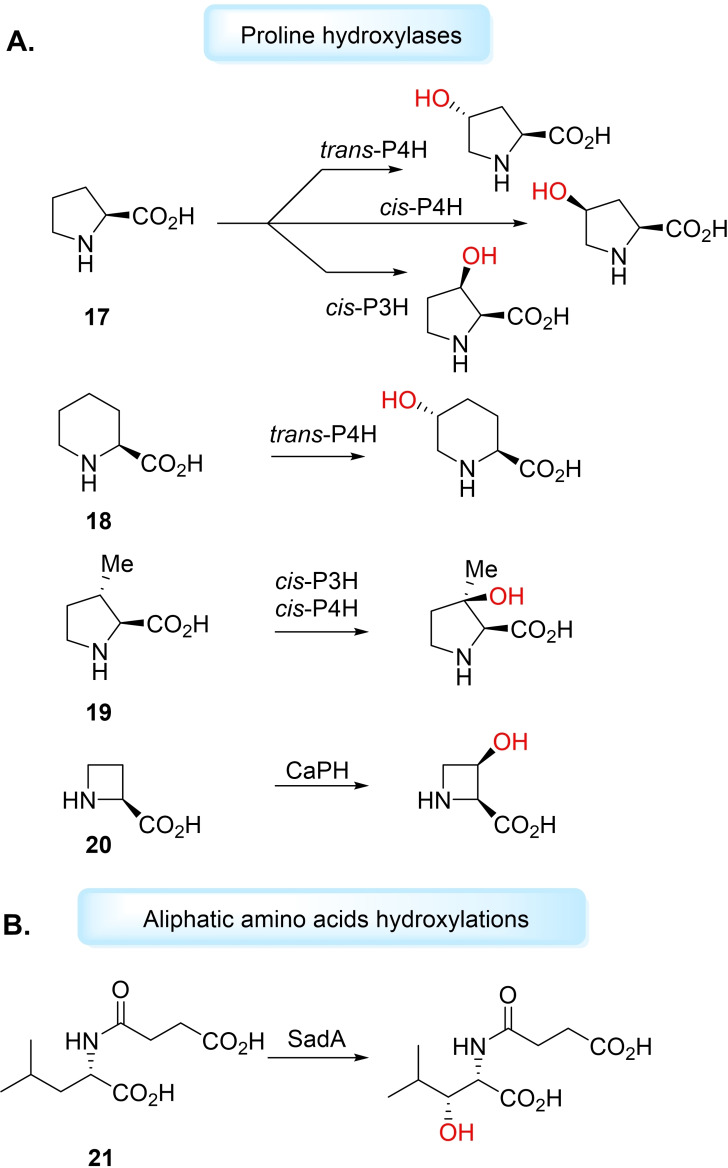
The remote hydroxylation of unactivated sp 3 C−H sites of amino acid substrates is a desired, but challenging, synthetic reaction. Proline hydroxylases (PHs) represent well‐studied and characterised Fe/αKGs for free amino acid hydroxyl functionalisation. Hüttel and Klein applied PHs for the selective hydroxylation of ‐proline (17) and its congener ‐pipecolic acid (18) ( ‐Pip), yielding building blocks for pharmaceutical synthesis. [111] The authors fed ‐proline to growing cultures of cis‐P3H, cis‐P4H and trans‐P4H in E. coli strains to produce the respective hydroxyprolines (Hyps) in isolated yields of 35–61 % after purification via ion‐exchange chromatography. It was also shown that trans‐P4H was capable of hydroxylating non‐natural substrate ‐Pip, producing trans‐5‐hydroxypipecolic acid (trans‐5‐Hypip) in good yield (68 %) in vitro. [111] Hüttel and Klein also demonstrated, in later work, cis‐P3H and cis‐P4H ability to accept non‐natural substrate trans‐3‐methyl‐ ‐proline (19) to produce the tertiary alcohol product, a biotransformation which had not yet been reported for Fe/αKGs. [112] Since these publications, other work has investigated PHs for oxygenation of other proline derivatives or for improved regio‐ and stereoselectivity.[ 113 , 114 ] Kino et al. showed a range of cis‐PHs MIP4H, SmP4H, SrPH and CaPH could selectively hydroxylate cis‐3‐Hyp to the 3,4‐cis‐dihydroxylated product and CaPH could also hydroxylate ‐azetidine‐2‐carboxylic acid (20) to the cis‐3‐hydroxy product (Figure 4A). [113] Higuchi et al. used protein engineering to improve the selectivity of WT cis‐P4H hydroxylation of ‐Pip. [114] After three rounds of directed evolution on SmP4H, the cis‐P4H triple‐mutant, V97F/V95W/E114G, catalysed hydroxyl functionalisation to produce cis‐5‐Hypip, in 95 % regioselectivity, and minor product cis‐3‐Hypip (60 : 40 cis‐5 : cis‐3 reported for WT). [114] Hüttel and Mattay have since found a clade of PHs with selectivity for ‐Pip as native substrate, rather than ‐proline, and therefore coined pipecolic acid hydroxylases (PiHs). [115] PiHs GetF and PiFa were found to catalyse hydroxylations on congeners 3,4‐dehydro‐ ‐proline and ‐azetidine‐2‐carboxylic acid. [115] Other work has seen recombinant strain improvement of P4H can increase the production of trans‐4‐Hyp from ‐proline.[ 116 , 117 ] Jing et al. have used genome mining to identify a new Fe/αKG from Kutzneria albida, KaPH1, that could convert ‐proline to trans‐4‐Hyp in vitro with 92.8 % yield. [118] Snajdrova et al. have also recently applied genome mining for the discovery of novel Fe/αKGs for the oxidation of ‐proline. Fe/αKG Ssp5PH was found to catalyse dihydroxylation, and following Fmoc protection of the diol, yielded (2S,3R,4S)‐N‐Fmoc‐3,4‐dihydroxyproline with 40 % selectivity. [119] Schofield and co‐workers have demonstrated a relaxed substrate tolerance of cis‐P3H, cis‐P4H and trans‐P4H to catalyse the hydroxylation of proline analogues with larger ring sizes, varying substituents, N‐methylations, fluorinated and hydroxylated rings and bicyclic substrates. [120]
Figure 4.
Hydroxylations catalysed by Fe/αKGs. A) Proline hydroxylases catalyse hydroxylation of ‐proline (17), ‐pipecolic acid (18), trans‐3‐methyl‐ ‐proline (19) and L‐azetidine‐2‐carboxylic acid (20). B) C−H hydroxyl functionalisation on other amino acids such as SadA used to hydroxylate N‐succinyl ‐leucine (21).
Other Fe/αKGs catalyse the hydroxylation of different aliphatic amino acids, forming important precursors for the synthesis of therapeutic compounds. Ogawa et al. have reported a ‐leucine‐5‐hydroxylase (LdoA) which catalyses regio‐ and stereoselective hydroxylation of ‐leucine and ‐norleucine into (2S,4S)‐5‐hydroxyleucine and (2S)‐5‐hydroxynorleucine, respectively. [121] Ogawa et al. have also obtained a novel Fe/αKG, SadA, from Burkholderia ambifaria AMMD for the production of N‐succinyl‐ ‐threo‐β‐hydroxyleucine, a target for the synthesis of cyclic depsipeptide antibiotics. [122] SadA was found to accept N‐formyl N‐acetyl ‐leucine, N‐succinyl‐ ‐leucine (Ns‐L‐Leu) and N‐carbamoyl ‐leucine as substrates. Recombinant SadA catalysed hydroxylation of Ns‐L‐Leu (21) with >99 % diastereoselectivity, in 93 % conversion, and was the first enzyme to catalyse β‐hydroxylation of amino acid‐related substrates (Figure 4B). [122] Kourist et al. have developed the SadA system by coupling the Fe/αKG with ‐glutamate oxidase (LGOX), for the in situ production of co‐substrate αKG from ‐glutamate. [123] Zaparucha et al. have used a genome‐mining approach to identify Fe/αKGs able to perform hydroxyl functionalisation with high regio‐ and stereoselectivity on basic amino acids ‐lysine, ‐arginine and ‐ornithine.[ 124 , 125 ] Kino et al. have studied a novel Fe/αKG from Sulfobacillus thermotolerans Y0017 and shown its ability to catalyse the regio‐ and stereoselective threo‐β‐hydroxylation of ‐histidine and ‐glutamine on a preparative scale. [126] Recently, work from Snajdrova and Buller has demonstrated structure‐guided semi‐rational mutagenesis of SmP4H could produce mutants capable of catalysing selective γ‐hydroxylation of nonproteinogenic amino acid ‐homophenylalanine to produce pharmacologically significant molecules. [127] These results attest the viability of Fe/αKGs to form optically active hydroxylated amino acids, important synthetic intermediates to pharmaceuticals.[ 122 , 128 ]
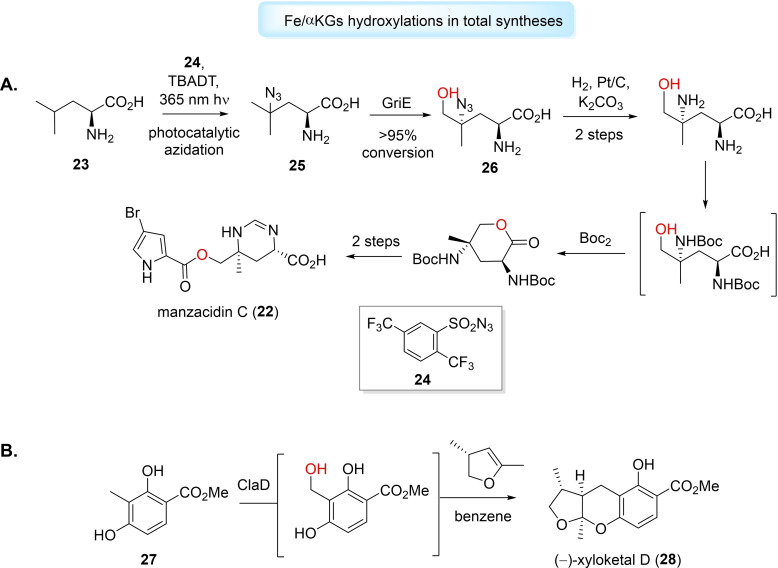
Hydroxyl functionalisation catalysed by Fe/αKGs have also been utilised for the total synthesis of complex natural products and bioactive molecules.[ 105 , 108 , 129 ] Renata et al. reported a formal synthesis of rare alkaloid, manzacidin C (22), and densely substituted amino acid derivatives, using a Fe/αKG that selectively catalysed δ‐hydroxylation of various aliphatic acids.[ 130 , 131 ] The group applied ‐leucine‐5‐hydroxylase, GriE, and inspired by work of Britton, [132] devised a strategy for ‐leucine (23) to undergo photo‐catalysed C−H azidation, with azide 24 as radical acceptor. The resulting azidoleucine (25) was converted by GriE in >95 % to the β‐hydroxylated derivative (26). The group implemented this enzymatic hydroxyl functionalisation into a five‐step formal synthesis of manzacidin C (Figure 5A), that was dramatically simplified and favourable compared to previously reported routes.[ 130 , 131 ] The Renata lab have also employed Fe/αKG‐catalysed hydroxyl functionalisation for the chemoenzymatic total syntheses of cepafungin I, [133] GE81112 B1 [134] and tambromycin, [135] among others. Finally, Narayan et al. have used Fe/αKGs, CitB and ClaD, for the benzylic hydroxylation of o‐cresol. [136] The alcohol product, under the aqueous reaction conditions, can convert to o‐quinone methide and be captured by nucleophiles or dienophiles to generate further functionalised species. The group employed this system with ClaD using methyl 2,4‐dihydroxy‐3‐methylbenzoate (27) to synthesise fungal metabolite (−)‐xyloketal D (28) in 64 % yield (Figure 5B). [136]
Figure 5.
