Abstract
A previous study of deletions in the protocatechuate (pca) region of the Acinetobacter sp. strain ADP1 chromosome revealed that genes required for utilization of the six-carbon dicarboxylic acid, adipic acid, are linked to the pca structural genes. To investigate the genes involved in adipate catabolism, a 33.8-kb SacI fragment, which corrects a deletion spanning this region, was cloned. In addition to containing known pca, qui, and pob genes (for protocatechuate, quinate, and 4-hydroxybenzoate dissimilation), clone pZR8000 contained 10 kb of DNA which was the subject of this investigation. A mutant strain of Escherichia coli DH5α, strain EDP1, was isolated that was able to utilize protocatechuate and 4-hydroxybenzoate as growth substrates when EDP1 cells contained pZR8000. Sequence analysis of the new region of DNA on pZR8000 revealed open reading frames predicted to be involved in β-oxidation. Knockouts of three genes implicated in β-oxidation steps were introduced into the chromosome of Acinetobacter sp. strain ADP1. Each of the mutants was unable to grow with adipate. Because the mutants were affected in their ability to utilize additional saturated, straight-chain dicarboxylic acids, the newly discovered 10 kb of DNA was termed the dca (dicarboxylic acid) region. Mutant strains included one with a deletion in dcaA (encoding an acyl coenzyme A [acyl-CoA] dehydrogenase homolog), one with a deletion in dcaE (encoding an enoyl-CoA hydratase homolog), and one with a deletion in dcaH (encoding a hydroxyacyl-CoA dehydrogenase homolog). Data on the dca region should help us probe the functional significance and interrelationships of clustered genetic elements in this section of the Acinetobacter chromosome.
Microbial β-oxidation of fatty acids has enjoyed prolonged research interest, yet the genetics and biochemistry of dicarboxylic acid catabolism have received minimal attention. The latter acids are of particular interest because they have the potential to play a significant role in the natural environment by serving as cross-linkers between other compounds. In addition, saturated, straight-chain dicarboxylic acids or their thioesters arise as intermediates in catabolic pathways for diverse compounds. Adipic acid is an intermediate in the metabolism of cyclohexanol (14), and other dicarboxylic acids form during oxidation of the corresponding cyclic alcohols. Additional catabolic pathways include ω-oxidation of fatty acids (31), alkane oxidation (29), aerobic degradation of cyclohexanecarboxylic acid (6), and anaerobic metabolism of aromatic compounds such as benzoate, which generates pimelyl coenzyme A (pimelyl-CoA) as an intermediate (22).
Straight-chain dicarboxylic acids of 6 to 10 carbon atoms in length serve as carbon sources for aerobic growth of diverse microbial strains (4, 37, 42). In Acinetobacter spp. (4), as in other bacteria characterized for the trait, the ability to utilize saturated dicarboxylic acids of this size range aerobically is often a unit characteristic (23). Experimental evidence with Pseudomonas fluorescens supported the hypothesis that this unit trait is a consequence of cyclic β-oxidation steps analogous to those of fatty acid degradation (23).
In the naturally transformable Acinetobacter sp. strain ADP1, also designated strain BD413 (28), there is a remarkable, extended cluster of genes for related function in one region of the chromosome, an “island of catabolic diversity” (35). Downstream from 10 genes required for protocatechuate catabolism are genes for conversion of diverse hydroaromatic and aromatic compounds to protocatechuate (Fig. 1). A positive selection strategy for mutations that protect against accumulation of a toxic intermediate in protocatechuate catabolism has been used to study Acinetobacter sp. strain ADP1 proteins and regulatory sequences which are required for generating the toxic compound. In one study, a quarter of the spontaneous mutations were deletions, and some of them extended into neighboring genes (18). The discovery that some of the deletions upstream of the pca structural genes eliminated the ability of strains to grow on the six-carbon dicarboxylic acid, adipic acid, provided the first evidence for linkage of adipate utilization genes and pca genes (10, 11).
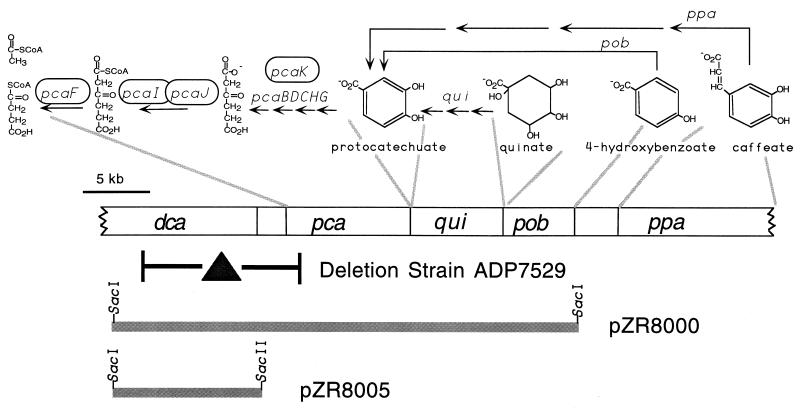
FIG. 1.
Relevant strain and plasmids used to clone and sequence DNA adjacent to the pca operon from strain ADP1. Gene designations are dca, dicarboxylic acid; pca, protocatechuate; qui, quinate and shikimate; pob, 4-hydroxybenzoate; and ppa, phenylpropenoid and phenylpropanoid. Above the layout of genetic sections, delineated by the gray lines, are the outlines of pathways for dissimilation of aromatic and hydroaromatic compounds. Genes required for the illustrated biochemical transformations are shown above the related arrows. Those encircled have homologs among the dca genes. The deletion within strain ADP7529 was identified as part of this study.
This communication describes the cloning and initial characterization of a cluster of open reading frames defined as dca genes because of their role in the dissimilation of an array of straight-chain, saturated dicarboxylic acids. Particular emphasis was placed on three genes that were predicted to be required for the central steps of β-oxidation.
MATERIALS AND METHODS
Source of dicarboxylic acids and their nomenclature.
Sigma Chemical Co. was the source of all dicarboxylic acids except tridecanedioic and dodecanedicarboxylic acids, which were obtained from Aldrich Chemical Co. Common names are used for the shorter dicarboxylic acids: adipic (6 carbons), pimelic (7 carbons), suberic (8 carbons), and sebacic (10 carbons) acids. Nomenclature for the less familiar acids is dodecanedioic (12 carbons), tridecanedioic (13 carbons), tetradecanedioic (14 carbons), and hexadecanedioic (16 carbons) acids. The dicarboxylic acids were acquired a short time before use, and all of the longer dicarboxylic acids were purported to be 99% pure except tridecanedioic acid, at 94%.
Strains, media, and growth of cells.
