Abstract
During the past several decades, numerous studies have provided insights into biological characteristics of cancer cells and identified various hallmarks of cancer acquired in the tumorigenic processes. However, it is still challenging to image these distinctive traits of cancer to facilitate the management of patients in clinical settings. The rapidly evolving field of positron emission tomography (PET) imaging has provided opportunities to investigate cancer's biological characteristics in vivo. This article reviews the current status of PET imaging on characterizing hallmarks of cancer and discusses the future directions of PET imaging strategies facilitating in vivo cancer phenotyping.
Keywords: biomarker, cancer, cancer hallmarks, molecular imaging, positron emission tomography (PET), translational medicine
Short abstract
Various direct and indirect imaging strategies have been developed in positron emission tomography. Positron emission tomography has shown great potential in characterizing cancer hallmarks in vivo.
Introduction
Cancer is a critical issue worldwide with high incidence and mortality, imposing a heavy burden on the global health care system. 1 To date, a critical issue facing cancer management is the spatiotemporal heterogeneity both inter‐ and intratumor lesions. 2 Developing imaging techniques and noninvasive biomarkers holds great promise in addressing this challenge across the spectrum of cancer management.
Positron emission tomography (PET) is a representative molecular imaging technique enabling the noninvasive visualization, characterization, and quantification of biologic processes at cellular and molecular levels. 3 By using various radiolabeled molecular probes, PET imaging has been widely applied in cancer diagnosis and treatment. Indeed, the development of PET imaging has shown great potential to reform traditional pathology and may lead to a new pattern of pathological practice termed transpathology. 4
In this review, we summarize principles of developing PET probes and discuss emerging strategies of PET imaging for in vivo cancer phenotyping with representative examples. The conceptual framework of cancer hallmarks is used to describe how PET imaging would help characterizing cancer phenotype noninvasively. 5
Molecular Recognition‐Based Radiopharmaceuticals for PET Imaging
Based on the principles of molecular recognition and radionuclide tracing, radiopharmaceuticals serve as a primary driving force of PET imaging (Fig. 1). Typically, a radiopharmaceutical comprises a targeting moiety, a radionuclide, and sometimes a linker connecting them.
Figure 1.
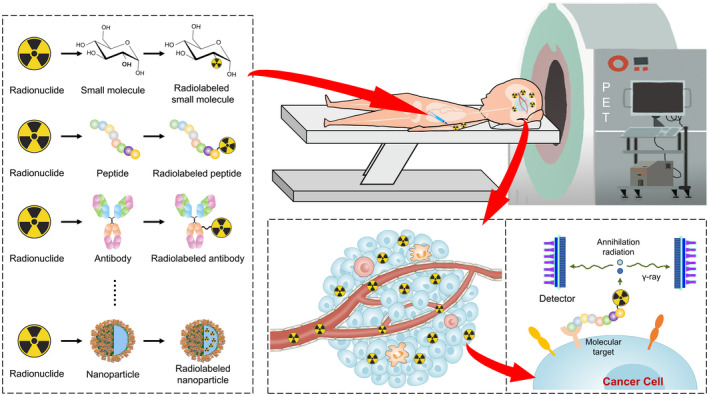
Principles of PET molecular imaging. To date, several ligands have been radiolabeled for PET imaging, including small molecules, peptides, antibodies, and nanoparticles. By using the radiopharmaceuticals developed, PET enables the whole‐body evaluation of cancer. Based on the principles of molecular recognition and radionuclide tracing, the assessment of biological processes at the molecular/cellular level can be achieved. PET indicates positron emission tomography.
According to imaging targets, numerous ligands can be used as the targeting moiety, including small molecules, peptides, antibodies, and nanoparticles. The targeting moiety determines the binding sensitivity, specificity, and in vivo pharmacokinetics of radiopharmaceuticals. To date, strategies to develop targeting moiety can be categorized into random and rational approaches. 6 For example, random compound‐making techniques (eg, combinatorial chemistry) could help generate compound libraries, and high‐throughput screen techniques may characterize a great many candidate probes in a short period. 7 Alternatively, targeting moiety could be previously characterized molecules, such as drug candidates, and established probes of other imaging modalities. 8
Positron‐emitting radionuclides serve as PET signal agents, of which the commonly used include 18F, 11C, 68Ga, and 89Zr. Currently, radiolabeling strategies are divided into direct and indirect approaches. 9 For example, 11C‐methylation reactions are commonly used for direct carbon labeling and 18F‐fluorination reactions can be used for substitutions of H or OH groups by fluorine‐18. In contrast, when pharmacophores lack suitable sites for direct labeling, labeling a linked prosthetic group is more applicable. Notably, the half‐lives of radionuclides should match the pharmacokinetics of targeting vectors. 10 For instance, monoclonal antibodies are generally labeled by radionuclides with long half‐lives (T1/2) (eg, 89Zr, T1/2 = 78.4 hours and 124I, T1/2 = 100.8 hours), whereas antibody fragments with relatively rapid pharmacokinetics are labeled by those with short T1/2 (eg, 18F, T1/2 = 109.8 minutes and 68Ga, T1/2 = 68.3 minutes).
Reporter gene strategies reduce the need for radiopharmaceuticals targeting each gene or protein of various signaling pathways. By tracking reporter gene products, which are expressed under the control of promoters, biological processes could be visualized (Fig. 2). The reporter protein can be expressed constitutively (eg, promoter cytomegalovirus) or inducibly (eg, promoter p53), enabling the imaging of gene expression, cell tracking, and protein‐protein interactions. 11 One of the most widely applied reporter gene is herpes simplex virus type 1 thymidine kinase (HSV1‐tk), with several probes, such as 18F‐FEAU, 131I‐FIAU, and 18F‐FHBG, being the commonly used radiopharmaceuticals.
Figure 2.
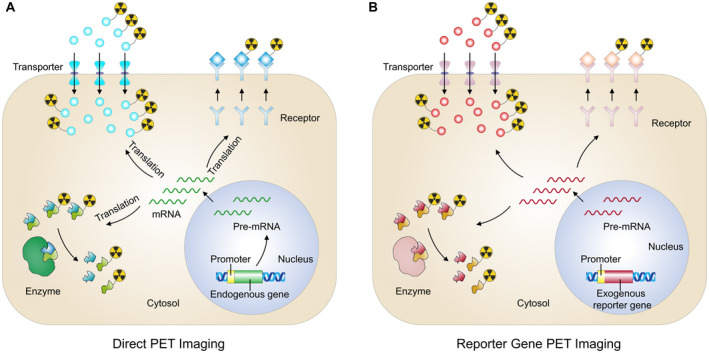
Principles of direct PET imaging and indirect reporter gene PET imaging. Targets of direct PET imaging (eg, transporters, enzymes, receptors) are translated from mRNA of endogenous genes (A), whereas the most commonly used imaging targets, transporters, enzymes, and receptors, are translated from transfected exogenous reporter gene (B). mRNA indicates messenger RNA; PET, positron emission tomography.
With the discovery of key mediators regulating cancer biological processes (Fig. 3), as well as novel imaging strategies, PET imaging has greatly facilitated in vivo cancer characterization (Table 1).
Figure 3.
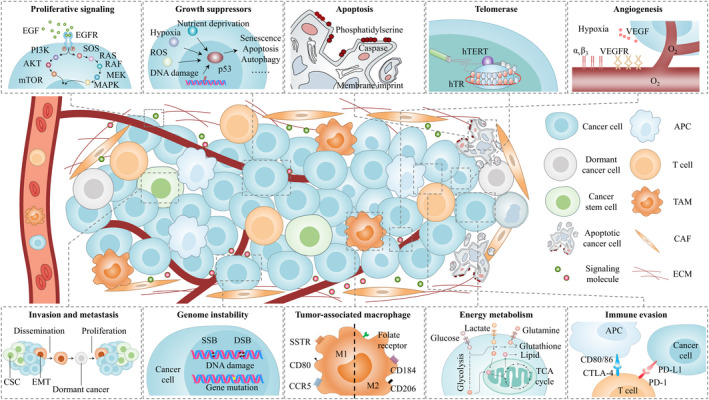
Imaging targets investigated for caner phenotyping. With the stunning progress in cancer biology, several distinctive mediators have been identified to drive cancer initiation and progression, ranging from proliferative signaling to immune evasion. Several examples of these key processes and corresponding imaging targets are depicted. To some extent, this depiction is simplistic because many molecules are also involved in other processes, and there are complex interactions among them. APC, antigen‐presenting cell; CAF, cancer associated fibroblasts; CSC, cancer stem cell; CTLA‐4, cytotoxic T lymphocyte antigen‐4; DSB, double‐strand break; ECM, extracellular matrix; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; EMT, epithelial–mesenchymal transition; hTERT, human telomerase reverse transcriptase; hTR, human telomerase RNA; PD‐1, programmed cell death protein 1; PD‐L1, programmed cell death ligand 1; SSB, single‐strand break; SSTR, somatostatin receptor; TAM, tumor associated macrophage; VEGF, vascular endothelial growth factor; VEGFR, vascular endothelial growth factor receptor.
