Abstract
Methanogenesis represents an important electron sink reaction in the hindgut of soil-feeding termites. This is the first comprehensive analysis of the archaeal community structure within the highly compartmentalized intestinal tract of a humivorous insect, combining clonal analysis and terminal restriction fragment (T-RF) length polymorphism (T-RFLP) fingerprinting of the archaeal communities in the different gut compartments of Cubitermes orthognathus. We found that the morphological and physicochemical heterogeneity of the gut is reflected in a large phylogenetic diversity and pronounced axial differences in the composition of the archaeal gut microbiota, notably among those clones or ribotypes that could be assigned to methanogenic taxa. Comparative analysis of the relative frequencies of different archaeal lineages among the small-subunit rRNA gene (SSU rDNA) clones and their corresponding T-RF indicated that the archaeal community in the anterior, extremely alkaline hindgut compartment (P1) consists mainly of members of the Methanosarcinaceae, whereas Methanobacteriaceae and Methanomicrobiales predominate in the subsequent, more posterior compartments (P3/4a and P4b). The relative abundance of Thermoplasmales increased towards the rectum (P5). SSU rDNA sequences representing Crenarchaeota, which have not yet been reported to occur in the intestinal tracts of arthropods, were detected in all gut sections. We discuss how the spatial distribution of methanogenic populations may be linked to axial heterogeneity in the physicochemical gut conditions and to functional adaptations to their respective ecological niches.
Termites are considered an important source of the climate-relevant greenhouse gas methane (for a review, see references 5 and 40), which is formed by methanogenic Archaea located in the enlarged hindguts of these insects. Methane emission rates differ strongly between termite species; these differences are closely correlated with the feeding habits of the respective taxa.
In the hindgut of most wood-feeding species, homoacetogenesis is the major hydrogen sink reaction (8, 9). The methane emission rates of the extremely abundant and globally important soil-feeding species, however, exceed those of wood-feeding termites considerably. In the humivorous Termitinae, the rates of reductive acetogenesis measured in gut homogenates were about 10-fold lower than the rates of methanogenesis (8). Methanogenesis represents a major electron sink in the hindgut metabolism of these termites; in Cubitermes orthognathus, methane production amounts to almost 10% of the respiratory activity of the insect (46).
Initially, the factors influencing the outcome of the competition of these CO2-reducing processes for hydrogen were quite enigmatic (9). However, several studies using microsensors and radiotracer analysis of intestinal CO2-reduction rates have provided evidence that the radial and axial distribution of the microbial populations involved in the production and consumption of hydrogen may play a key role in controlling the fluxes of reducing equivalents in the gut (18, 41, 47).
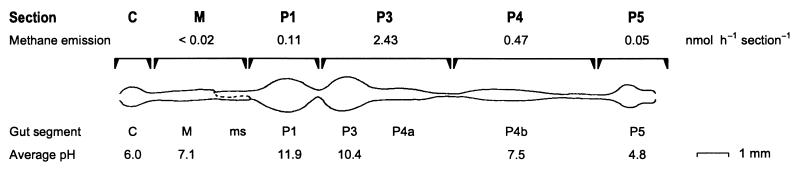
In soil-feeding Termitinae, the hindgut is highly compartmentalized and characterized by an unusually high pH in the anterior region (4) (Fig. 1). The luminal pH increases sharply in the mixed segment, a gut region located between the neutral midgut and the first proctodeal dilation (P1), which possesses the highest alkalinity ever observed in biological systems (around pH 12; [14]). Significant amounts of hydrogen accumulate only in the mixed segment and in the P3 (41). Conversely, methane is formed mainly in the posterior gut compartments P3/4a and P4b (Fig. 1); only small amounts of methane are emitted by the anterior, alkaline compartments (41). The close contact of hydrogen-producing and hydrogen-consuming gut compartments in the abdomen and the stimulation of methanogenesis in the isolated posterior hindgut compartments by the addition of external H2 and formate (41) indicate that a cross-epithelial transfer of reducing equivalents and the contribution of electron donors other than hydrogen also have to be considered when analyzing the functional interactions between the different microbial populations (13, 41).
FIG. 1.
Gut morphology of a Cubitermes sp. worker termite. The gut was drawn in its unraveled state to illustrate the various segments: C, crop; M, midgut; ms, mixed segment; P1 to P5, proctodeal segments 1 to 5, respectively. Gut sections were separated at the indicated positions. Methane emission rates are for individual gut sections of C. orthognathus incubated under N2 atmosphere with 20% H2 (41); the average luminal pH of the major gut segments was determined with glass pH microelectrodes for Cubitermes speciosus (14). (Modified after reference 41).
Unfortunately, little is known about the microbial community involved in these processes, the numerical abundance and spatial distribution of the different hydrogen-producing and hydrogen-consuming populations, and their metabolic properties. Despite the ubiquitous presence of methanogenic activities in termites, the only methanogens isolated in pure culture from termites are three Methanobrevibacter spp. from the wood-feeding Reticulitermes flavipes (27, 28); most of the current knowledge of the methanogen diversity in termite guts is based on cultivation-independent molecular studies using PCR-based 16S rRNA gene (rDNA) retrieval from total gut homogenates. All clones obtained from lower termites represent Methanobacteriaceae (33, 33b, 49) and are closely related to the Methanobrevibacter species isolated from R. flavipes. Also, the archaeal 16S rDNA clones obtained by Shinzato et al. (42) from the gut of Reticulitermes speratus mainly clustered with the Methanobacteriaceae. A few clones, however, fell into the nonmethanogenic order of the Thermoplasmales (42). The archaeal clones obtained from higher termites also include members of the Methanosarcinaceae and Methanomicrobiales; those from the soil-feeding Pericapritermes nitobei, the only humivorous species tested to date, represent the highest methanogen diversity among all termites so far investigated (34). Interestingly, all archaeal clones so far recovered from termite guts form termite-specific clusters.