Strains and plasmids are listed in Table 1. Cells were cultured in Luria-Bertani medium (41) or minimal medium (36). Solidified minimal medium contained adipate at 5 mM or succinate at 10 mM. In some instances, modified gradient plates were used to screen Acinetobacter cells for substrate utilization patterns (36): cells were spread on agar-solidified minimal medium, and the carbon source was applied to one spot at the edge of the plate, providing a concentration gradient.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Genotype | Reference or source |
|---|---|---|---|
| E. coli strains | |||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK−) supE44 λ−thi-1 gyrA96 relA1 | Gibco-BRL | |
| EDP1 | Mutant of strain DH5α which degrades 4-hydroxybenzoate when it carries pZR8000 | This study | |
| Acinetobacter sp. strains | |||
| ADP1 | Wild-type strain (BD413) | 28 | |
| ADP992 | ADP1 with a 90-bp deletion in pobA, affected in 4-hydroxybenzoate hydroxylase | ΔpobA992 | D. D'Argenio, unpublished data |
| ADP7529 | 11.6-kb deletion with endpoints in dcaE and the intergenic region between pcaU and pcaI | ΔdcaI | 10; this study |
| ADP8018 | 0.33-kb dcaH deletion created in pZR8044 and introduced into the chromosome of ADP992 | ΔpobA992 ΔdcaH1 | This study |
| ADP8023 | Kmr; dcaA::Kmr from pZR8053 in the chromosome of ADP1 | dcaA1::Kmr | This study |
| ADP8061 | 812-bp deletion in dcaA created in pZR8075 and introduced into the chromosome of ADP1 | ΔdcaA2 | This study |
| ADP8062 | 100-bp deletion in dcaE constructed in pZR8077 and introduced into the chromosome of ADP1 | ΔdcaE2 | This study |
| Plasmids | |||
| pBBR1MCS | Chlr; broad-host-range cloning vector | 30 | |
| pBKS | Apr; narrow-host-range cloning vector | Stratagene | |
| pBSK | Apr; narrow-host-range cloning vector | Stratagene | |
| pUC4K | Apr Kmr; carries antibiotic resistance cassette | Amersham Pharmacia Biotech | |
| pUC19 | Apr; narrow-host-range cloning vector | 46 | |
| pZR8000 | Chlr; 34.8-kb Acinetobacter sp. strain ADP1 SacI insertion in pBBR1MCS containing dca genes as well as pca, qui, and pob genes | This study | |
| pZR8005 | Apr; 11.8-kb SacI-to-SacII insertion encompassing dca genes from pZR8000 in pBSK | This study | |
| pZR8006 | Apr; 11.8-kb SacI-to-SacII insertion with dca genes from pZR8000 in pBKS; insertion orientation is flipped relative to that of pZR8005 | This study | |
| pZR8030 | Apr; 5.5-kb KpnI deletion of pZR8006 | This study | |
| pZR8031 | Apr; 8.8-kb HindIII deletion of pZR8006 | This study | |
| pZR8038 | Apr; 8.9-kb HindIII insertion of pZR8006 in pUC19 | This study | |
| pZR8044 | Apr; 0.34-kb BglII-Eco47III deletion of pZR8031 | This study | |
| pZR8053 | Apr; Kmr cassette from pUC4K in XhoI site of pZR8030 | This study | |
| pZR8054 | Apr; 4-kb HindIII-to-EcoRI insertion of pZR8038 in pUC19 | This study | |
| pZR8061 | Apr; 1.68-kb HindIII-SphI insertion of pZR8006 in pUC19 | This study | |
| pZR8075 | 812-bp HincII-NruI deletion of pZR8054, which removes much of the dcaA gene | This study | |
| pZR8077 | 100-bp MscI-BsgI deletion of pZR8061, which removes a central piece of the dcaE gene | This study |
To screen Acinetobacter cells for antibiotic resistance markers, a kanamycin concentration of 15 μg ml−1 was used in Luria-Bertani medium, and ampicillin at 110 μg ml−1 was used in minimal medium. When Escherichia coli cells were under selection, Luria-Bertani medium was supplemented with chloramphenicol, kanamycin, or ampicillin at 20, 25, or 90 μg ml−1, respectively.
For all tests involving relative yields of Acinetobacter strains in liquid medium, stock solutions of substrates were prepared in dimethyl sulfoxide (DMSO) at a concentration of 0.5 M or 0.25 M, as required by solubility. Adipic acid was added to liquid minimal medium at a final concentration of 2 mM. For each of the longer-chain dicarboxylic acids, the final concentration of carbon atoms was equivalent to that provided by 2 mM adipic acid. Controls contained only DMSO, provided at the maximum amount added with any carbon source.
Comparative growth tests of Acinetobacter strains were carried out by growing cells overnight at 37°C in minimal medium containing succinate as the sole carbon source. A 50-μl aliquot of an overnight culture of cells was added to 5 ml of fresh minimal medium containing a carbon source, with the inoculum size designed to minimize the contribution of possible revertants to growth. Cultures were incubated at 37°C and 250 rpm. Growth of the wild-type strain was monitored, and the density of each mutant strain on a particular substrate was measured when wild-type cells, grown in parallel on the same substrate, attained their maximum level of growth.
Doubling times on particular substrates were determined by measuring turbidity after inoculating an overnight culture into 10 ml of minimal medium in a 50-ml Erlenmeyer flask and shaking at 37°C and 250 rpm.
Analysis of the ability of E. coli cells bearing pZR8000 to grow at the expense of various compounds was tested on solidified medium and in liquid minimal medium, both supplemented with 0.1% yeast extract and chloramphenicol. In this case, substrates were prepared in water and neutralized with sodium hydroxide. A positive control for growth was glucose, and negative controls were sodium chloride or no addition. Since some tested compounds might be toxic at high concentrations, they were provided on gradient plates. Cells were streaked for single colonies across half of each plate, and 70 to 100 μmol of substrate was distributed evenly in a line in the middle of the other half of the plate, perpendicular to the bacterial streaks. Liquid cultures contained substrate at a concentration of 5 mM, and turbidity was determined after 2 days in a 37°C shaker at 250 rpm. Turbidity was measured at 600 nm, and the correlation between increased turbidity and cell growth was confirmed by performing viable counts, particularly important with protocatechuate-supplemented medium, which had a colored tint.
Analysis of revertant frequencies.
The apparent revertant frequency of each mutant strain was measured by spreading aliquots of an overnight culture onto minimal medium plates containing adipate as the carbon source. Viable counts of the original cultures were determined on nonselective medium. The fraction of the total viable count that appeared as CFU on the selective medium was taken as the presumptive revertant frequency.
Construction of plasmids and mutant strains.
Standard techniques were used in molecular biology manipulations (2, 39). Natural transformation of Acinetobacter strains followed published methods (27). The ability of mutant strains of ADP1 to take up DNA carried on plasmids in E. coli by replica plating has been demonstrated (3). Competent E. coli DH5α cells (26) were transformed with a SacI library of Acinetobacter sp. strain ADP1 in vector pBBR1MCS. E. coli transformants were replica plated onto a lawn of strain ADP7529 which had been made competent for natural transformation. An E. coli colony that transformed the deletion strain to an adipate-positive phenotype was isolated off a master plate, and plasmid pZR8000 was purified from the E. coli cells. DH5α(pZR8000) (Fig. 1) also transformed ADP992, a pobA-defective strain, to a 4-hydroxybenzoate-positive phenotype.
Strains ADP8018 (ΔdcaH1), ADP8023 (dcaA1::Kmr), ADP8061 (ΔdcaA2), and ADP8062 (ΔdcaE2) were created by transformation of the competent parental strain with plasmid pZR8044, pZR8053, pZR8075, or pZR8077, respectively (Table 1; Fig. 2 and 3), followed by phenotype assessment on minimal medium plates containing adipate; the strains were also screened for the absence of the vector antibiotic resistance marker. Because Klenow fragment was used to construct the deletions of pZR8044 (in dcaH) and pZR8077 (in dcaE), with the potential for unintended removal of exposed single-stranded DNA, the deletions of pZR8044 and pZR8077 were sequenced to verify their specificities to those genes.
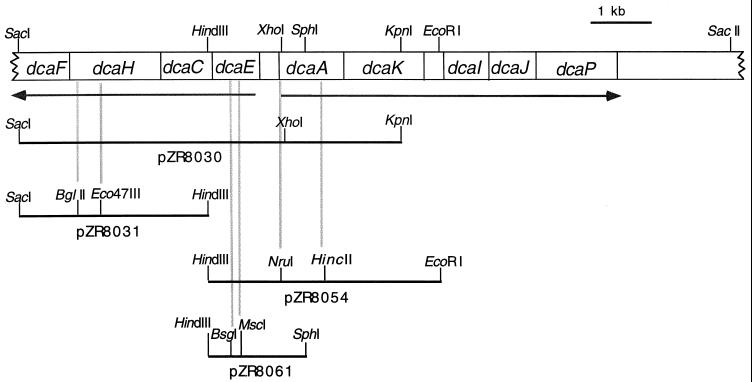
FIG. 2.
