Abstract
Burkholderia sp. strain JS150 is able to metabolize a wide range of alkyl-and chloroaromatic hydrocarbons through multiple, apparently redundant catabolic pathways. Previous research has shown that strain JS150 is able to synthesize enzymes for multiple upper pathways as well as multiple lower pathways to accommodate variously substituted catechols that result from degradation of complex mixtures of monoaromatic compounds. We report here the genetic organization and functional characterization of a gene cluster, designated tbc (for toluene, benzene, and chlorobenzene utilization), which has been cloned as a 14.3-kb DNA fragment from strain JS150 into vector pRO1727. The cloned DNA fragment expressed in Pseudomonas aeruginosa PAO1c allowed the recombinant to grow on toluene or benzene and to transform chlorobenzene, trichloroethylene, phenol, and cresols. The tbc genes are organized into two divergently transcribed operons, tbc1 and tbc2, each comprised of six open reading frames. Similarity searches of databases revealed that the tbc1 and tbc2 genes showed significant homology to multicomponent cresol and phenol hydroxylases and to toluene and benzene monooxygenases, respectively. Deletion mutagenesis and product analysis were used to demonstrate that tbc2 plays a role in the initial catabolism of the unactivated alkyl- or chloroaromatic substrate and that the tbc1 gene products play a role in the catabolism of the first metabolite that results from transformation of the initial substrate. Phylogenetic analysis was used to compare individual components of these tbc monooxygenases with similar sequences in the databases. These results provide further evidence for the existence of multiple, functionally redundant alkyl- and chloroaromatic monooxygenases in strain JS150.
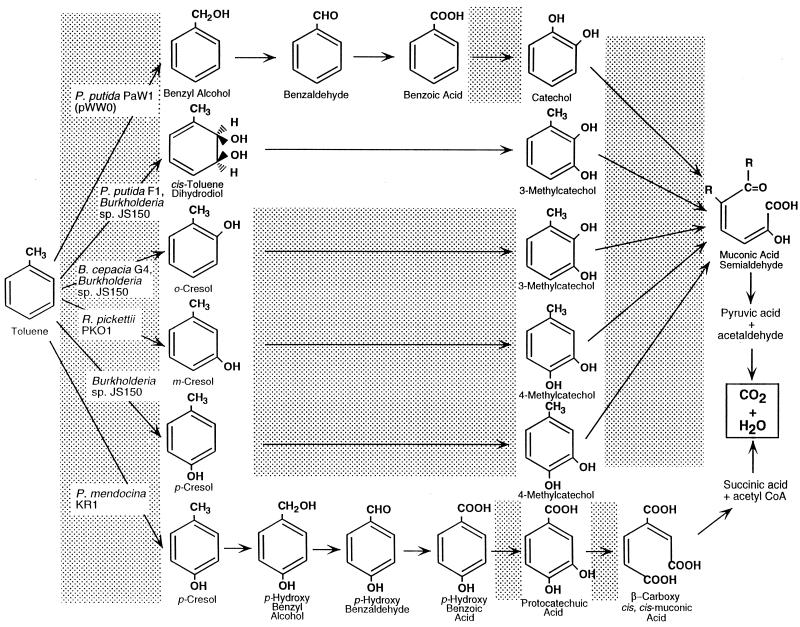
Biodegradation of the monoaromatic hydrocarbons, benzene, toluene, ethylbenzene, and the xylenes (collectively designated BTEX) has been extensively investigated as a basis for understanding the intrinsic biodegradation potential of these fuel hydrocarbons when they occur as groundwater contaminants (17, 21, 39, 57, 61). Toluene has been studied as a model compound representative of this group of aromatic hydrocarbons (14), and its biodegradation under aerobic conditions has been found to proceed by the six pathways shown in Fig. 1. Implicit in much of the literature on biodegradation of toluene is the assumption that each toluene degrader elaborates a single pathway for toluene degradation. This is seen, for example, for Pseudomonas putida mt-2 (PaW1), which carries the TOL plasmid pWW0 (64); for P. putida F1 (13); for Burkholderia cepacia G4 (52); for Ralstonia pickettii PKO1 (25); or for Pseudomonas mendocina KR-1 (63). However, Burkholderia sp. strain JS150 seems to be an exception to this rule. Previous research conducted with this strain by Haigler and coworkers (15) has demonstrated that a broad-substrate-range toluene dioxygenase is at least partly responsible for the extended aromatic substrate range of JS150. Subsequently, the studies of Johnson and Olsen (23, 24) have shown that this strain also produces an ortho- and a para-monooxygenase that are used in the degradation of toluene. We have continued to investigate strain JS150 in order to learn more about the physiological significance of its unusual ability to produce apparently redundant oxygenases for dissimilation of monoaromatic hydrocarbons. In this study we report the cloning and characterization of additional genes encoding multicomponent monooxygenases that allow strain JS150 to transform toluene, as well as several related monoaromatic substrates.
FIG. 1.
Pathways for aerobic catabolism of toluene. The shaded areas indicate steps where oxygen is consumed for substrate-level oxygenation. R, a methyl group. CoA, coenzyme A.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. P. aeruginosa PAO1c containing cloned fragments in vector pRO1727 was maintained on plate count medium (TNA [36]) containing carbenicillin (500 μg/ml). Escherichia coli JM109 was used for routine maintenance and construction of plasmids and for construction of fragments used for DNA sequence analysis. E. coli cells containing recombinant plasmids were maintained on Luria-Bertani (LB) medium (48) supplemented with ampicillin (50 μg/ml). When used for enzyme assays or for high-performance liquid chromatography (HPLC) analyses of metabolites, cells were routinely grown in a basal salts medium (BM [32]) containing (per liter) 2.49 g of Na2HPO4, 3.05 g of KH2PO4, 0.1995 g of MgSO4, 0.995 g of CaCl2 · 2H2O, 0.00005 g of FeSO4 · 7H2O, 0.00025 g of NaMoO4 · 2H2O, 1.0 ml of Hunter's trace metal solution (7), 1.0 g of (NH4)2SO4, and 1.0 g of KNO3. When needed, Casamino Acids (Difco Laboratories, Detroit, Mich.) or glucose was added to BM to a final concentration of 0.1 or 0.25%, respectively. When used for enzyme induction experiments, liquid toluene, benzene, chlorobenzene, trichloroethylene (TCE), phenol, or o-cresol was added directly to BM to a final concentration of 1.0 mM. Cultures maintained in liquid or solid media were incubated at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | recA1 relA1 thi-1 Δ(lac-proAB) gyrA96 hsdR17 endA1 supE44 (rK− mK+) [F′ traD36 proAB lacIqZΔM15] | Promega |
| E. coli BL21 | F−dcm ompT hsdR (rB− mB+) gal | Stratagene |
| Burkholderia sp. strain JS150 | Tol+ Ben+ Chlben+ Crl+ Phl+ | 15 |
| P. aeruginosa PAO1c | Prototroph | 20 |
| Plasmids | ||
| pBluescript II KS+(SK+) | lacZ Apr; 2.96-kb cloning vector | Stratagene |
| pRO1727 | Tcr Cbr; 3.77-kb Pseudomonas cloning vector | 8 |
| pHYK2000 | Cbr; pRO1727::14.3-kb HindIII-BamHI fragment from JS150 containing tbc1 and tbc2 | This study |
| pHYK2001 | Cbr; pRO1727::11.3-kb HindIII-BamHI fragment of pHYK2000, with a 3-kb HindIII deletion | This study |
| pHYK2002 | Cbr; pRO1727::10.1-kb HindIII-BamHI fragment of pHYK2000, with a 4.2-kb internal NotI deletion | This study |
| pHYK2003 | Cbr; pRO1727::8.9-kb HindIII-BamHI fragment of pHYK2000, with a 5.4-kb internal XhoI deletion | This study |
| pHYK2004 | Cbr; pRO1727::10.5-kb HindIII-XhoI fragment of pHYK2000, with a 3.8-kb deletion fusing an internal XhoI site with a vector SalI site | This study |
| pHYK2005 | Cbr; pRO1727::9.2-kb BamHI-XhoI fragment of pHYK2000 ligated to BamHI- and XhoI-digested pRO1727 | This study |
| pJCM1000 | Apr; pBluescript::5.4-kb XhoI fragment of pHYK2000 | This study |
| pJCM1001 | Apr; pBluescript::6.2-kb HindIII-NotI fragment of pHYK2000 | This study |
Abbreviations: Tol, toluene; Ben, benzene; Chlben, chlorobenzene; Crl, cresol; Phl, phenol; Ap, ampicillin; Tc, tetracycline; Cb, carbenicillin.
Clone isolation, deletion mutagenesis, and genetic techniques.