Nevertheless, the implications of the morphologically and physicochemically highly structured hindgut of the soil-feeding species on the diversity and distribution of archaeal populations remain to be addressed. In this study, we investigated the composition of the archaeal microbiota in the individual gut sections of the soil-feeding termite C. orthognathus and compared it to the archaeal community structure in the nest and the surrounding soil.
MATERIALS AND METHODS
Termites.
Nests of C. orthognathus Emerson (Termitidae: Termitinae) were collected near Busia, Kenya. The termites together with nest fragments and soil from the original collection site were brought to the laboratory in polypropylene containers. Only worker caste termites were used in this study.
DNA extraction and purification.
Termites were dissected with sterile, fine-tipped forceps, and guts were separated into six sections, comprising the crop, the midgut, and the major hindgut compartments (Fig. 1). Between 10 and 20 gut sections each were placed in sterile 2-ml tubes filled with 1 ml of buffered saline solution (48) and were stored frozen at −80°C. DNA was extracted following a direct lysis protocol modified after that of Moré et al. (33) as previously described in detail (23). Aliquots (1 g) of soil samples collected at about 3 m from the nest and of the inner nest material were extracted using the same procedure. DNA was purified from the supernatant by consecutive ammonium acetate, isopropanol, and ethanol precipitation steps. To remove humic substances, the extracts were passed through spin columns filled with polyvinylpolypyrrolidone (3) as previously described (36). Extraction efficiency and quality of the extracted DNA were verified by standard gel electrophoresis. DNA concentrations were determined fluorometrically using Hoechst dye 33258 and a DyNA Quant 200 fluorometer (Amersham Pharmacia Biotech, Freiburg, Germany) as recommended by the manufacturer.
PCR amplification of archaeal 16S rRNA genes.
PCR amplification was carried out as described previously (21) with modifications. Oligonucleotide primers specific for archaeal 16S rDNAs were Ar109F (21) and Ar912R (CTC CCC CGC CAA TTC CTT TA) (Escherichia coli 16S rRNA numbering [11]: positions 109 to 125 and 912 to 931, respectively.). PCR (30 cycles) was carried out at an annealing temperature of 52°C, and 1 μl of undiluted (for P4 and P5) or 10-fold-diluted DNA preparation (all others) was used as template. Aliquots of the 16S rDNA amplicons (5 μl) were analyzed by electrophoresis on a 1% agarose gel and visualized after staining with ethidium bromide.
Fragments of crenarchaeotal SSU rDNA were specifically amplified using primers Ar109F and Cren752R (5′-ACG GTG AGG GAT GAA AGC TG-3′), which were modified from those of Buckley et al. (15). All other PCR conditions were as described above.
SSU rDNA libraries.
Clone libraries were created from archaeal small-subunit (SSU) rDNA amplicons obtained from termite gut community DNA. PCR products (∼800 bp [Ar109F-Ar912R] or ∼750 bp [Arch21F-Cren752R] long) were ligated into the pGEM-T Easy plasmid vector (Promega, Mannheim, Germany), and E. coli JM109 (Promega) was transformed with the recombinant plasmids according to the manufacturer's instructions. Randomly picked clones were further analyzed as described previously by Rotthauwe et al. (37). Clones were checked for the correct insert size by vector-targeted PCR and standard agarose gel electrophoresis. Clones were designated P1 (gut section P1; 30 clones), P3 (gut section P3, representing the P3/4a double compartment; 30 clones), P4b (gut section P4; 30 clones), P5 (gut section P5; 42 clones), or Cren (whole gut; PCR product amplified with Arch21F-Cren752R; 30 clones).
T-RFLP analysis.
Terminal restriction fragment (T-RF) length polymorphism (T-RFLP) analysis was performed as described by Lueders and Friedrich (31) using TaqI restriction digestion. Archaeal SSU rDNAs were amplified using primer Ar109F and 6-carboxyfluorescein-labeled primer Ar912R following the PCR protocol described above, except that only 100-μl PCR mixtures and 28 cycles were used. Standards (GeneScan-1000 ROX; Applied Biosystems), sample preparation, electrophoretic separation on an ABI 373 sequencer, and data analysis using GeneScan analysis software were as previously described (16). T-RFLP electropherograms were analyzed by peak area integration of the different T-RFs. In order to obtain a measure of the relative SSU rDNA frequency, the relative fluorescence intensity of each individual band representing a single T-RF was compared to the total fluorescence intensity of all T-RF bands (31).
Diversity analysis.
Clones with similar (>97% similarity) SSU rDNA sequences were assigned to the same operational taxonomic unit (OTU). Diversity coverage of SSU rDNA clone libraries was analyzed by using the analytical approximation algorithm of Hurlbert (24), and 95% confidence intervals were estimated as described by Heck et al. (22). Rarefaction curves were produced using the Analytic Rarefaction software (version 1.2; S. M. Holland, University of Georgia, Athens [http://www.uga.edu/strata/Software.html]).
Phylogenetic analysis.