Fe/aKG‐catalysed hydroxyl functionalisations aid the total synthesis of complex compounds. A) GriE employed for manzacidin C (22) total synthesis. B) ClaD used in (−)‐xyloketal D (28) total synthesis.
2.2.2. Unspecific Peroxygenases
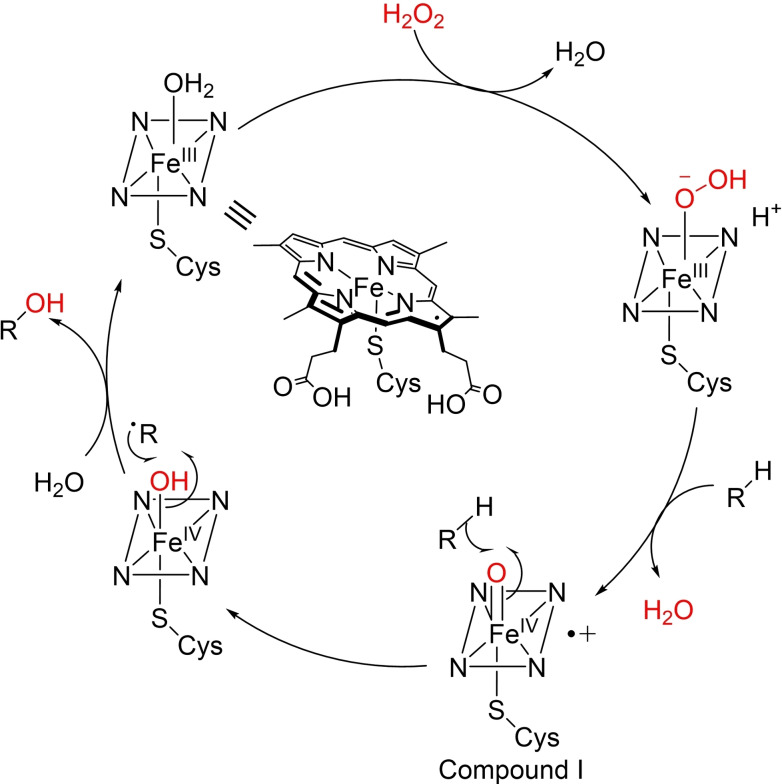
Unspecific peroxygenases (UPOs) have been identified as an emerging enzyme class for key C−H oxidation reactions, producing valuable compounds for the pharmaceutical industry.[ 137 , 138 ] UPOs are structurally‐similar to cytochrome P450s, in that they are haem‐thiolate proteins, but instead rely on inexpensive H2O2 as oxidant, while independent of NAD(P)H cofactors. [139] Additionally, UPOs are extracellular secreted enzymes and display tolerance towards pH and organic solvents, [140] providing attractive versatility and simplicity. In the catalytic cycle, H2O2 is activated when bound to a Fe(III)‐species, followed by expulsion of water, to form a Fe(III)‐peroxo complex. After heterolytic O−O bond cleavage, the oxoiron (IV) compound I species is formed which can facilitate substrate hydrogen abstraction and subsequent oxidation (Scheme 4). [141]
Scheme 4.
Simplified catalytic cycle for C−H hydroxylation catalysed by unspecific peroxygenases.
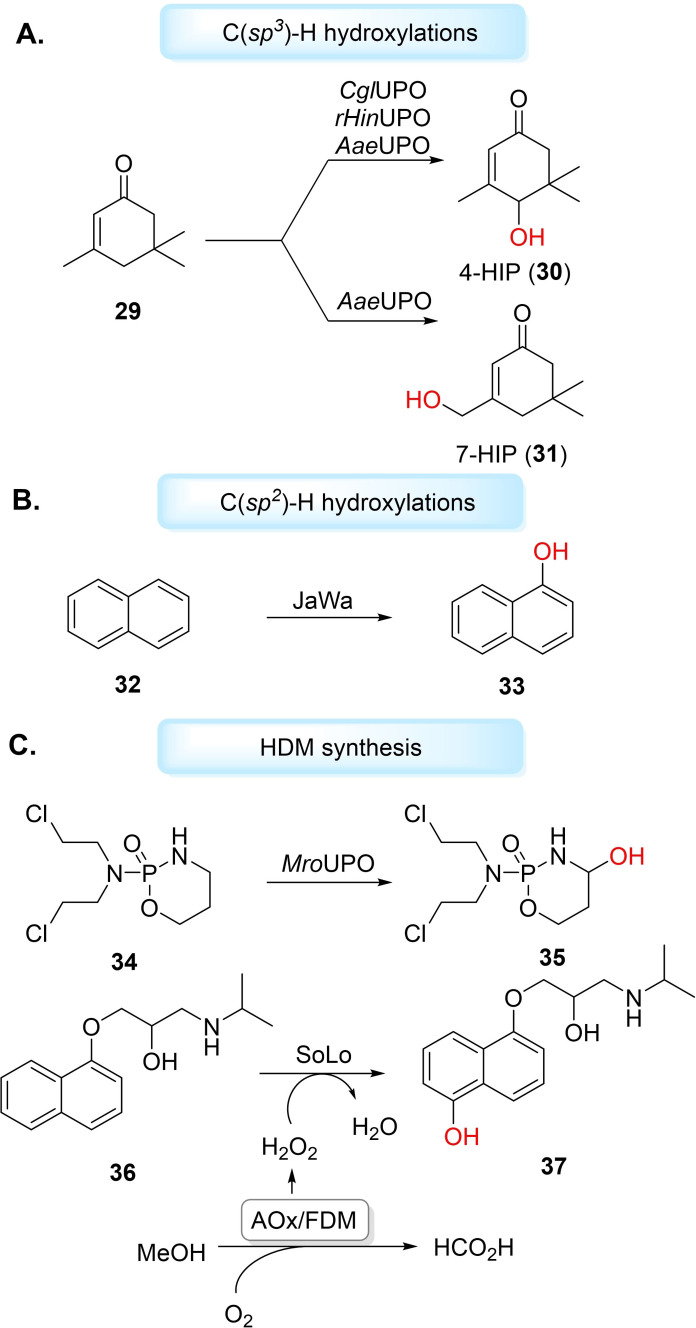
Over the past decade, UPOs have gained attention for catalysing aromatic and aliphatic hydroxylation [142] reactions. [139] Río and co‐workers used model UPO from Agrocybe aegerita (AaeUPO) for the regioselective hydroxylation of fatty acids, fatty alcohols and other aliphatic compounds. [143] For the fatty acid substrates, monohydroxylation was observed to produce predominately ω‐1 and ω‐2 hydroxy fatty acids, except for myristoleic acid, which only produced the ω‐2 derivative. Linear alkanes dodecane, tetradecane and hexadecane, yielded a mixture of 2‐alkanols, 3‐alkanols and dehydroxylated derivates and demonstrated the lowest substrate conversion by the UPO. [143] Hofrichter et al. have also studied AaeUPO hydroxylation of linear, branched and cyclic alkanes. [144] AaeUPO catalysed monohydroxylation of n‐heptane and n‐octane with high enantioselectivity to the corresponding 3‐alcohol (>99 % ee). [144] Nevertheless, both Río and Hofrichter reported subsequent over‐oxidation of the alcohol products by the UPOs,[ 143 , 144 ] which is a reported problem for UPO alkane hydroxylations. AaeUPO has more recently been applied for the hydroxyl functionalisation of isophorone (29), along with recombinant UPOs from Chaetomium globosum DSM‐62110 (CglUPO) and Humicola insolens (rHinUPO). [145] AaeUPO catalysed formation of 4‐hydroxyisophorone (30) (4‐HIP) and 7‐hydroxyisophorone (31) (7HIP), while CglUPO and rHinUPO produced only 4HIP (Figure 6A). All UPOs showed a tendency to over‐oxidise the alcohol products to their respective keto‐forms. [145] UPOs hydroxylate aromatic rings by the formation of an epoxide intermediate, which can rearrange to the corresponding phenol. [139] Hofrichter and co‐workers reported the hydroxylation of benzene using AaeUPO. As well as the phenol product, the group observed a mixture of hydroquinone, o‐ and p‐benzoquinone, catechol and traces of 1,2,4‐trihydroxybenzene and hydroxy‐p‐benzoquinone because of further oxidation. [146] The variety of products reflects the ability of AaeUPO to hydroxylate unactivated aromatic compounds such as benzene and its derivatives under mild conditions. Additionally, protein engineering may improve selectivity of UPO aromatic hydroxyl functionalisations and avoid quinone formation from peroxidative activity. AaeUPO has been engineered by directed evolution to produce a double mutant, JaWa (G241D‐R257K), for the hydroxylation of naphthalene (32) to 1‐napthol (33) with high regioselectivity (97 %) and diminished peroxidative activity (Figure 6B). [147] However, UPO production and engineering in recombinant strains remains a substantial bottleneck for industrial application. Indeed, most UPO engineering studies have been through eukaryotic expression systems,[ 148 , 149 ] with only a few successful examples in heterologous hosts.[ 139 , 150 ] Alcade et al. have demonstrated that directed evolution on AaeUPO, to produce mutant PaDa‐I, could lead to enhanced heterologous expression and activity in yeast. [149] Mutant PaDa‐I has since been used to produce novel UPO chimeras by combining different sequence blocks with other UPOs Galerina marginata (GmaUPO) and Coprinopsis cinerea (CciUPO), followed by a multiple‐injection GC‐MS high‐throughput screening for tetralin hydroxyl functionalisation. [151] Weissenborn et al. have recently applied a modular Golden Gate‐based secretion system to successfully produce four UPOs from Marasmius rotula (MroUPO), Chaetomium globosum (CglUPO), Myceliophthora thermophila (MthUPO) and Thielavia terrestris (TteUPO), respectively. [152] These enzymes were expressed in Saccharomyces cerevisiae and found to catalyse the benzylic hydroxylation of various phenylalkanes. Additionally, after transferring expression to Pichia pastoris, MthUPO was used on a preparative scale to catalyse the hydroxylation of N‐phthaloyl‐phenethylamine to produce 2‐N‐phthaloyl‐1‐phenylethanol in 57 % yield, 99 % ee. [152] The group have extended this methodology for the engineering of MthUPO in S. cerevisiae.[ 153 , 154 ] Consequently, MthUPO variant A161L was found to catalyse octane hydroxylation, with selectivity towards terminal 1‐octanol (38 %). The only other reported UPO‐catalysed terminal hydroxylation of linear alkanes demonstrated over‐oxidation to the corresponding carboxylic acid. [155] No aldehyde or carboxylic acid products were found with A161L, making it an interesting case for future work on terminal hydroxyl‐functionalisation of linear alkanes. [153] In a similar study, engineered MthUPO mutant L60F was found to catalyse the benzylic hydroxylation of indane and tetralin to produce (R)‐1‐indanol (95 % ee) and (R)‐1‐tetralol (74 % ee), respectively. [154] Babot et al. have studied AaeUPO, MroUPO, CglUPO, CciUPO and HinUPO for the oxyfunctionalisation of α‐ and β‐ionones and their respective isomers, α‐ and β‐damascones, to produce hydroxylated products with high significance as synthetic building blocks for pharmaceuticals. [156]
Figure 6.
Hydroxyl functionalisations catalysed by UPOs. A) C(sp 3 )−H hydroxylation of isophorone (29) to produce 4‐HIP (30) and 7‐HIP (31) with AaeUPO, while CglUPO and rHinUPO produce only 4‐HIP. B) C(sp 2 )−H hydroxylation of naphthalene (32) catalysed by AaeUPO mutant to produce 1‐napthol (33). C) UPOs used for metabolite synthesis such as forming HDM 4‐OH‐CPA (35) and 5’‐hydroxypropranolol (37). SoLo‐catalysed hydroxylation uses bienzymatic cascade alcohol oxidase (AOx) and formaldehyde dismutase (FDM) for the in situ generation of H2O2.
