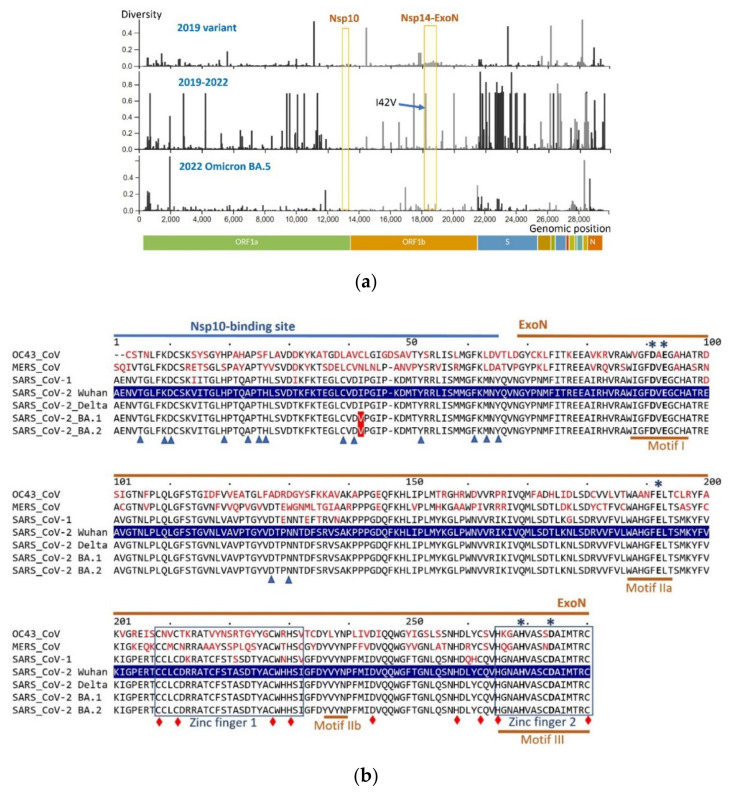
Figure 1.
(a) Diversity of the encoded amino acids along genomic sequences of SARS-CoV-2. Profiles for three genomic sets are shown—top: diversity within ~2000 genomic sequences of the early SARS-CoV-2 sampled between December 2019 and March 2020; middle: diversity within ~2000 genomes collected in December 2019–May 2022; bottom: diversity within ~900 genomes of the recent Omicron BA.5 variant collected from January–May of 2022. The Nsp14-ExoN boundaries include only the first 280 aa positions involved in ExoN function (the same region shown in the sequence alignment). The arrow points to the position of the ubiquitous I42V aa substitution in Nsp14. Profiles are constructed by the Nextstrain webtool using genomic sequences from GISAID (https://www.gisaid.org/; Nextstrain datasets: “early-19A” and “22B Omicron”; webtool accessed on 25 May 2022 at https://nextstrain.org [61]). (b) Protein sequence conservation in the ExoN domain of Nsp14 in representative beta Coronaviridae strains. Partial Nsp14 protein alignment spans the Nsp10-binding site (amino acid positions 1–64) and the adjacent ExoN domain (aa positions 68–280). Nsp14 of the SARS-CoV-2 Wuhan-Hu-1 variant (highlighted in blue) was used as a reference: amino acid differences in other variants compared to the reference sequence are shown in red. Functional motifs comprising the ExoN active site are underlined, and the zinc finger regions are boxed [12,14,36]. ExoN catalytic residues are indicated with asterisks; residues interacting with Nsp10 are marked by blue triangles; and positions critical for zinc fingers, protein solubility or ExoN activity are shown as red diamonds [12,14]. The sequences were retrieved from GenBank: OC43-CoV (common cold; accession number YP_009924328), MERS-CoV (YP_009047225), SARS-CoV-1 (JX163928), and variants of SARS-CoV-2: Wuhan-Hu-1 (YP_009725309, original strain isolated in 2019), Delta (OM990852), and Omicron BA.1 (ON141240) and BA.2 (ON553707). Protein alignment was built using Clustal Omega [62,63], visualized by MView [63,64], and then annotated.