Sequence data were analyzed and trees were constructed using the ARB software package with its database (version 2.5b; O. Strunk and W. Ludwig, Technische Universität München, Munich, Germany [http://www.biol.chemie.tu-muenchen.de/pub/ARB/]) as described previously in detail (20). SSU rDNA sequences were added to the database and aligned by using the Fast Aligner tool (version 1.03). Alignments were corrected manually if necessary. Sequences were phylogenetically placed by comparing them to reference sequences for the main lines of descent within the archaeal kingdoms Euryarchaeota and Crenarchaeota (53), as well as Korarchaeota (2). For tree construction, nearly full-length SSU rDNA sequences (>1,300 bases) from the ARB database were selected to construct an archaeal base frequency filter (50 to 100% similarity), and an initial tree was generated by using the neighbor-joining algorithm (39). Sequences from termite gut clone libraries were added to this tree by using the ARB parsimony tool, which allows the addition of short sequences to phylogenetic trees without changing global tree topologies (30). Clonal SSU rDNA sequences were compared to sequences in public databases by using BLAST (1), and closely related SSU rDNA sequences from databases were retrieved and added to the alignment. To detect chimeric rDNA primary structures prior to phylogenetic analysis, the terminal 300 sequence positions of the 5′ and 3′ ends of the archaeal SSU rDNA sequences were used in separate treeing analyses (“fractional treeing” [29]), and significant differences in phylogenetic placement of these terminal sequence fragments were indicative of chimera formation. Also, sequences with unusual mismatches of the rRNA secondary structure were checked for chimera formation.
Nucleotide sequence accession numbers.
SSU rDNA sequences of clones from the termite gut were deposited in GenBank under accession numbers AF293481 to AF293613.
RESULTS
DNA was successfully extracted from all gut sections. Final yields in purified extracts ranged between 50 and 100 ng of DNA per gut section. Whole-gut preparations yielded about 280 ng of DNA per gut, which corresponded to approximately 130 μg of DNA per g (dry weight); yields from purified soil and nest material were 2.0 and 2.5 μg of DNA per g (dry weight), respectively. All DNA extracts except those obtained from the crop yielded amplicons with the Archaea-specific primer pair. Since SSU rDNA was successfully amplified from all extracts, including the crop, with a Bacteria-specific primer pair (27F-1492R [23, 52]) (data not shown), we concluded that the purification protocol had removed PCR-inhibiting compounds (e.g., humic substances) to such an extent that a high degree of inhibition was unlikely.
SSU rDNA clone libraries.
The DNA extracts obtained from gut sections P1, P3, P4, and P5 were used to construct individual clone libraries of archaeal SSU rDNA. From each clone library, 30 clones (section P5 contained 42 clones) were randomly selected and sequenced. Sequence analysis revealed that nearly all clones were affiliated with known taxa of Archaea. While most clones clustered with Euryarchaeota (Fig. 2), a small number (5%) of the clones in the libraries were more closely related to so-far-uncultured Crenarchaeota (Fig. 3). Separate phylogenetic analysis of the terminal 400 nucleotide sequence positions at the 5′ and the 3′ ends of the SSU rDNA clones identified three clones as chimeras (Table 1). These and four clones in the clone libraries that were not affiliated with the Archaea were excluded from further analysis (Table 1).
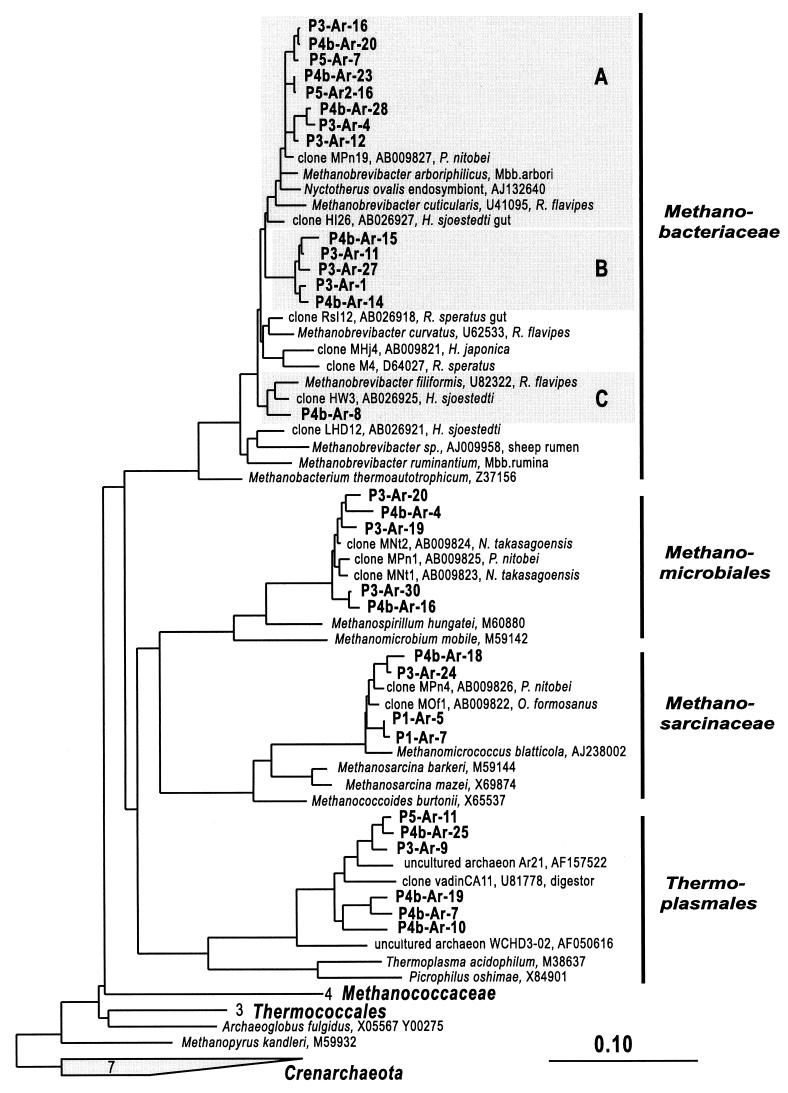
FIG. 2.