The oxygenation pattern of UPOs can mimic that of human liver cytochrome P450s, and thus may be exploited for HDM synthesis. [157] Hofrichter et al. used AaeUPO and a UPO from Coprinellus radians (CraUPO) for the aromatic hydroxylation of pharmaceuticals including propranolol, carbamazepine, diclofenac and tamoxifen. [157] Both UPOs were also found to catalyse aliphatic hydroxylation of ibuprofen and tolbutamide, with all hydroxylated products reported as HDMs. [157] Recently, Scheibner et al. used MroUPO for hydroxylation of the cytostatic drug cyclophosphamide (34) (CPA), to produce HDM 4‐hydroxycyclophosphamide (35) (4‐OH‐CPA) in 32 % yield (>97.6 % purity). [158] CglUPO has been studied for the oxyfunctionalisation of testosterone, producing 90 % of the 4,5‐epoxide product and 10 % HDM 16α‐hydroxytestosterone, both in high diastereoselectivities (>98 %). [159] Gomez De Santos et al. have engineered a highly active and stable AaeUPO mutant SoLo, using earlier mutant JaWa as a starting point, in yeast for the synthesis of propranolol (36) HDM 5’‐hydroxypropranolol (37) (5’‐OHP), in 99 % regioselectivity. [160] Notably, the group generated H2O2 in situ utilising a two‐enzyme cascade of alcohol oxidase (AOx) and formaldehyde dismutase (FDM) for the double oxidation of methanol, as sacrificial electron donor, to formic acid. The gentle in situ supply of H2O2 can avoid oxidative inactivation of the UPO caused by excess concentrations of H2O2. Previous efforts to maintain optimal H2O2 concentrations levels had used a glucose/glucose oxidase system for in situ generation, but has suffered poor atom‐efficiency before Hollmann et al. presented the oxidation of methanol as a solution. [161] Moreover, Alcade et al. have demonstrated that the AaeUPO WT and mutants could be applied to catalyse the hydroxylation of Na+‐channel blocker tolbutamide to produce HDM 4‐hydroxymethyl‐tolbutamide, albeit with some UPOs producing minor amounts of over‐oxidised product, 4‐formyl‐tolbutamide, additionally. Recently, photochemical [162] and electrochemical [163] methods have been investigated for the generation H2O2 in recombinant AaeUPO‐catalysed hydroxylations.
UPOs have proven to be viable oxygenating biocatalysts for hydroxyl functionalisation of a diverse range of substrates. The simplicity of their application, such as cofactor‐independence and reliance on inexpensive H2O2, stands them in good stead to support the practical shortcomings of cytochrome P450s. Their applicability will likely surge with increasing developments in heterologous expression systems and protein engineering efforts, increasing their biocatalytic potential. [164] Screening kits of phylogenetically diverse UPOs, both WT and mutants, have recently become more widely available from commercial suppliers such as Aminoverse and EvoEnzyme, [165] providing a simplified route to implement these enzymes in drug discovery settings.
3. Applications of Oxygenating Biocatalysts in Drug Discovery and Development
3.1. Late‐stage functionalisation
Late‐stage functionalisation (LSF) of (drug‐like) compounds offers diversification of chemical space without laborious synthetic routes and may only consume milligram‐quantities of material. [166] We wish to clarify that LSF can be distinguished from HDM synthesis, where other goals such as characterisation or assessing drug toxicity are desired, whereas LSF can be utilised solely as a synthetic tool and may not result in HDMs. Hydroxylations on a drug‐scaffold can improve activity, selectivity and solubility, and therefore accelerate lead compound generation, or offer a handle for further functionalisation. [166] Romero et al. have recently exhaustively covered enzymatic LSFs. [167] This section will only summarise the aforesaid oxygenating biocatalysts’ applications in late‐stage hydroxyl functionalisations, and the benefit this brings to drug discovery and development.
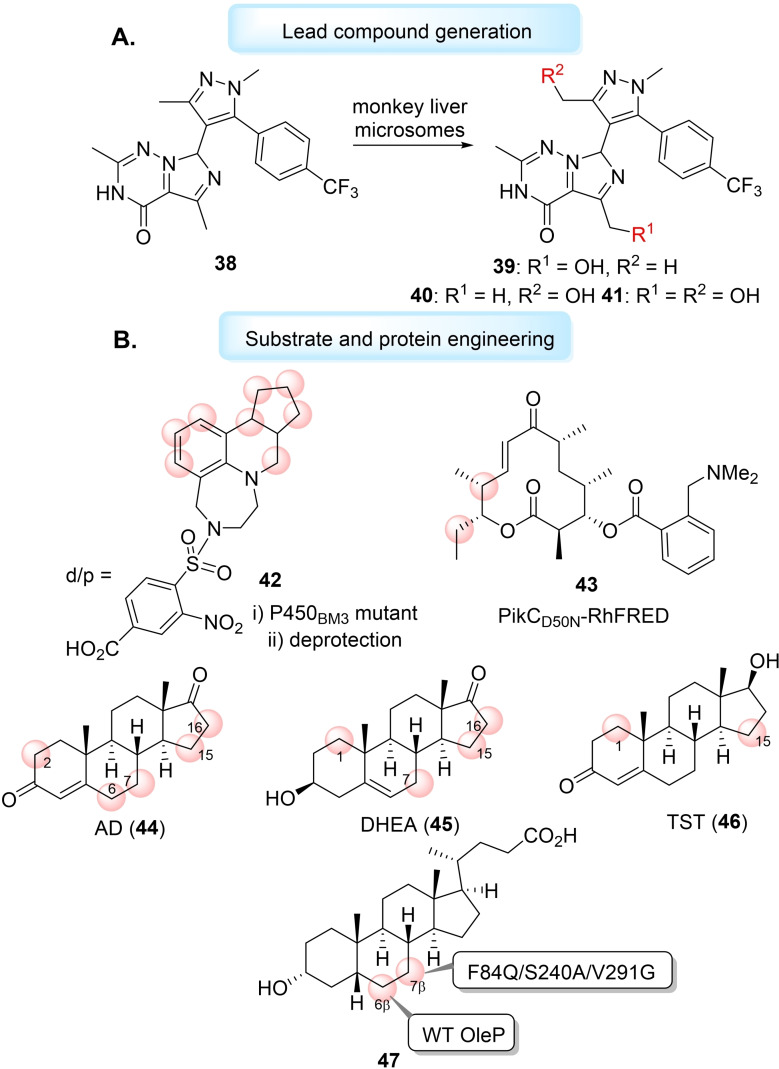
Late‐stage hydroxylations, catalysed by cytochrome P450s, of complex molecule scaffolds can generate analogues of existing bioactive molecules or produce new lead compounds. [168] Obach et al. used liver microsomes, a mixture of cytochrome P450s, to generate a new lead candidate for a phosphodiesterase 2 (PDE2) inhibitor (38), which had improved metabolic clearance and minimised drug‐drug interactions. Lead candidate 38 (PDE2 IC50=0.6 nM), was metabolised by monkey liver microsomes to produce three new products, two monohydroxy products (39 and 40) and one dihydroxy product (41) (Figure 7A). New analogue 38 had improved potency (PDE IC50=0.4 nM), metabolic stability and lower lipophilicity. [169] Recently, Fessner et al. have repurposed the human liver cytochrome P450 3A4 for LSF utility, pushing the oxygenating biocatalyst to beyond its traditional use in inhibition and toxicology studies. [170] The group exploited the enzyme's natural lack of substrate selectivity to oxyfunctionalise a range of natural products including terpenes, steroids and alkaloids, generating hydroxylated derivatives of pharmacological interest, without the need for protein engineering. [170] However, a combination of protein and substrate engineering can still be a valuable route to improve other cytochrome P450 LSF applicability. Lange et al. designed docking/protecting (d/p) groups on vabicaserin (42), a serotonin (5‐HT)2C receptor agonist, to steer the regioselectivity of a WT and mutant P450BM3‐mediated late‐stage monohydroxylation. [171] Sherman et al. used synthetic N,N‐dimethylamino anchoring groups on the macrocyclic YC‐17 aglycone (43) through an ester linkage to control the regioselectivity of hydroxylation catalysed by the engineered self‐sufficient P450 PikCD50N‐RhFRED (Figure 7B). [172] The group found the size, stereochemistry and rigidity of the anchoring group influenced the regioselectivity of the enzymatic C−H bond hydroxylation. [172] In seminal work, Sherman et al. have shown that engineering the iterative cytochrome P450 TamI could generate a toolbox of TamI mutants capable of multiple late‐stage oxidations on the antibiotic, tirandamycin. [173] One such mutant, L101A_L295I, was found to catalyse epoxidation and hydroxyl functionalisation to generate new congener tirandamycin N, with comparable antimicrobial activity to erythromycin. [173] Work has also been conducted towards the random mutagenesis of cytochrome P450 MycG to produce mutant V135G/E355K that can selectively hydroxylate potent antibiotic mycinamicin to produce analogue mycinamicin V. [174] Urlacher and co‐workers have shown active site mutagenesis on P450BM3 could generate mutants capable of catalysing hydroxylation of β‐cembrenediol, a 14‐membered macrocycle with a breadth of biological activity, in high regio‐ and stereoselectivity. [175] Urlacher et al. have followed this work by developing chemoenzymatic syntheses of structurally‐related cembranoid‐ols and cembranoid‐diols using P450BM3 mutants. [176] Wong et al. have engineered P450BM3, to hydroxylate unreactive sites of androstenedione (AD) (44), dehydroepiandrosterone (DHEA) (45) and testosterone (TST) (46). After analysis of the steroid C19‐demethylase CYP19A1 active site, with AD‐bound, and comparing this to the active site of P450BM3, the group combined scanning glycine mutagenesis with a second round of mutations to elect a library of mutants able to selectively hydroxylate (up to 97 %) of AD, DHEA and TST at the widest range of positions reported by a bacterial cytochrome P450. [177] Recently, Bornscheuer et al. have engineered the P450 CYP107D1 (OleP) for the regio‐ and stereoselective 7β‐hydroxylation of lithocholic acid (LCA) (47) to produce therapeutic agent ursodeoxycholic acid (UDCA). [178] WT OleP exclusively hydroxylates LCA at the 6β‐position to generate murideoxycholic acid (MDCA), but the group used enzyme structural analysis for the rational design of triple mutant F84Q/S240A/V291G for the near complete inversion of regioselectivity to 7β‐hydroxylation (Figure 7B). [178]
Figure 7.
A) Late‐stage hydroxylation of PDE2 inhibitor 38 to produce new lead candidate 39 using monkey liver microsomes. B) Protein and substrate engineering for late‐stage hydroxylations, different hydroxylation sites shown with red circles. Docking/protecting (d/p) groups used on vabicaserin 42 to control regioselectivity of hydroxylations. Anchoring groups used on macrocycle YC‐17 43 direct site of hydroxylations using PikCD50N‐RhFRED. Sites of hydroxylations using P450BM3 engineered by combining scanning glycine mutations and mutagenesis on AD (44), DHEA (45) and TST (46). Engineering the regioselectivity of OleP for the late‐stage hydroxylation of LCA (47).