TABLE 1.
PET Molecular Imaging to Study Cancer Hallmarks
| Hallmarks | Studied Aspects | Imaging Targets a | PET Imaging Tools a | References |
|---|---|---|---|---|
| Sustaining proliferative signaling | Signaling | EGFR | 89Zr‐cetuximab, 89Zr‐panitumumab, 89Zr‐nimotuzumab | 12, 13 |
| PI3K | 18F‐FMTA‐2, 11C‐pictilisib and 18F‐PEG3‐GDC‐0941 | 14 | ||
| Evading growth suppressors | Specific gene | P53 | p53‐TKGFP system, p53‐TAg‐TK‐GFP system | 15, 16 |
| Transduction pathway | TGF‐β | 89Zr‐fresolimumab, 64Cu‐NOTA‐TRC105 | 17, 18 | |
| Resisting cell death | Apoptosis | Phosphatidylserine exposure | 18F‐annexin V, 18F‐FBAM, 18F‐C2Am | 19 |
| Apoptotic membrane imprint | 18F‐ML‐10 | 20 | ||
| Caspase | 18F‐ICMT‐11, 18F‐CP18, caspase‐3‐cTK system | 21, 22, 23 | ||
| Enabling replicative immortality | Telomerase function | hTERT | hTERT‐reporter systems, radiolabeled ASON, radiolabeled siRNA, 64Cu‐hTERT IgM | 24, 25, 26, 27, 28 |
| hTR | hTR‐NIS system | 24 | ||
| Inducing angiogenesis | Direct angiogenetic processes | VEGF/VEGFR | 18F‐AlF‐NODA‐scVR1, 89Zr‐bevacizumab, 89Zr‐ranibizumab, 11C‐erlotinib | 29, 30 |
| Integrin | 18F‐galacto‐RGD, 18F‐fluciclatide, 18F‐RGD‐K5, and 68Ga‐NOTA‐RGD | 31 | ||
| Indirect angiogenetic state | Hypoxia | 18F‐FMISO, 18F‐FAZA, 18F‐HX4, and 64Cu‐ATSM | 32, 33, 34, 35 | |
| Activating invasion and metastasis | Metastasis‐initiating processes | CSCs | 64Cu‐NOTA‐AC133 mAb, 64Cu‐T140‐2D | 36, 37 |
| Cancer dormancy | 18F‐NFTG | 38 | ||
| Phenotypic plasticity | EMT and MET | 11C‐SU11274 | 39 | |
| Genome instability and mutation | DNA damage | Single‐strand break | 18F‐FTT and 18F‐PARPi | 40, 41 |
| Double‐strand break | 89Zr‐anti‐γH2AX‐TAT | 42 | ||
| Gene mutation | Nucleic acid | Radiolabeled ASON | 43 | |
| Tumor‐promoting inflammation | Cellular components of tumor microenvironment | Macrophages | 68Ga‐pentixafor, 64Cu‐MAN‐LIPs, 3′‐Aza‐2′‐[18F]fluorofolic acid, 18F‐FDR‐NOC, 11C‐AM7, 64Cu‐DOTA‐DAPTA | 44, 45 |
| Enzymes of tumor microenvironment | MMPs | 64Cu‐DOTA‐CTT, 18F‐CGS27023A | 46, 47 | |
| COX‐2 | 18F‐desbromo‐Dup‐697, 18F‐SC58125, 11C‐celecoxib, and 11C‐rofecoxib | 48 | ||
| Reprogramming energy metabolism | Glucose metabolism | Glucose | 18F‐FDG | 49 |
| Amino acid metabolism | Various amino acids | 11C‐MET, 18F‐FET, 18F‐DOPA, 18F‐FGln, 11C‐glutamine, | 50 | |
| Metabolism of other nutrients | Fatty acids, choline, etc. | 11C‐acetate, 11C‐choline, 18F‐choline, 18F‐fluoroethylcholine | 51, 52 | |
| Evading immune destruction | Immune cell infiltration | CD8+ T cell | 89Zr‐DFO‐CD3, 89Zr‐malDFO‐GK1.5 cDb, 89Zr‐Df‐IAB22M2C | 53 |
| Cancer checkpoint | PD‐1, PD‐L1, and CTLA‐4 | 64Cu‐NOTA‐PD‐1 mAb, 89Zr‐Df‐nivolumab, 64Cu‐NOTA‐PD‐L1 mAb, 64Cu‐DOTA‐anti‐CTLA‐4, 64Cu‐DOTA‐ipilimumab | 54, 55, 56 | |
| Other tumor‐associated immune cells | TAM, MDSC, neutrophils, natural killer cell, etc. | Radiopharmaceuticals targeting macrophages, 64Cu‐NOTA‐αCD11b‐mAb, 18F‐MAPP, 89Zr‐NKp30Ab | 57, 58, 59 |
Abbreviations: ASON, antisense oligonucleotide; COX‐2, cyclooxygenase‐2; CSCs, cancer stem cells; CTLA‐4, cytotoxic T‐lymphocyte‐associated antigen 4; EGFR, epidermal growth factor receptor; EMT, epithelial‐to‐mesenchymal transition; hTERT, human telomerase reverse transcriptase; hTR, human telomerase RNA; IgM, immunoglobulin M; mAb, monoclonal antibody; MDSC, myeloid‐derived suppressor cells; MET, mesenchymal‐to‐epithelial transition; MMPs, matrix metalloproteinases; PD‐1, programmed cell death protein 1; PD‐L1, programmed death‐ligand 1; PET, positron emission tomography; PI3K, phosphoinositide 3‐kinase; TAM, tumor associated macrophages; TGF‐β, transforming growth factor β; VEGF, vascular endothelial growth factor; VEGFR, vascular endothelial growth factor receptor.
Examples of imaging targets and corresponding PET imaging tools.
Sustaining Proliferative Signaling
Arguably, the most distinctive trait of cancer is the ability to sustain uncontrolled cell proliferation. 5 Specifically, the gain‐of‐function mutation, gene amplification and recombination, and overexpressed tumorigenic receptor and ligand could be key players that maintain the self‐sufficient proliferative ability.
PET imaging has emerged as a powerful tool to visualize cell‐surface receptors triggering proliferation signaling circuits. 12 For example, epidermal growth factor receptor (EGFR) is a critical imaging target because of its wide expression in epithelial malignancies as well as its crucial role in promoting cancer proliferation. A number of radiopharmaceuticals, especially antibody‐based probes, including 89Zr‐cetuximab, 89Zr‐panitumumab, and 89Zr‐nimotuzumab, have been developed to visualize EGFR expression. 12 , 13 Several radiopharmaceuticals were investigated in clinical settings to select patients suitable for EGFR‐targeted therapies. 60 , 61 Unfortunately, not all patients with high EGFR expression are sensitive to EGFR‐targeted therapy because the anti‐EGFR effects could be bypassed by other EGFR family members, mutations in downstream signaling cascades (eg, phosphatidylinositol 3‐kinase [PI3K]), and tumor suppressor proteins (eg, p53). 62 The complexity in tumor proliferation biology underlines the importance to image‐related compensatory mechanisms.
Estrogen receptor (ER) is another representative receptor regulating the growth and development for both healthy tissue and hormone‐regulated cancers (eg, cancers originating in the breast and ovary). To date, 2 subtypes of ER, ERα and ERβ, have been discovered: ERα is the primary subtype in hormone‐regulated cancers, triggering proliferation and survival of cancer cells, whereas ERβ functions as a proliferative “brake” against ERα, with a declined expression level in tumor progression. 63 For ER imaging, 18F‐fluoroestradiol, a radiolabeled estrogen analog with ERα selectivity (ERα/ERβ = 6.3), is the most widely used PET agent, showing great value in assessing tumor ER status and informing therapeutic decision‐making. 64 Additionally, attempts have been made in developing Erβ‐selective probes, including 18F‐FHNP, 18F‐FEDPN, and 18F‐PVBO, with an ERβ/ERα selectivity of 3.5 to 12.46. 65 However, further studies are warranted to characterize and improve the in vivo targeting ability as well as to evaluate the potential application in patients with ERβ‐positive cancer.