Phylogenetic tree showing the positions of SSU rDNA sequences recovered from termite hindgut gut compartments P1, P3(/4a), P4b, and P5 relative to members of the Euryarchaeota. Selected sequences (nearly full-length rDNA) of cultivated representatives from euryarchaeotal lineages were used as references to construct an evolutionary distance dendrogram. SSU rDNA sequences of members of Crenarchaeota and Korarchaeota were used as outgroup references. Partial SSU rDNA sequences (>500 bp) of clones from the guts of the termites Nasutitermes takasagoensis (34), Pericapritermes nitobei (34), Odontotermes formosanus (34), Hodotermopsis sjoestedti, Hodotermopsis japonica (34, 49), Reticulitermes flavipes, and Reticulitermes speratus (49) related to archaeal clones from C. orthognathus were added to the tree using the ARB parsimony tool. Methanobacterial subclusters were designated as follows: A, M. cuticularis; B, C. orthognathus termite cluster; C, M. filiformis. The termite hosts from which SSU rDNA sequences were retrieved are indicated. The scale bar represents 10% sequence difference. Accession numbers of reference sequences are indicated.
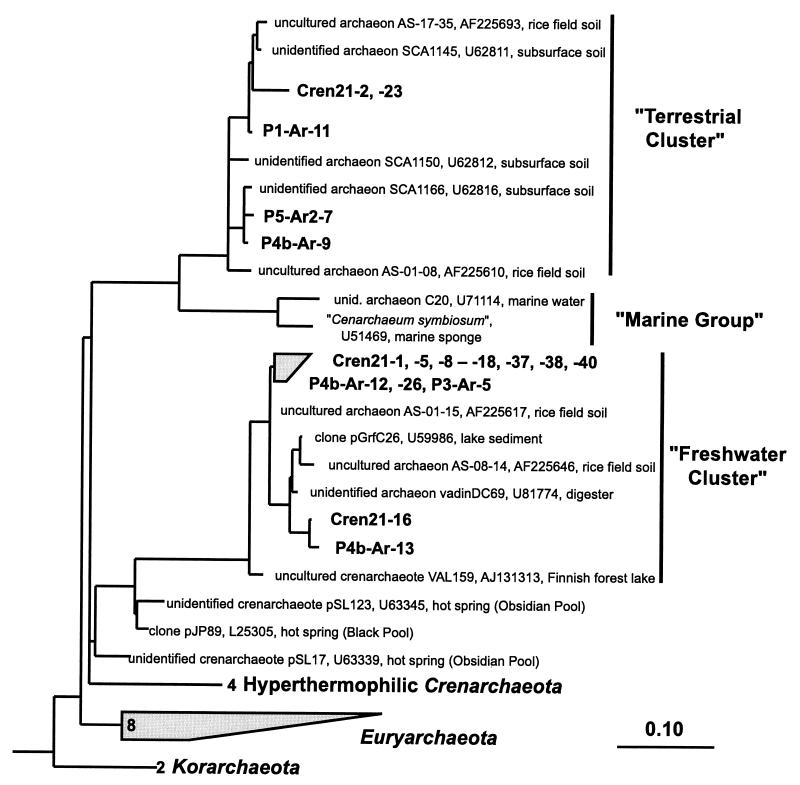
FIG. 3.
Phylogenetic tree showing the positions of SSU rDNA sequences recovered from termite hindgut compartments P1, P3(/4a), P4b, and P5 as well as from total gut homogenates (Cren21-) relative to members of the Crenarchaeota. The tree was rooted with SSU rDNA sequences of Aquifex pyrophilus and Thermotoga maritima, and sequences of members of Euryarchaeota and Korarchaeota were used as additional outgroup references. The habitat from which isolates and SSU rDNA clones were retrieved is indicated. The scale bar represents 10% sequence difference. Accession numbers of reference sequences are indicated.
TABLE 1.
Lengths of TaqI-specific T-RFs of SSU rDNA clonesc
| T-RF [bp] | Clones per compartment
|
Phylogenetic group | |||
|---|---|---|---|---|---|
| P1 | P3(/4a) | P4(b) | P5 | ||
| 88 | P4b-Ar-11, -17, -23, -27 | P5-Ar-5 to -7, -9, -14, -15; P5-Ar2-1, -4, -8, -10, -11, -15 to -19, -21 to -23, -25 to -27, -30 | Methanobacteriaceae | ||
| 181 | P1-Ar-11 | P3-Ar-5 | P4b-Ar-26 | P5-Ar2-7 | Crenarchaeota |
| 220 | P5-Ar2-2 | Thermoplasmales | |||
| 280 | Unknown | ||||
| 341 | P4b-Ar-7, -19 | Thermoplasmales | |||
| 375 | P3-Ar-9 | P4b-Ar-10, -22, -25 | P5-Ar-1 to -3, -10 to -12; P5-Ar2, -3, -6, -9, -13, -14, -20, -24, -28 | Thermoplasmales | |
| 388 | P3-Ar-3, -7, -18, -19, -20, -25, -30 | P4b-Ar-4, -29, -16 | Methanomicrobiales | ||
| 389 | P4b-Ar-13 | Crenarchaeota | |||
| 457 | Unknown | ||||
| 611 | P4b-Ar-5, -8 | Methanobacteriaceae | |||
| ∼800a | P3-Ar-1, -2, -4, -6, -10 to -14, -16, -17, -21 to -23, -26 to -29 | P4b-Ar-1, -3, -6, -14, -15, -20, -21, -28, -30 | Methanobacteriaceae | ||
| ∼800a | P1-Ar-1 to -8, -10, -12 to -30 | P3-Ar-24 | P4b-Ar-18 | Methanosarcinaceae | |
| ∼800a | P4b-Ar-9, -12 | Crenarchaeota | |||
| NDb | P1-Ar-9 | P3-Ar-8 | P4b-Ar-2 | P5-Ar2-12, -29 | Unidentified/chimera |
T-RFs of ∼800 bp represent PCR fragments without a TaqI restriction site.