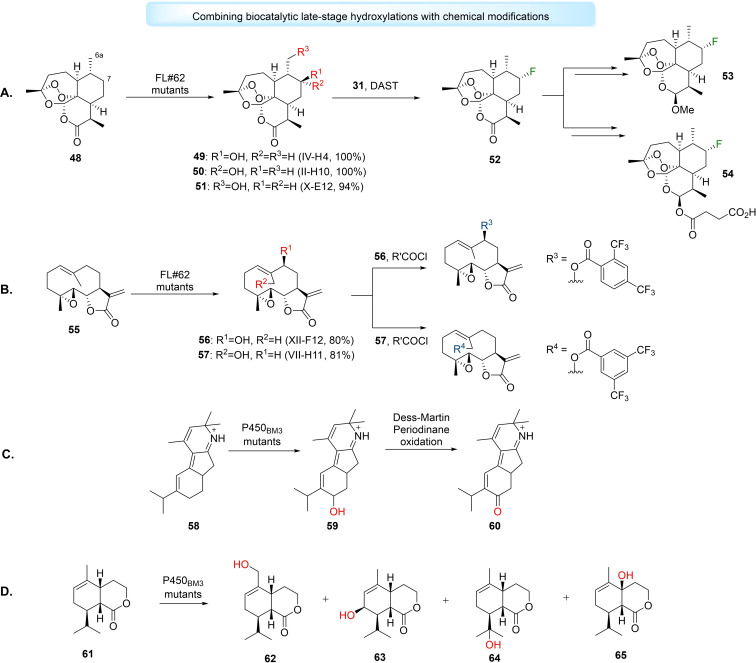
Approaches which combine LSF with chemical modifications may be utilised to diversify a bioactive molecule's architecture. Fasan et al. used cytochrome P450‐catalysed hydroxylations to elaborate unactivated C−H sites in artemisinin (ART) (48). [179] The group used an engineered P450BM3, FL#62, and utilised fingerprinting methods to create three efficient mutants, IV‐H4, II‐H10 and X‐E12, that catalysed selective hydroxylation of ART to 7(S)‐hydroxy‐ART (49) (100 %), 7(R)‐hydroxy‐ART (50) (100 %) and 6a‐hydroxy‐ART (51) (94 %), respectively. Subsequently, 49 was subjected to deoxyfluorination by diethylaminosulfur trifluoride (DAST) to produce enantiopure 7(R)‐fluoro‐ART (52), in 82 % yield, which could be converted to analogues of clinical antimalarial drugs artemether (53) and artesunate (54), in two steps, in high yields (Figure 8A). [179]
Figure 8.
Combined cytochrome P450 late‐stage hydroxylations with chemical transformations. A) Generation of artemisinin (48) analogues using FL#62 mutants engineered utilising cytochrome P450 fingerprinting methods, the generated alcohol product 49 was subjected to deoxyfluorination and the fluorinated analogue 52 was used to synthesise fluorinated derivates of antimalarial drugs artemether (53) and artesunate (54). B) Generation of parthenolide analogues using late‐stage hydroxylations catalysed by FL#62 mutants and then benzoylation using acid chlorides. C) Enantioselective total synthesis of nigelladine A (60) using hydroxylation catalysed by engineered P450BM3 variant followed by Dess‐Martin Periodinane oxidation. D) Synthesis of hydroxylated eleuthoside synthetic intermediates using P450BM3 variants with the aim to synthesise eleutherobin.
Similarly, Obach et al. have also used a combination of cytochrome P450‐catalysed hydroxylation with chemical halogenation via DAST to enable the late‐stage replacement of hydrogen with metabolically stable fluorine in existing drugs such as midazolam, celecoxib, ramelteon and risperidone. [180] Fasan et al. have also used FL#62 and cytochrome P450 fingerprinting methods for the chemoenzymatic functionalisation of aliphatic sites on parthenolide (PTL) (55). [181] Mutants XII‐F12 and VII‐H11 catalysed the selective hydroxylation of PTL to form 9(S)‐hydroxy‐PTL 56 (80 %) and 14‐hydroxy‐PTL 57 (81 %), respectively. 56 and 57 were chemically benzoylated using acid chlorides to generate a panel of 9‐ and 14‐substituted PTL analogues, with some analogues possessing improved antileukemic properties (Figure 8B). [181]
Late‐stage hydroxylations can be used to aid the total syntheses of advanced synthetic intermediates or natural products by diversifying complex precursors in high efficiency when compared to conventional linear approaches. Stolz et al. devised the first enantioselective total synthesis of nigelladine A (60), a norditerpenoid alkaloid, via a regioselective late‐stage allylic hydroxylation of intermediate 58 catalysed by an engineered P450BM3, followed by oxidation with Dess‐Martin periodinane, in collaboration with the Arnold group (Figure 8C). [182] Robertson et al. used a panel of engineered P450BM3 mutants for the synthesis of hydroxylated analogues of eleutherobin, a cytotoxic compound with comparable activity to taxol, through LSF. [183] By screening eleutherobin precursor lactone 61 against the P450BM3 panel, the authors could produce five oxidised products and a sixth unidentified compound. The monohydroxylated products observed included the primary alcohol 62, allylic secondary alcohol 63 and two tertiary alcohols 64 and 65. Mutants such as RP/HL/IG catalysed highly regioselective hydroxylations of 61 to form only 62 (100 %). [183] Recently, Renata et al. have designed the syntheses of numerous α‐pyrone meroterpenoids utilising the direct C‐3 hydroxyl functionalisation of sclareol and sclareolide catalysed by P450BM3 mutant (BM3 MERO1), followed by further synthetic steps. [184]
There are fewer examples of FMOs applied for late‐stage hydroxylations. Kino et al. have demonstrated that two‐component FMO HpaBC could not only selectively hydroxylate chemically‐complex resveratrol, [185] but also the hydroxylated‐product, piceatannol. [186] The latter reaction afford 3,4,5,3’,5’‐pentahydroxy‐trans‐stilbene (PHS), a biologically‐active stilbene derivative in 1.8 g L−1 product titre, and represented the first example of this enzymatic hydroxylation. [186] Hong et al. also studied the regioselective hydroxylation of stilbene compounds but with a different FMO, Sam5. [187] The group studied Sam5 3’‐hydroxylase activities on a series of methylated‐resveratrol analogues (pinostilbene and pterostilbene) and hydroxylated‐resveratrol (oxyresveratrol), which produced the 3’‐hydroxylated products in 37–54 % conversions. [187] These examples demonstrate the LSF‐potential for FMO‐catalysed hydroxylations to generate a variety of hydroxylated‐stilbene derivatives with attractive pharmaceutical uses.
While the majority of LSF examples have relied on the well‐studies cytochrome P450s, emerging oxygenating biocatalysts may be utilised more frequently in the future. After their initial report of hydroxyl functionalisation with SadA, [122] Ogawa et al. used rational structure‐based protein engineering to improve the hydroxylation activity of Fe/αKG SadA on N‐succinyl‐threo‐3,4‐dimethyoxyphenylalanine (NSDOPA) to produce N‐succinyl‐ ‐threo‐3,4‐dimethoxyphenylserine (NSDOPS), a precursor to the psychoactive drug, Droxidopa. [188] Recently, Abe et al. have shown Fe/αKG SptF to display remarkable promiscuity, and can catalyse a series of oxidative biotransformations, including hydroxyl functionalisation, on meroterpenoid substrates. [189] Encouraged by these findings, the authors tested SptF's ability to catalyse hydroxylation of steroids including androsterone, testosterone and progesterone, and found SptF exhibited good conversion rates (34–53 %) with these substrates. [189] UPOs have also been studied to catalyse hydroxyl functionalisation on pharmaceutical substrates.[ 157 , 159 , 190 ] Babot et al. employed three different UPOs AaeUPO, MroUPO and recombinant UPO CciUPO, to catalyse regioselective hydroxylation on a variety of steroids. [191] In one example, the antiviral therapeutic 25‐hydroxycholestrol was produced from cholesterol using CciUPO. [191] CciUPO has also been studied for the hydroxylation of ergocalciferol (vitamin D2) and cholecalciferol (vitamin D3) to produce therapeutic agents 25‐hydroxyergocalciferol and 25‐hydroxycholecalciferol, in high yields of 90 % and 100 %, respectively. [192] Aranda et al. have tested UPOs including AaeUPO and MroUPO for the hydroxyl functionalisation of (E)‐stilbene to produce (E)‐4,4’‐dihydroxystilbene (DHS), a compound with antiproliferative and antioxidant properties. [190] AaeUPO and MroUPO achieved complete conversion of the substrate with reported DHS molar yields of 94 % and 96 %, respectively, considerably higher than previous yields for DHS chemical synthesis (25 %). [190] Notably, a cytochrome P450 mutant CYP154E1 has also catalysed late‐stage hydroxylation to produce (E)‐stilbene derivatives, including DHS, [193] with comparable regioselectivity and conversion albeit lower turnovers (20 000) than AaeUPO (200 000) and MroUPO (25 000). [190]
3.2. Biocatalytic Cascades
Access to diverse biotransformations, like those described above, have enabled the generation of multi‐enzyme, multi‐step pathways in biocatalytic cascades. Cascades may rely on the combination of multiple enzymatic reactions, such as the remarkable recent work to synthesise islatravir, [194] or depend on biocatalytic and chemical steps to operate synergistically. [195] Often aided by biocatalytic retrosynthesis, which may offer disconnections without chemical alternatives, [196] cascades shift equilibria to the formation of product while obviating product inhibition and efficiently converting unstable intermediates.[ 197 , 198 ] We wish to highlight how the aforesaid oxygenating biocatalysts can be utilised for the design of such biocatalytic cascades, providing hydroxyl functionalisation which may be required in the final product or used for further manipulations. The adoption of biocatalytic cascades will only increase in the future, providing a sustainable strategy for the synthesis of complex molecules in drug discovery and development.
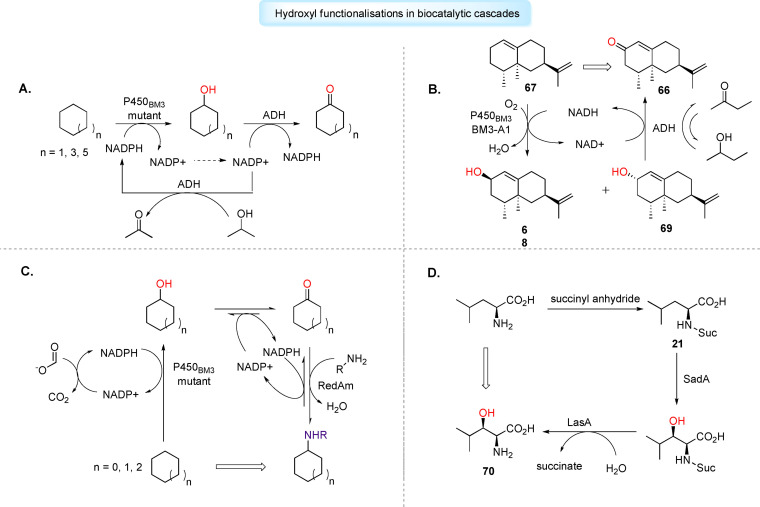
The implementation of cytochrome P450s in artificial cascades remains a relatively unexplored research area, but recent uses which couple cytochrome P450s with alcohol dehydrogenases (ADH) show considerable potential. [199] In such two‐step cascades, the cytochrome P450 catalyses hydroxylation of the starting substrate, with the resulting alcohol oxidised to the corresponding aldehyde or ketone by ADH. Conveniently, NAD(P)H consumed as a cofactor by cytochrome P450 can be reformed by the ADH‐catalysed oxidation step, providing facile cofactor regeneration in situ. [200] Gröger et al. have used the P450‐ADH cascade for the direct conversion of cycloalkanes to the corresponding ketones. [201] The group selected two P450BM3 mutants through screening studies, and an ADH from Lactobacillus kefir to perform double oxidation on inert substrates cyclohexane, cyclooctane and cyclodecane. Additionally, the group used the oxidised cofactor NADP+ which was reduced to NADPH by ADH using 2‐propanol as a sacrificial substrate, to initiate the reaction (Figure 9A). [201] Urlacher et al. extended use of the P450‐ADH cascade to synthesise ( )‐nootkatone (66), a high‐value sesquiterpenoid. [202] The group selected P450BM3 mutant BM3‐A1 to catalyse the allylic hydroxylation of ( )‐valencene (67) to produce cis‐nootkatol (68) and trans‐nootkatol (69), which were subsequently oxidised to 66 by an unselective ADH mutant ADH‐21, in yields reaching 360 mg L−1 after 20 hours. [202] Similarly to work by Gröger et al., the authors used the oxidised cofactor NADP+, which was reduced in situ by ADH using 2‐butanol as sacrificial substrate (Figure 9B). [202] Turner et al. have applied a P450‐ADH cascade involving chimeric P450cam‐RhFRed mutants and Cm‐ADH10 for the one‐pot biocatalytic synthesis of ketoisophorone, a key building block for vitamins and pharmaceuticals. [203] Furthermore, the P450‐ADH cascade may be extended with additional enzymes to produce chiral amines, a ubiquitous motif in pharmaceuticals. Turner et al. have employed a P450BM3 mutant, an ADH variant and a reductive aminase (RedAm) for the amination of unfunctionalised cycloalkanes. [204] Unlike Gröger and Urlacher, the authors used a FDH for the cofactor regeneration in the cytochrome P450‐catalysed step (Figure 9C). [204] Recently, work by Schwaneberg et al. has demonstrated an engineered P450BM3 and cpADH5 coupled cascade can be utilised for the generation of hydroxyl‐ or keto‐functionalised fatty acid methyl esters, such as methyl 3‐hydroxyhexanoate and methy‐3‐oxohexanoate, important pharmaceutical precursors. [205] Other cytochrome P450‐based cascades may not require additional ADH enzyme activity. Continuing their work with the iterative cytochrome P450, TamI, Sherman et al. engineered mutant L244A_L295V capable of catalysing an unprecedent four‐step oxidative biocatalytic cascade on tirandamycin C to generate biologically‐active tirandamycin B, overriding the enzyme's innate need for its oxidative partner, TamL. [173]
Figure 9.