Subclones used to create mutant strains and to verify their genotypes. The physical map of the dca genes shows the location of restriction sites used to create the subclones. Subclones used to create deletion or insertion mutations have additional, internal restriction sites. The gray lines show the sites' locations within the gene that was altered.
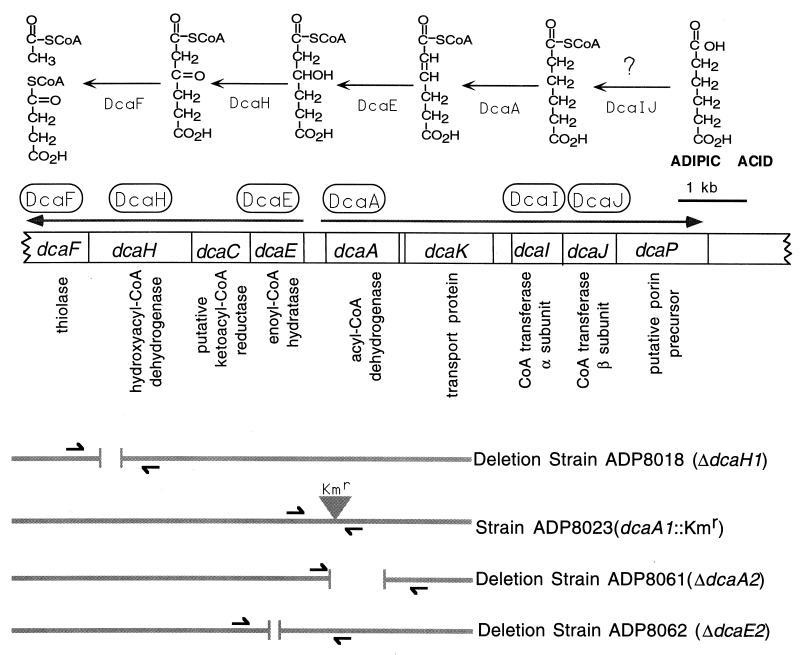
FIG. 3.
Proposed roles of dca genes and physical maps of dca mutations in Acinetobacter strains. The dcaP gene is proximal to the pca genes shown in Fig. 1. Beneath each gene is the likely function of its product, deduced from data presented in Tables 3 and 5. A putative catabolic pathway for adipic acid, which enlists the circled gene products, is shown at the top. Activation of adipic acid is marked by a question mark because it is possible that an as-yet-unidentified ligase carries out the reaction. Small arrows flanking the sites of mutations, aligned beneath the genetic map, denote the locations of primers used to verify mutations.
Insertion or deletion mutations in the Acinetobacter chromosome were analyzed by PCR (Fig. 3). In addition, verification that an introduced, defined mutation was responsible for an adipate-negative phenotype was tested by transformation of a mutant strain with a subclone containing the wild-type gene (Fig. 2). ADP8018 (ΔdcaH1) was transformed to the adipate-positive phenotype by pZR8031 but not by pZR8054; ADP8061 (ΔdcaA2) and ADP8023 (dcaA1::Kmr) were corrected to the adipate-positive phenotype by pZR8054 but not by pZR8031; and ADP8062 (ΔdcaE2) was made adipate positive by pZR8061 but not by pZR8031 (Fig. 2 and 3).
PCRs.
For shorter PCR products, preparation of template DNA followed the instructions provided with InstaGene Matrix from Bio-Rad; longer products were produced using crude chromosomal preparations as the template. The usual PCR conditions were 94°C for 3 min followed by 30 cycles of denaturing at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for an appropriate length of time. Primer sets were as follows (Fig. 3): MGZR80T31 and MGZR80F15 for analysis of ΔdcaH1 in ADP8018; MGZR80T36 and MGZR80F9 for ΔdcaE2 in ADP8062; MGZR80T37 and MGZR80F9 for dcaA1::Kmr in ADP8023; and MGZR80T38 and MGZR80F7 for ΔdcaA2 in ADP8061. The oligonucleotide sequences of these primers are as follows: MGZR80F7, 5′-CCT GCT TGT AAT CCA GTG AGA TGA-3′; MGZR80F9, 5′-CAA TAA TTT CTC CAC TGG CAT AAC G-3′; MGZR80F15, 5′-GAA AAT GTC ACA GAT ATT AGT GAA ATT GAC-3′; MGZR80T31, 5′-CGG TAC AGT CAC AGG CAA ACC TG-3′; MGZR80T36, 5′-AGC TGC CAT CAT TTC TTC CTA GAT A-3′; MGZR80T37, 5′-TTA CGG GCT TGG GAC ATT GTG-3′; and MGZR80T38, 5′-ATA GTA GAT TGC TAT AGC GAA ATA TAG AGA-3′.
The primer pair I8pcaU1 (5′-GAT AAC TCC AAT GTG CAT CTA GC-3′) and MGZR80T34 (5′-GCT GCA TCT GCG TAT TCA GTC GT-3′) was used to amplify DNA covering the dca-pca deletion of strain ADP7529 by PCR.
DNA sequencing.
The Acinetobacter DNA insertion of pZR8005 (Fig. 1) was sequenced by primer walking at the Yale Keck Biotechnology Resource Laboratory. Standard ABI PRISM terminator cycle sequencing with AmpliTaq DNA polymerase was used.
Nucleotide sequence accession number.
The DNA sequence for the dca genes from Acinetobacter sp. strain ADP1 may be found under accession no. LO5770 in the GenBank database.
RESULTS
Phenotype of E. coli carrying pZR8000.
The 33.8-kb SacI insertion of pZR8000 contains pca, qui, and pob genes as well as putative adipate dissimilation genes (Fig. 1). pZR8000 had a low yield on isolation, probably due to the size of its insertion and the low copy number of the vector. Low copy number likely contributed to its stability in E. coli. When tested on gradient plates, E. coli DH5α(pZR8000) did not show enhanced growth in the presence of protocatechuate, 4-hydroxybenzoate, quinate, or adipate. Its inability to utilize adipate was not surprising given that it lacked at least one necessary dca gene, as discussed below.
A mutant strain of E. coli DH5α(pZR8000), designated EDP1(pZR8000′), grew with 4-hydroxybenzoate. To determine if the mutation conferring this phenotype was in the plasmid or in the bacterium, growth properties of the two bacterial strains (DH5α and EDP1) and two plasmids (pZR8000 and pZR8000′) in different combinations were examined. As shown in Table 2, either plasmid conferred upon strain EDP1 the ability to grow with 4-hydroxybenozate, whereas neither plasmid allowed growth of strain DH5α with the compound. Therefore, it is evident that a mutation giving rise to strain EDP1 allowed the strain to use genes in either plasmid to support growth with 4-hydroxybenzoate. Such strains also grew with protocatechuate but not with quinate.
TABLE 2.
Growth phenotypes of E. coli strains with respect to 4-hydroxybenzoate
| E. coli strain and plasmid | Growth at the expense of 4-hydroxybenzoatea |
|---|---|
| DH5α | − |
| DH5α(pZR8000) | − |
| DH5α(pZR8000′)b | − |
| EDP1 | − |
| EDP1(pZR8000) | + |
| EDP1(pZR8000′) | + |
Cells were grown in minimal medium supplemented with 0.1% yeast extract and chloramphenicol to maintain plasmids. Tubes with 5 mM sodium 4-hydroxybenzoate were compared to those containing no additional carbon source as well as those with 5 mM sodium chloride.
pZR8000′ is the plasmid isolated from mutant EDP1(pZR8000), which grows at the expense of 4-hydroxybenzoate.
Sequence analysis of the new chromosomal region captured in pZR8000.