Alternatively, PET has been used in imaging intracellular proliferative signaling pathways. For example, the PI3K/protein kinase B/mammalian target of rapamycin pathway, a commonly activated pathway in cancer, critically regulates cell growth and proliferation. 66 Correspondingly, PI3K inhibitors have be radiolabeled for PET imaging, such as 18F‐FMTA‐2, 11C‐pictilisib, and 18F‐PEG3‐GDC‐0941. 14 It is noteworthy that inhibition of the PI3K/protein kinase B/mammalian target of rapamycin pathway could also stimulate feedback loops that lead to receptor tyrosine kinase expression and drug resistance. Interestingly, the pattern of receptor tyrosine kinase expression depends on the inhibited signaling node and can be evaluated through receptor PET imaging. 67
Evading Growth Suppressors
Cancer can accomplish aberrant growth by circumventing tumor‐suppressive programs. 5 To date, numerous tumor suppressors have been discovered, with the role of specific genes and transduction pathways regulating cellular quiescence being gradually revealed. 68 PET molecular imaging is powerful in visualizing key processes of tumor suppressors.
p53 is an extensively investigated gene controlling cell growth. Although no radiopharmaceuticals directly targeting p53 have been reported, imaging of the transcriptional regulation of p53 is feasible. By placing the HSV1‐tk/GFP (TKGFP) dual reporter gene under the control of a cis‐acting p53‐specific enhancer, the expression of TKGFP could be transcriptionally imaged and activate the p53 protein. 15 Once transgenic mice carrying cis‐p53/TKGFP were generated, these animals have been valuable to longitudinally monitoring the dynamic activity of p53 in oncogenesis processes. 15 Another indirect strategy was developed to image protein‐protein interactions of p53 in vivo. By fusing p53 with Gal4‐BD (from Saccharomyces cerevisiae) and T‐antigen with the VP16‐AD (from HSV1), the interaction of p53‐T‐antigen is able to regulate the expression of reporter gene (HSV1‐TK and GFP). 16 This imaging system may facilitate the evaluation of pharmacokinetics, and overall efficacy of drugs targeting protein‐protein interactions.
Another tumor suppressor explored is transforming growth factor‐β (TGF‐β). Notably, TGF‐β arrests growth in many premalignant lesions, whereas it promotes growth in advanced tumors. 69 Imaging techniques are important in understanding the dynamic alternation of TGF‐β. To date, 2 components of the TGF‐β signaling pathway, the cytokine TGF‐β and the accessory receptor endoglin, have been imaged. 17 Among them, 89Zr‐labeled fresolimumab, a monoclonal antibody neutralizing TGF‐β, was charactered in different cancer models in a preclinical study. 18 Increased 89Zr‐fresolimumab uptake was observed in tumor ulceration and scar tissue, in which TGF‐β is known to be highly active. For endoglin imaging, multiple antibodies have been radiolabeled as PET tracers, such as 64Cu‐NOTA‐TRC105 and 64Cu‐TRC105‐Fab. 17 , 70 The activation of TGF‐β could also be visualized using TGF‐β‐inducible reporter genes (HSV1‐tk, GFP, and luciferase). Interestingly, in mouse xenografting metastases models, reporter gene expression was observed in bone metastases but not in adrenal metastases, indicating different mechanisms are mediating these metastases. 71
Resisting Cell Death
Programmed cell death serves as a fundamental cellular program for tissue homeostasis. Over the past several decades, several types of programmed cell death have been described, such as apoptosis, necroptosis, and pyroptosis, among which apoptosis represents the most extensively investigated.
To date, various strategies have been developed to image apoptosis processes, involving cellular membrane composition, protein synthesis, and enzyme activation. 19 Membrane phosphatidylserine exposure was a widely explored dying cell target, with commonly used radiopharmaceuticals including radiolabeled annexins, phosphatidylserine‑binding peptides, and synaptotagmin I derivatives. 19 These probes have been extensively assessed in preclinical and early‐phase clinical studies but may not meet clinical expectations for several reasons (eg, the variable tracer uptake, low signal‐to‐noise ratio, nonspecific accumulation in liver and kidneys). 72
The cell marker of phosphatidylserine exposure is shared by apoptotic and necrotic cells 73 ; therefore, strategies for imaging specific apoptotic mediators were developed. For example, a family of cysteine‐aspartate specific proteases, caspase, critically involved in programmed cell death, was targeted to specifically visualize apoptosis. 72 Because different apoptotic pathways ultimately converge in caspase‐3 and caspase‐7, these proteases have been identified as the key executors of apoptosis. 74 Currently, imaging activated caspase can be achieved by using radiolabeled caspase inhibitors (eg, 18F‐ICMT‐11) or substrates (eg, 18F‐CP18), 21 , 22 among which the best validated probe is 18F‐CP18. 19 However, no significant 18F‐CP18 uptake was observed in bone marrow, where apoptotic blood cells were removed, so further specific evaluations are warranted. 21
Besides, most caspase radiopharmaceuticals suffer from low cellular penetration and high background signal because of nonspecific cleavage. 75 To circumvent this issue, another interesting strategy is apoptosis‐responsive reporter gene imaging, in which a cyclic HSV1‐TK reporter was designed with a caspase‐3 recognition domain as the switch. The caspase‐3 cleavage in apoptotic cells could restore the activity of thymidine kinase to enable PET detection. This imaging system showed low background noise and high sensitivity in response to caspase‐3 activation and therefore presented significant value in both high‐throughput apoptosis‐inducing drug screening in vitro and the therapeutic efficacy assessment in vivo. 23
Enabling Replicative Immortality
Another trait of cancer is the ability to enable replicative immortality. 5 Commonly, the ends of linear eukaryotic chromosomes, telomeres, progressively shorten with cell divisions and ultimately reach a critical length, leading to cell death. However, a specialized DNA polymerase activated in cancer cells, telomerase, elongates or maintains telomeres by adding sequence to the telomeres. Telomerase is minimally composed of RNA (hTR), reverse transcriptase (hTERT), and telomerase‐associated proteins. 76 Both hTR and hTERT have been visualized by PET imaging.
Reporter gene methods are commonly used for imaging telomerase. For example, the Na/I symporter has been placed under the control of hTR and hTERT to measure the telomerase activity in vivo. Both the hTR and hTERT promoter could drive the expression of the Na/I symporter. Interestingly, imaging results highlighted the difference of hTR and hTERT expression pattern: the hTERT promoter was inactive in normal tissues, with a weak expression in cancer cells, whereas the hTR promoter expression was less restricted in cancer, but its expression in cancer cells was higher. 24 In another study, by introducing a trimodality fusion reporter (luciferase, fluorescence protein, and TK) under the control of hTERT promoter fragments, a highly expressed reporter system could be imaged in hTERT‐positive cells, providing a potential multimodality system for imaging telomerase in vivo. 25
In contrast, direct imaging of telomerase activity is more challenging. Using an 99mTc‐radiolabeled 18‐mer antisense oligonucleotide targeting hTERT messenger RNA significantly increased radiopharmaceutical uptake in MCF‐7 xenografts and the tracer delivery and normal tissue clearance improved. 26 Similar probe design has also been applied in small interference RNA. 27 Recently, the cell‐penetrating peptide Tat was conjugated with the hTERT‐antibody to improve cell membrane permeability, enabling the visualization of intracellular and intranuclear hTERT protein after 64Cu radiolabeling. 28
Inducing Angiogenesis
Angiogenesis is crucial for meeting the vast demand of oxygen and nutrients in tumorigenesis. 5 Overexpressed proangiogenetic factors in a cancer microenvironment drive aberrant neovascularization, typically branching, distorted, enlarged, and leaky vessels. 77 Imaging targets of angiogenesis can be broadly categorized as 1) direct targets, which mediate the angiogenetic processes directly (eg, vascular endothelial growth factor [VEGF], integrins, CD105); and 2) indirect targets, which reflect the vascular formation indirectly (eg, hypoxia, glucose metabolism). 78
Vascular endothelial growth factor pathways are critical in initiating a signaling cascade of proliferation, migration, and survival of endothelial cells. To date, 3 categories of probes have been developed for imaging VEGF and VEGF receptors. Category 1 is the radiolabeled isoforms of VEGF (eg, VEGF121, VEGF165), with site‐specific labeling and protective sequence insertion technologies were developed to minimize the affinity loss due to random radiolabeling. 29 , 79 Category 2 is the radiolabeled antibody or antibody fragments against VEGF, such as 89Zr‐bevacizumab and 89Zr‐ranibizumab, which help measure the VEGF level and predict antiangiogenic therapeutic efficiency. 30 However, the exact correlation between the probe uptake and VEGF expression remains to be established. 80 Category 3 is the radiolabeled VEGF receptor inhibitors, either for the protein (eg, aflibercept) or small molecular inhibitors (eg, erlotinib). 12 , 30 Notably, tyrosine kinase inhibitor–based radiopharmaceuticals are more likely to form radioactive metabolites than monoclonal antibodies (mAbs); thus, an in vivo stability evaluation is essential. 81