Total genomic DNA of Burkholderia sp. strain JS150 was isolated using a Nucleospin tissue kit (Clontech). Genomic DNA was digested with BamHI and HindIII and ligated with similarly digested vector plasmid pRO1727. Ligation products were transformed into cells of P. aeruginosa PAO1c using the CaCl2 method (28), with initial selection on TNA medium containing carbenicillin (500 μg/ml). Plating the primary transformants onto BM with toluene as the sole carbon source yielded no colonies capable of growth on this substrate. Therefore, a multistep selection and screening protocol was used. Primary transformants were first screened for the ability to grow on BM (supplemented with carbenicillin) containing Casamino Acids together with either toluene, benzene, or chlorobenzene. This was designed to select for transformants capable of growth in the presence of the aromatic carbon source. Several hundred transformants selected at random from this primary screen were then further screened individually for the ability to transform the aromatic carbon source, using the substrate conversion assay described below. Using such a protocol, three transformants capable of converting the aromatic carbon source to a more polar product, as deduced from HPLC analysis described below, were obtained. One of these transformants was selected for detailed analysis, owing to its ability to rapidly convert the aromatic carbon sources. The recombinant plasmid, purified from this isolate using a Qiagen kit, was designated pHYK2000.
For functional mapping of the cloned DNA fragment from strain JS150, plasmid pHYK2000 (Table 1) was digested with HindIII, NotI, XhoI, and BamHI. The resultant HindIII, NotI, XhoI, HindIII-XhoI, and BamHI-XhoI DNA fragments were extracted following electrophoretic separation in a 1.2% agarose gel, and each fragment was then ligated with vector plasmid pRO1727 digested with the same restriction endonucleases. The ligation mixture was introduced into P. aeruginosa PAO1c by electroporation using the method of Smith and Iglewski (55), and electrotransformants were selected on TNA medium containing carbenicillin (500 μg/ml). Plasmids with the expected deletion were confirmed by restriction endonuclease digestion patterns of purified DNAs obtained from selected clones. The deletion constructs were designated pHYK2001, pHYK2002, pHYK2003, pHYK2004, and pHYK2005 (Table 1; Fig. 2). Each of the deletion subclones was also ligated to vector pBluescript SK+ digested with the same restriction enzyme for DNA sequencing. The pBluescript clones were introduced into E. coli JM109 by the CaCl2 method (28), with selection on LB medium containing ampicillin (50 μg/ml).
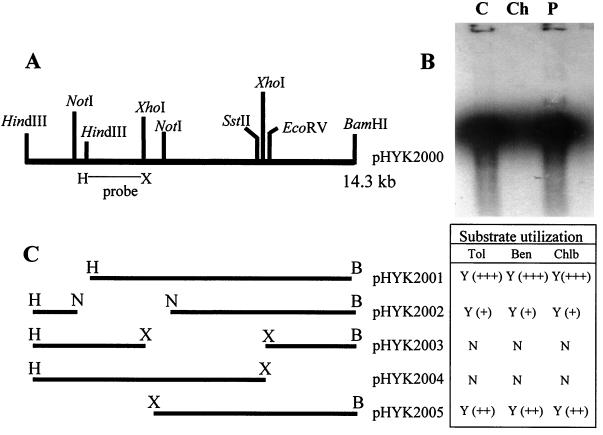
FIG. 2.
Physical and functional map of pHYK2000. (A) Restriction map of the 14.3-kb BamHI-HindIII DNA fragment from Burkholderia sp. strain JS150 cloned into vector pRO1727 as pHYK2000. The XhoI-HindIII fragment used as a probe is indicated. (B) Southern hybridization results for DNA digested with XhoI and HindIII. Lanes: C, control (cloned) DNA; Ch, chromosomal DNA; P, plasmid DNA. (C) Construction of deletion mutants of pHYK2000 and assays for substrates oxidized by each construct. For functional mapping of pHYK2000, HindIII, NotI, XhoI, BamHI-XhoI, and HindIII-XhoI deletions were constructed, and the constructs were used to assay for oxidation (+, weak; ++, intermediate; +++, strong) of toluene (Tol), benzene (Ben), and chlorobenzene (Chlb).
Isolation of plasmid DNA from the indigenous plasmid of strain JS150 was accomplished using the method of Olsen and Hansen (36). This plasmid DNA was purified using cesium chloride-ethidium bromide density gradient centrifugation.
Substrate conversion assays.
Cultures were grown in 50 ml of BM with 0.1% Casamino Acids, 0.25% glucose, and a 1 mM concentration of either toluene, benzene, chlorobenzene, TCE, o-cresol, or phenol in tightly stoppered 500 ml-bottles at 30°C with shaking until they reached the late log phase. Cells were harvested by centrifugation at 10,000 × g at 4°C and were washed twice with 40 mM potassium-sodium phosphate buffer (pH 6.8). Washed cells were transferred to 10 ml of the same buffer containing either toluene, benzene, chlorobenzene, TCE, o-cresol, or phenol (1.0 mM) to produce an A600 of 1.0. These washed cell suspensions were incubated with shaking at 30°C for 24 h, and 500-μl samples were taken every 4 h, mixed with 500 μl of methanol in 1.5-ml microcentrifuge tubes, and then centrifuged at 4°C for 10 min to remove cells. The resulting supernatants were carefully transferred to autosampler vials and were analyzed by reverse-phase HPLC. Uninoculated bottles, which served as controls, were incubated under the same conditions, and results were corrected for substrate losses from the controls (which were never more than 10% of the initial hydrocarbon concentration). Reverse-phase chromatography was performed with a PhaseSep H4726 column (4.6 by 250 mm) filled with Spherisorb ODS2 (particle diameter, 5 μm) preceded by a Whatman CSKI guard column (6.5 by 65 mm) coupled to a Shimadzu SCL-6B solvent delivery system and a CR501 Chromatopac computing integrator. A methanol-water solvent was used at a flow rate of 1 ml/min. The methanol/water ratio was adjusted between 90:10 and 70:30, depending on the target analyses. Each substrate was detected by monitoring at A254, and concentrations were calculated by comparison with a standard curve as described previously (27, 38).
Analysis of tbc gene expression in E. coli.
In order to analyze separately the functions encoded by tbc1 and tbc2, the genes were cloned into pBluescript under transcriptional control of the vector's lac promoter for analysis in E. coli. For this, tbc2 was excised from pHYK2000 as an XhoI fragment (see Fig. 4) and was subcloned into XhoI-digested pBluescript SK. Selection of a clone with the proper orientation of the inserted fragment was verified by restriction digest analyses. This plasmid was designated pJCM1000. Subcloning of tbc1 into pBluescript was done in a three-step process. First, the small HindIII fragment of pHYK2000 (see Fig. 4) was subcloned into pBluescript, with selection of a clone with the proper orientation (relative to the lac promoter) made by restriction digest analyses. Then a HindIII-NotI deletion was made of this fragment to an adjacent NotI site in the vector. This produced a unique NotI restriction site. Finally, the NotI fragment from pHYK2000 that partially overlaps the small HindIII fragment was subcloned into the unique NotI site of the pBluescript recombinant, yielding a fusion that would regenerate the original HindIII-NotI DNA fragment of pHYK2000. Selection of a clone with the proper orientation of the NotI insert was made by restriction digest analyses. This plasmid was designated pJCM1001.
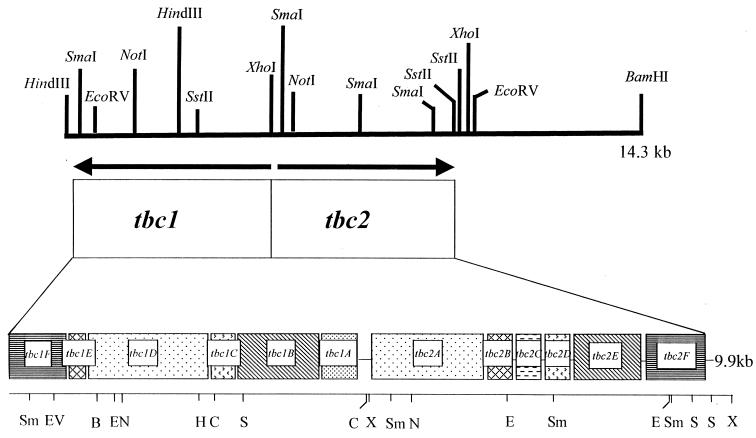
FIG. 4.
Schematic showing the organization on pHYK2000 of the genes encoding the Tbc1 and Tbc2 monooxygenases, as deduced from DNA sequence analysis. Nucleotide sequence analyses revealed that tbc genes are organized into two divergent operons, tbc1 and tbc2, each comprised of six open reading frames, designated tbc1ABCDEF and tbc2ABCDEF, respectively. Abbreviations: EV, EcoRV; B, BglII; E, EcoRI; N, NotI; H, HindIII; C, ClaI; S, SstII; P, PstI; X, XhoI; Sm, SmaI.