Applications of oxygenating biocatalysts in biocatalytic cascades for drug discovery and development. A) Enzymatic one‐pot synthesis of cycloalkanones using P450BM3‐ADH cascade. Oxidised cofactor NADP+ is used in ADH‐catalysed oxidation of propanol as sacrificial substrate to generate NADPH in situ. B) P450BM3‐ADH cascade employed for selective allylic oxidation of (+)‐valence (67) to high‐value (+)‐nootkatone (66), an additive used in pharmaceuticals. Simultaneous butanol oxidation catalysed by ADH used to generate NADH. C) Three‐enzyme cascade used for the amination of unfunctionalised cycloalkanes. D) Fe/αKG SadA used in cascade with LasA for the synthesis L‐threo‐βOH‐Leu (70), a precursor for cyclic depsipeptide.
Recently, Sewald et al. have shown that FMOs can be successfully integrated in biocatalytic cascades to form an array of indigoids. [206] Starting with ‐tryptophan, the group employed crosslinked enzyme aggregates for C‐5, C‐6 or C‐7 bromination of the indole moiety, followed by cleavage of the amino acid backbone catalysed by tryptophanase (TnaA), to give bromoindole. FMO mFMO from Methlophaga sp., genetically fused to phosphite dehydrogenase (PTDH) for NADPH regeneration, was then used to catalyse C‐3 hydroxylation on the bromoindole, reaching a total conversion of 96 % for 6‐bromoindole. The hydroxylated product was found to spontaneously dimerise to produce dibromoindingo, that could then be subjected to cross‐coupling chemistries. [206]
Fe/αKGs have been incorporated in biocatalytic cascades to form key building blocks for pharmaceuticals. Ogawa et al. coupled SadA with N‐succinyl ‐amino acid desuccinylase (LasA) to produce β‐hydroxy α‐amino acids. [207] Based off their previous studies with SadA, [122] the group had shown SadA's hydroxylation activity towards Ns‐L‐Leucine (21) but not L‐Leucine. Consequently, the group developed a two‐step production of ‐threo‐β‐hydroxyleucine (70) ( ‐threo‐βOH‐Leu), a precursor for cyclic depsipeptides, from chemically‐prepared 21, utilising hydroxylation catalysed by SadA and the desuccinylation of the resulting alcohol by LasA. Through this cascade, ‐threo‐βOH‐Leu was prepared in 93 % conversion and >99 % diastereomeric excess. [207] Zaparucha et al. designed an enzymatic cascade utilising Fe/αKGs and pyridoxal phosphate (PLP)‐dependent amino acid decarboxylases to synthesise aliphatic chiral β‐ and γ‐amino alcohols from amino acids. [125] By retrosynthetic analysis, the group devised enzymatic cascades which started from either ‐ornithine, ‐lysine or (5R)‐hydroxy‐ ‐lysine, by one or two hydroxylation steps and cleavage of the carboxylic acid moiety. In three semi‐preparative syntheses starting from ‐lysine, three amino alcohols were formed with full conversion and isolated in excellent yields, respectively (93 to >95 %). [125] UPO‐catalysed hydroxyl functionalisations have been less adopted in biocatalytic cascades, but increasing work on developing multienzymatic cascades for in situ generation of H2O2[ 208 , 209 ] could, in the future, incorporate additional enzymes to incite further transformations on intermediate species.
4. Conclusions and Outlook
The past decade has witnessed an explosion of research interest in oxygenating biocatalysts for C−H hydroxyl functionalisation reactions. In drug discovery and development, cytochrome P450s have been the dominant oxygenating biocatalyst to produce chiral alcohols or phenols as pharmaceutical building blocks, or to synthesise HDMs for drug toxicity and characterisation. Several bottlenecks to cytochrome P450 application exist, such as cofactor dependence, multi‐component systems and instability. Ongoing efforts in protein engineering to produce novel chimeric cytochrome P450s, improve stability and enzymatic cofactor regeneration will undoubtedly ease some of these challenges. Likewise, the adoption of NCBs for FMO‐catalysed reactions will facilitate cheaper industrial applications of these enzymes on larger scales. However, in light of the preparative limitations of cytochrome P450s, emerging oxygenating biocatalysts such as Fe/αKGs and UPOs, which do not rely on expensive cofactor systems and demonstrate stability and a wide substrate scope, may soon be more widely adopted.
Enzymatic processes will inevitably be increasingly incorporated in drug discovery and development as we strive for greener and more efficient drug syntheses. Hydroxyl functionalisation in LSF and biocatalytic cascades is proving an invaluable tool for the diversification of drug‐like molecules and synthesis of precursors. Oxygenating biocatalysts described in this work will play a key role in the future of these processes. Advances in protein engineering and genome mining will improve and expand the current repertoire of oxygenating biocatalysts with chemistries so far undiscovered.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Sacha N. Charlton received her Master of Chemistry (MChem), with industrial experience, degree from the University of Warwick in 2021. Her industrial placement year was working at AstraZeneca, Gothenburg in the iLAB's Biocatalysis team. In this role, she was responsible for conducting high‐throughput screening of P450BM3 enzyme libraries for metabolite synthesis and late‐stage functionalisation of drug‐like compounds. She is now completing her Ph.D. studies at the University of Bristol under the co‐supervision of Prof. Chris Willis and Prof. Paul Race, working towards the engineering of biosynthetic pathways and biocatalysts to deliver new antimicrobial compounds.

Biographical Information
Martin A. Hayes is currently Biocatalysis Leader in the iLAB, Discovery Sciences at AstraZeneca, Gothenburg. He completed his Ph.D. with Prof. T. J. Simpson FRS at the University of Bristol. After postdoctoral studies with J. Bryan Jones at the University of Toronto, he started his industrial career. He has contributed to the discovery of small molecule therapeutics including Brilinta and the FLAP inhibitor AZD5718, currently in Phase2 clinical trials. His research interests include biocatalysis, drug design and high‐throughput experimentation.

S. N. Charlton, M. A. Hayes, ChemMedChem 2022, 17, e202200115.
References
- 1. Devine P. N., Howard R. M., Kumar R., Thompson M. P., Truppo M. D., Turner N. J., Nat. Chem. Rev. 2018, 2, 409–421. [Google Scholar]
- 2. Patel R. N., Coord. Chem. Rev. 2008, 252, 659–701. [Google Scholar]
- 3. Panke S., Wubbolts M., Curr. Opin. Chem. Biol. 2005, 9, 188–194. [DOI] [PubMed] [Google Scholar]
- 4. Hughes G., Lewis J. C., Chem. Rev. 2018, 118, 1–3. [DOI] [PubMed] [Google Scholar]
- 5. Woodley J. M., Trends Biotechnol. 2008, 26, 321–327. [DOI] [PubMed] [Google Scholar]
- 6. Patel R. N., in Org. Synth. Using Biocatal. (Eds.: Goswami A., Stewart J. D.), Elsevier Inc., 2016, pp. 339–411. [Google Scholar]
- 7. Sheldon R. A., Woodley J. M., Chem. Rev. 2018, 118, 801–838. [DOI] [PubMed] [Google Scholar]
- 8. Hanefeld U., Hollmann F., Paul C. E., Chem. Soc. Rev. 2022, 51, 594–627. [DOI] [PubMed] [Google Scholar]
- 9. Bornscheuer U. T., Philos. Trans. R. Soc. London 2018, 376, 1–7. [Google Scholar]
- 10. Truppo M. D., ACS Med. Chem. Lett. 2017, 8, 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arnold F. H., Q. Rev. Biophys. 2015, 48, 404–410. [DOI] [PubMed] [Google Scholar]
- 12. Brustad E. M., Arnold F. H., Curr. Opin. Chem. Biol. 2011, 15, 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Foley A. M., Maguire A. R., Eur. J. Org. Chem. 2019, 2019, 3713–3734. [Google Scholar]
- 14. Chen K., Arnold F. H., Nat. Catal. 2020, 3, 203–213. [Google Scholar]
- 15. Fryszkowska A., Devine P. N., Curr. Opin. Chem. Biol. 2020, 55, 151–160. [DOI] [PubMed] [Google Scholar]
- 16. Li G., bo Wang J., Reetz M. T., Bioorg. Med. Chem. 2018, 26, 1241–1251. [DOI] [PubMed] [Google Scholar]
- 17. Bornscheuer U. T., Hauer B., Jaeger K. E., Schwaneberg U., Angew. Chem. Int. Ed. 2019, 58, 36–40; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 36–41. [Google Scholar]
- 18. Simić S., Zukić E., Schmermund L., Faber K., Winkler C. K., Kroutil W., Chem. Rev. 2022, 122, 1052–1126. [DOI] [PubMed] [Google Scholar]
- 19. Ortiz De Montellano P. R., Chem. Rev. 2010, 110, 932–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chakrabarty S., Wang Y., Perkins J. C., Narayan A. R. H., Chem. Soc. Rev. 2020, 49, 8137–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hollmann F., Arends I. W. C. E., Buehler K., Schallmey A., Bühler B., Green Chem. 2011, 13, 226–265. [Google Scholar]
- 22. Holtmann D., Fraaije M. W., Arends I. W. C. E., Opperman D. J., Hollmann F., Chem. Commun. 2014, 50, 13180–13200. [DOI] [PubMed] [Google Scholar]
- 23. Dong J. J., Fernández-Fueyo E., Hollmann F., Paul C. E., Pesic M., Schmidt S., Wang Y., Younes S., Zhang W., Angew. Chem. Int. Ed. 2018, 57, 9238–9261; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 9380–9404. [Google Scholar]
- 24. Cramer J., Sager C. P., Ernst B., J. Med. Chem. 2019, 62, 8915–8930. [DOI] [PubMed] [Google Scholar]
- 25. Islam S., Leissing T. M., Chowdhury R., Hopkinson R. J., Schofield C. J., Annu. Rev. Biochem. 2018, 87, 585–620. [DOI] [PubMed] [Google Scholar]
- 26. Romero E., Gómez Castellanos J. R., Gadda G., Fraaije M. W., Mattevi A., Chem. Rev. 2018, 118, 1742–1769. [DOI] [PubMed] [Google Scholar]
- 27. Münch J., Püllmann P., Zhang W., Weissenborn M. J., ACS Catal. 2021, 11, 9168–9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fasan R., ACS Catal. 2012, 2, 647–666. [Google Scholar]
- 29. Liang Y., Wei J., Qiu X., Jiao N., Chem. Rev. 2018, 118, 4912–4945. [DOI] [PubMed] [Google Scholar]
- 30. Ener M. E., Lee Y. T., Winkler J. R., Gray H. B., Cheruzela L., Proc. Natl. Acad. Sci. USA 2010, 107, 18783–18786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z., Jiang Y., Guengerich F. P., Ma L., Li S., Zhang W., J. Biol. Chem. 2020, 295, 833–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Di Nardo G., Gilardi G., Trends Biochem. Sci. 2020, 45, 511–525. [DOI] [PubMed] [Google Scholar]