Angiogenesis can also be visualized by indirect targets, such as hypoxia. Because hypoxia is an important factor driving angiogenesis and resistance to therapy, it makes sense to image hypoxia as a pseudo‐target of angiogenesis. The most extensively studied PET probe for hypoxia is 18F‐fluorinated radiosensitizer nitroimidazole, which binds to cellular components permanently when it cannot be oxidized in hypoxic cells. However, the suboptimal pharmacokinetics of 18F‐fluorinated radiosensitizer nitroimidazole limit its application. 32 Novel nitroimidazole‐based radiopharmaceuticals, 18F‐FAZA and 18F‐HX4, have shown improved pharmacokinetics and biodistribution. 33 , 34 Another interesting ligand is 64Cu‐ATSM. 35 After the reduction of Cu(II) to Cu(I), the intracellular Cu(I)‐ATSM reoxidizes and outflows from normoxic cells, whereas in hypoxic cells, it is dissociated into Cu(I) (and then trapped) and ATSM. 64Cu‐ATSM showed favorable imaging performance for clinical translation. 78
Activating Invasion and Metastasis
Metastasis remains the major cause of cancer‐related death and closely correlates with poor prognosis and patient survival. Advances in cancer biology have provided valuable insights into metastatic cascade processes, involving invasive migration, circulation, extravasation, and colonization. 82
During the past decade, cancer stem cells (CSCs) have been identified in many malignancies, with properties of self‐renewal, tumor initiation, and clonal long‐term replication. Metastatic cancers are hypothesized to be initiated by a small number of CSCs. Imaging CSCs is therefore conceptually attractive. A representative radiopharmaceutical imaging CSCs is the 64Cu‐NOTA‐AC133 mAb, targeting one of the most investigated CSC markers, the AC133 epitope of CD133. Notably, imaging intracerebral xenografts with a low density of AC133+ glioblastoma stem cells was accomplished. 36 Additionally, another CSC marker, CXCR4, was visualized by the Cu‐64–labeled CXCR4 peptide antagonist, despite nonspecific accumulation in red blood cells and high accumulation in both liver and kidneys. 37 Many other biomarkers of CSCs (eg, EpCAM) have been characterized and various potential ligands (eg, EpCAM RNA aptamer) are available 83 ; these will also contribute to the development of CSC PET imaging.
Epithelial–mesenchymal transition (EMT) is a critical phenotypic plasticity process mediating tumor metastasis by enhancing the abilities of mobility, invasion, and resistance to apoptosis upon cancer cells. Although the EMT process has not been visualized by PET, other imaging modalities can be potential strategies for PET translation. For example, by using cancer cells derived from the MMTV‐PyMT, Rosa26‐RFP‐GFP, and Fsp1‐Cre transgenic mouse model, the conversion of RFP+ epithelial cells to GFP+ mesenchymal cells under the control of the Fsp1 promoter (a gatekeeper of EMT initiation) can be imaged. 84 Another canonical biomarker for EMT, vimentin, has also been targeted in direct imaging (eg, vimentin‐traced antibodies) and indirect imaging (vimentin promoter). 85 , 86 Interestingly, radiopharmaceuticals imaging a mesenchymal‐epithelial transition receptor have been developed. 39
Genome Instability and Mutation
Cancer cells may acquire random mutations and chromosomal rearrangements, contributing to spatiotemporal tumor heterogeneity. Genome instability results from increased sensitivity to mutagenic events or the dysfunction of genomic maintenance machinery. 87 Several proteins mentioned previously, such as p53 and telomerase, also play critical roles. 5 , 87
DNA damage serves as a source of genomic instability, whereas defects in the defensive mechanism could cause genomic instability and drives tumorigenesis. Poly(ADP‐ribose) polymerase 1 is a critical sensor in repairing single‐strand breaks and represents a imaging biomarker for PET imaging. Notably, this enzyme has also been targeted in the therapeutic strategy of synthetic lethality. Several radionuclide probes, commonly based on PARP inhibitors (eg, olaparib, rucaparib), have been developed, 88 among which 18F‐FTT and 18F‐PARPi have been tested in clinical trials. 40 , 41 For double‐strand breaks (DSBs), an extensively explored imaging biomarker is the protein γH2AX, and antibody‐based radiopharmaceuticals (eg, 89Zr‐anti‐γH2AX‐TAT) have been developed. 42 Notably, γH2AX is a secondary marker of DSBs, so it is essential to take the biology of γH2AX into account when quantifying the numbers of DSBs from PET images.
The representative method for PET to detect a specific gene mutation is antisense gene imaging. By using a radiolabeled oligonucleotide (15‐20 base in length) specifically complementary to targeted nucleic acids, monitoring gene expression in cancer cells is feasible. 26 To date, a series of radiolabeled antisense oligonucleotides have been developed that target different gene mutations such as MYCC, CCND1, BCL2, and KRAS, although the applications in clinical conditions are still scare. 43 Future studies should optimize the in vivo properties of antisense oligonucleotide, including stability, retention in target tissues, and clearance in nontarget tissues. Additionally, fluorescence imaging with modular proteins enabling specific DNA recognition (eg, ZFP, CRISPR‐Cas9) has been developed recently. 89 This radionuclide‐based transformation could be an important direction for clinical translation.
Tumor‐Promoting Inflammation
Accumulating evidence has indicated that inflammation possesses protumorigenic effects. 5 A group of tumor‐promoting inflammation cells have been identified, including tumor‐associated macrophages, neutrophils, and lymphocytes. These inflammatory cells may release various signaling molecules (eg, EGF, matrix metalloproteinases [MMPs], chemokines, cytokines) that can shape the tumor microenvironment (TME) toward a more tumor‐permissive state. 5 , 90
Macrophages constitute the largest population of tumor‐promoting components in the TME, driving carcinogenetic processes in a variety of ways, such as promoting genetic instability, supporting invasion and metastasis, and taming protective adaptive immunity. 91 During the past several decades, 2 major subtypes, M1 and M2, of macrophages have been identified. The M2‐polarized subtype represents the predominant subtype of macrophages within TME, so many efforts were made to develop M2‐targeted imaging agents, such as 68Ga‐pentixafor targeting CD184, 64Cu‐MAN‐LIPs targeting CD206, and 3′‐Aza‐2′‐18F‐fluorofolic acid targeting folate receptor. 44 Besides, the increased interest in macrophage‐modulating therapies implies a need for radiopharmaceuticals targeting M1‐subtypes, including 18F‐FDR‐NOC targeting somatostatin receptor, 11C‐AM7 targeting CD80, and 64Cu‐DOTA‐DAPTA targeting CCR5. 45 Notably, because differentiation of macrophage subtypes with precision generally need 2 or 3 cell markers, and the interconversion between M1 and M2 subtypes may occur in response to TME signals, the precise identification of macrophage subtypes using PET is still challenging.
Chemokines mediate the activation and migration of inflammatory cells. A family of enzyme reported with the function‐regulating chemokine gradient is the MMPs. Inspired by the MMP inhibitory activity of CGS27023A, several MMP inhibitors have been radiolabeled and used for cancer detection, such as 64Cu‐DOTA‐CTT, 18F‐CGS27023A, and 11C‐methyl‐halo‐CGS27023A analogs. 46 , 47 Because MMPs are able to degrade the basal membrane and extracellular matrix and provide space for neovascularization, they also serve as imaging targets for cancer invasion and angiogenesis.
Reprogramming Energy Metabolism
Tumorigenesis relies on the reprogrammed energy metabolism to fuel the cell growth and division, 5 exerting extensive effects on gene expression, cellular differentiation, and tumor microenvironment. 92 PET is powerful in investigating altered cancer metabolism through radiolabeled nutrients.
One of the most widely known reprogrammed metabolism is the ability of cancer cells to produce energy through glycolysis even under aerobic conditions, known as the Warburg effect. Cancer cells upregulate glucose transporters, notably GLUT1, in compensation for ATP's lower efficiency production afforded by glycolysis. Markedly enhanced glucose uptake has been documented in many tumor types in clinical settings by PET imaging using 18F‐FDG, a radiolabeled glucose analog. Indeed, 18F‐FDG remains the most used radiopharmaceutical in evaluating cancer, yielding applications in both cancer diagnosis and treatment. Interestingly, a recent study has revealed impressive results that it is myeloid cells rather than cancer cells that showed the highest uptake of intratumoral glucose across a range of cancer models. 49 These findings may lead to a change of perception on cancer biology and glucose PET imaging and contribute an explanation for intratumoral regional variability in glucose avidity.