The recombinant plasmids were introduced into E. coli BL21 for expression analysis. For this, cells were grown overnight at 37°C with shaking in 25 ml of LB medium with ampicillin in 250-ml conical flasks. Cells harvested by centrifugation were washed once in phosphate-buffered saline (48) and were resuspended in 3 ml of phosphate-buffered saline to a final A600 of 15 in stoppered 25-ml flasks. Toluene or o-cresol was added to a final concentration of 1 mM. These resting cell suspensions were incubated with shaking at 37°C for 19 h. Aliquots were removed for reverse-phase HPLC analysis (described above) at 0, 6, and 19 h. Control cultures consisted of E. coli BL21 carrying the pBluescript vector, as well as pHYK2000 carried in P. aeruginosa PAO1c. For the latter, cells were grown under inducing conditions as described above.
RNA preparation and Northern hybridization.
In order to analyze expression of Tbc monooxygenases, cells of P. aeruginosa PAO1c containing pHYK2000 were grown overnight in 50 ml of BM containing 1 mM toluene, benzene, chlorobenzene, TCE, or o-xylene with 0.1% Casamino Acids. Cells were harvested by centrifugation at 4°C and were suspended in 50 ml of protoplasting buffer (15 mM Tris-Cl, pH 8.0; 0.45 M sucrose; 8 mM EDTA), which contained 100 μl of lysozyme (10 mg/ml). Following centrifugation the cell pellet was resuspended in 2.5 ml of lysis buffer (30 mM Tris-Cl, pH 7.4; 100 mM NaCl; 5 mM EDTA; 1% sodium dodecyl sulfate [SDS]), to which 50 μl of diethyl pyrocarbonate (DEPC) was added. The suspension was incubated at 37°C for 5 min and cooled on ice, and then 1.25 ml of 6.8 M NaCl was added to precipitate proteins which were sedimented by centrifugation. Nucleic acids were precipitated by addition of 2 to 3 volumes of LiCl to the supernatant, which was kept at −70°C overnight. The pellet, which had been washed with 70% ethanol, was dissolved in 450 μl of DNase buffer (50 mM Tris-Cl, pH 7.4; 10 mM CaCl2) to which was added 50 μl of DNase preheated to 25°C. DNase was removed by phenol treatment, and the RNA was precipitated at −70°C in a sodium acetate (pH 5.2) solution with ethanol. RNA was recovered by centrifugation, and the pellet was washed twice with 75% ethanol and was dried under vacuum. The RNA pellet was dissolved in DEPC-treated water and was stored at −70°C with RNase Block RNase inhibitor (Stratagene, La Jolla, Calif.). All the chemicals, glassware, and plasticware for RNA isolation were treated with DEPC and were sterilized before use. Total RNA (5 μg) was separated in a formamide-containing 1% agarose gel and then blotted to a Hybond-N membrane (Amersham) for 6 h.
Hybridization was done using methods essentially as described by Thomas (60). Briefly, the blotted membrane was washed and air dried, and then DNA cross-linking was accomplished with a Spectrolinker (Spectronics Corporation, Westbury, N.Y.). The membrane was prehybridized in hybridization solution (30% formamide, 5× Denhardt's solution, 5× SSPE [4% NaCl, 2.2% sodium phosphate, 0.2% EDTA], 100 μg of salmon sperm DNA per ml) for 2 h at 42°C and then hybridized with the denatured probe in the same solution for 20 h at 42°C. The 2-kb XhoI-HindIII fragment of pHYK2000 which was used as the probe (see Fig. 2), was end labeled with [α-32P]dCTP using T4 polynucleotide kinase according to the protocol provided by Promega. The hybridized membrane was washed three times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.05% SDS and then twice in 0.2× SSC with 0.1% SDS for 10 min each at 42°C. The membrane was then washed twice in 0.2× SSC with 0.1% SDS for 10 min each at 68°C. The washed membrane was then air dried and exposed to X-ray film at −85°C.
Nucleotide sequence analysis.
Nucleotide sequencing was carried out using an ABI 373A automated sequencer based on the method of Sanger et al. (49), with T7, T3, and specific synthetic oligonucleotide primers. For PCR amplification of fragments to be sequenced, a total 10-μl reaction mixture containing 0.2 μg of template DNA, 1.6 pmol of primer, and 1 U of Amplitaq FS (Gibco BRL, Gaithersburg, Md.) was employed for 25 cycles of 10 s at 96°C, 5 s at 50°C, and 4 min at 60°C. Sequence analysis was done with Lasergene software (DNA Star, Inc., Madison, Wis.), MacVector version 4.5.3 (Oxford Molecular, Campbell, Calif.), and the Genetics Computer Group (GCG) (University of Wisconsin, Madison) software package, version 8.1. Searches of the GenBank database and pairwise sequence comparisons were carried out with GCG programs TFASTA and BESTFIT, respectively. Similarity searches were also performed with the BLAST program and the NCBI databases. Nucleotide sequence alignments were done by using the GCG multiple sequence alignment program PILEUP or the Lasergene program MegAlign.
Chemicals.
All chemicals used in this study were of the highest purity commercially available. Aromatic hydrocarbons were purchased from Sigma Chemical Co. (St. Louis, Mo.). Components for cell growth were purchased from Difco, Aldrich Chemical Co. (St. Louis, Mo.), and Gibco BRL. Enzymes and reagents used for nucleic acid manipulations were purchased from Promega, Gibco BRL, and Stratagene.
Nucleotide sequence accession numbers.
The sequence data obtained in this study have been deposited in the GenBank data library under accession numbers AF282897 and AF282898.
RESULTS AND DISCUSSION
Cloning of a DNA fragment from Burkholderia sp. strain JS150 that allowed P. aeruginosa PAO1c to transform toluene, benzene, and chlorobenzene.
Previous research conducted on Burkholderia sp. strain JS150 had revealed that this organism is capable of utilizing a wide range of alkyl- as well as chloroaromatic hydrocarbons (15) and that this degradative ability was linked, in part, to the presence of multiple dioxygenases and monooxygenases of broad substrate specificity (15, 23, 24). In order to gain a better understanding of the physiological significance of multiple and apparently redundant oxygenases in this strain, we have continued to clone additional genes allowing for catabolism of alkyl- and chloroaromatic substrates. For this, total genomic DNA from strain JS150 digested with HindIII and BamHI was randomly ligated into cloning vector pRO1727 and was transformed into the heterologous recipient, P. aeruginosa PAO1c, which itself is unable to transform toluene, benzene, or chlorobenzene. From the isolation and screening protocol described in Materials and Methods, we obtained a recombinant plasmid that allowed P. aeruginosa PAO1c to transform toluene, benzene and chlorobenzene. This clone was designated pHYK2000. Restriction digest analysis of the recombinant plasmid demonstrated that it contained a 14.3-kb HindIII-BamHI insert. The clone was mapped with restriction endonucleases, and the results are shown in Fig. 2A.
In order to determine whether the cloned DNA fragment originated from the chromosome or from one of the plasmids of strain JS150, Southern hybridization analyses were performed (56). For this, an arbitrarily chosen, 2-kb HindIII-XhoI fragment from pHYK2000 was labeled with [α-32P]dCTP. The hybridization results, presented in Fig. 2B, showed a strong positive signal for HindIII-XhoI-digested plasmid DNA. However, a similarly strong signal was not detected for HindIII-XhoI-digested total genomic DNA, which would have a preponderance of chromosomal DNA. The intensity of the signals from the control and plasmid hybridizations slightly obscures the total genomic DNA digest lane such that a weakly positive signal from a similarly sized band (as would be expected since the total genomic DNA preparation should include some plasmid DNA) would not be readily detected. Nevertheless, these results clearly indicate that the source of the cloned DNA in pHYK2000 was the indigenous plasmid of strain JS150. Johnson and Olsen (23), in their previous study on characterization of a toluene or benzene monooxygenase from strain JS150, also found that these tbm genes were located on a plasmid in this strain. Interestingly, they detected two hybridizing fragments of different sizes in their Southern blots, only one of which was the tbm locus. Based on our results, it is likely that the other hybridizing fragment corresponds to the genes that we have cloned here.
Functional characterization of pHYK2000.