- 33. Urlacher V. B., Girhard M., Trends Biotechnol. 2019, 37, 882–897. [DOI] [PubMed] [Google Scholar]
- 34. Whitehouse C. J. C., Bell S. G., Wong L. L., Chem. Soc. Rev. 2012, 41, 1218–1260. [DOI] [PubMed] [Google Scholar]
- 35. Pflug S., Richter S. M., Urlacher V. B., J. Biotechnol. 2007, 129, 481–488. [DOI] [PubMed] [Google Scholar]
- 36. Weber E., Seifert A., Antonovici M., Geinitz C., Pleiss J., Urlacher V. B., Chem. Commun. 2011, 47, 944–946. [DOI] [PubMed] [Google Scholar]
- 37. Agudo R., Roiban G. D., Reetz M. T., ChemBioChem 2012, 13, 1465–1473. [DOI] [PubMed] [Google Scholar]
- 38. Tang W. L., Li Z., Zhao H., Chem. Commun. 2010, 46, 5461–5463. [DOI] [PubMed] [Google Scholar]
- 39. Sarkar M. R., Dasgupta S., Pyke S. M., Bell S. G., Chem. Commun. 2019, 55, 5029–5032. [DOI] [PubMed] [Google Scholar]
- 40. Hammerer L., Winkler C. K., Kroutil W., Catal. Lett. 2018, 148, 787–812. [Google Scholar]
- 41. Kim K. R., Oh D. K., Biotechnol. Adv. 2013, 31, 1473–1485. [DOI] [PubMed] [Google Scholar]
- 42. Neufeld K., Henßen B., Pietruszka J., Angew. Chem. Int. Ed. 2014, 53, 13253–13257; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 13469–13473. [Google Scholar]
- 43. Ma D., Zhang L., Yin Y., Wang Q., Chin. Chem. Lett. 2020, 9–12. [Google Scholar]
- 44. Chen H., Huang M., Yan W., Bai W. J., Wang X., ACS Catal. 2021, 11, 10625–10630. [Google Scholar]
- 45. Zong L., Zhang Y., Shao Z., Wang Y., Guo Z., Gao R., Eser B. E., Catalysts 2021, 11, 665. [Google Scholar]
- 46. Dennig A., Lülsdorf N., Liu H., Schwaneberg U., Angew. Chem. Int. Ed. 2013, 52, 8459–8462; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 8617–8620. [Google Scholar]
- 47. O'Hanlon J. A., Ren X., Morris M., Wong L. L., Robertson J., Org. Biomol. Chem. 2017, 15, 8780–8787. [DOI] [PubMed] [Google Scholar]
- 48. Shoji O., Yanagisawa S., Stanfield J. K., Suzuki K., Cong Z., Sugimoto H., Shiro Y., Watanabe Y., Angew. Chem. Int. Ed. 2017, 56, 10324–10329; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 10460–10465. [Google Scholar]
- 49. Zhou H., Wang B., Wang F., Yu X., Ma L., Li A., Reetz M. T., Angew. Chem. Int. Ed. 2019, 58, 764–768; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 774–778. [Google Scholar]
- 50. Thistlethwaite S., Jeffreys L. N., Girvan H. M., McLean K. J., Munro A. W., Int. J. Mol. Sci. 2021, 22, 11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guengerich F. P., Chem. Res. Toxicol. 2001, 14, 611–650. [DOI] [PubMed] [Google Scholar]
- 52. Butler C. F., Peet C., McLean K. J., Baynham M. T., Blankley R. T., Fisher K., Rigby S. E. J., Leys D., Voice M. W., Munro A. W., Biochem. J. 2014, 460, 247–259. [DOI] [PubMed] [Google Scholar]
- 53. Venkataraman H., Verkade-Vreeker M. C. A., Capoferri L., Geerke D. P., Vermeulen N. P. E., Commandeur J. N. M., Bioorg. Med. Chem. 2014, 22, 5613–5620. [DOI] [PubMed] [Google Scholar]
- 54. Ren X., Yorke J. A., Taylor E., Zhang T., Zhou W., Wong L. L., Chem. A Eur. J. 2015, 21, 15039–15047. [DOI] [PubMed] [Google Scholar]
- 55. Klenk J. M., Nebel B. A., Porter J. L., Kulig J. K., Hussain S. A., Richter S. M., Tavanti M., Turner N. J., Hayes M. A., Hauer B., Flitsch S. L., Biotechnol. J. 2017, 12, 1600520. [DOI] [PubMed] [Google Scholar]
- 56. Worsch A., Eggimann F. K., Girhard M., von Bühler C. J., Tieves F., Czaja R., Vogel A., Grumaz C., Sohn K., Lütz S., Kittelmann M., Urlacher V. B., Biotechnol. Bioeng. 2018, 115, 2156–2166. [DOI] [PubMed] [Google Scholar]
- 57. Rinnofner C., Kerschbaumer B., Weber H., Glieder A., Winkler M., Biocatal. Agric. Biotechnol. 2019, 17, 525–528. [Google Scholar]
- 58. O'Reilly E., Köhler V., Flitsch S. L., Chem. Commun. 2011, 47, 2490–2501. [DOI] [PubMed] [Google Scholar]
- 59. Di Nardo G., Gilardi G., Int. J. Mol. Sci. 2012, 13, 15901–15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Julsing M. K., Cornelissen S., Bühler B., Schmid A., Curr. Opin. Chem. Biol. 2008, 12, 177–186. [DOI] [PubMed] [Google Scholar]
- 61. Kaluzna I., Schmitges T., Straatman H., Van Tegelen D., Müller M., Schürmann M., Mink D., Org. Process Res. Dev. 2016, 20, 814–819. [Google Scholar]
- 62. Zhang Z., Li F., Cao Y., Tian Y., Li J., Zong Y., Song H., Catal. Sci. Technol. 2019, 9, 4877–4887. [Google Scholar]
- 63. Shalan H., Kato M., Cheruzel L., Biochim. Biophys. Acta Proteins Proteomics 2018, 1866, 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kato M., Melkie M., Li J., Foley B., Nguyen H. T., Leti L., Cheruzel L., Arch. Biochem. Biophys. 2019, 672, 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Robin A., Köhler V., Jones A., Ali A., Kelly P. P., O'Reilly E., Turner N. J., Flitsch S. L., Beilstein J. Org. Chem. 2011, 7, 1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kulig J. K., Spandolf C., Hyde R., Ruzzini A. C., Eltis L. D., Grönberg G., Hayes M. A., Grogan G., Bioorg. Med. Chem. 2015, 23, 5603–5609. [DOI] [PubMed] [Google Scholar]
- 67. Lundemo M. T., Notonier S., Striedner G., Hauer B., Woodley J. M., Appl. Microbiol. Biotechnol. 2016, 100, 1197–1208. [DOI] [PubMed] [Google Scholar]
- 68. Luan Z. J., Yin Y. C., Li A. T., Yu H. L., Xu J. H., J. Mol. Catal. B 2015, 116, 78–82. [Google Scholar]
- 69. Correddu D., Di Nardo G., Gilardi G., Trends Biotechnol. 2021, 39, 1184–1207. [DOI] [PubMed] [Google Scholar]
- 70. Zhang K., El Damaty S., Fasan R., J. Am. Chem. Soc. 2011, 133, 3242–3245. [DOI] [PubMed] [Google Scholar]
- 71. Haatveit K. C., Garcia-Borràs M., Houk K. N., Front. Chem. 2019, 7, 1–6.30778383 [Google Scholar]
- 72. Reis R. A. G., Li H., Johnson M., Sobrado P., Arch. Biochem. Biophys. 2021, 699, 108765. [DOI] [PubMed] [Google Scholar]
- 73. Paul C. E., Eggerichs D., Westphal A. H., Tischler D., van Berkel W. J. H., Biotechnol. Adv. 2021, 51, 107712. [DOI] [PubMed] [Google Scholar]
- 74. Deng Y., Zhou Q., Wu Y., Chen X., Int. J. Mol. Sci. 2022, 23, 2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Holtmann D., Hollmann F., ChemBioChem 2016, 17, 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Toplak M., Matthews A., Teufel R., Arch. Biochem. Biophys. 2021, 698, 108732. [DOI] [PubMed] [Google Scholar]
- 77. Saleem-Batcha R., Teufel R., Curr. Opin. Chem. Biol. 2018, 47, 47–53. [DOI] [PubMed] [Google Scholar]
- 78. Teufel R., Stull F., Meehan M. J., Michaudel Q., Dorrestein P. C., Palfey B., Moore B. S., J. Am. Chem. Soc. 2015, 137, 8078–8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Teufel R., Miyanaga A., Michaudel Q., Stull F., Louie G., Noel J. P., Baran P. S., Palfey B., Moore B. S., Nature 2013, 503, 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chenprakhon P., Wongnate T., Chaiyen P., Protein Sci. 2019, 28, 8–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Palfey B. A., McDonald C. A., Arch. Biochem. Biophys. 2010, 493, 26–36. [DOI] [PubMed] [Google Scholar]
- 82. Huijbers M. M. E., Montersino S., Westphal A. H., Tischler D., Van Berkel W. J. H., Arch. Biochem. Biophys. 2014, 544, 2–17. [DOI] [PubMed] [Google Scholar]
- 83. Romero E., Gómez Castellanos J. R., Gadda G., Fraaije M. W., Mattevi A., Chem. Rev. 2018, 118, 1742–1769. [DOI] [PubMed] [Google Scholar]
- 84. Montersino S., te Poele E., Orru R., Westphal A. H., Barendregt A., Heck A. J. R., van der Geize R., Dijkhuizen L., Mattevi A., van Berkel W. J. H., Front. Microbiol. 2017, 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yu H., Zhao S., Lu W., Wang W., Guo L., Appl. Microbiol. Biotechnol. 2018, 102, 4843–4852. [DOI] [PubMed] [Google Scholar]
- 86. Meyer A., Held M., Schmid A., Kohler H. P. E., Witholt B., Biotechnol. Bioeng. 2003, 81, 518–524. [DOI] [PubMed] [Google Scholar]
- 87. Lutz J., Mozhaev V. V., Khmelnitsky Y. L., Witholt B., Schmid A., J. Mol. Catal. B 2002, 19–20, 177–187. [Google Scholar]
- 88. Tomaszewski B., Schmid A., Buehler K., Org. Process Res. Dev. 2014, 18, 1516–1526. [Google Scholar]
- 89. Bregman-Cohen A., Deri B., Maimon S., Pazy Y., Fishman A., ChemBioChem 2018, 19, 583–590. [DOI] [PubMed] [Google Scholar]
- 90. Shen X., Zhou D., Lin Y., Wang J., Gao S., Kandavelu P., Zhang H., Zhang R., Wang B. C., Rose J., Yuan Q., Yan Y., Sci. Rep. 2019, 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Furuya T., Kino K., Appl. Microbiol. Biotechnol. 2014, 98, 1145–1154. [DOI] [PubMed] [Google Scholar]
- 92. Nakagawa A., Nakamura S., Matsumura E., Yashima Y., Takao M., Aburatani S., Yaoi K., Katayama T., Minami H., Appl. Microbiol. Biotechnol. 2021, 105, 5433–5447. [DOI] [PubMed] [Google Scholar]
- 93. Dhammaraj T., Phintha A., Pinthong C., Medhanavyn D., Tinikul R., Chenprakhon P., Sucharitaku J., Vardhanabhuti N., Jiarpinitnun C., Chaiyen P., ACS Catal. 2015, 5, 4492–4502. [Google Scholar]
- 94. Dhammaraj T., Pinthong C., Visitsatthawong S., Tongsook C., Surawatanawong P., Chaiyen P., ACS Chem. Biol. 2016, 11, 2889–2896. [DOI] [PubMed] [Google Scholar]
- 95. Chenprakhon P., Dhammaraj T., Chantiwas R., Chaiyen P., Arch. Biochem. Biophys. 2017, 620, 1–11. [DOI] [PubMed] [Google Scholar]
- 96. Hashimoto T., Nozawa D., Mukai K., Matsuyama A., Kuramochi K., Furuya T., RSC Adv. 2019, 9, 21826–21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Deng Y., Faivre B., Back O., Lombard M., Pecqueur L., Fontecave M., ChemBioChem 2020, 21, 163–170. [DOI] [PubMed] [Google Scholar]
- 98. Herrmann S., Dippe M., Pecher P., Funke E., Pietzsch M., Wessjohann L. A., ChemBioChem 2022, 23, e202100480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Feng L., Wang W., Cheng J., Ren Y., Zhao G., Gao C., Tang Y., Liu X., Han W., Peng X., Liu R., Wang L., Proc. Natl. Acad. Sci. USA 2007, 104, 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dong Y., Yan J., Du H., Chen M., Ma T., Feng L., Appl. Microbiol. Biotechnol. 2012, 94, 1019–1029. [DOI] [PubMed] [Google Scholar]