Dysregulated catabolism of amino acids also plays critical roles in the tumorigenesis, such as supplying carbons to tricarboxylic acid cycle, nitrogen to nucleobase synthesis, and mediating redox balance. 93 By using radiolabeled amino acids (eg, 11C‐MET, 18F‐FET, 18F‐DOPA), PET enables the characterization of tumor amino acid metabolism in vivo. 50 Notably, nonessential amino acids, such as glutamine, could become essential in determining rapid growth or other stresses of cancer. Accordingly, through radiolabeled glutamine analogues (eg, 18F‐FGln, 11C‐glutamine), glutamine transport and kinetics in various cancers have been evaluated. 94 , 95 Besides, because distinct aspects of glutamine metabolism are controlled by the balance of oncogenes and tumor suppressors, glutamine PET imaging has the potential to evaluate transforming tumor mutations and assess the sensitivity of therapeutic agents targeting glutamine utilization. 96
Evading Immune Destruction
Another critical cancer hallmark involves the trait of cancer cells to evade immune destruction 5 via strategies regulating tumor antigen expression, releasing immune suppressive cytokines, and inducing T‐cell tolerance and immune deviation. 97
In recent years, infiltration of T cells, in particular CD8+ T cells, has been reported to influence the therapeutic effect of immune checkpoint blockade therapy. Several immune infiltration patterns of tumors have been described: immune‐inflamed, immune‐excluded, and immune‐desert. 98 Accordingly, imaging T cells could be powerful for an efficiency evaluation. Representative imaging targets for T cells are CD3, CD4, and CD8, with radiopharmaceuticals being derived from an antibody, antibody fragment, and small molecules, such as 89Zr‐DFO‐CD3, 89Zr‐malDFO‐GK1.5 cDb, and 89Zr‐Df‐IAB22M2C. 53 Very recently, the first‐in‐human imaging study with radiolabeled anti‐CD8 minibody 89Zr‐Df‐IAB22M2C has been reported, demonstrating increased uptake of radiopharmaceutical in CD8+ T cell–rich tissues (eg, spleen, bone marrow) and tumor lesions. 99 Further results are highly anticipated.
To date, 3 primary targets of checkpoint inhibition including the programmed death protein‐1 receptor, its ligand programmed death ligand‐1 (PD‐L1), and the cytotoxic T‐lymphocyte–associated antigen‐4 receptor (CTLA‐4) have the widest clinical applications in many cancer types. 100 PET may help assess tumor programmed death protein‐1/PD‐L1/CTLA‐4 expression in vivo. Radiolabeled mAb tracers (eg, 89Zr‐Df‐nivolumab, 64Cu‐NOTA‐PD‐L1 mAb, 64Cu‐DOTA‐anti‐CTLA‐4) provide an elegant solution to obtaining quantitative whole‐body biodistribution and kinetic information of these antibodies, including parameters such as tumor accumulation and blood T1/2. 54 , 55 , 56 After full evaluation, PET has the potential to select patients who are most likely to benefit from immune checkpoint therapies and monitor dynamic checkpoint expression during treatment.
Also, PET enables the imaging of some other immune‐oncology components, including the tumor‐associated macrophages mentioned, myeloid‐derived suppressor cells, neutrophils, and natural killer cells. 57 , 58 , 59 The noninvasive characterization of the tumor microenvironment by PET molecular imaging is of significance because it depicts the cell‐autonomous properties of various cells and relative regulatory signaling in the TME, which may provide major mechanisms of immune evasion in specific cancer patients.
PET Imaging–Based Phenotyping and Future Perspectives
The past few decades have witnessed stunning progress in cancer biology, identifying a roster of distinctive features in the processes of cancer initiation and progression. 5 We have sought here to provide a framework of PET imaging for cancer phenotyping, as a generalized evaluation system in transpathology, to help investigators in various fields better understand PET imaging tools available in cancer research. 4 The field of PET imaging for cancer is in rapid flux, showing extensive applications in imaging for all the cancer hallmarks proposed. With advances in molecular biology and radiolabeling technologies, emerging novel vectors, such as antibody derivatives, protein scaffolding, and small molecule drugs, have been used for direct PET imaging. Alternatively, through specific reporter constructs, either protein expression or protein‐protein interaction could be dynamically assessed.
Yet, the development of PET imaging lags behind cancer biology research. Compared with the substantial number of in‐depth molecular mechanisms of tumorigenesis, only a few pathophysiological processes are visualized and translated, and a great many interesting and important biomarkers of cancer remain to be investigated through PET imaging. For example, imaging metastasis‐initiating cells and cellular dormancy would further provide opportunities for treating metastatic relapse. 101 , 102 Similarly, imaging the interaction between cancer cells and some other novel factors such as nerves and the microbiome would provide insights into developing cancer and potential therapeutic targets. 103 Significant effort is still required to better integrate oncology and PET molecular imaging.
Looking ahead, the field of PET imaging would continue to benefit from a broad range of disciplines. Notably, with advances in genetic, transcriptomic, proteomic, and metabolomic research, as well as the growth in big data and artificial intelligence, remarkable progress would be made in understanding cancer complexity, its relationship with tumor microenvironment, and the whole body, providing mounting novel imaging targets. Similarly, development of chemical and radiochemical techniques would contribute to the discovery of targeting vectors with favorable binding abilities, as well as the optimization of available imaging agents, and lead to a newer generation of more sensitive and specific molecular probes. Exhaustive assessment of in vivo characteristics, including pharmacokinetics, toxicity, and some other properties (eg, off‐target effects, immune response), would help to determine the optimal agent and accelerate clinical translation.
The imaging potential of radiopharmaceuticals has not remained the same as technological advances. For example, advances in detectors, electronics, and processing algorithms have fueled the emergence of total‐body PET devices, which strengthen the ability to detect lesions with very low levels of radioactivity. 104 The generation of bispecific or multispecific imaging agents would enhance specificity in assessing target status or processes. 105
Moreover, rigorous translation and validation procedures are essential for the imaging agent to be accepted in clinical settings. A few concerns need to be addressed, including technical validation (eg, repeatability, reproducibility), clinical validation (eg, value in informing decision‐making), and cost‐effectiveness evaluation. 106 Imaging procedures and evaluation criteria should also be standardized to allow comparability of data across centers. Besides, before widespread distribution and clinical use, consultations with the US Food and Drug Administration and Centers for Medicare & Medicaid Services are important to ensuring continued reimbursement. All these issues require broad communication and extensive effort by scientists and clinicians.
Taken together, PET has gained increasing importance in characterizing cancer features both in preclinical and clinical settings. As oncology and PET imaging become more closely integrated, PET imaging would further improve cancer evaluation methods, achieving a mode for in vivo cancer phenotyping. To move the field forward, both cancer and nuclear medicine researchers should collaborate with people who have different areas of expertise, including but not limited to clinicians, molecular biologists, statisticians, chemical engineers, technologists, and computational biologists, with the common goal of improving the management of patients with cancer.
Funding Support
This work was supported by the National Natural Science Foundation of China (81725009 and 21788102 to Mei Tian, 32027802 to Hong Zhang), the National Key R&D Program of China (2021YFE0108300 to Hong Zhang), and the Fundamental Research Funds for the Central Universities (2021FZZX002‐05 to Hong Zhang).
Conflict of Interest Disclosure
The authors made no disclosures.
Jin C, Luo X, Li X, Zhou R, Zhong Y, Xu Z, Cui C, Xing X, Zhang H, Tian M. Positron emission tomography molecular imaging‐based cancer phenotyping. Cancer. 2022. 10.1002/cncr.34228
Contributor Information
Hong Zhang, Email: hzhang21@zju.edu.cn.
Mei Tian, Email: meitian@zju.edu.cn.