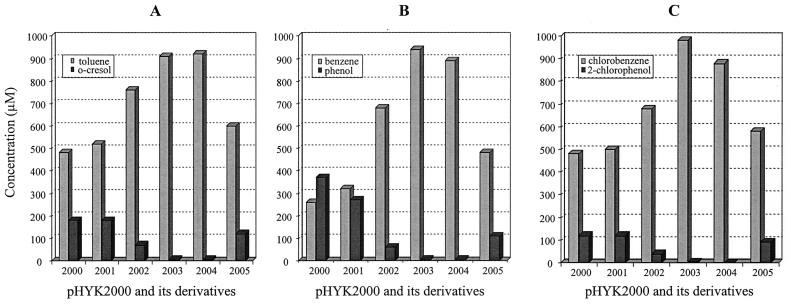
Deletion mutagenesis was used to determine the location of the genes on pHYK2000 that allow for oxidation of toluene, benzene, and chlorobenzene. For this, plasmid pHYK2000 was digested with the restriction endonucleases shown in Fig. 2C. The deletants as well as pHYK2000 were inserted into P. aeruginosa, and cells were grown in the presence of toluene, benzene, or chlorobenzene. These induced cells were then washed and were used as resting cells to determine the products from the initial oxidation of the homologous substrate. Analysis of metabolites using reverse-phase HPLC revealed that P. aeruginosa PAO1c containing pHYK2000 was able to oxidize toluene to o-cresol, benzene to phenol, and chlorobenzene to 2-chlorophenol (Fig. 3). In addition, we determined that TCE-induced cells were able to convert TCE to more-polar products (data not shown). In order to determine whether the genes allowing for transformation of toluene, benzene, or chlorobenzene were associated with a single locus or with separate loci on pHYK2000, the various deletants shown in Fig. 2C were also tested for metabolite production. Reverse-phase HPLC analysis demonstrated that cells of PAO1c containing pHYK2001, pHYK2002, or pHYK2005 were able to transform toluene, benzene, and chlorobenzene. However, cells containing pHYK2003 or pHYK2004 had no ability to transform the substrates (Fig. 3). The results also showed that there was no physical or functional separation in the ability to transform toluene, benzene, or chlorobenzene among the various deletants, suggesting that there is only a single locus for this function. Since the pHYK2002, -2005, and -2001 deletants retained progressively greater amounts of the original clone, the product yield results suggested to us that the additional DNA provided enhanced functionality, indicating that some additional catabolic functions were located on these fragments.
FIG. 3.
Functional analysis of pHYK2000 and deletion constructs made from pHYK2000. Cells were grown and induced as described in the text. (A) Conversion of toluene by cells containing pHYK2000 or its derivatives. The light shaded bars indicate the amount of toluene remaining; the dark shaded bars indicate the amount of o-cresol formed from degradation of toluene. (B) Conversion of benzene. The light shaded bars indicate the amount of benzene remaining; the dark shaded bars indicate the amount of phenol formed from degradation of benzene. (C) Conversion of chlorobenzene. The light shaded bars indicate the amount of chlorobenzene remaining; the dark shaded bars indicate the amount of 2-chlorophenol formed from degradation of chlorobenzene.
Previous studies conducted by Haigler et al. (15) had found that toluene and chlorobenzene could be attacked by a dioxygenase in strain JS150. Subsequent research by Johnson and Olsen (23) demonstrated that a toluene monooxygenase was also present in this strain, yielding a mixture of o- and p-cresol as the initial oxidation products. The oxygenase(s) present on pHYK2000 was clearly different from these previously described enzymes in that o-cresol was the sole product detected from oxidation of toluene.
Organization of genes encoding the duplex Tbc monooxygenases.
Nucleotide sequence analysis of the 14.3-kb DNA fragment of pHYK2000 revealed two gene clusters arranged in divergent orientation (Fig. 4). Each cluster was comprised of six open reading frames that appeared to be operonic in organization. We have designated these gene clusters tbc1 and tbc2.
The deduced products of the tbc1 gene cluster showed significant similarity to a group of multicomponent monooxygenases that have been shown to function in hydroxylation of phenol, cresol, and related hydroxylated aromatic substrates. Table 2 shows the homologs that have at least 70% overall sequence identity to the comparable Tbc1 components. In addition to these highly homologous sequences, other polypeptide components of multicomponent monooxygenases with lower overall similarity to the Tbc1 enzyme were found in the phenol hydroxylases of Ralstonia eutropha E2 (19), Ralstonia sp. strain KN1 (33). P. putida P35X (35), P. putida H (18), Acinetobacter calcoaceticus NCIB8250 (9), and Pseudomonas sp. strain BH (58). Also included in this group is the well-characterized phenol and dimethylphenol hydroxylase from Pseudomonas sp. strain CF600 (53). Interestingly, Tbc1 was also found to have a low (∼45%) degree of overall similarity to a multicomponent oxygenase associated with the ability to oxidize dimethyl sulfoxide in Acinetobacter sp. strain 20B (22).
TABLE 2.
Gene product homologs of the tbc1 and tbc2 gene clusters
| Gene and homologs | Size (amino acids) | % Identity | % Similarity | Function | Organism | Accession no. | Reference |
|---|---|---|---|---|---|---|---|
| tbc1A | 59 | ||||||
| tbmA | 69 | 84 | 85 | Toluene | Burkholderia sp. strain JS150 | L40033 | 23 |
| crpA | 69 | 84 | 85 | Cryptic | Ralstonia pickettii PKO1 | AF012632 | 37 |
| tbc1B | 335 | ||||||
| crpB | 335 | 99 | 99 | Cryptic | Ralstonia pickettii PKO1 | AF012632 | 37 |
| tbmB | 336 | 91 | 91 | Toluene | Burkholderia sp. strain JS150 | L40033 | 23 |
| tbc1C | 89 | ||||||
| crpC | 89 | 100 | 100 | Cryptic | Ralstonia pickettii PKO1 | AF012632 | 37 |
| tbmC | 89 | 98 | 98 | Toluene | Burkholderia sp. strain JS150 | L40033 | 23 |
| tbc1D | 514 | ||||||
| tbmD | 513 | 94 | 94 | Toluene | Burkholderia sp. strain JS150 | L40033 | 23 |
| aphN | 536 | 84 | 89 | Phenol | Comamonas testosteroni TA441 | AB006479 | 1 |
| tomA3 | 519 | 82 | 87 | Toluene, TCE | Burkholderia cepacia PR123 | I24403 | 51 |
| phcN | 540 | 79 | 85 | Phenol | Comamonas testosteroni R5 | AB024741 | 59 |
| tbc1E | 66 | ||||||
| tomA4 | 93 | 51 | 54 | Toluene, TCE | Burkholderia cepacia PR123 | I24403 | 51 |
| tbc1F | 354 | ||||||
| tbmF | 355 | 91 | 92 | Toluene | Burkholderia sp. strain JS150 | L40033 | 23 |
| phcP | 357 | 71 | 78 | Phenol | Comamonas testosteroni R5 | AB024741 | 59 |
| aphP | 357 | 70 | 78 | Phenol | Comamonas testosteroni TA441 | AB006479 | 1 |
| tbc2A | 501 | ||||||
| tbuA1 | 501 | 99 | 99 | Toluene, benzene | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlK | 501 | 93 | 97 | Phenol | Ralstonia eutropha JMP134 | AF065891 | 2 |
| bmoA | 500 | 71 | 82 | Benzene | Pseudomonas aeruginosa JI104 | D83068 | 26 |
| tbc2B | 86 | ||||||
| tbuU | 86 | 100 | 100 | Toluene, benzene | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlL | 86 | 90 | 98 | Phenol | Ralstonia eutropha JMP134 | AF065891 | 2 |
| tbc2C | 111 | ||||||
| tbuB | 111 | 98 | 98 | Toluene, benzene | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlM | 111 | 92 | 95 | Phenol | Ralstonia eutropha JMP134 | AF065891 | 2 |
| tbc2D | 104 | ||||||
| tbuV | 104 | 97 | 97 | Toluene, benzene | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlN | 105 | 89 | 92 | Phenol | Ralstonia eutropha JMP134 | AF065891 | 2 |
| tbc2E | 352 | ||||||
| tbuA2 | 329 | 92 | 92 | Toluene, benzene | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlO | 332 | 81 | 86 | Phenol | Ralstonia eutropha JMP134 | AF065891 | 2 |
| tbc2F | 334 | ||||||
| tbuC | 334 | 99 | 99 | Toluene, benzene | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlP | 332 | 77 | 86 | Phenol | Ralstonia eutropha JMP134 | AF065891 | 2 |
Based on overall sequence similarity, as well as similarity in gene order to this family of enzymes, it is reasonable to conclude that Tbc1D, Tbc1B, and Tbc1E would comprise, respectively, the α, β, and γ subunits of a putative α2β2γ2 hexameric nonheme iron monooxygenase. Further support for this conclusion comes from the presence in Tbc1D, the putative α subunit of the monooxygenase, of the highly conserved motif Asp-Glu-X-Arg-His, which occurs twice in Tbc1D at positions 139 and 234 (data not shown). These ligands form the dinuclear iron binding site in the large subunits of this family of monooxygenases (11, 12). In addition, the spacing of 94 amino acids between these amino acid repeats was conserved in Tbc1D.
Tbc1F shared homology with a very large group of iron-sulfur flavoproteins which function as oxidoreductases to transfer electrons from reduced pyridine nucleotides to a terminal electron acceptor via a flavin and [2Fe-S] center (29). Supportive of this deduced function was the presence near the N terminus (residues 37 to 77, data not shown) of Tbc1F of the conserved sequence Cys-X3-Cys-X2-Cys-X29-Cys, which is found in chloroplast-type ferredoxins (40). The C-terminal portion (residues 149 to 155 and 207 to 236 [data not shown]) of Tbc1F contained motifs characteristic of the binding domains for the isoalloxazine ring of flavin adenine dinucleotide and NADP ribose (44).