- 101. Baker Dockrey S. A., Narayan A. R. H., Tetrahedron 2019, 75, 1115–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Heine T., van Berkel W. J. H., Gassner G., van Pée K. H., Tischler D., Biology (Basel) 2018, 7, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Guarneri A., Westphal A. H., Leertouwer J., Lunsonga J., Franssen M. C. R., Opperman D. J., Hollmann F., van Berkel W. J. H., Paul C. E., ChemCatChem 2020, 12, 1368–1375. [Google Scholar]
- 104. Paul C. E., Arends I. W. C. E., Hollmann F., ACS Catal. 2014, 4, 788–797. [Google Scholar]
- 105. Zwick C. R., Renata H., Nat. Prod. Rep. 2020, 37, 1065–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Martinez S., Hausinger R. P., J. Biol. Chem. 2015, 290, 20702–20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hüttel W., Chemie (Ingenieur-Technik) 2013, 85, 809–817. [Google Scholar]
- 108. King-Smith E., Zwick C. R., Renata H., Biochemistry 2018, 57, 403–412. [DOI] [PubMed] [Google Scholar]
- 109. Peters C., Buller R. M., Catalysts 2019, 9, 221. [Google Scholar]
- 110. Sangster J. J., Marshall J. R., Turner N. J., Mangas-Sanchez J., ChemBioChem 2021, 23, e2021004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Klein C., Hüttel W., Adv. Synth. Catal. 2011, 353, 1375–1383. [Google Scholar]
- 112. Klein C., Hüttel W., Beilstein J. Org. Chem. 2011, 7, 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hara R., Uchiumi N., Okamoto N., Kino K., Biosci. Biotechnol. Biochem. 2014, 78, 1384–1388. [DOI] [PubMed] [Google Scholar]
- 114. Structure X. C., Koketsu K., Shomura Y., Moriwaki K., Hayashi M., Mitsuhashi S., ACS Synth. Biol. 2015, 4, 383–392. [DOI] [PubMed] [Google Scholar]
- 115. Mattay J., Hüttel W., ChemBioChem 2017, 18, 1523–1528. [DOI] [PubMed] [Google Scholar]
- 116. Zhang H. L., Zhang C., Pei C. H., Han M. N., Xu Z. D., Li C. H., Li W., Lett. Appl. Microbiol. 2018, 66, 400–408. [DOI] [PubMed] [Google Scholar]
- 117. Zhao T. X., Li M., Zheng X., Wang C. H., Zhao H. X., Zhang C., Xing X. H., J. Biosci. Bioeng. 2017, 123, 109–115. [DOI] [PubMed] [Google Scholar]
- 118. Jing X., Wang X., Zhang W., An J., Luo P., Nie Y., Xu Y., ACS Omega 2019, 4, 8350–8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tassano E., Moore C., Dussauge S., Vargas A., Snajdrova R., Org. Process Res. Dev. 2022, doi.org/10.1021/acs.oprd.1c00405. [Google Scholar]
- 120. Smart T. J., Hamed R. B., Claridge T. D. W., Schofield C. J., Bioorg. Chem. 2020, 94, 103386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hibi M., Kawashima T., Sokolov P. M., Smirnov S. V., Kodera T., Sugiyama M., Shimizu S., Yokozeki K., Ogawa J., Appl. Microbiol. Biotechnol. 2013, 97, 2467–2472. [DOI] [PubMed] [Google Scholar]
- 122. Hibi M., Kawashima T., Kasahara T., Sokolov P. M., Smirnov S. V., Kodera T., Sugiyama M., Shimizu S., Yokozeki K., Ogawa J., Lett. Appl. Microbiol. 2012, 55, 414–419. [DOI] [PubMed] [Google Scholar]
- 123. Busch F., Busch F., Brummund J., Calderini E., Schürmann M., Kourist R., ACS Sustainable Chem. Eng. 2020, 8, 8604–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Baud D., Saaidi P. L., Monfleur A., Harari M., Cuccaro J., Fossey A., Besnard M., Debard A., Mariage A., Pellouin V., Petit J. L., Salanoubat M., Weissenbach J., De Berardinis V., Zaparucha A., ChemCatChem 2014, 6, 3012–3017. [Google Scholar]
- 125. Baud D., Peruch O., Saaidi P. L., Fossey A., Mariage A., Petit J. L., Salanoubat M., Vergne-Vaxelaire C., de Berardinis V., Zaparucha A., Adv. Synth. Catal. 2017, 359, 1563–1569. [Google Scholar]
- 126. Hara R., Nakajima Y., Yanagawa H., Gawasawa R., Hirasawa I., Kino K., Appl. Environ. Microbiol. 2021, 87, e01335–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Meyer F., Frey R., Ligibel M., Sager E., Schroer K., Snajdrova R., Buller R., ACS Catal. 2021, 11, 6261–6269. [Google Scholar]
- 128. Jing X., Liu H., Nie Y., Xu Y., Syst. Microbiol. Biomanufacturing 2021, 1, 275–290. [Google Scholar]
- 129. Stout C. N., Renata H., Acc. Chem. Res. 2021, 54, 1143–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zwick C. R., Renata H., J. Am. Chem. Soc. 2018, 140, 1165–1169. [DOI] [PubMed] [Google Scholar]
- 131. Zwick C. R., Renata H., J. Org. Chem. 2018, 83, 7407–7415. [DOI] [PubMed] [Google Scholar]
- 132. Nodwell M. B., Yang H., Čolović M., Yuan Z., Merkens H., Martin R. E., Bénard F., Schaffer P., Britton R., J. Am. Chem. Soc. 2017, 139, 3595–3598. [DOI] [PubMed] [Google Scholar]
- 133. Amatuni A., Shuster A., Adibekian A., Renata H., Cell Chem. Biol. 2020, 27, 1318–1326.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zwick C. R., Sosa M. B., Renata H., J. Am. Chem. Soc. 2021, 143, 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhang X., King-Smith E., Renata H., Angew. Chem. Int. Ed. 2018, 57, 5037–5041; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 5131–5135. [Google Scholar]
- 136. Doyon T. J., Perkins J. C., Baker Dockrey S. A., Romero E. O., Skinner K. C., Zimmerman P. M., Narayan A. R. H., J. Am. Chem. Soc. 2019, 141, 20269–20277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Adams J. P., Brown M. J. B., Diaz-Rodriguez A., Lloyd R. C., Roiban G. D., Adv. Synth. Catal. 2019, 361, 2421–2432. [Google Scholar]
- 138. Grogan G., JACS 2021, 1, 1312–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hobisch M., Holtmann D., Gomez de Santos P., Alcalde M., Hollmann F., Kara S., Biotechnol. Adv. 2021, 51, 107615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Martin-Diaz J., Molina-Espeja P., Hofrichter M., Hollmann F., Alcalde M., Biotechnol. Bioeng. 2021, 118, 3002–3014. [DOI] [PubMed] [Google Scholar]
- 141. Wang Y., Lan D., Durrani R., Hollmann F., Curr. Opin. Chem. Biol. 2017, 37, 1–9. [DOI] [PubMed] [Google Scholar]
- 142. Aranda C., Carro J., González-Benjumea A., Babot E. D., Olmedo A., Linde D., Martínez A. T., Gutiérrez A., Biotechnol. Adv. 2021, 51, 107615. [DOI] [PubMed] [Google Scholar]
- 143. Gutiérrez A., Babot E. D., Ullrich R., Hofrichter M., Martínez A. T., Del Río J. C., Arch. Biochem. Biophys. 2011, 514, 33–43. [DOI] [PubMed] [Google Scholar]
- 144. Peter S., Kinne M., Wang X., Ullrich R., Kayser G., Groves J. T., Hofrichter M., FEBS J. 2011, 278, 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Aranda C., Municoy M., Guallar V., Kiebist J., Scheibner K., Ullrich R., Del Río J. C., Hofrichter M., Martínez A. T., Gutiérrez A., Catal. Sci. Technol. 2019, 9, 1398–1405. [Google Scholar]
- 146. Karich A., Kluge M., Ullrich R., Hofrichter M., AMB Express 2013, 3, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Molina-Espeja P., Cañellas M., Plou F. J., Hofrichter M., Lucas F., Guallar V., Alcalde M., ChemBioChem 2016, 17, 341–349. [DOI] [PubMed] [Google Scholar]
- 148. Molina-Espeja P., Ma S., Mate D. M., Ludwig R., Alcalde M., Enzyme Microb. Technol. 2015, 73–74, 29–33. [DOI] [PubMed] [Google Scholar]
- 149. Molina-Espeja P., Garcia-Ruiz E., Gonzalez-Perez D., Ullrich R., Hofrichter M., Alcalde M., Appl. Environ. Microbiol. 2014, 80, 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Linde D., Olmedo A., González-Benjumea A., Estévez M., Renau-Mínguez C., Carro J., Fernández-Fueyo E., Gutiérrez A., Martínez A. T., Appl. Environ. Microbiol. 2020, 86, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Knorrscheidt A., Püllmann P., Schell E., Homann D., Freier E., Weissenborn M. J., ChemCatChem 2020, 12, 4788–4795. [Google Scholar]
- 152. Püllmann P., Knorrscheidt A., Münch J., Palme P. R., Hoehenwarter W., Marillonnet S., Alcalde M., Westermann B., Weissenborn M. J., Commun. Biol. 2021, 4, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Knorrscheidt A., Soler J., Hünecke N., Püllmann P., Garcia-Borràs M., Weissenborn M. J., Catal. Sci. Technol. 2021, 11, 6058–6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Knorrscheidt A., Soler J., Hünecke N., Püllmann P., Garcia-Borràs M., Weissenborn M. J., ACS Catal. 2021, 11, 7327–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Olmedo A., Aranda C., del Río J. C., Kiebist J., Scheibner K., Martínez A. T., Gutiérrez A., Angew. Chem. Int. Ed. 2016, 55, 12248–12251; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 12436–12439. [Google Scholar]
- 156. Babot E. D., Aranda C., Del Rĺo J. C., Ullrich R., Kiebist J., Scheibner K., Hofrichter M., Martĺnez A. T., Gutiérrez A., J. Agric. Food Chem. 2020, 68, 5375–5383. [DOI] [PubMed] [Google Scholar]
- 157. Poraj-Kobielska M., Kinne M., Ullrich R., Scheibner K., Kayser G., Hammel K. E., Hofrichter M., Biochem. Pharmacol. 2011, 82, 789–796. [DOI] [PubMed] [Google Scholar]
- 158. Steinbrecht S., Kiebist J., König R., Thiessen M., Schmidtke K. U., Kammerer S., Küpper J. H., Scheibner K., AMB Express 2020, 10, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kiebist J., Schmidtke K. U., Zimmermann J., Kellner H., Jehmlich N., Ullrich R., Zänder D., Hofrichter M., Scheibner K., ChemBioChem 2017, 18, 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Gomez De Santos P., Cañellas M., Tieves F., Younes S. H. H., Molina-Espeja P., Hofrichter M., Hollmann F., Guallar V., Alcalde M., ACS Catal. 2018, 8, 4789–4799. [Google Scholar]
- 161. Ni Y., Fernández-Fueyo E., Baraibar A. G., Ullrich R., Hofrichter M., Yanase H., Alcalde M., VanBerkel W. J. H., Hollmann F., Angew. Chem. Int. Ed. 2016, 55, 798–801; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 809–812. [Google Scholar]
- 162. Zhang W., Burek B. O., Fernández-Fueyo E., Alcalde M., Bloh J. Z., Hollmann F., Angew. Chem. Int. Ed. 2017, 56, 15451–15455; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 15654–15658. [Google Scholar]
- 163. Horst A. E. W., Bormann S., Meyer J., Steinhagen M., Ludwig R., Drews A., Ansorge-Schumacher M., Holtmann D., J. Mol. Catal. B 2016, 133, S137–S142. [Google Scholar]
- 164. Beltrán-Nogal A., Sánchez-Moreno I., Méndez-Sánchez D., Gómez de Santos P., Hollmann F., Alcalde M., Curr. Opin. Struct. Biol. 2022, 73, 102342. [DOI] [PubMed] [Google Scholar]
- 165.T. N. Karsten, Mäder; Verena Weiss, Jörg Kressler, Mutants of Unspecific Peroxygenase with High Monooxygenase Activity and Their Uses, 2017.