ND, not determined.
Archaeal SSU rDNA clones related to methanogens.
The majority of the clones affiliated with the Euryarchaeota clustered within different methanogenic lineages (Fig. 2). One group of clones was related to the family Methanobacteriaceae and grouped closely with Methanobrevibacter filiformis and Methanobrevibacter cuticularis (94 to 96% and 95 to 97% similarity, respectively), which have been isolated from the gut of the wood-feeding termite Reticulitermes flavipes (Kollar) (27), and to sequences of clones retrieved from the guts of other wood-, grass-, and soil-feeding termite species.
A second group of clones was related to members of the Methanomicrobiales (7) and clustered with SSU rDNA sequences retrieved from the guts of Nasutitermes takasagoensis and Pericapritermes nitobei (34) (98 to 99% similarity; 550 bp of overlapping sequence information). Clones of this cluster were 95 to 100% similar to each other, and their closest cultivated relative was Methanospirillum hungatei (86 to 89% similarity [Fig. 2]).
A third group of clones was related to members of the family Methanosarcinaceae and grouped closely with SSU rDNA sequences retrieved from the guts of Pericapritermes nitobei and Odontotermes formosanus (34) (96 to 98% similarity; 550 bp overlapping sequence information). Clones of this cluster were 94 to 100% similar to each other, and their closest cultivated relative was Methanomicrococcus blatticola (93.5 to 97.5% similarity), isolated from the gut of the cockroach Periplaneta americana (43).
Each of these three clone groups contained distinct subclusters within their respective clade (Fig. 2). For example, at least three distinct subclusters of Methanobrevibacter spp. (i.e., an M. cuticularis cluster, an M. filiformis cluster, and a distinct C. orthognathus clone group, labeled A to C in Fig. 2) were retrieved from the clone libraries that displayed intersubcluster sequence differences of 95 to 96%, whereas sequence similarities within these subclusters were between 97.5 and 100% similarity. Some of these clones even differed in their TaqI restriction sites (Table 1).
Archaeal SSU rDNA clones related to members of the Thermoplasmales.
A significant proportion of clones affiliated with the Euryarchaeota, especially those recovered from the P5, clustered with members of the Thermoplasmales (Fig. 2). The clones of this cluster were 92 to 100% similar to each other and formed two distinct subclusters. They were closely related to a clone from a fluidized-bed reactor fed with vinasses (19) (92 to 95% sequence similarity) but only distantly related to cultivated members of the Thermoplasmales (between 75 to 78% similarity).
Crenarchaeotal SSU rDNA sequences.
A smaller proportion (5%) of the clones in the clone library carried sequences affiliated with the Crenarchaeota (Fig. 3). Seven of these clones grouped closely with several clones of “rice cluster IV” (16, 20) and from agricultural soil (6); both clone groups are affiliated with the “freshwater cluster” of the kingdom Crenarchaeota. Four clones were affiliated with rice cluster VI (16, 20), a clone group within the “terrestrial cluster” of the Crenarchaeota (95 to 96%, 99%, and 98% similarity, respectively). Clones retrieved from termite gut sections within the freshwater cluster and the terrestrial cluster were 94 to 100% similar to each other.
To further explore the diversity of Crenarchaeota in the gut of C. orthognathus, we specifically amplified only crenarchaeotal SSU rDNA from total community DNA extracted from whole guts. A total of 24 clones from this clone library were analyzed, one of which was a potential chimera. All other clones were closely related to the crenarchaeotal clones in the archaeal clone libraries obtained from the individual gut sections, which indicated that further crenarchaeotal genotypes were not to be expected. The largest proportion of the new clones (21 clones) fell into rice cluster IV, and the remainder fell into the terrestrial cluster (Fig. 3).
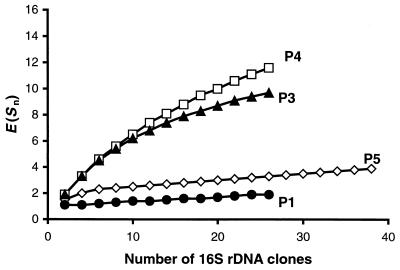
Since the cloning analysis may not reflect the total community diversity due to the method-inherent undersampling bias (45), we checked the diversity coverage of the individual clone libraries by rarefaction analysis (22). We defined sequences as belonging to the same species at >97% sequence similarity, which is in agreement with current microbial species concepts (44). Results for gut sections P1 and P5 revealed a good diversity coverage, as indicated by the slopes of the rarefaction curves, which approached saturation for the 97% sequence similarity level (Fig. 4). In the case of the P3 and P4 sections, however, additional archaeal species would likely be detected if more clones were analyzed (Fig. 4).
FIG. 4.
Rarefaction analysis of archaeal SSU rDNA clones as retrieved from termite gut sections P1, P3, P4, and P5. The expected number of clones [E(Sn)] was calculated from the number of clones analyzed at the species level using 97% sequence similarity. The slope of the curves indicates whether the diversity was covered (zero or low slope) or whether further clones with <97% sequence similarity may be expected by analyzing additional clones (steep slope).
T-RFLP analyses.