Tbc1C was found to be homologous to the small polypeptides that are believed to play a role in regulating monooxygenase activity. The only member of this group of polypeptides among the phenol and cresol hydroxylases that has been studied in detail is DmpM from Pseudomonas sp. strain CF600. DmpM binds to the DmpNLO hydroxylase, but it does not participate directly in redox reactions. Rather, its role appears to be one of increasing product yield from the phenol hydroxylase, possibly by controlling entrance of substrate and exit of products to and from the active site (46). Tbc1C retained the conserved amino acids Glu54 and Gly57, which were identified as conserved components of the functionally important helix 2 of DmpM (46); however, Leu56 of DmpM was replaced by Thr in both Tbc1C and in TbmC of strain JS150 (data not shown).
Unlike the other Tbc1 components which exhibited homology to polypeptide components from a variety of enzymes associated with phenol, toluene, alkene, and methane oxidation, Tbc1A was unique in that it only shared significant similarity with a small group of 11 polypeptides, all of which are only found as components of enzymes associated with phenol or cresol oxidation. The functional role of Tbc1A can be inferred from studies on DmpK from Pseudomonas sp. strain CF600, where it has been shown that DmpK binds to both DmpN and DmpL and plays an essential role in assembly of the active oxygenase, possibly by posttranslational insertion of iron into the α subunit (45).
Analysis of the six deduced protein products of the tbc2 gene cluster revealed significant similarity to a group of multicomponent monooxygenases that have been shown to function in initial hydroxylation of a range of unactivated hydrocarbons, including aromatic substrates such as benzene, toluene, and o-xylene; alkenes such as propene, isoprene, butene, butadiene, and pentene; and halogenated alkenes such as trichloroethylene. Table 2 shows the homologs that have at least 70% overall sequence identity to the comparable Tbc2 components. In addition to these highly homologous sequences, other polypeptide components of multicomponent monooxygenases with lower overall similarity to the Tbc2 enzyme were found in the toluene and o-xylene monooxygenase of Pseudomonas stutzeri OX1 (3), the toluene monooxygenase of P. mendocina KR1 (66, 67), the isoprene monooxygenase of Rhodococcus sp. strain AD45 (62), the propene monooxygenases from Xanthobacter sp. strain Py2 (68) and Rhodococcus rhodochrous B276 (47), and the carbazole monooxygenases of Pseudomonas sp. strain CA10 (50) and P. stutzeri OMI (41). Enzymes of this family are comprised of four dissociable components, three of which constitute a short electron transfer chain made of an oxidoreductase, a ferredoxin, and a terminal oxygenase. Based on overall sequence similarity, as well as similarity in gene order to this family of monooxygenases, it is reasonable to assign Tbc2F as the oxidoreductase, Tbc2C as the ferredoxin, and Tbc2AEB as the terminal hydroxylase in the electron transfer chain of the Tbc2 monooxygenase. Support for these assignments comes from analyses similar to those conducted on the Tbc1 components described above.
Tbc2F, the putative oxidoreductase, was similar to the proteins that comprise the very large family of iron-sulfur flavoproteins that function as oxidoreductases for most mono- and dioxygenase systems. Consistent with this assignment was the presence in Tbc2F of conserved Cys (Cys37, -42, -45, and -77) and Gly (Gly40 and -52) residues (data not shown) necessary for coordination of the two iron atoms of the [2Fe-2S] cluster (29), as well as conserved motifs near the C terminus of the protein (residues 147 to 155 and 202 to 233 [data not shown]) characteristic of the binding domains for flavin adenine dinucleotide and NADPH (44). It is of interest that the homologs of Tbc2F (Table 2) include polypeptides associated with both mono- and dioxygenases associated with dissimilation of a diverse array of substrates, including monoaromatics such as benzene and toluene, alkenes such as propene and isoprene, polycyclics such as phenanthrene, and N-containing polycyclics such as carbazole. It appears that the oxidoreductase function is not as substrate specific as that found for the other components of these multicomponent enzymes. In fact, it has been previously shown that TmoF, the oxidoreductase associated with the toluene 4-monooxygenase of P. mendocina KR1, can be replaced by the reductase component from naphthalene dioxygenase and vice versa (66). Moreover, recent biochemical studies with purified protein components have shown that TmoF can be replaced by spinach ferredoxin reductase and NADPH as the oxidoreductase components, in combination with TmoC (the ferredoxin) and the TmoAEB hydroxylase for multiple turnover oxidation of toluene in vitro (42). These results indicate that there is a certain amount of interchangeability among these oxidoreductases.
Tbc2C was highly homologous to the ferredoxin components associated with multicomponent monooxygenases involved in degradation of benzene, toluene, o-xylene, and alkenes (Table 2). These polypeptides were, in turn, related to a large family of Rieske-type ferredoxins that function as soluble electron carriers for a variety of bacterial oxygenases. Tbc2C contained the metal-binding motif Cys-X-His-X15–21-Cys-X2-His at positions 44 to 66 (data not shown) that is characteristic of all Rieske proteins (29). The Tbc2C homolog among the multicomponent aromatic monooxygenases that has been investigated in greatest detail is TmoC from P. mendocina KR1. Spectroscopic analysis of purified TmoC has revealed that this protein exhibits absorption bands similar to ferredoxins with mixed Cys and His ligation (42). In addition, 15N nuclear magnetic resonance studies have shown that four residues, His47, Gln48, Ala66, and His67, likely provide the peptidyl N-H bonds to the inorganic sulfides (65). These residues are conserved in Tbc2C (data not shown).
A high degree of similarity to the components of the terminal hydroxylases of multicomponent monooxygenases (Table 2) suggests that Tbc2AEB would comprise the α, β, and γ subunits, respectively, of a putative (αβγ)2 dimeric nonheme iron monooxygenase. Consistent with this is the presence in Tbc2A, the putative α subunit of the monooxygenase, of two copies of the amino acid sequence motif (Asp/Glu)-X−30-Asp-Glu-X-Arg-His at positions 104 to 137 and 197 to 234 (data not shown), which are the ligands for a diiron center in the active site of this family of enzymes (10). Site-directed mutagenesis studies have been conducted on the active site of the α subunit of the toluene 4-monooxygenase hydroxylase (T4MOH) component from P. mendocina KR1 (43). Using a model of the T4MOH active site constructed from the crystal structure of soluble methane monooxygenase, with which T4MOH is homologous, it has been proposed that the diiron center is surrounded by two hydrophobic regions. Site-directed mutagenesis of selected amino acid residues in this hydrophobic region has shown that product distribution from oxidation of toluene can be altered. However, it is clear that these amino acids (Ile100, Ala107, Gln141, Phe176, Leu179, Phe180, Leu192, Phe196, Thr210, Phe205, Ile224, and Ile227) alone cannot be the sole determinants of regiospecificity for toluene oxidation since these residues are identical between the toluene 2-monooxygenase (tbc2 gene product) of JS150 and the toluene 3-monooxygenase of R. pickettii PKO1.
Tbc2D was found to be homologous to the small polypeptides that are believed to play a role in regulating monooxygenase activity. The members of this group of polypeptides among the toluene and alkene monooxygenases that have been studied in detail include TmoD from P. mendocina KR1 and XamoD (encoded by aamD) from Xanthobacter sp. strain Py2. In the KR1 system, TmoD, which appears to be present normally as a substoichiometric constituent of the TmoAEB hydroxylase, can mildly stimulate the rate of toluene hydroxylation when added to separately purified hydroxylase (42). In the alkene oxidation system of strain Py2, it has been shown that XamoD is essential for steady-state alkene epoxidation (54). Moreover, Tbc2D retained the conserved amino acids Glu54, Leu56, and Gly57, which were identified as conserved components of the functionally important helix 2 of DmpM, which provides a similar regulatory function for the phenol and cresol hydroxylase of Pseudomonas sp. strain CF600 (46). From this it is reasonable to conclude that Tbc2D might function as a regulatory component of the Tbc2 monooxygenase complex.