The G+C content of the 33.8-kb SacI insertion of pZR8000 was 40.9%, typical of strain ADP1. Translation of the DNA sequence of its subclone pZR8005 (Fig. 1) in all six possible reading frames revealed the open reading frames shown in Fig. 3. Note that the SacI end of the pZR8000 clone truncated dcaF 516 nucleotides short of its 3′ end (D. Parke, unpublished data); however, the complete sequence of the gene was used in the tables and discussion below.
For each protein encoded by the dca region, the highest-scoring homolog revealed by a BLAST search (1) of the nonredundant NCBI database is listed in Table 3. Data from sequence analysis and Table 3 form the basis of the physical map of the dca genes shown in Fig. 3. That dcaECHF and dcaAK form two divergent transcripts is unambiguous. What is less certain, given the distance between dcaK and dcaI, is whether the genetic unit dcaIJP is part of a dcaAKIJP transcript or is subject to independent control. At least four of the dca genes encoding the products listed in Table 3 have homologs in adjacent regions of the ADP1 chromosome. The relationships between these genes and their products are shown in Table 4.
TABLE 3.
Dicarboxylic acid pathway genes and their products and relationships to other proteinsa
| Gene | Size of gene product (aa)b | Related gene products
|
||||
|---|---|---|---|---|---|---|
| Function (source) | GenBank accession no. | Size of gene product (aa) | % Identityc | % Similarityd | ||
| dcaF | 401 | Probable acyl-CoA thiolase (Pseudomonas aeruginosa PAO1) | AAG06977 | 401 | 67 | 82 |
| dcaH | 505 | Probable 3-hydroxyacyl-CoA DHe (Pseudomonas aeruginosa PAO1) | AAG05017 | 509 | 52 | 66 |
| dcaC | 253 | Ketoreductase (Streptomyces fradiae) | S54815 | 261 | 34 | 51 |
| dcaE | 262 | Enoyl-CoA dehydratase (Acinetobacter sp. strain SE19) | AAG10018 | 258 | 61 | 80 |
| dcaA | 384 | Acyl-CoA DH (Acinetobacter sp. strain SE19) | AAG10019 | 384 | 82 | 89 |
| dcaK | 435 | cis,cis-Muconate transport (Acinetobacter sp. strain ADP1) | AAC27117 | 413 | 55 | 72 |
| dcaI | 223 | β-Kaf succinyl-CoA transferase, α chain (Pseudomonas putida) | AAA25922 | 231 | 65 | 74 |
| dcaJ | 224 | β-Ka succinyl-CoA transferase, β chain (Acinetobacter sp. strain ADP1) | AAC46433 | 217 | 69 | 81 |
| dcaPg | 438 | Sucrose porin precursor (plasmid pUR400, Salmonella and E. coli) | AAA98417 | 505 | 25 | 39 |
Based on BLAST program (1) using sequences in the nonredundant protein databases.
aa, number of amino acids.
Percentage of amino acids that are identical between the Dca protein and its aligned homolog.
Percentage of amino acids that are identical or conserved between the Dca protein and its aligned homolog.
DH, dehydrogenase.
β-Ka, β-ketoadipate.
The 3′ end of dcaP was sequenced previously (M. Lee and D. Young, unpublished data).
TABLE 4.
Dicarboxylic acid pathway genes and their homologs in flanking regions of the ADP1 chromosome
| dca gene of homolog(s) | % G+C
content
|
% nta identity | Distance (kb) between dca gene and homolog | Function(s) of gene product | % aab identity (% similarity) | GenBank accession no. or reference | |
|---|---|---|---|---|---|---|---|
| dca gene | Homolog | ||||||
| dcaI | 40 | Putative Dcac succinyl-CoA transferase, α chain | This study | ||||
| pcaI | 55 | 61 | 8.4 | β-Ketoadipate succinyl-CoA transferase, α chain | 61 (75) | AAC46432 | |
| dcaJ | 41 | Putative Dca adipate succinyl-CoA transferase, β chain | This study | ||||
| pcaJ | 54 | 65 | 8.4 | β-Ketoadipate succinyl-CoA transferase, β chain | 68 (80) | AAC37147 | |
| dcaF | 44 | Putative Dca-CoA thiolase | This study | ||||
| pcaF | 56 | 59 | 16 | β-Ketoadipyl-CoA thiolase | 56 (70) | AAC37148 | |
| dcaK | 41 | Putative Dca transport | This study | ||||
| mucK | 40 | 63 | 12d | cis,cis-Muconate transport; possible adipate transport | 55 (72) | AAC27117 | |
| pcaK | 42.5 | Divergent | 13.5 | Protocatechuate transport | 21 (37) | AAC37151 | |
nt, nucleotide.
aa, amino acid.
Dca, dicarboxylic acid.
Based on sequence analysis of DNA between mucK and dcaF (D. Parke, unpublished data).
Most of the deduced protein sequences possess amino acid sequence motifs characteristic of the functional family to which each belongs (Table 5) (25). DcaK, being closely related to MucK, is a member of the aromatic acid transporter subgroup within the major facilitator superfamily (34). The prediction of 12 transmembrane helices in DcaK is in accord with the number predicted for other members of this group of transporters. Although DcaC shares a relatively low level of identity with its closest homolog in the database compared to the other enzymes encoded by the dca region (Table 3), it preserves the short-chain dehydrogenase/reductase family signature (Table 5).
TABLE 5.
Motifs conserved in dca gene productsa
| Gene product | Motif peptide in Dca protein | Location in proteinb | Family for which motif is characteristic |
|---|---|---|---|
| DcaF | VNRLCASGLAAIIDSARAI | 85–103 | Thiolase acyl-enzyme intermediate |
| NPNGGAIAVGHPLGASG | 346–362 | Thiolase signature 2 | |
| AVVSLCIGVGQGLA | 381–394 | Thiolase active-site motif | |
| DcaC | SIAGRMGYPFRLAYSTSKWGIVGFTKTLS | 135–163 | Short-chain DHc/reductase signature |
| DcaE | IAAVNGYALGGGCELAMHTDI | 103–123 | Enoyl-CoA hydratase/isomerase signature |
| DcaA | CLTEPEAGSDAAS | 123–135 | Acyl-CoA DH signature 1 |
| QIHGGAGYISEYSIERFYRD | 337–356 | Acyl-CoA DH signature 2 | |
| DcaK | AGMIADKLGRRAVFAFGT | 304–321 | Sugar transport protein signature 1 |
| FFGFLYGIPYAINATYMTESFPTSIR | 346–371 | Sugar transport protein signature 2 | |
| DcaJ | LHSENGVLA | 47–55 | CoA transferase signature 2 |
| DcaP precursor | MKKLILAVACATASGTLLA | 1–19 | Signal peptide motif |
Two dca gene products that did not register a motif were DcaI and DcaH. The region from residues 18 to 33 of DcaI corresponds to the CoA transferase 1 signature with the exception of two amino acid changes. At residue 24 of DcaI, Thr is present rather than one of the motif amino acids (Leu, Ile, Val, Met, Phe, or Ala), and position 25 has Ser rather than Gly. Analogously, the peptide in DcaH that correlates to the 3-hydroxyacyl-CoA dehydrogenase signature region of the protein, from residues 184 to 209, has several differences from the conserved motif: His192 rather than Arg, Gly198 rather than a motif amino acid (Leu, Ile, Val, Met, Phe, or Tyr), and Asn at 209 rather than the Gly or Val found in the motif sequence. Since some other members of the hydroxyacyl-CoA dehydrogenase family in the NCBI database also fail to elicit the Prosite motif, it is likely that the signature sequence is based on too narrow a database.
DcaP elicited no convincing homologs from a database search (Table 3). The weakness of the match with ScrY, a sucrose-specific porin, is reflected in an alignment restricted to 185 residues at the N termini of the proteins. Moreover, the DcaP and ScrY match had an Expect value of 0.34, which underlines the weakness of their relationship compared with the low Expect values obtained with the other homolog pairs shown in Table 3. When ScrY is aligned with its closest homolog, the maltoporin LamB, the overall identity is only 20%; however, their relationship is revealed convincingly in their similar structures (15, 40). Circumstantial evidence, which includes the prediction that DcaP has a leader sequence (33) (Table 5), suggests that DcaP may be a porin precursor.