- 166. Guillemard L., Kaplaneris N., Ackermann L., Johansson M. J., Nat. Chem. Rev. 2021, 5, 522–545. [DOI] [PubMed] [Google Scholar]
- 167. Romero E., Jones B. S., Hogg B. N., Rué Casamajo A., Hayes M. A., Flitsch S. L., Turner N. J., Schnepel C., Angew. Chem. Int. Ed. 2021, 60, 16824–16855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Fessner N. D., ChemCatChem 2019, 11, 2226–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Stepan A. F., Tran T. P., Helal C. J., Brown M. S., Chang C., O'Connor R. E., De Vivo M., Doran S. D., Fisher E. L., Jenkinson S., Karanian D., Kormos B. L., Sharma R., Walker G. S., Wright A. S., Yang E. X., Brodney M. A., Wager T. T., Verhoest P. R., Obach R. S., ACS Med. Chem. Lett. 2018, 9, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Fessner N. D., Grimm C., Srdič M., Weber H., Kroutil W., Schwaneberg U., Glieder A., ChemCatChem 2022, 14, e202101564. [Google Scholar]
- 171. Vickers C., Backfisch G., Oellien F., Piel I., Lange U. E. W., Chem. A Eur. J. 2018, 24, 17936–17947. [DOI] [PubMed] [Google Scholar]
- 172. Negretti S., Narayan A. R. H., Chiou K. C., Kells P. M., Stachowski J. L., Hansen D. A., Podust L. M., Montgomery J., Sherman D. H., J. Am. Chem. Soc. 2014, 136, 4901–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Espinoza R. V., Haatveit K. C., Grossman S. W., Tan J. Y., McGlade C. A., Khatri Y., Newmister S. A., Schmidt J. J., Garcia-Borràs M., Montgomery J., Houk K. N., Sherman D. H., ACS Catal. 2021, 11, 8304–8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Iizaka Y., Arai R., Takahashi A., Ito M., Sakai M., Fukumoto A., Sherman D. H., Anzai Y., J. Ind. Microbiol. Biotechnol. 2022, 49, kuab069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Le-Huu P., Heidt T., Claasen B., Laschat S., Urlacher V. B., ACS Catal. 2015, 5, 1772–1780. [Google Scholar]
- 176. Le-Huu P., Rekow D., Krüger C., Bokel A., Heidt T., Schaubach S., Claasen B., Hölzel S., Frey W., Laschat S., Urlacher V. B., Chem. A Eur. J. 2018, 24, 12010–12021. [DOI] [PubMed] [Google Scholar]
- 177. Chen W., Fisher M. J., Leung A., Cao Y., Wong L. L., ACS Catal. 2020, 10, 8334–8343. [Google Scholar]
- 178. Grobe S., Badenhorst C. P. S., Bayer T., Hamnevik E., Wu S., Grathwol C. W., Link A., Koban S., Brundiek H., Großjohann B., Bornscheuer U. T., Angew. Chem. Int. Ed. 2021, 60, 753–757; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 764–768. [Google Scholar]
- 179. Zhang K., Shafer B. M., Demars M. D., Stern H. A., Fasan R., J. Am. Chem. Soc. 2012, 134, 18695–18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Obach R. S., Walker G. S., Brodney M. A., Drug Metab. Dispos. 2016, 44, 634–646. [DOI] [PubMed] [Google Scholar]
- 181. Kolev J. N., O'Dwyer K. M., Jordan C. T., Fasan R., ACS Chem. Biol. 2014, 9, 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Loskot S. A., Romney D. K., Arnold F. H., Stoltz B. M., J. Am. Chem. Soc. 2017, 139, 10196–10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Syntrivanis L. D., Wong L. L., Robertson J., Eur. J. Org. Chem. 2018, 2018, 6369–6378. [Google Scholar]
- 184. Li J., Li F., King-Smith E., Renata H., Nat. Chem. 2020, 12, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Furuya T., Kino K., Tetrahedron Lett. 2014, 55, 2853–2855. [Google Scholar]
- 186. Furuya T., Sai M., Kino K., Biosci. Biotechnol. Biochem. 2016, 80, 193–198. [DOI] [PubMed] [Google Scholar]
- 187. Heo K. T., Lee B., Son S., Ahn J. S., Jang J. H., Hong Y. S., J. Microbiol. Biotechnol. 2018, 28, 1105–1111. [DOI] [PubMed] [Google Scholar]
- 188. Qin H. M., Miyakawa T., Nakamura A., Hibi M., Ogawa J., Tanokura M., Biochem. Biophys. Res. Commun. 2014, 450, 1458–1461. [DOI] [PubMed] [Google Scholar]
- 189. Tao H., Mori T., Chen H., Lyu S., Nonoyama A., Lee S., Abe I., Nat. Commun. 2022, 13, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Aranda C., Ullrich R., Kiebist J., Scheibner K., Del Río J. C., Hofrichter M., Martínez A. T., Gutiérrez A., Catal. Sci. Technol. 2018, 8, 2394–2401. [Google Scholar]
- 191. Babot E. D., del Río J. C., Cañellas M., Sancho F., Lucas F., Guallar V., Kalum L., Lund H., Gröbe G., Scheibner K., Ullrich R., Hofrichter M., Martínez A. T., Gutiérrez A., Appl. Environ. Microbiol. 2015, 81, 4130–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Babot E. D., Del Río J. C., Kalum L., Martínez A. T., Gutiérrez A., ChemCatChem 2015, 7, 283–290. [Google Scholar]
- 193. Rühlmann A., Antovic D., Müller T. J. J., Urlacher V. B., Adv. Synth. Catal. 2017, 359, 984–994. [Google Scholar]
- 194. Huffman M. A., Fryszkowska A., Alvizo O., Borra-garske M., Campos K. R., Canada K. A., Devine P. N., Duan D., Forstater J. H., Grosser S. T., Halsey H. M., Hughes G. J., Jo J., Joyce L. A., Kolev J. N., Liang J., Maloney K. M., Mann B. F., Marshall N. M., Mclaughlin M., Moore J. C., Murphy G. S., Nawrat C. C., Nazor J., Novick S., Patel N. R., Rodriguez-granillo A., Science 2019, 1259, 1255–1259. [DOI] [PubMed] [Google Scholar]
- 195. Schmidt-Dannert C., Lopez-Gallego F., Microb. Biotechnol. 2016, 9, 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Green A. P., Turner N. J., Perspect. Sci. 2016, 9, 42–48. [Google Scholar]
- 197. Bell E. L., Finnigan W., France S. P., Hepworth L. J., Lovelock S. L., Hayes M. A., Osuna S., Romero E., Ryan K. S., Turner N. J., Flitsch S. L., Nat. Rev. Methods Prim. 2021, 1, 1–21. [Google Scholar]
- 198. McIntosh J. A., Owens A. E., Curr. Opin. Green Sustain. Chem. 2021, 29, 100448. [Google Scholar]
- 199. Urlacher V. B., Schulz S., in Cascade Biocatal. Integr. Stereoselective Environ. Friendly React. (Eds.: Riva S., Fessner W.-D.), John Wiley & Sons, Incorporated, 2014, pp. 87–132. [Google Scholar]
- 200. Urlacher V. B., Girhard M., Trends Biotechnol. 2019, 37, 882–897. [DOI] [PubMed] [Google Scholar]
- 201. Staudt S., Burda E., Giese C., Müller C. A., Marienhagen J., Schwaneberg U., Hummel W., Drauz K., Gröger H., Angew. Chem. Int. Ed. 2013, 52, 2359–2363; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 2415–2419. [Google Scholar]
- 202. Schulz S., Girhard M., Gaßmeyer S. K., Jäger V. D., Schwarze D., Vogel A., Urlacher V. B., ChemCatChem 2015, 7, 601–604. [Google Scholar]
- 203. Tavanti M., Parmeggiani F., Castellanos J. R. G., Mattevi A., Turner N. J., ChemCatChem 2017, 9, 3338–3348. [Google Scholar]
- 204. Tavanti M., Mangas-Sanchez J., Montgomery S. L., Thompson M. P., Turner N. J., Org. Biomol. Chem. 2017, 15, 9790–9793. [DOI] [PubMed] [Google Scholar]
- 205. Ensari Y., de Almeida Santos G., Ruff A. J., Schwaneberg U., Enzyme Microb. Technol. 2020, 138, 109555. [DOI] [PubMed] [Google Scholar]
- 206. Schnepel C., Dodero V. I., Sewald N., Chem. A Eur. J. 2021, 27, 5404–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Hibi M., Kasahara T., Kawashima T., Yajima H., Kozono S., Smirnov S. V., Kodera T., Sugiyama M., Shimizu S., Yokozeki K., Ogawa J., Adv. Synth. Catal. 2015, 357, 767–774. [Google Scholar]
- 208. Pesic M., Willot S. J. P., Fernández-Fueyo E., Tieves F., Alcalde M., Hollmann F., Z. Naturforsch. C 2019, 74, 101–104. [DOI] [PubMed] [Google Scholar]
- 209. Carro J., Fernández-Fueyo E., Fernández-Alonso C., Cañada J., Ullrich R., Hofrichter M., Alcalde M., Ferreira P., Martínez A. T., Biotechnol. Biofuels 2018, 11, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]