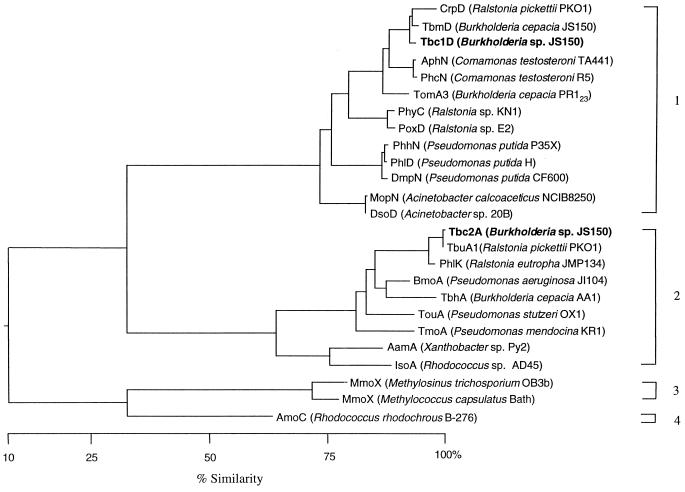
Phylogenetic as well as biochemical analyses have revealed that the diiron monooxygenases form an enzyme family. Figure 5 shows a phenogram constructed for the α subunits of the hydroxylase component of these multicomponent diiron monooxygenases. Since the α subunit contains the diiron center and is the site of substrate hydroxylation, it is reasonable to assume that amino acids in this subunit essential for function would have been conserved, whereas variation in other amino acids might indicate phylogenetic lineage. It is apparent from Fig. 5 that there are four major subfamilies of diiron monooxygenases. Subfamily 1, represented by Tbc1D, is comprised of three-component enzymes that contain an αβγ-hydroxylase, a coupling protein, and an oxidoreductase. These enzymes also contain a noncatalytic Tbc1A-like homolog that apparently functions as an assembly polypeptide. In addition to sharing similar components, the genes encoding the polypeptide components of the enzymes of this subfamily also conserve a similar gene order. Subfamily 2, represented by Tbc2A, is comprised of four-component enzymes that contain an αβγ-hydroxylase, a coupling protein, a Rieske-type ferredoxin, and an oxidoreductase. The order of genes that encode the constituent monooxygenase polypeptides is also conserved within this subfamily. Subfamily 3 contains the soluble methane monooxygenases. These enzymes are comprised of an αβγ-hydroxylase, a coupling protein, and an oxidoreductase, which are encoded by genes that have an organization unique to this subfamily of enzymes (6, 31). Subfamily 4 contains only a single member, the alkene monooxygenase of R. rhodochrous (formerly Nocardia corallina) B-276. This enzyme is comprised of an αβ-hydroxylase, a coupling protein, and an oxidoreductase.
FIG. 5.
Phylogenetic tree for α subunits of multicomponent diiron monooxygenases. The brackets indicate the four subfamilies that are discussed in the text.
The enzymes in subfamily 1 are primarily phenol and cresol monooxygenases, which appears to be the role of Tbc1 in aromatic hydrocarbon oxidation in strain JS150. The exceptions to this group trait are the toluene and benzene 2-monooxygenase previously isolated from strain JS150 (23) and the toluene 2-monooxygenase from B. cepacia PR123, which is a derivative of strain G4 (51). However, for both of these enzymes it has been found that the o-cresol produced from the initial hydroxylation of toluene can be further oxidized to 3-methylcatechol by the same enzyme (24, 34), supporting the description of this subfamily as a group largely associated with oxidation of monohydroxylated aromatic substrates. Subfamily 2, in contrast, is comprised of enzymes that oxidize nonhydroxylated substrates. In this subfamily there are clearly two subgroups: a group represented by Tbc2 that preferentially hydroxylate aromatic substrates and a group comprised of the Aam hydroxylase of Xanthobacter sp. strain Py2 and the Iso hydroxylase of Rhodococcus sp. strain AD45 that preferentially oxidize alkenes. Biochemical characterization of at least two members of this subfamily (the Tmo hydroxylase of P. mendocina KR1 [30] and the Aam hydroxylase of Xanthobacter sp. strain Py2 [68]) has demonstrated that the aromatic oxygenases have some ability to oxidize alkenes and vice versa, further supporting the designation of this as a phylogenetically and functionally cohesive subfamily. Among the aromatic monooxygenases of subfamily 2, the Phl monooxygenase of R. eutropha JMP134 appears to be a singular exception to the group trait in that it has been characterized as a phenol monooxygenase (16). However, other enzymes in this subfamily have also been shown to be capable of oxidation of phenol and cresols (3, 68), although to a lesser extent than their ability to oxidize unactivated aromatic substrates; therefore, it would be of interest to determine whether the Ph1 monooxygenase of strain JMP134 also has the ability to function as an oxygenase for nonhydroxylated aromatic substrates.
Analysis of Tbc1 and Tbc2 expressed from a heterologous promoter in E. coli.
Sequence analysis suggested that Tbc1 and Tbc2 might catalyze different steps in the initial oxidation of toluene and related aromatic substrates. In order to test this hypothesis, tbc1 and tbc2 were separately cloned into pBluescript under the control of a lac promoter, as described in Materials and Methods. E. coli cells carrying these clones were evaluated for the ability to transform toluene and o-cresol. E. coli DH5α carrying pJCM1000, which expressed the Tbc2 monooxygenase, was able to convert 75% of an initial concentration of 1.0 mM toluene into o-cresol following 6 h of incubation, whereas cells carrying pJCM1001, which expressed the Tbc1 monooxygenase, produced no detectable product from toluene in 6 or in 19 h of incubation. When o-cresol was provided as the initial substrate, cells expressing Tbc1 converted 10% of the initial 1 mM concentration to 3-methylcatechol in 19 h of incubation. Interestingly, cells expressing the Tbc2 monooxygenase were also able to convert o-cresol to 3-methylcatechol, with approximately 10% of 1 mM o-cresol being converted to the methylcatechol product in 19 h of incubation.
These results provide functional analysis and support for the conclusions drawn from the sequence inference, namely, that Tbc1 functions as a phenol- and cresol-oxidizing enzyme, and Tbc2 functions as a toluene-oxidizing enzyme. The ability of Tbc2 to convert both toluene and cresol is interesting and is similar to the broad substrate transformation abilities reported previously for the toluene 2-monooxygenase from B. cepacia G4 (52). The relative contribution of the Tbc1 and Tbc2 monooxygenases to aromatic hydrocarbon utilization in strain JS150 is currently being investigated.
Analysis of tbc transcripts.
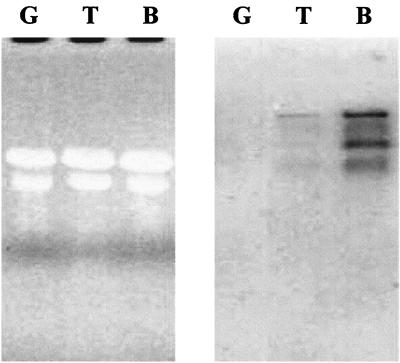
Northern hybridization analysis was used to detect the number and size of mRNA transcripts produced from the pHYK2000 clone for cells that had been induced with toluene and benzene. For this, a 2-kb XhoI-HindIII fragment of pHYK2000 (Fig. 2) was used as a probe. From sequence analysis we knew that this fragment would contain genes encoding an assembly protein, a hydroxylase β subunit, and a coupling protein. The Northern blot results (Fig. 6), which demonstrated that benzene was a stronger inducer of transcription than was toluene, also showed that there were two major transcripts produced as a consequence of toluene or benzene induction. This result is consistent with analysis of the sequence upstream of the tbc2A and tbc1A genes, which showed the presence of elements of two ς54-dependent promoters (data not shown). Homology between the tbc1BC genes on the probe would be expected to allow for hybridization to a tbc1 message as well as a tbc2 message. Surprisingly, though, the two major mRNA transcripts were not of the same size, even though the coding regions for tbc1 and tbc2 are approximately equal. One plausible interpretation for this size difference is that the tbc2 genes might be cotranscribed as part of a polycistronic operon together with the gene for an adjacent transcriptional activator that was detected from DNA sequence analysis. Such a transcriptional organization has been previously documented for the tbuA1UBVA2C-tbuT operon in R. pickettii PKO1 (5).
FIG. 6.
Transcriptional analysis of Tbc monooxygenases. RNA was isolated from cells grown in BM in the presence of toluene (T) or benzene (B). Transcripts were analyzed using a 2-kb HindIII-XhoI probe (Fig. 2). The left panel shows an agarose gel profile of total RNA. The right panel shows an autoradiogram of transcripts.
Analysis of transcripts, taken together with the results from the sequence analysis, provide a plausible basis for interpreting the results of the functional characterization of the pHYK2000 clone (Fig. 2 and 3). It is clear that the pHYK2001 deletant has little or no effect on toluene, benzene, or chlorobenzene transformation, because the initial oxygenase, Tbc2, remains intact. The slight reduction in transformation ability seen for pHYK2005 is most likely the result of a deletion of part of an upstream activating sequence that affects tbc2 transcription. The complete absence of transformation ability in pHYK2003 is clearly due to the deletion of tbc2, whereas the absence of activity associated with pHYK2004 is a consequence of the transcriptional activator having been deleted. This gene has been identified from sequence analysis of the region immediately downstream of tbc2 (data not shown). The results from the pHYK2002 deletant, however, cannot be explained based on our current understanding of the tbc system. The NotI deletion removes almost all of Tbc1 as well as the promoter and amino-terminal third of the α subunit of the Tbc2 oxygenase. Nevertheless, the ability to produce a small amount of product from toluene, benzene, or chlorobenzene has been found to be reproducible from independent experiments. This phenomenon is currently being investigated.
Conclusions.