Sequence of the dca-pca deletion of strain ADP7529.
The nucleotide sequence of ADP7529, the recipient used to identify the E. coli clone carrying pZR8000, revealed that the strain contains a deletion of 11.6 kb with endpoints in the dcaE gene and in the pcaU-pcaI intergenic region (Fig. 4).
FIG. 4.
Analysis of deletion Δdca1 of ADP7529. The sequence of the relevant region of dcaE is shown aligned above that of the pcaU-pcaI intergenic region, followed by that of the deletion strain. Nucleotide numbers refer to the current listing for accession no. LO5770 in the GenBank database. The repeated element present in all three sequences is underlined. Overlining designates hypothetical parent sequences that may have contributed to the deletion juncture; they were arbitrarily chosen to clarify the subtle changes at the juncture.
Adipic acid phenotypes of Acinetobacter dcaH,dcaE, and dcaA mutants.
The ability of Acinetobacter sp. strain ADP1 to undergo natural transformation allowed introduction of defined deletions and of a Kmr marker into genes located in divergently transcribed regions (Fig. 3). Sequence analysis indicated that the large dcaA deletion in ADP8061 should cause a frameshift resulting in only an 18-residue peptide being produced rather than the wild-type protein. DcaE is ordinarily a 262-amino-acid polypeptide; the ADP8062 mutant retains the first 93 residues of the wild-type protein, but the mutant protein is presumed to be only 100 residues long due to a frameshift caused by the deletion. The dcaH mutation of ADP8018 results in retention of the original reading frame but loss of a 111-amino-acid segment of the translated product.
Note that the Kmr insertion marker used in ADP8023 has a G+C content higher than that typical of most Acinetobacter genes. Such a disparity may cause polar effects. This possibility in ADP8023 led to the creation of dcaA2 deletion strain ADP8061.
Mutant strains ADP8018 (ΔdcaH1), ADP8023 (dcaA1::Kmr), ADP8061 (ΔdcaA2), and ADP8062 (ΔdcaE2) grew with doubling times similar to that of the parental strain when the carbon source was succinate, but all failed to grow on adipate (Fig. 5). The phenotype of ADP8062 (ΔdcaE2) appeared to be slightly leaky on adipate plates; the effect was heat sensitive, being minimal at 37°C. As expected, the phenotype of ADP992, the parental strain of ADP8018 (Table 1), was similar to that of ADP1 in response to adipate and other dicarboxylic acids provided on gradient plates.
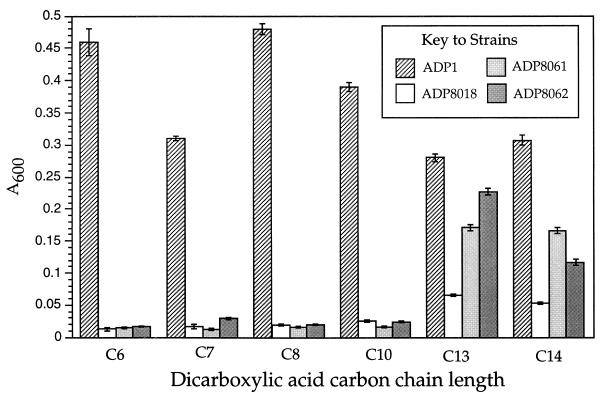
FIG. 5.
Relative yields of wild-type and dca mutant Acinetobacter strains grown at the expense of dicarboxylic acids of different chain length. Strains represented in the legend are ADP1, wild type; ADP8018, ΔdcaH1; ADP8062, ΔdcaE2; and ADP8061, ΔdcaA2. Dicarboxylic acids of different chain length are C6, adipic acid; C7, pimelic acid; C8, suberic acid; C10, sebacic acid; C13, tridecanedioic acid; and C14, tetradecanedioic acid. Adipic acid was supplied at 2 mM, and other dicarboxylic acids were supplied at a concentration that provided an equivalent molarity of carbon atoms. The optical density of a mutant culture on each substrate was measured at the time that wild-type cells, grown in parallel on the same substrate, reached a plateau in density. For two of the dicarboxylic acids (chain length C7 and C14), this required an extra overnight period of growth.
Selection of adipate-positive second-site suppressor mutations.
Since ADP8018 (ΔdcaH1), ADP8061 (ΔdcaA2), and ADP8062 (ΔdcaE2) each contained a significant dca deletion mutation that eliminated the ability to grow at the expense of adipate, their adipate-negative phenotypes were expected to be very stable. Indeed, the reversion frequency of ΔdcaH2 mutant strain ADP8018 at 30 or 37°C was below 4 × 10−10, the limit of detection. By contrast, adipate-positive colonies of ADP8023 (dcaA1::Kmr), ADP8061 (ΔdcaA2), and ADP8062 (ΔdcaE2) did arise on selective plates. When 2 × 108 cells were plated on adipate medium, ADP8061 formed adipate-positive colonies at a frequency of 10−7 or lower after 7 days at 30°C. For ADP8023, under similar conditions, the frequency was 3 × 10−7. The frequency of adipate-positive colonies of ADP8062 at 37°C was 4 × 10−7 or less.
A reduced growth rate on adipate differentiated the adipate-positive mutant derivatives from ADP1 and indicated that they were not true revertants. Strains carrying putative second-site suppressor mutations were purified, and chromosomal lysates of the strains were prepared. Purified, representative adipate-positive derivatives of ADP8061 and ADP8062 were analyzed by PCR and were shown to have retained their original deletions.
Because ADP8023 had an easily monitored Kmr marker, its phenotypic revertants received the greatest attention. Their phenotypes with respect to growth rate on adipate varied, and 36% of them were temperature sensitive, failing to grow on adipate at 37°C. All of them carried the Kmr marker. Moreover, when the Kmr marker of representative phenotypic revertant strains was transformed into ADP1, it conferred an adipate-negative phenotype. In addition, DNA from the ADP8023 suppressor mutants was able to transform two mutant strains to an adipate-positive phenotype: ADP8023 as well as ADP8061, which contains a deletion that flanks the Kmr marker in ADP8023 (Fig. 3). One class of ADP8023 suppressor mutants proved to be unstable, throwing off adipate-negative colonies on subculture; its phenotype was interpreted to be caused by unstable amplification of a genetic region encoding low-level suppressor activity.
Utilization of fatty acids, pimelic acid, and longer-chain dicarboxylic acids.
A gradient plate method was used to assess the ability of mutant strains to utilize medium-chain fatty acids. The fatty acids caprylic and capric acid (8 and 10 carbons, respectively) were metabolized by all, indicating that the dca genes under investigation are not involved in ω-oxidation of these particular fatty acids.
Strain ADP1 and the three deletion strains were also screened for their ability to utilize longer-chain dicarboxylic acids. Figure 5 shows that ADP1 grew at the expense of all of the tested compounds. The doubling time of adipate-grown cultures of ADP1 at 37°C was about 30 min. The doubling time of dodecanedioic acid- or tetradecanedioic acid-grown cultures was 2.6 or 4.7 times longer, respectively. ADP1 cells were very slow to grow at the expense of pimelic acid; as noted in the legend to Fig. 5, an extra day was allowed for growth of cells at the expense of pimelic acid as well as tetradecanedioic acid.