Archaeal community structure and diversity in the different gut sections were also assessed directly by T-RFLP analysis. Typical fingerprints of the gut sections P1 and P3 are shown as examples (Fig. 5). The T-RFs obtained from the different profiles and the corresponding clones were grouped according to their phylogenetic affiliations with major lineages of the domain Archaea (Table 1). Relative frequencies of SSU rDNAs, deduced from the T-RF fingerprints of the different sections, the parent soil, and the nest material, were compared to those obtained by analysis of the respective clone libraries (Fig. 6).
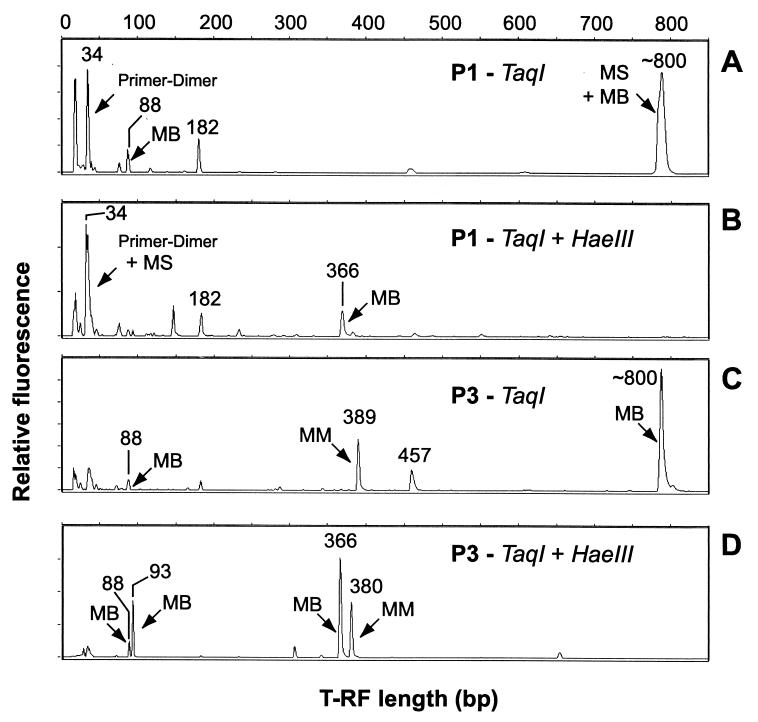
FIG. 5.
T-RFLP analyses of archaeal SSU rDNA PCR products amplified from DNA extracts of gut sections P1 and P3. T-RFLP profiles after TaqI digestion of 6-carboxyfluorescein-labeled PCR products from P1 (A) and P3 (C) and after double digestion with TaqI and HaeIII from P1 (B) and P3 (D). The x axis shows the length (base pairs) of the terminal restriction fragments, and the y axis shows the intensity of fragments in arbitrary units. Designations of archaeal lineages are derived from in silico analysis of sequence data (see also Table 1).
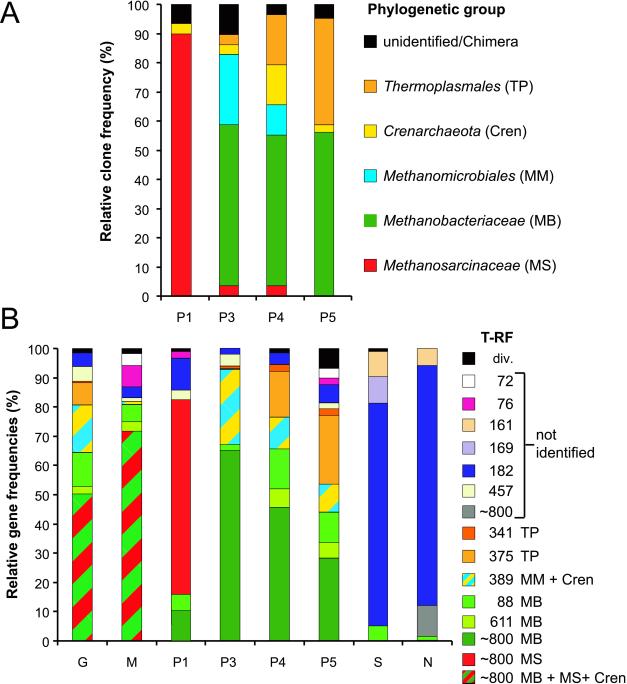
FIG. 6.
Relative clone frequencies (A) and/or T-RFLP-derived SSU rDNA frequencies (B) of archaeal ribotypes in different gut sections (M, P1, P3, P4, and P5), total guts (G), the inner nest material (N), and soil collected at a 3-m distance from the nest (S). Relative SSU rDNA frequencies (B) were determined by integrating peak areas of T-RFs. For designations of ribotypes, see Table 1; “div.” summarizes unidentified T-RFs with <1% relative gene frequency. In the case of the gut sections P1 to P5, the relative gene frequencies of the ∼800-bp OTUs are based on the combined results of TaqI and double-digestion analysis (Fig. 5). With the other data sets, the ∼800-bp OTUs were not further resolved.
Most of the major T-RFs observed in the T-RFLP profiles of the different gut sections matched with OTUs predicted by in silico analysis of clone sequence data. Clones belonging to the Methanobacteriaceae were characterized by three T-RFs of 88, 611, and 800 bp. The third of these fragments did not have a TaqI restriction site and was also characteristic for clones belonging to members of the Methanosarcinaceae and the Crenarchaeota. Clones falling into the Methanomicrobiales and the Crenarchaeota were characterized by T-RF lengths of 388 and 389 bp, which is a fragment size difference below the resolution limit of the T-RFLP analysis (35). However, only one crenarchaeotal clone with a T-RF of 389 bp was detected in the library of the P4 section, which indicated the low frequency of these sequences. All other T-RF sizes predicted were unique for one phylogenetic group. Clones belonging to the Thermoplasmales could even be differentiated by distinct T-RF sizes of 220, 341, and 375 bp (Table 1).