References
- 1. Soerjomataram I, Bray F. Planning for tomorrow: global cancer incidence and the role of prevention 2020‐2070. Nat Rev Clin Oncol. 2021;18:663‐672. doi: 10.1038/s41571-021-00514-z [DOI] [PubMed] [Google Scholar]
- 2. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613‐628. doi: 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 3. Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat Rev Cancer. 2002;2:683‐693. doi: 10.1038/nrc882 [DOI] [PubMed] [Google Scholar]
- 4. Tian M, He X, Jin C, et al. Transpathology: molecular imaging‐based pathology. Eur J Nucl Med Mol Imaging. 2021;48:2338‐2350. doi: 10.1007/s00259-021-05234-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646‐674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 6. Chen K, Chen X. Design and development of molecular imaging probes. Curr Top Med Chem. 2010;10:1227‐1236. doi: 10.2174/156802610791384225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gagnon MK, Hausner SH, Marik J, Abbey CK, Marshall JF, Sutcliffe JL. High‐throughput in vivo screening of targeted molecular imaging agents. Proc Natl Acad Sci U S A. 2009;106:17904‐17909. doi: 10.1073/pnas.0906925106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Waaijer SJH, Kok IC, Eisses B, et al. Molecular imaging in cancer drug development. J Nucl Med. 2018;59:726‐732. doi: 10.2967/jnumed.116.188045 [DOI] [PubMed] [Google Scholar]
- 9. Lau J, Rousseau E, Kwon D, Lin KS, Benard F, Chen X. Insight into the development of PET radiopharmaceuticals for oncology. Cancers (Basel). 2020;12:1312. doi: 10.3390/cancers12051312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dammes N, Peer D. Monoclonal antibody‐based molecular imaging strategies and theranostic opportunities. Theranostics. 2020;10:938‐955. doi: 10.7150/thno.37443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Serganova I, Blasberg RG. Molecular imaging with reporter genes: has its promise been delivered? J Nucl Med. 2019;60:1665‐1681. doi: 10.2967/jnumed.118.220004 [DOI] [PubMed] [Google Scholar]
- 12. Wei W, Ni D, Ehlerding EB, Luo QY, Cai W. PET imaging of receptor tyrosine kinases in cancer. Mol Cancer Ther. 2018;17:1625‐1636. doi: 10.1158/1535-7163.MCT-18-0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rinne SS, Orlova A, Tolmachev V. PET and SPECT imaging of the EGFR family (RTK class I) in oncology. Int J Mol Sci. 2021;22:3363. doi: 10.3390/ijms22073663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Altine B, Gai Y, Han N, et al. Preclinical evaluation of a fluorine‐18 ((18)F)‐labeled phosphatidylinositol 3‐kinase inhibitor for breast cancer imaging. Mol Pharm. 2019;16:4563‐4571. doi: 10.1021/acs.molpharmaceut.9b00690 [DOI] [PubMed] [Google Scholar]
- 15. Doubrovin M, Ponomarev V, Beresten T, et al. Imaging transcriptional regulation of p53‐dependent genes with positron emission tomography in vivo. Proc Natl Acad Sci U S A. 2001;98:9300‐9305. doi: 10.1073/pnas.161091198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luker GD, Sharma V, Pica CM, Prior JL, Li W, Piwnica‐Worms D. Molecular imaging of protein‐protein interactions: controlled expression of p53 and large T‐antigen fusion proteins in vivo. Cancer Res. 2003;63:1780‐1788. [PubMed] [Google Scholar]
- 17. Rotteveel L, Poot AJ, Bogaard HJ, Ten Dijke P, Lammertsma AA, Windhorst AD. In vivo imaging of TGFbeta signalling components using positron emission tomography. Drug Discov Today. 2019;24:2258‐2272. doi: 10.1016/j.drudis.2019.08.011 [DOI] [PubMed] [Google Scholar]
- 18. Oude Munnink TH, Arjaans ME, Timmer‐Bosscha H, et al. PET with the 89Zr‐labeled transforming growth factor‐beta antibody fresolimumab in tumor models. J Nucl Med. 2011;52:2001‐2008. doi: 10.2967/jnumed.111.092809 [DOI] [PubMed] [Google Scholar]
- 19. Van de Wiele C, Ustmert S, De Spiegeleer B, De Jonghe PJ, Sathekge M, Alex M. Apoptosis imaging in oncology by means of positron emission tomography: a review. Int J Mol Sci. 2021;22:2753. doi: 10.3390/ijms22052753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoglund J, Shirvan A, Antoni G, et al. 18F‐ML‐10, a PET tracer for apoptosis: first human study. J Nucl Med. 2011;52:720‐725. doi: 10.2967/jnumed.110.081786 [DOI] [PubMed] [Google Scholar]
- 21. Doss M, Kolb HC, Walsh JC, et al. Biodistribution and radiation dosimetry of 18F‐CP‐18, a potential apoptosis imaging agent, as determined from PET/CT scans in healthy volunteers. J Nucl Med. 2013;54:2087‐2092. doi: 10.2967/jnumed.113.119800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dubash SR, Merchant S, Heinzmann K, et al. Clinical translation of [(18)F]ICMT‐11 for measuring chemotherapy‐induced caspase 3/7 activation in breast and lung cancer. Eur J Nucl Med Mol Imaging. 2018;45:2285‐2299. doi: 10.1007/s00259-018-4098-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang F, Wang Z, Hida N, et al. A cyclic HSV1‐TK reporter for real‐time PET imaging of apoptosis. Proc Natl Acad Sci U S A. 2014;111:5165‐5170. doi: 10.1073/pnas.1321374111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Riesco‐Eizaguirre G, De la Vieja A, Rodriguez I, et al. Telomerase‐driven expression of the sodium iodide symporter (NIS) for in vivo radioiodide treatment of cancer: a new broad‐spectrum NIS‐mediated antitumor approach. J Clin Endocrinol Metab. 2011;96:E1435‐E1443. doi: 10.1210/jc.2010-2373 [DOI] [PubMed] [Google Scholar]
- 25. Padmanabhan P, Otero J, Ray P, et al. Visualization of telomerase reverse transcriptase (hTERT) promoter activity using a trimodality fusion reporter construct. J Nucl Med. 2006;47:270‐277. [PMC free article] [PubMed] [Google Scholar]
- 26. Liu M, Wang RF, Zhang CL, et al. Noninvasive imaging of human telomerase reverse transcriptase (hTERT) messenger RNA with 99mTc‐radiolabeled antisense probes in malignant tumors. J Nucl Med. 2007;48:2028‐2036. doi: 10.2967/jnumed.107.042622 [DOI] [PubMed] [Google Scholar]
- 27. Kang L, Wang R‐f, Yan P. Cellular uptake study on radiolabeled telomerase‐targeted small interference RNA in hepatocarcinoma cells. J Oncol. 2010;16(6): 22–25. [Google Scholar]
- 28. Jung KO, Youn H, Kim SH, Kim YH, Kang KW, Chung JK. A new fluorescence/PET probe for targeting intracellular human telomerase reverse transcriptase (hTERT) using Tat peptide‐conjugated IgM. Biochem Biophys Res Commun. 2016;477:483‐489. doi: 10.1016/j.bbrc.2016.06.068 [DOI] [PubMed] [Google Scholar]
- 29. Mason CA, Carter LM, Mandleywala K, et al. Imaging early‐stage metastases using an (18)F‐labeled VEGFR‐1‐specific single chain VEGF mutant. Mol Imaging Biol. 2021;23:340‐349. doi: 10.1007/s11307-020-01555-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaufman NEM, Dhingra S, Jois SD, Vicente M. Molecular targeting of epidermal growth factor receptor (EGFR) and vascular endothelial growth factor receptor (VEGFR). Molecules. 2021;26:1076. doi: 10.3390/molecules26041076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Debordeaux F, Chansel‐Debordeaux L, Pinaquy JB, Fernandez P, Schulz J. What about alphavbeta3 integrins in molecular imaging in oncology? Nucl Med Biol. 2018;62‐63:31‐46. doi: 10.1016/j.nucmedbio.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 32. Kobayashi K, Manabe O, Hirata K, et al. Influence of the scan time point when assessing hypoxia in (18)F‐fluoromisonidazole PET: 2 vs. 4 h. Eur J Nucl Med Mol Imaging. 2020;47:1833‐1842. doi: 10.1007/s00259-019-04626-8 [DOI] [PubMed] [Google Scholar]
- 33. Savi A, Incerti E, Fallanca F, et al. First evaluation of PET‐based human biodistribution and dosimetry of (18)F‐FAZA, a tracer for imaging tumor hypoxia. J Nucl Med. 2017;58:1224‐1229. doi: 10.2967/jnumed.113.122671 [DOI] [PubMed] [Google Scholar]
- 34. Sanduleanu S, Wiel A, Lieverse RI, et al. Hypoxia PET imaging with [18F]‐HX4—a promising next‐generation tracer. Cancers. 2020;12:1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lopci E, Grassi I, Rubello D, et al. Prognostic evaluation of disease outcome in solid tumors investigated with 64Cu‐ATSM PET/CT. Clin Nucl Med. 2016;41:e87‐e92. doi: 10.1097/RLU.0000000000001017 [DOI] [PubMed] [Google Scholar]