At a carbon chain length of 14, solubility of the saturated dicarboxylic acids in minimal medium starts to fall off, and at 16 atoms, insolubility obscures differences in the amount of growth. In the case of hexadecanedioic acid, the amount of growth was determined by viable cell counts on minimal medium plates containing succinate. Under the conditions described in the legend to Fig. 5, an ADP1 culture yielded 108 cells ml−1 on hexadecanedioic acid compared to the DMSO control of 7 × 106 cells ml−1. Although the same number of carbon atoms was provided to cultures with each substrate, decreasing solubility and the tendency of cells to clump onto insoluble crystals may account for the apparent reduced growth of ADP1 on the longer-chain acids shown in Fig. 5.
When cultured on adipic, pimelic, suberic, or sebacic acid, strains mutated in dcaE, dcaA, and dcaH failed to grow (Fig. 5). Strain ADP8023 has a similar phenotype on these substrates (data not shown). Clearly, the dcaA, dcaE, and dcaH gene products are specific for at least the medium-chain dicarboxylic acid thioesters. Growth at the expense of homologs longer than sebacic acid was reduced in the mutant strains compared to those of ADP1 but to a lesser extent (Fig. 5). Viable counts from tetradecanedioic acid cultures confirmed that the optical density at 600 nm reflected the number of cells present. Finally, growth at the expense of hexadecanedioic acid yielded about 27% of the viable count of ADP1 for ADP8061 and ADP8062; it was 16% of the wild-type level in ADP8018 (data not shown).
Reversion of mutant strains can be ruled out as an explanation for the disproportionately high relative yields of ADP8061 and ADP8062 on the longer-chain dibasic acids. After exposure to tridecanedioic or tetradecanedioic acid, cultures of ADP8061 or ADP8062, which showed an elevated amount of growth, continued to show the same adipate-negative phenotype as the original strains. Moreover, the relatively elevated amount of growth of the two strains on the higher-carbon-number homologs versus that on adipate was observed on solidified medium containing tetradecanedioic acid as the sole carbon source, where suppressor mutants or revertants could be observed directly.
Ability of mutant strains to dissimilate glutarate.
Dissimilation of even- and odd-chain dicarboxylic acids is postulated to diverge at the shorter thioesters: even-numbered compounds would be converted to succinyl-CoA following β-oxidation steps (Fig. 3), whereas odd-numbered ones would be converted to the C5 derivative glutaryl-CoA. Indeed, in another strain of Acinetobacter, glutarate was found to accumulate when chloramphenicol-inhibited cells were incubated with pimelic acid or its C9, C11, or C13 homologs (8). Evidence for metabolism of pimelate via a glutaryl-CoA intermediate under anaerobic conditions also exists (16, 20).
Although strain ADP1 may well metabolize odd-chain-length dicarboxylic acids through glutaryl-CoA, it does not grow with externally supplied glutarate alone. However, growth with external glutarate is observed if 0.1 mM adipate is added to the medium (D. A. D'Argenio, personal communication). Strain ADP7529 was shown to have a large deletion that extends into the dcaE gene (Fig. 1 and 4). Because the strain lacks the intergenic region between the dcaECHF transcript and the divergently transcribed dcaA, it is unlikely to express dcaH as well as dcaA and dcaE. Although unable to utilize pimelate, ADP7529 maintained its ability to grow at the expense of glutarate in the presence of 0.1 mM adipate (data not shown). This rules out any role for DcaA, DcaE, or DcaH in the metabolism of glutaryl-CoA.
Indeterminate role of other genes identified on pZR8005.
The functions of the other open reading frames identified on pZR8005 (Fig. 3 and Table 3) are under investigation. Initial analysis of knockout mutations in several of these genes has revealed that they do not lead to simple adipate-negative phenotypes. Nevertheless, dcaK and dcaC are located on transcripts which encode genes shown to be required for adipic acid dissimilation. Induction of dcaP::lacZ-Kmr by adipate in Acinetobacter (D. Parke, unpublished data) demonstrates that the putative transcript dcaIJP, which may be part of a longer dcaAKIJP transcript, also encodes genes which participate in dicarboxylic acid metabolism.
DISCUSSION
E. coli mutant with improved potential to dissimilate phenolics.
Isolation of a large clone, pZR8000, that contained not only genes that corrected an adipate-negative mutant but also suites of genes involved in aromatic and hydroaromatic catabolism led us to examine the phenotypes of E. coli carrying the clone. The full potential of DH5α(pZR8000) to utilize protocatechuate and 4-hydroxybenzoate required a mutation giving rise to E. coli strain EDP1 (Table 2). Although the scope of our research was limited to a single, genetically uncharacterized mutant strain, it opens the door to expanding the catabolic potential of E. coli as an experimental vehicle. It is not clear why the hydroaromatic compound quinate was not readily catabolized by the mutant E. coli harboring genes for all of the requisite enzymes and transporter, but this may be related to the periplasmic conversion of quinate to protocatechuate in Acinetobacter sp. strain ADP1 (12).
Analysis of the dca-pca deletion of strain ADP7529.
It is an indication of the remarkable natural competence of the “Juni” strain of Acinetobacter (28) that our cloning efforts were successful in spite of the hurdle of screening clones against a recipient strain that turned out to have a relatively large, 11.6-kb deletion. The sequences of the large deletions found in previous studies of Acinetobacter (13, 18) have not been determined, and it is not clear why they arise at high frequency upon application of the ΔpcaBDK positive selection method (21). An explanation might be that they are mediated by crossing over between distant copies of a transposon or insertion sequence such as IS1236, which occurs seven times in the ADP1 chromosome (17). However, the sequence of Δdca1 in ADP7529, isolated in a ΔpcaBDK positive selection (10), gives no evidence that IS1236 played a role in this deletion. Although the molecular events that created the deletion are unknown, it is noteworthy that the sequence ATAATA, itself a direct repetition, which is separated by 11.6 kb, occurs at the deletion juncture (Fig. 4).
Phenotypic evidence for overlapping functions of β-oxidation genes in Acinetobacter.
Data based on sequence analysis and mutant phenotypes support the conclusion that DcaA, DcaE, and DcaH act on thioesters of adipic, pimelic, and suberic acids as depicted in Fig. 3. Growth studies and analysis of pseudorevertants suggest that homologs of the cloned dca genes exist in strain ADP1. It remains to be determined whether such homologs are required for the conversion of longer-chain thioesters to medium-chain ones, which are then subject to catalysis by DcaA, DcaE, and DcaH. Results with the ΔdcaH1 mutant ADP8018 are consistent with this hypothesis, showing an elevated growth yield on the longer dibasic acids that would be predicted by liberation of only one or two acetyl units per chain (Fig. 5). It is difficult to explain the elevated growth yields of ADP8061 and ADP8062 at the expense of tridecanedioic or tetradecanedioic acid. It is possible that a putative DcaA or DcaH homolog that acts preferentially on longer-chain acids may possess some activity towards certain shorter-chain derivatives, which could be enhanced by amplification of the coding segment of the chromosome. Although the physiological data do not allow us to determine the absolute limits of specificity for the three cloned dca gene products, they do indicate a role for them in at least part of the dissimilation of the longer-chain acids.
It is also possible that other enzymes, not directly engaged in straight-chain dicarboxylic acid dissimilation, may act nonspecifically on the longer-chain acids. Isolation of an array of phenotypic revertants of dcaA and dcaE mutants provides genetic evidence for the presence of related β-oxidation genes in the Acinetobacter chromosome. The inference that other genes with functions similar to those of dcaE or dcaA are present is not surprising given the array of DcaA, DcaE, and DcaH homologs from single bacterial strains pulled up in database queries. Further characterization of the suppressor mutant strains and the dca genetic region afford synergistic lines of investigation.
Interpretation of functions of other open reading frames in the dca region of pZR8000.