Archaeal community structure in different gut sections.
Both T-RFLP analysis and cloning analysis revealed significant differences in the archaeal community structure among the individual gut sections of C. orthognathus. In general, the relative gene frequencies of OTUs derived from T-RFLP analyses were in good agreement with the frequencies of clones falling into the respective T-RF size classes (Table 1; Fig. 6). T-RFLP analysis detected a few additional, less-frequent ribotypes (<4%) with T-RFs of 72, 76, and 457 bp, which were not detected by cloning analysis (Fig. 6; Table 1).
The highest diversity was found for the whole termite gut (Fig. 6B), as indicated by 10 different, mostly less-frequent ribotypes, and two frequent groups, represented by T-RFs of 389 and ∼800 bp (see Table 1 and the legend of Fig. 6 for affiliation of T-RFs to phylogenetic groups). All major T-RFs, however, could be traced back to the individual gut compartments, where they were represented with a different, compartment-specific gene frequency (Fig. 6).
Midgut section.
The midgut section, which included the increasingly alkaline mixed segment, exhibited a T-RFLP profile that resembled in the relative frequencies and size classes those observed in the following hindgut compartment (P1; see below) with a predominance of the ∼800-bp OTU (72% [Fig. 6B]). Since no clone library was generated for this section, it remains open whether the ±800-bp OTU represents members of the Methanosarcinaceae or of the Methanobacteriaceae prevailing in this section.
Hindgut section P1.
In the case of the extremely alkaline P1, we observed a high proportion of clones (90%) related to members of the Methanosarcinaceae, which all had a T-RF of ∼800 bp (Fig. 6A), and only one crenarchaeotal clone. Direct T-RFLP analysis revealed the presence of a slightly lower relative frequency of 78% for the ∼800-bp OTU, and in addition T-RFs with 88 bp (5%; OTU specific for members of the Methanobacteriaceae) (Table 1) and 181 bp (11%), which probably represented members of the Crenarchaeota (Table 1).
Hindgut section P3.
In the more posterior P3/4a double compartment, the archaeal community changed distinctly and was dominated by Methanobrevibacter spp. (55% of clones, 67% of T-RFs), followed by members of the Methanomicrobiales (24% of clones, 26% of T-RFs). However, only one clone each was closely related to Methanosarcinaceae, Thermoplasmales, and Crenarchaeota, which is in agreement with the relative gene frequencies of <5% for the 341-, 375-, and 182-bp T-RFs, respectively (Fig. 6A). Using a modified T-RFLP assay, we were able to differentiate between Methanosarcinaceae-like, Methanobrevibacter-like, and crenarchaeotal amplicons, none of which had a restriction site for TaqI in the PCR fragment analyzed (∼800-bp OTU [Table 1]). This approach involved a double digestion with HaeIII and TaqI, which resulted in a characteristic 34-bp T-RF (Fig. 5B) for clones falling into the Methanosarcinaceae and a 366-bp T-RF for Methanobrevibacter-like sequences (Fig. 5B and D). Unfortunately, the 34-bp T-RF comigrated with the primer-dimer artifact (34 bp), which rendered quantification impossible. Nevertheless, we were able to show that the Methanobrevibacter-like gene frequencies were low in the P1 section (Fig. 5B), whereas Methanosarcina-like genes were virtually not present in the adjacent P3/4a section (Fig. 5D).
Gut sections P4 and P5.
Towards the posterior end of the gut, the relative frequencies of the Methanobrevibacter-like sequences (88- and 611-bp T-RFs and ∼800-bp OTU) decreased from 64% in the P4 section to 45% in the P5, whereas cloning analyses gave similar clone frequencies of 52 and 56% for these sections. Sequences falling into the Methanomicrobiales represented 10% relative gene (389-bp T-RF) and clone frequency in both sections. The proportion of Thermoplasmales-like sequences increased to 18 and 28% (341- and 375-bp T-RFs), and that of the Crenarchaeota (182-bp T-RF) increased to 4 and 7% in the P4 and P5 sections, respectively. Interestingly, Methanobrevibacter-like sequences with a T-RF of 88 bp represented a larger proportion of archaeal OTUs in the P4 and P5 sections (14 and 11%, respectively) than in the P3 section (2%). This may indicate that the phylogenetically distinct subclusters (A to C [Fig. 2]) represent Methanobrevibacter spp. which occupy specific niches within the respective gut segments.
Archaeal community composition in termite nest and adjacent soil.
Both the T-RFLP fingerprints of the nest material and the surrounding soil were characterized by a predominant 182-bp T-RF, which represented 82 and 76% of the ribotypes in the respective samples (Fig. 6B) but which was not present in the fingerprints of any of the gut sections. Also, two other, less-frequent OTUs with T-RFs of 161 and 169 bp were observed in soil, but not in the termite gut samples. OTUs present both in soil and gut samples had relative frequencies in the fingerprints of the soil samples that were significantly lower than those of the gut samples. Only 10% of the predominant ∼800-bp OTU was present in nest material, and only 5% of the 88-bp T-RF was present in the surrounding soil. The distinct differences in the community composition did not require the assignment of the OTUs to a specific archaeal group; therefore, no clonal analysis was performed for the soil and nest material samples.
DISCUSSION
This is the first comprehensive analysis of the archaeal community structure within the highly compartmentalized intestinal tract of a soil-feeding termite. We could show that the morphological and physicochemical heterogeneity of the gut (14, 25, 41) is paralleled by a high phylogenetic diversity and pronounced axial differences in the composition of the archaeal gut microbiota.
Phylogenetic diversity of termite gut Archaea.