From our investigation we can conclude that an additional set of monooxygenases is present in strain JS150 that allow this organism to carry out the initial oxidation of toluene and a variety of related alkyl- and chloro-substituted hydrocarbons. The tbc1 and tbc2 gene cluster cloned and analyzed in this study represents a set of monooxygenases that are somewhat similar to the toluene 2- and toluene 4-monooxygenases previously cloned from this strain (23, 24); however, the differences in Southern blot hybridization patterns, DNA sequence, and product distribution profiles clearly indicate that the tbc genes are a new and distinctly separate set of functions. The presence of multiple and apparently functionally redundant oxygenases in strain JS150 is intriguing. It has been previously noted by Haigler and coworkers (15) that this isolate is capable of dissimilating a remarkably broad range of substrates, and this catabolic versatility is consistent with the presence of a wide array of oxygenases. The apparent functional redundancy of these oxygenases could allow for utilization of related substrates under different physiological conditions, depending on the regulation patterns controlling expression of these genes. This is currently being investigated in our laboratory.
It is worthwhile noting that the presence in a single bacterial strain of multiple oxygenases with similar catabolic potential may not be a singularity for strain JS150. Many of the strains that have been investigated genetically or biochemically for aromatic hydrocarbon catabolism have generally been studied for the presence of a single oxygenase function, but multiple and functionally related oxygenases may be present in some of these organisms. Support for this argument comes from recent mutagenesis studies conducted with strain KR1 (30). In this case, toluene 4-monooxygenase-deficient mutants of strain KR1 were still able to oxidize five- to eight-carbon alkenes. These results suggest the presence of alternate oxygenases in strain KR1 with catabolic capabilities similar to those of toluene 4-monooxygenase. It would be of interest to analyze other well-studied aromatic hydrocarbon-degrading bacteria for the presence of multiple mono- and dioxygenases.
ACKNOWLEDGMENTS
This research was supported by the National Institute of Environmental Health Sciences through Superfund Basic Research Program grant P42-ES-04911.
Technical assistance provided by Andrew Berger and Cecilia Lim is also gratefully acknowledged.
REFERENCES
- 1.Arai H, Akahira S, Ohishi T, Maeda M, Kudo T. Adaptation of Comamonas testosteroni TA441 to utilize phenol: organization and regulation of the genes involved in phenol degradation. Microbiology. 1998;144:2895–2903. doi: 10.1099/00221287-144-10-2895. [DOI] [PubMed] [Google Scholar]
- 2.Ayoubi P J, Harker A R. Whole-cell kinetics of trichloroethylene degradation by phenol hydroxylase in a Ralstonia eutropha JMP134 derivative. Appl Environ Microbiol. 1998;64:4353–4356. doi: 10.1128/aem.64.11.4353-4356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertoni G, Martino M, Galli E, Barbieri P. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl Environ Microbiol. 1998;64:3626–3632. doi: 10.1128/aem.64.10.3626-3632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrne A M, Kukor J J, Olsen R H. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii PKO1. Gene. 1995;27:65–70. doi: 10.1016/0378-1119(94)00844-i. [DOI] [PubMed] [Google Scholar]
- 5.Byrne A M, Olsen R H. Cascade regulation of the toluene-3-monooxygenase operon (tbuA1UBVA2C) of Burkholderia pickettii PKO1: role of the tbuA1 promoter (PtbuA1) in the expression of its cognate activator, TbuT. J Bacteriol. 1996;178:6327–6337. doi: 10.1128/jb.178.21.6327-6337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardy D L, Laidler V, Salmond G P, Murrell J C. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991;5:335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 7.Cohen-Bazaire G, Sistrom W R, Stainer R Y. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Comp Physiol. 1987;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 8.Cuskey S M, Pecoraro V, Olsen R H. Initial catabolism of aromatic biogenic amines by Pseudomonas aeruginosa PAO1: pathway description, mapping of mutations, and cloning of essential genes. J Bacteriol. 1987;169:2398–2404. doi: 10.1128/jb.169.6.2398-2404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrt S, Schirmer F, Hillen W. Genetic organization, nucleotide sequence and regulation of expression of genes encoding phenol hydroxylase and catechol 1,2-dioxygenase in Acinetobacter calcoaceticus NCIB8250. Mol Microbiol. 1995;18:13–20. doi: 10.1111/j.1365-2958.1995.mmi_18010013.x. [DOI] [PubMed] [Google Scholar]
- 10.Fox B G, Shanklin J, Ai J, Loehr T M, Sanders-Loehr J. Resonance Raman evidence for an Fe-O-Fe center in stearoyl-ACP desaturase: primary sequence identity with other diiron-oxo proteins. Biochemistry. 1994;33:12776–2786. doi: 10.1021/bi00209a008. [DOI] [PubMed] [Google Scholar]
- 11.Fox B G, Shanklin J, Somerville C, Munck E. Stearoyl-acyl carrier protein Δ9 desaturase from Ricinus communis is a diiron-oxo protein. Proc Natl Acad Sci USA. 1993;90:2486–2490. doi: 10.1073/pnas.90.6.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox B G, Surerus K K, Munck E, Lipscomb J D. Evidence for a μ-oxobridged binuclear iron cluster in the hydroxylase component of methane monooxygenase. J Biol Chem. 1988;263:10553–10556. [PubMed] [Google Scholar]
- 13.Gibson D T, Hensley M, Yoshioka H, Mabry T J. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from Pseudomonas putida. Biochemistry. 1970;9:1626–1630. doi: 10.1021/bi00809a023. [DOI] [PubMed] [Google Scholar]
- 14.Gibson D T, Subramanian V. Microbial degradation of aromatic hydrocarbons. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc; 1984. pp. 187–212. [Google Scholar]
- 15.Haigler B E, Pettigrew C A, Spain J C. Biodegradation of mixtures of substituted benzenes by Pseudomonas sp. strain JS150. Appl Environ Microbiol. 1992;58:2237–2244. doi: 10.1128/aem.58.7.2237-2244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harker A R, Kim Y. Trichloroethylene degradation by two independent aromatic-degrading pathways in Alcaligenes eutrophus JMP134. Appl Environ Microbiol. 1990;56:1179–1181. doi: 10.1128/aem.56.4.1179-1181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heitzer A, Sayler G S. Monitoring the efficacy of bioremediation. Trends Biotechnol. 1993;11:334–343. doi: 10.1016/0167-7799(93)90156-4. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann H, Muller C, Schmidt I, Mahnke J, Petruschka L, Hahnke K. Localization and organization of phenol degradation genes of Pseudomonas putida strain H. Mol Gen Genet. 1995;247:240–246. doi: 10.1007/BF00705655. [DOI] [PubMed] [Google Scholar]
- 19.Hino S, Watanabe K, Takahashi N. Phenol hydroxylase cloned from Ralstonia eutropha strain E2 exhibits novel kinetic properties. Microbiology. 1998;144:1765–1772. doi: 10.1099/00221287-144-7-1765. [DOI] [PubMed] [Google Scholar]
- 20.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooker B, Skeen R S. Intrinsic bioremediation: an environmental restoration technology. Curr Opin Biotechnol. 1996;7:317–320. doi: 10.1016/s0958-1669(96)80037-1. [DOI] [PubMed] [Google Scholar]
- 22.Horinouchi M, Kasuga K, Nojiri H, Yamane H, Omori T. Cloning and characterization of genes encoding an enzyme which oxidizes dimethyl sulfide in Acinetobacter sp. strain 20B. FEMS Microbiol Lett. 1997;155:99–105. doi: 10.1111/j.1574-6968.1997.tb12692.x. [DOI] [PubMed] [Google Scholar]
- 23.Johnson G R, Olsen R H. Nucleotide sequence analysis of genes encoding a toluene/benzene-2-monooxygenase from Pseudomonas sp. strain JS150. Appl Environ Microbiol. 1995;61:3336–3346. doi: 10.1128/aem.61.9.3336-3346.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson G R, Olsen R H. Multiple pathways for toluene degradation in Burkholderia sp. strain JS150. Appl Environ Microbiol. 1997;63:4047–4052. doi: 10.1128/aem.63.10.4047-4052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaphammer B J, Kukor J J, Olsen R H. Cloning and characterization of a novel toluene degradative pathway from Pseudomonas pickettii PKO1. In: Rossmore H W, editor. Biodeterioration and biodegradation. London, United Kingdom: Elsevier Applied Science; 1991. pp. 571–572. [Google Scholar]
- 26.Kitayama A, Suzuki E, Kawakami Y, Nagamune T. Gene organization and low regiospecificity in aromatic-ring hydroxylation of a benzene monooxygenase of Pseudomonas aeruginosa JI104. J Ferment Bioeng. 1996;82:421–425. [Google Scholar]