In addition to the three open reading frames that were the focus of this study, six other genes were identified as part of the dca region on pZR8000. The dcaF gene, encoding a thiolase homolog, is presumed to be required for thiolytic cleavage of acetyl-CoA from acyl-CoA. The role of the dcaC gene is unknown. The products of dcaK and dcaP are being investigated for their roles in the control of dicarboxylic acid traffic into the cell. Genetic linkage of dcaP with dcaIJ is seen as a tantalizing hint of the functional relationship of these genes' products. The dcaIJ genes are located not far from the homologous pcaIJ genes, which encode a β-ketoadipate succinyl-CoA transferase involved in protocatechuate catabolism (Table 4). The dcaIJ genes undoubtedly encode the third enzyme (in addition to PcaIJ and CatIJ) with β-ketoadipate succinyl-CoA transferase activity previously detected in Acinetobacter; this broader-specificity enzyme was induced by adipate, and unlike PcaIJ and CatIJ, its activity was inhibited by adipate (7). Activity of this presumed adipate succinyl-CoA transferase towards longer dicarboxylic acids remains a subject for future investigation. In P. fluorescens, an adipate succinyl-CoA transferase did not appear to act on dicarboxylic acids longer than pimelate; a separate ligase was responsible for activation of longer-chain acids (23, 24). If this is the case in Acinetobacter, a critical ligase gene remains to be discovered.
Relationships of dca gene products to homologous Acinetobacter proteins.
The closest homologs of DcaE and DcaA elicited by BLAST queries are those from Acinetobacter sp. strain SE19 (Table 3). The homologous SE19 genes, termed fadB and fadE, respectively, lie next to a cluster of genes required for the conversion of cyclohexanol to adipic acid (9). Only 147 residues of the deduced N-terminal sequence of FadC, the DcaH homolog, from strain SE19 have been deposited in GenBank, and therefore this enzyme was not pulled up in a database query; however, alignment of the N-terminal sequences of FadC and DcaH demonstrated a sequence identity of 55%. Organization of the three fad genes from strain SE19 is similar to that of the dca homologs, with the exception of the intervening dcaC in ADP1. Given their location adjacent to genes required for generating adipic acid from cyclohexanol and their close relationship to the dca homologs, the SE19 fad genes probably encode enzymes for adipic acid dissimilation.
In Acinetobacter sp. strain ADP1, the region between dcaK and mucK spans 12 kb of DNA which encodes additional genes involved in dicarboxylic acid metabolism (D. Parke and G. Peterson, unpublished data). The location of and close relationship between DcaK and MucK (Tables 3 and 4) suggest that phenotypic masking by the DcaK transporter may have obscured adipic or other dicarboxylic acid carrier functions of the cis,cis-muconate transporter in a mucK mutant (44).
Organization of putative dca genes in Pseudomonas aeruginosa.
Like Acinetobacter sp. strain ADP1, P. aeruginosa strains are able to grow at the expense of dicarboxylic acids containing from 6 to 10 carbon atoms (42). As mentioned above, early work established the presence of two genes that contributed to activating dicarboxylic acids in the closely related P. fluorescens (23, 24). In light of that work and the identification of P. aeruginosa homologs of the Dca proteins from strain ADP1 (Table 3), the P. aeruginosa PAO1 genome (43) was examined to determine the organization and location of putative dca gene homologs.
One likely dca genetic region in the P. aeruginosa PAO1 genome lies at position 1771122 to 1775757. Giving genes the dca designations, a putative dcaEH transcript is divergently transcribed from a likely dcaRA transcript. Identification numbers for the PAO1 genes are indicated in parentheses: dcaE (PA1629), dcaH (PA1628), dcaR (PA1630), and dcaA (PA1631). In Acinetobacter, dcaR is linked to dcaF and has been identified tentatively as one of the regulatory genes for dicarboxylic acid dissimilation (D. Parke and G. Peterson, unpublished data). In P. aeruginosa, this gene cluster lies between sets of genes associated with transport. As discussed below, it seems likely that they play a role in medium-chain dicarboxylic acid dissimilation.
A second cluster of putative dca genes, at position 4021918 to 4028538 in the PAO1 genome, forms a unit of transcription that includes dcaA, a gene of unknown function, dcaE, dcaH, dcaF, and a probable porin gene. These genes have gene identification numbers as indicated parenthetically: dcaA homolog (PA3593), dcaE homolog (PA3591), dcaH homolog (PA3590), dcaF homolog (PA3589), and the possible porin gene (PA3588).
In all cases, the DcaE, DcaH, and DcaA homologs which correspond to the genes of the first cluster at position 1771122 to 1775757 are more similar to the ADP1 proteins deduced in this study than are those in the second cluster. The fact that the dcaF gene product is very similar to that of the ADP1 homolog (Table 3) is taken as evidence supporting the hypothesis that the second cluster is related to dicarboxylic acid degradation as well.
The two PAO1 DcaH homologs bear a close similarity to each other, as do the two DcaE homologs; the aligned proteins possess amino acid identities of 66 and 72%, respectively. By contrast, the two PAO1 DcaA homologs are quite divergent, sharing only 29% amino acid identity in the aligned portion of the proteins. Only DcaA homolog PA1631 is very similar to its ADP1 homolog. In addition, the DcaA homolog PA3593 is 191 amino acids longer than DcaA homolog PA1631. The size difference suggests that the longer protein may be adapted for longer-chain substrates, as occurs in fatty acid degradation. The genes in the region of position 4021918 to 4028538 of the PAO1 chromosome may have evolved to play a role in long-chain dicarboxylic acid dissimilation, and those in the region of position 1771122 to 1775757 may be adapted to shorter compounds such as adipate. Curiously, there is no close homolog for dcaIJ in the PAO1 genome, and different transporter genes appear to be associated with the putative dca genes.
Phylogenetic trees based on rRNA sequences place Acinetobacter and Pseudomonas species on neighboring twigs of the same branch (45). The common elements in dicarboxylic acid degradation appear to form a mere skeleton, which was fleshed out by appropriating distinct genes in new contexts.
Genomic context of dca genes and intimations of host bacterial niche.
In Acinetobacter sp. strain ADP1, linkage of genes for associated functions is a striking feature of the pca-qui-pob-ppa region (Fig. 1). Proximity of the dca genes to the pca region raises the question of whether the dca-pca linkage provides a selective benefit. In addition to allowing horizontal transfer of a suite of genes, linkage enables them to be amplified (10, 38) when the concentrations of their specific, related substrates are low. Suberin is a complex, natural polyester that could potentially yield substrates for dca, pca, and ppa gene products. Synthesized by plants as a protective barrier in response to wounding, its components include hydroxycinnamic acids esterified to dicarboxylic acids (5, 19, 32). Typically, dicarboxylic acids found in suberin networks are comprised of at least 16 carbon atoms. As we have demonstrated in this paper, Acinetobacter sp. strain ADP1 is capable of degrading the long-chain hexadecanedioic acid. Given these elements, it is tempting to term the dca-pca-qui-pob-ppa region of the ADP1 genome a “suberon.”
ACKNOWLEDGMENTS
We are indebted to Wayne Coco, David D'Argenio, and David Young for their helpful suggestions as well as J. Hanrahan for technical contributions in the initial stages of this project. We acknowledge the Pseudomonas Genome Project, which was the source of information on organization of putative dca gene homologs in P. aeruginosa PAO1.
The research was funded by grants DAAG55-98-1-0232 from the Army Research Office and MCB-9603980 from the National Science Foundation to L.N.O.
Footnotes
This article is publication number 28 from the Biological Transformation Center in the Yale Institute for Biospheric Studies.