- 36. Gaedicke S, Braun F, Prasad S, et al. Noninvasive positron emission tomography and fluorescence imaging of CD133+ tumor stem cells. Proc Natl Acad Sci U S A. 2014;111:E692‐E701. doi: 10.1073/pnas.1314189111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jacobson O, Weiss ID, Szajek LP, et al. PET imaging of CXCR4 using copper‐64 labeled peptide antagonist. Theranostics. 2011;1:251‐262. doi: 10.7150/thno/v01p0251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Witney TH, Carroll L, Alam IS, et al. A novel radiotracer to image glycogen metabolism in tumors by positron emission tomography. Cancer Res. 2014;74:1319‐1328. doi: 10.1158/0008-5472.CAN-13-2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu C, Tang Z, Fan W, et al. In vivo positron emission tomography (PET) imaging of mesenchymal‐epithelial transition (MET) receptor. J Med Chem. 2010;53:139‐146. doi: 10.1021/jm900803q [DOI] [PubMed] [Google Scholar]
- 40. Chen D, Dyroff S, Michel L, et al. First‐in‐human studies characterizing a poly (ADP‐ribose) polymerase (PARP) targeted tracer, 18F‐FluorThanatrace (18F‐FTT) for cancer imaging. J Nuclear Med. 2016;57(suppl 2):582. [Google Scholar]
- 41. Schoder H, Franca PDS, Nakajima R, et al. Safety and feasibility of PARP1/2 imaging with (18)F‐PARPi in patients with head and neck cancer. Clin Cancer Res. 2020;26:3110‐3116. doi: 10.1158/1078-0432.CCR-19-3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knight JC, Topping C, Mosley M, et al. PET imaging of DNA damage using (89)Zr‐labelled anti‐gammaH2AX‐TAT immunoconjugates. Eur J Nucl Med Mol Imaging. 2015;42:1707‐1717. doi: 10.1007/s00259-015-3092-8 [DOI] [PubMed] [Google Scholar]
- 43. Mukherjee A, Wickstrom E, Thakur ML. Imaging oncogene expression. Eur J Radiol. 2009;70:265‐273. doi: 10.1016/j.ejrad.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mukherjee S, Sonanini D, Maurer A, Daldrup‐Link HE. The yin and yang of imaging tumor associated macrophages with PET and MRI. Theranostics. 2019;9:7730‐7748. doi: 10.7150/thno.37306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jiemy WF, Heeringa P, Kamps J, van der Laken CJ, Slart R, Brouwer E. Positron emission tomography (PET) and single photon emission computed tomography (SPECT) imaging of macrophages in large vessel vasculitis: current status and future prospects. Autoimmun Rev. 2018;17:715‐726. doi: 10.1016/j.autrev.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 46. Sprague JE, Li WP, Liang K, Achilefu S, Anderson CJ. In vitro and in vivo investigation of matrix metalloproteinase expression in metastatic tumor models. Nucl Med Biol. 2006;33:227‐237. doi: 10.1016/j.nucmedbio.2005.10.011 [DOI] [PubMed] [Google Scholar]
- 47. Wagner S, Breyholz HJ, Holtke C, et al. A new 18F‐labelled derivative of the MMP inhibitor CGS 27023A for PET: radiosynthesis and initial small‐animal PET studies. Appl Radiat Isot. 2009;67:606‐610. doi: 10.1016/j.apradiso.2008.12.009 [DOI] [PubMed] [Google Scholar]
- 48. Wu C, Li F, Niu G, Chen X. PET imaging of inflammation biomarkers. Theranostics. 2013;3:448‐466. doi: 10.7150/thno.6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reinfeld BI, Madden MZ, Wolf MM, et al. Cell‐programmed nutrient partitioning in the tumour microenvironment. Nature. 2021;593:282‐288. doi: 10.1038/s41586-021-03442-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang C, McConathy J. Radiolabeled amino acids for oncologic imaging. J Nucl Med. 2013;54:1007‐1010. doi: 10.2967/jnumed.112.113100 [DOI] [PubMed] [Google Scholar]
- 51. Spick C, Herrmann K, Czernin J. Evaluation of prostate cancer with 11C‐acetate PET/CT. J Nucl Med. 2016;57(suppl 3):30S‐37S. doi: 10.2967/jnumed.115.169599 [DOI] [PubMed] [Google Scholar]
- 52. Giovacchini G, Giovannini E, Leoncini R, Riondato M, Ciarmiello A. PET and PET/CT with radiolabeled choline in prostate cancer: a critical reappraisal of 20 years of clinical studies. Eur J Nucl Med Mol Imaging. 2017;44:1751‐1776. doi: 10.1007/s00259-017-3700-x [DOI] [PubMed] [Google Scholar]
- 53. Wei W, Jiang D, Ehlerding EB, Luo Q, Cai W. Noninvasive PET imaging of T cells. Trends Cancer. 2018;4:359‐373. doi: 10.1016/j.trecan.2018.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hettich M, Braun F, Bartholoma MD, Schirmbeck R, Niedermann G. High‐resolution PET Imaging with therapeutic antibody‐based PD‐1/PD‐L1 checkpoint tracers. Theranostics. 2016;6:1629‐1640. doi: 10.7150/thno.15253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ehlerding EB, England CG, Majewski RL, et al. ImmunoPET imaging of CTLA‐4 expression in mouse models of non‐small cell lung cancer. Mol Pharm. 2017;14:1782‐1789. doi: 10.1021/acs.molpharmaceut.7b00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Broos K, Lecocq Q, Raes G, Devoogdt N, Keyaerts M, Breckpot K. Noninvasive imaging of the PD‐1:PD‐L1 immune checkpoint: embracing nuclear medicine for the benefit of personalized immunotherapy. Theranostics. 2018;8:3559‐3570. doi: 10.7150/thno.24762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoffmann SHL, Reck DI, Maurer A, et al. Visualization and quantification of in vivo homing kinetics of myeloid‐derived suppressor cells in primary and metastatic cancer. Theranostics. 2019;9:5869‐5885. doi: 10.7150/thno.33275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang C, Keliher E, Zeller MWG, et al. An activatable PET imaging radioprobe is a dynamic reporter of myeloperoxidase activity in vivo. Proc Natl Acad Sci U S A. 2019;116:11966‐11971. doi: 10.1073/pnas.1818434116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shaffer TM, Aalipour A, Schurch CM, Gambhir SS. PET imaging of the natural killer cell activation receptor NKp30. J Nucl Med. 2020;61:1348‐1354. doi: 10.2967/jnumed.119.233163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lindenberg L, Adler S, Turkbey IB, et al. Dosimetry and first human experience with (89)Zr‐panitumumab. Am J Nucl Med. Mol Imaging. 2017;7:195‐203. [PMC free article] [PubMed] [Google Scholar]
- 61. van Helden EJ, Elias SG, Gerritse SL, et al. [(89)Zr]Zr‐cetuximab PET/CT as biomarker for cetuximab monotherapy in patients with RAS wild‐type advanced colorectal cancer. Eur J Nucl Med Mol Imaging. 2020;47:849‐859. doi: 10.1007/s00259-019-04555-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van Dijk LK, Boerman OC, Kaanders JH, Bussink J. PET Imaging in head and neck cancer patients to monitor treatment response: a future role for EGFR‐targeted imaging. Clin Cancer Res. 2015;21:3602‐3609. doi: 10.1158/1078-0432.CCR-15-0348 [DOI] [PubMed] [Google Scholar]
- 63. Antunes IF, van Waarde A, Dierckx RA, de Vries EG, Hospers GA, de Vries EF. Synthesis and evaluation of the estrogen receptor beta‐selective radioligand 2‐(18)F‐fluoro‐6‐(6‐hydroxynaphthalen‐2‐yl)pyridin‐3‐ol: comparison with 16alpha‐(18)F‐fFluoro‐17beta‐estradiol. J Nucl Med. 2017;58:554‐559. doi: 10.2967/jnumed.116.180158 [DOI] [PubMed] [Google Scholar]
- 64. Liao GJ, Clark AS, Schubert EK, Mankoff DA. 18F‐fluoroestradiol PET: current status and potential future clinical applications. J Nucl Med. 2016;57:1269‐1275. doi: 10.2967/jnumed.116.175596 [DOI] [PubMed] [Google Scholar]
- 65. Zhou Y, Lei P, Han J, et al. Development of a novel 18F‐labeled probe for PET imaging of estrogen receptor β. Eur J Nucl Med Mol Imaging. Forthcoming 2022. doi: 10.21203/rs.3.rs-1334238/v1 [DOI] [PubMed] [Google Scholar]
- 66. Janku F, Yap TA, Meric‐Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol. 2018;15:273‐291. doi: 10.1038/nrclinonc.2018.28 [DOI] [PubMed] [Google Scholar]