- 27.Leahy J G, Byrne A M, Olsen R H. Comparison of factors influencing trichloroethylene degradation by toluene-oxidizing bacteria. Appl Environ Microbiol. 1996;62:825–833. doi: 10.1128/aem.62.3.825-833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 29.Mason J R, Cammack R. The electron transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 30.McClay K, Fox B G, Steffan R J. Toluene monooxygenase-catalyzed epoxidation of alkenes. Appl Environ Microbiol. 2000;66:1877–1882. doi: 10.1128/aem.66.5.1877-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald I R, Uchiyama H, Kambe S, Yagi O, Murrell J C. The soluble methane monooxygenase gene cluster of the trichloroethylene-degrading methanotroph Methylocystis sp. strain M. Appl Environ Microbiol. 1997;63:1898–1904. doi: 10.1128/aem.63.5.1898-1904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikesell M D, Kukor J J, Olsen R H. Metabolic diversity of aromatic hydrocarbon-degrading bacteria from a petroleum-contaminated aquifer. Biodegradation. 1993;4:249–259. doi: 10.1007/BF00695973. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura K, Ishida H, Iizumi T. Constitutive trichloroethylene degradation led by tac promoter chromosomally integrated upstream of phenol hydroxylase genes of Ralstonia sp. KN1 and its nucleotide sequence analysis. J Biosci Bioeng. 2000;89:47–54. doi: 10.1016/s1389-1723(00)88049-4. [DOI] [PubMed] [Google Scholar]
- 34.Newman L M, Wackett L P. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry. 1995;34:14066–14076. doi: 10.1021/bi00043a012. [DOI] [PubMed] [Google Scholar]
- 35.Ng L C, Shingler V, Sze C C, Poh C L. Cloning and sequences of the first eight genes of the chromosomally encoded (methyl) phenol degradation pathway from Pseudomonas putida P35X. Gene. 1994;151:29–36. doi: 10.1016/0378-1119(94)90629-7. [DOI] [PubMed] [Google Scholar]
- 36.Olsen R H, Hansen J. Evolution and utility of a Pseudomonas aeruginosa drug resistance factor. J Bacteriol. 1976;125:837–844. doi: 10.1128/jb.125.3.837-844.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen R H, Kukor J J, Byrne A M, Johnson G R. Evidence for the evolution of a single component phenol/cresol hydroxylase from a multicomponent toluene monooxygenase. J Ind Microbiol Biotechnol. 1997;19:360–368. doi: 10.1038/sj.jim.2900453. [DOI] [PubMed] [Google Scholar]
- 38.Olsen R H, Kukor J J, Kaphammer B. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J Bacteriol. 1994;176:3749–3756. doi: 10.1128/jb.176.12.3749-3756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsen R H, Mikesell M D, Kukor J J. Enumeration and characterization of BTEX-degrading bacteria from hypoxic environments functional with mixed electron acceptors. Res Microbiol. 1994;145:47–49. doi: 10.1016/0923-2508(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 40.Otaka E, Ooi T. Examination of protein sequence homologies. V. New perspectives on evolution between bacterial and chloroplast-type ferredoxins inferred from sequence evidence. J Mol Evol. 1989;29:246–254. doi: 10.1007/BF02100208. [DOI] [PubMed] [Google Scholar]
- 41.Ouchiyama N, Miyachi S, Omori T. Cloning and nucleotide sequence of carbazole catabolic genes from Pseudomonas stutzeri strain OM1, isolated from activated sludge. J Gen Appl Microbiol. 1998;44:57–63. doi: 10.2323/jgam.44.57. [DOI] [PubMed] [Google Scholar]
- 42.Pikus J D, Studts J M, Achim C, Kauffmann K E, Münck E, Steffan R J, McClay K, Fox B G. Recombinant toluene-4-monooxygenase: catalytic and Mössbauer studies of the purified diirion and Rieske components of a four-protein complex. Biochemistry. 1996;35:9106–9119. doi: 10.1021/bi960456m. [DOI] [PubMed] [Google Scholar]
- 43.Pikus J D, Studts J M, McClay K, Steffan R J, Fox B G. Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron enzyme toluene 4-monooxygenase. Biochemistry. 1997;36:9283–9289. doi: 10.1021/bi971049t. [DOI] [PubMed] [Google Scholar]
- 44.Porter T D, Kasper C B. NADPH-cytochrome P-450 oxidoreductase: flavin mononucleotide and flavin adenine dinucleotide domains evolved from different flavoproteins. Biochemistry. 1986;25:1682–1687. doi: 10.1021/bi00355a036. [DOI] [PubMed] [Google Scholar]
- 45.Powlowski J, Sealy J, Shingler V, Cadieux E. On the role of DmpK, an auxiliary protein associated with multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Biol Chem. 1997;272:945–951. doi: 10.1074/jbc.272.2.945. [DOI] [PubMed] [Google Scholar]
- 46.Qian H, Edlund U, Powlowski J, Shingler V, Sethson I. Solution structure of phenol hydroxylase protein component P2 determined by NMR spectroscopy. Biochemistry. 1997;36:495–504. doi: 10.1021/bi9619233. [DOI] [PubMed] [Google Scholar]
- 47.Saeki H, Furuhashi K. Cloning and characterization of a Nocardia corallina B-276 gene cluster encoding alkene momnoxygenase. J Ferment Bioeng. 1994;78:399–406. [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain- terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato S I, Ouchiyama N, Kimura T, Nojiri H, Yamane H, Omori T. Cloning of genes involved in carbazole degradation of Pseudomonas sp. strain CA10: nucleotide sequences of genes and characterization of meta-cleavage enzymes and hydrolase. J Bacteriol. 1997;179:4841–4849. doi: 10.1128/jb.179.15.4841-4849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shields M S, Francesconi S C. Microbial degradation of trichloroethylene, dichloroethylenes and aromatic pollutants. U.S. patent 5,543,317. 6 August 1996. [Google Scholar]
- 52.Shields M S, Montgomery S O, Chapman P J, Cuskey S M, Pritchard P H. Novel pathway for toluene catabolism in the trichloroethylene-degrading bacterium G4. Appl Environ Microbiol. 1989;55:1624–1629. doi: 10.1128/aem.55.6.1624-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shingler V, Powlowski J, Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethyl phenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992;174:711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Small F J, Ensign S A. Alkene monooxygenase from Xanthobacter strain Py2: purification and characterization of a four-component system central to the bacterial metabolism of aliphatic alkenes. J Biol Chem. 1997;272:24913–24920. doi: 10.1074/jbc.272.40.24913. [DOI] [PubMed] [Google Scholar]
- 55.Smith A W, Iglewski B H. Transformation of Pseudomonas aeruginosa by electroporation. J Mol Biol. 1989;17:10509. doi: 10.1093/nar/17.24.10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 57.Spain J. Synthetic chemicals with potential for natural attenuation. Bioremed J. 1997;1:1–9. [Google Scholar]
- 58.Takeo M, Maeda Y, Okada H, Miyama K, Mori K, Ike M, Fujita M. Molecular cloning and sequencing of the phenol hydroxylase gene from Pseudomonas putida BH. J Ferment Bioeng. 1995;79:485–488. [Google Scholar]
- 59.Teramoto M, Futamata H, Harayama S, Watanabe K. Characterization of a high-affinity phenol hydroxylase from Comamonas testosteroni R5 by gene cloning, and expression in Pseudomonas aeruginosa PAO1c. Mol Gen Genet. 1999;262:552–558. doi: 10.1007/s004380051117. [DOI] [PubMed] [Google Scholar]
- 60.Thomas P S. Hybridization of denatured RNA transferred or dotted to nitrocellulose paper. Methods Enzymol. 1983;100:255. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- 61.van Agteren M H, Keuning S, Janssen D B. Handbook on biodegradation and biological treatment of hazardous organic compounds. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. [Google Scholar]
- 62.van Hylckama Vlieg J E, Leemhuis H, Spelberg J H, Janssen D B. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45. J Bacteriol. 2000;182:1956–1963. doi: 10.1128/jb.182.7.1956-1963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whited G, Gibson D T. Toluene-4-monooxygenase: a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991;173:3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Worsey M J, Williams P A. Metabolism of toluene and the xylenes by Pseudomonas ([sic] putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia B, Pikus J D, Xia W, McClay K, Steffan R J, Chae Y C, Westler W M, Markley J L, Fox B G. Detection and classification of hyperfine-shifted 1H, 2H, and 15N resonances of the Rieske ferredoxin component of toluene 4-monooxygenase. Biochemistry. 1999;38:727–739. doi: 10.1021/bi981851a. [DOI] [PubMed] [Google Scholar]
- 66.Yen K-M, Karl M R. Identification of a new gene, tmoF, for the Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1992;174:7253–7261. doi: 10.1128/jb.174.22.7253-7261.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yen K-M, Karl M R, Blatt L M, Simon M J, Winter R B, Fausset P R, Lu H S, Harcourt A A, Chen K. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1991;173:5315–5327. doi: 10.1128/jb.173.17.5315-5327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou N-Y, Jenkins A, Chan Kwo Chion C K N, Leak D J. The alkene monooxygenase from Xanthobacter strain Py2 is closely related to aromatic monooxygenases and catalyzes aromatic monohydroxylation of benzene, toluene, and phenol. Appl Environ Microbiol. 1999;65:1589–1595. doi: 10.1128/aem.65.4.1589-1595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]