REFERENCES
- 1.Altschul S F, Madden T L, Schafer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1991. [Google Scholar]
- 3.Averhoff B, Gregg-Jolly L, Elsemore D, Ornston L N. Genetic analysis of supraoperonic clustering by use of natural transformation in Acinetobacter calcoaceticus. J Bacteriol. 1992;174:200–204. doi: 10.1128/jb.174.1.200-204.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann P, Doudoroff M, Stanier R Y. A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter) J Bacteriol. 1968;95:1520–1541. doi: 10.1128/jb.95.5.1520-1541.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernards M A, Lewis N G. The macromolecular aromatic domain in suberized tissue: a changing paradigm. Phytochemistry. 1998;47:915–933. doi: 10.1016/s0031-9422(98)80052-6. [DOI] [PubMed] [Google Scholar]
- 6.Blakely E R. The microbial degradation of cyclohexanecarboxylic acid by a β-oxidation pathway with simultaneous induction to the utilization of benzoate. Can J Microbiol. 1978;24:847–855. doi: 10.1139/m78-141. [DOI] [PubMed] [Google Scholar]
- 7.Canovas J L, Stanier R Y. Regulation of the enzymes of the β-ketoadipate pathway in Moraxella calcoacetica. 1. General aspects. Eur J Biochem. 1967;1:289–300. doi: 10.1007/978-3-662-25813-2_40. [DOI] [PubMed] [Google Scholar]
- 8.Chapman P J, Dugglesby R G. Dicarboxylic acid catabolism by bacteria. Biochem J. 1967;103:7c–9c. doi: 10.1042/bj1030007c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Q, Thomas S M, Kostichka K, Valentine J R, Nagarajan V. Genetic analysis of a gene cluster for cyclohexanol oxidation in Acinetobactersp. strain SE19 by in vitro transposition. J Bacteriol. 2000;182:4744–4751. doi: 10.1128/jb.182.17.4744-4751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Argenio D A. Ph.D. thesis. New Haven, Conn: Yale University; 1999. [Google Scholar]
- 11.D'Argenio D A, Segura A, Bünz P V, Ornston L N. Spontaneous mutations affecting transcriptional regulation by protocatechuate in Acinetobacter. FEMS Microbiol Lett. 2001;201:15–19. doi: 10.1111/j.1574-6968.2001.tb10726.x. [DOI] [PubMed] [Google Scholar]
- 12.D'Argenio D A, Segura A, Coco W M, Bünz P V, Ornston L N. The physiological contribution of AcinetobacterPcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK. J Bacteriol. 1999;181:3505–3515. doi: 10.1128/jb.181.11.3505-3515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Argenio D A, Vetting M W, Ohlendorf D H, Ornston L N. Substitution, insertion, deletion, suppression, and altered substrate specificity in functional protocatechuate 3,4-dioxygenases. J Bacteriol. 1999;181:6478–6487. doi: 10.1128/jb.181.20.6478-6487.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donoghue N A, Trudgill P W. The metabolism of cyclohexanol by AcinetobacterNCIB 9871. Eur J Biochem. 1975;60:1–7. doi: 10.1111/j.1432-1033.1975.tb20968.x. [DOI] [PubMed] [Google Scholar]
- 15.Forst D, Welte W, Wacker T, Diederichs K. Structure of the sucrose-specific porin ScrY from Salmonella typhimuriumand its complex with sucrose. Nat Struct Biol. 1998;5:37–46. doi: 10.1038/nsb0198-37. [DOI] [PubMed] [Google Scholar]
- 16.Gallus C, Schink B. Anaerobic degradation of pimelate by newly isolated denitrifying bacteria. Microbiology. 1994;140:409–416. doi: 10.1099/13500872-140-2-409. [DOI] [PubMed] [Google Scholar]
- 17.Gerischer U, D'Argenio D A, Ornston L N. IS1236, a newly discovered member of the IS3 family, exhibits varied patterns of insertion into the Acinetobacter calcoaceticuschromosome. Microbiology. 1996;142:1825–1831. doi: 10.1099/13500872-142-7-1825. [DOI] [PubMed] [Google Scholar]
- 18.Gerischer U, Ornston L N. Spontaneous mutations in pcaH and -G, structural genes for protocatechuate 3,4-dioxygenase in Acinetobacter calcoaceticus. J Bacteriol. 1995;177:1336–1347. doi: 10.1128/jb.177.5.1336-1347.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graca J, Pereira H. Suberin structure in potato periderm: glycerol, long-chain monomers, and glyceryl and feruloyl dimers. Agric Food Chem. 2000;48:5476–5483. doi: 10.1021/jf0006123. [DOI] [PubMed] [Google Scholar]
- 20.Härtel U, Eckel E, Koch J, Fuchs G, Linder D, Buckel W. Purification of glutaryl-CoA dehydrogenase from Pseudomonassp., an enzyme involved in the anaerobic degradation of benzoate. Arch Microbiol. 1993;159:174–181. doi: 10.1007/BF00250279. [DOI] [PubMed] [Google Scholar]
- 21.Hartnett G, Averhoff B, Ornston L N. Selection of Acinetobacter calcoaceticus mutants deficient in the p-hydroxybenzoate hydroxylase gene (pobA) a member of a supraoperonic cluster. J Bacteriol. 1990;172:6160–6161. doi: 10.1128/jb.172.10.6160-6161.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harwood C S, Gibson J. Shedding light on anaerobic benzene ring degradation: a process unique to prokaryotes? J Bacteriol. 1997;179:301–309. doi: 10.1128/jb.179.2.301-309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoet P P, Stanier R Y. The dissimilation of higher dicarboxylic acids by Pseudomonas fluorescens. Eur J Biochem. 1970;13:65–70. doi: 10.1111/j.1432-1033.1970.tb00899.x. [DOI] [PubMed] [Google Scholar]
- 24.Hoet P P, Stanier R Y. Existence and functions of two enzymes with β-ketoadipate:succinyl-CoA transferase activity in Pseudomonas fluorescens. Eur J Biochem. 1970;13:71–76. doi: 10.1111/j.1432-1033.1970.tb00900.x. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coliwith plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 27.Juni E. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J Bacteriol. 1972;112:917–931. doi: 10.1128/jb.112.2.917-931.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juni E, Janick A. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kester A S, Foster J W. Diterminal oxidation of long-chain alkanes by bacteria. J Bacteriol. 1963;85:859–869. doi: 10.1128/jb.85.4.859-869.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. A broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 31.Kusunose M, Kusunose E, Coon M J. Enzymatic ω-oxidation of fatty acids. I. Products of octanoate, decanoate, and laurate oxidation. J Biol Chem. 1964;239:1374–1380. [PubMed] [Google Scholar]
- 32.Moire L, Schmutz A, Buchala A, Yan B, Stark R E, Ryser U. Glycerol is a suberin monomer. New experimental evidence for an old hypothesis. Plant Physiol. 1999;119:1137–1146. doi: 10.1104/pp.119.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parke D, D'Argenio D A, Ornston L N. Bacteria are not what they eat: that is why they are so diverse. J Bacteriol. 2000;182:257–263. doi: 10.1128/jb.182.2.257-263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parke D, Ornston L N. Nutritional diversity of Rhizobiaceaerevealed by auxanography. J Gen Microbiol. 1984;130:1743–1750. [Google Scholar]
- 37.Pichinoty F, Veron M, Mandel M, Durand M, Job C, Garcia J L. Physiological study and taxonomy of Alcaligenes species: A. denitrificans, A. odorans and A. faecalis. Can J Microbiol. 1978;24:743–753. doi: 10.1139/m78-123. [DOI] [PubMed] [Google Scholar]
- 38.Roth J R, Benson N, Galitski T, Haack K, Lawrence J G, Miesel L. Rearrangements of the bacterial chromosome: formation and applications. In: Neidhart F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D. C.: American Society for Microbiology; 1996. pp. 2256–2276. [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schirmer T, Keller T A, Wang Y, Rosenbusch J P. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 41.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 42.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic Pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 43.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L. Complete genome sequence of Pseudomonas aeruginosaPAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 44.Williams P A, Shaw L E. mucK a gene in Acinetobacter calcoaceticus ADP1 (BD413), encodes the ability to grow on exogenous cis,cis-muconate as the sole carbon source. J Bacteriol. 1997;179:5935–5942. doi: 10.1128/jb.179.18.5935-5942.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]