- 67. Wehrenberg‐Klee E, Turker NS, Heidari P, et al. Differential receptor tyrosine kinase PET imaging for therapeutic guidance. J Nucl Med. 2016;57:1413‐1419. doi: 10.2967/jnumed.115.169417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sever R, Brugge JS. Signal transduction in cancer. Cold Spring Harb Perspect Med. 2015;5:a006098. doi: 10.1101/cshperspect.a006098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Principe DR, Doll JA, Bauer J, et al. TGF‐beta: duality of function between tumor prevention and carcinogenesis. J Natl Cancer Inst. 2014;106:djt369. doi: 10.1093/jnci/djt369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sier VQ, van der Vorst JR, Quax PHA, et al. Endoglin/CD105‐based imaging of cancer and cardiovascular diseases: a systematic review. Int J Mol Sci. 2021;22:4804. doi: 10.3390/ijms22094804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Serganova I, Moroz E, Vider J, et al. Multimodality imaging of TGFbeta signaling in breast cancer metastases. FASEB J. 2009;23:2662‐2672. doi: 10.1096/fj.08-126920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rybczynska AA, Boersma HH, de Jong S, et al. Avenues to molecular imaging of dying cells: focus on cancer. Med Res Rev. 2018;38:1713‐1768. doi: 10.1002/med.21495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Blankenberg FG. In vivo detection of apoptosis. J Nucl Med. 2008;49(suppl 2):81S‐95S. doi: 10.2967/jnumed.107.045898 [DOI] [PubMed] [Google Scholar]
- 74. Chen KW, Demarco B, Heilig R, et al. Extrinsic and intrinsic apoptosis activate pannexin‐1 to drive NLRP3 inflammasome assembly. EMBO J. 2019;38:e101638. doi: 10.15252/embj.2019101638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. De Saint‐Hubert M, Prinsen K, Mortelmans L, Verbruggen A, Mottaghy FM. Molecular imaging of cell death. Methods. 2009;48:178‐187. doi: 10.1016/j.ymeth.2009.03.022 [DOI] [PubMed] [Google Scholar]
- 76. Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5:577‐584. doi: 10.1038/nrd2081 [DOI] [PubMed] [Google Scholar]
- 77. De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer. 2017;17:457‐474. doi: 10.1038/nrc.2017.51 [DOI] [PubMed] [Google Scholar]
- 78. Florea A, Mottaghy FM, Bauwens M. Molecular imaging of angiogenesis in oncology: current preclinical and clinical status. Int J Mol Sci. 2021;22:5544. doi: 10.3390/ijms22115544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Blankenberg FG, Backer MV, Levashova Z, Patel V, Backer JM. In vivo tumor angiogenesis imaging with site‐specific labeled (99m)Tc‐HYNIC‐VEGF. Eur J Nucl Med Mol Imaging. 2006;33:841‐848. doi: 10.1007/s00259-006-0099-1 [DOI] [PubMed] [Google Scholar]
- 80. Scheer MG, Stollman TH, Boerman OC, et al. Imaging liver metastases of colorectal cancer patients with radiolabelled bevacizumab: lack of correlation with VEGF‐A expression. Eur J Cancer. 2008;44:1835‐1840. doi: 10.1016/j.ejca.2008.05.026 [DOI] [PubMed] [Google Scholar]
- 81. van Dongen GA, Poot AJ, Vugts DJ. PET imaging with radiolabeled antibodies and tyrosine kinase inhibitors: immuno‐PET and TKI‐PET. Tumour Biol. 2012;33:607‐615. doi: 10.1007/s13277-012-0316-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ganesh K, Massague J. Targeting metastatic cancer. Nat Med. 2021;27:34‐44. doi: 10.1038/s41591-020-01195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shigdar S, Lin J, Li Y, et al. Cancer stem cell targeting: the next generation of cancer therapy and molecular imaging. Ther Deliv. 2012;3:227‐244. doi: 10.4155/tde.11.148 [DOI] [PubMed] [Google Scholar]
- 84. Zhao Z, Zhu X, Cui K, et al. In vivo visualization and characterization of epithelial‐mesenchymal transition in breast tumors. Cancer Res. 2016;76:2094‐2104. doi: 10.1158/0008-5472.CAN-15-2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Maier J, Traenkle B, Rothbauer U. Real‐time analysis of epithelial‐mesenchymal transition using fluorescent single‐domain antibodies. Sci Rep. 2015;5:13402. doi: 10.1038/srep13402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ieda T, Tazawa H, Okabayashi H, et al. Visualization of epithelial‐mesenchymal transition in an inflammatory microenvironment‐colorectal cancer network. Sci Rep. 2019;9:16378. doi: 10.1038/s41598-019-52816-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ferguson LR, Chen H, Collins AR, et al. Genomic instability in human cancer: molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Semin Cancer Biol. 2015;35(suppl):S5‐S24. doi: 10.1016/j.semcancer.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ambur Sankaranarayanan R, Kossatz S, Weber W, Beheshti M, Morgenroth A, Mottaghy FM. Advancements in PARP1 targeted nuclear imaging and theranostic probes. J Clin Med. 2020;9:2130. doi: 10.3390/jcm9072130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chen B, Guan J, Huang B. Imaging specific genomic DNA in living cells. Annu Rev Biophys. 2016;45:1‐23. doi: 10.1146/annurev-biophys-062215-010830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51:27‐41. doi: 10.1016/j.immuni.2019.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour‐associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14:399‐416. doi: 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27‐47. doi: 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lieu EL, Nguyen T, Rhyne S, Kim J. Amino acids in cancer. Exp Mol Med. 2020;52:15‐30. doi: 10.1038/s12276-020-0375-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhou R, Pantel AR, Li S, et al. [(18)F](2S,4R)4‐Fluoroglutamine PET detects glutamine pool size changes in triple‐negative breast cancer in response to glutaminase inhibition. Cancer Res. 2017;77:1476‐1484. doi: 10.1158/0008-5472.CAN-16-1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cohen AS, Grudzinski J, Smith GT, et al. First‐in‐human PET imaging and estimated radiation dosimetry of L‐[5‐(11)C]‐glutamine in patients with metastatic colorectal cancer. J Nucl Med. 2021;63:36‐43. doi: 10.2967/jnumed.120.261594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mates JM, Di Paola FJ, Campos‐Sandoval JA, Mazurek S, Marquez J. Therapeutic targeting of glutaminolysis as an essential strategy to combat cancer. Semin Cell Dev Biol. 2020;98:34‐43. doi: 10.1016/j.semcdb.2019.05.012 [DOI] [PubMed] [Google Scholar]
- 97. Vinay DS, Ryan EP, Pawelec G, et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;35(suppl):S185‐S198. doi: 10.1016/j.semcancer.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 98. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133‐150. doi: 10.1038/s41568-019-0116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pandit‐Taskar N, Postow MA, Hellmann MD, et al. First‐in‐humans imaging with (89)Zr‐Df‐IAB22M2C anti‐CD8 minibody in patients with solid malignancies: preliminary pharmacokinetics, biodistribution, and lesion targeting. J Nucl Med. 2020;61:512‐519. doi: 10.2967/jnumed.119.229781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. O’Donnell JS, Teng MW, Smyth MJ. Cancer immunoediting and resistance to T cell‐based immunotherapy. Nat Rev Clin Oncol. 2019;16:151‐167. [DOI] [PubMed] [Google Scholar]
- 101. Sosa MS, Bragado P, Aguirre‐Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14:611‐622. doi: 10.1038/nrc3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Massague J, Ganesh K. Metastasis‐initiating cells and ecosystems. Cancer Discov. 2021;11:971‐994. doi: 10.1158/2159-8290.CD-21-0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Monje M, Borniger JC, D'Silva NJ, et al. Roadmap for the emerging field of cancer neuroscience. Cell. 2020;181:219‐222. doi: 10.1016/j.cell.2020.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Berg E, Gill H, Marik J, et al. Total‐body PET and highly stable chelators together enable meaningful (89)Zr‐antibody PET studies up to 30 days after injection. J Nucl Med. 2020;61:453‐460. doi: 10.2967/jnumed.119.230961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ehlerding EB, Sun L, Lan X, Zeng D, Cai W. Dual‐targeted molecular imaging of cancer. J Nucl Med. 2018;59:390‐395. doi: 10.2967/jnumed.117.199877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. O'Connor JP, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14:169‐186. doi: 10.1038/nrclinonc.2016.162 [DOI] [PMC free article] [PubMed] [Google Scholar]