Our study reveals a remarkable diversity of Archaea in the gut of C. orthognathus. Both the results of the cloning analysis and the T-RFLP profiles demonstrated considerable differences in the composition of the archaeal community in the different gut segments. It should be pointed out that many of the phylogenetic groups were detected only by a separate analysis of the individual gut sections (Fig. 6A) and by sequencing a sufficiently large sample of clones (Fig. 4 and 6A). For example, clones related to the Methanosarcinaceae were barely represented in the clone library of the P3 and P4 sections and were not detected at all in the P5 section, whereas they predominated in the P1 section.
Besides detecting sequences grouping with methanogenic Archaea, we also detected SSU rRNA sequences that grouped with members of the Thermoplasmales (Fig. 2). To date, only three SSU rRNA clones (sequence of <500 bp) related to the Thermoplasmales have been recovered from the wood-feeding termite R. speratus (42). It was not possible to determine their phylogenetic relationship with the clones retrieved from the guts of C. orthognathus since the overlapping region of the sequences (∼20 bp) was too small for comparison. One can only speculate whether the Thermoplasmales-like clones represent non-methanogenic Archaea.
In addition to these euryarchaeotal clones, SSU rDNAs representing Crenarchaeota were also detected in all hindgut sections. Phylogenetic analysis indicates a close relationship of the termite gut clones to other clones with unknown phenotypes retrieved from terrestrial or aquatic environments (Fig. 3). There is an increasing body of evidence that Crenarchaeota, which were originally considered to be confined to habitats characterized by high temperature, high salinity, or an extreme pH, also seem to occur ubiquitously in temperate or cold aquatic (26) and terrestrial environments (for a review, see reference 17). Our findings represent the first report on the occurrence of Crenarchaeota in the digestive tract of arthropods and, together with the distantly related clones recovered from the midgut of the holothurian Oneirophanta mutabilis (32), also one of the first reports on the presence of Crenarchaeota in intestinal tracts. In the case of a crenarchaeotal clone recovered from the gut of a flounder (Platichthys flesus) (50), the presence of a specific intestinal population remains to be established.
Although both soil and nest material contained archaeal SSU rDNA, T-RFLP profiles of parent soil and nest material differed strongly from those of the different gut sections. It can be ruled out that the microorganisms whose DNA was amplified from the gut extracts stem from ingested soil. Rather, they seem to be specific members of the termite gut microbiota. T-RFs characteristic of Crenarchaeota and Methanomicrobiales were not even detected in the DNA amplified from the parent soil (Fig. 6B [389 bp]; Table 1), and the frequency of crenarchaeotal clones increased towards the P4b section (Fig. 6A). Interestingly, shedding of intestinal archaea seems to have little impact on the composition of the archaeal community in the nest material, despite the fact that the latter is constructed largely from feces.
Spatial distribution and functional implications.
The environmental factors which should be most decisive for the distribution of archaeal populations in the different gut sections of C. orthognathus are the intestinal pH and—at least in the case of methanogenic Archaea—the availability of reducing equivalents. In a previous study, we found that methane is formed mainly in the P3/4a and P4b compartments (41, 46) (Fig. 1). The results of the molecular analysis performed in the present study, however, indicate that the anterior gut sections (M/ms to P1) also harbor specific, probably autochthonous populations of methanogenic Archaea (Fig. 6).
Based on the extreme alkalinity of the P1 compartment of all Cubitermes spp. (pH 11 to 12.5 [14]), one might carefully conclude that the methanogenic populations specific for this gut section are alkaliphilic. More than 60% of the T-RFs and 90% of the archaeal clones recovered from the P1 section were members of the Methanosarcinaceae, which were almost completely replaced by Methanobrevibacter-related sequences in the subsequent sections consisting of less-alkaline to neutral gut segments (Fig. 6). All Methanosarcinaceae-related clones from the P1 compartment clustered within a clade comprising only clones from the intestinal tracts of insects, which may reflect coevolution with their host, and were closely related to clones recovered from other termites with alkaline gut segments (Nasutitermes takasagoensis and Pericapritermes nitobei). However, the same clade comprised also clones or pure cultures obtained from hosts without an elevated intestinal pH (Odontotermes formosanus and Periplaneta americana) (Fig. 2), and presently it cannot be excluded that the methanogenic population in the P1 compartment is located in microhabitats characterized by a less-alkaline pH than that reported for its bulk volume. Cultivation or in situ localization of the cells within the gut will be necessary to clarify whether these methanogenic symbionts are indeed adapted to extremely alkaline microenvironments.
Hydrogen appears to be a key substrate of methanogenesis in termite guts (for reviews, see references 10 and 12). In Cubitermes spp., however, a utilization of endogenous hydrogen is feasible only in the anterior gut regions, including the P3/4a compartment (41). Based on the large methanogenic potential of the posterior hindgut and the juxtaposition of hydrogen-forming and hydrogen-consuming gut regions in the abdomen of the termite, we have proposed a cross-epithelial hydrogen transfer from hydrogen-forming gut segments (ms, P1, and P3) to hydrogen-consuming gut segments (P4a and P4b) in situ (41), which would create additional microniches for H2-consuming populations.
Nevertheless, it is possible that different methanogenic populations use different electron donors in situ. An alternative, exogenous electron donor for methanogenesis in the posterior hindgut compartments could be formate. Formate, most probably a product of microbial fermentations in the anterior gut compartments (46), is present in the hemolymph of C. orthognathus in appreciable concentrations (2.6 mM [A. Tholen and A. Brune, unpublished results]). The stimulation of methane emission of isolated P3/4a compartments by formate is even stronger than that by exogenous H2 (41), and H2-oxidizing and formate-oxidizing methanogens seem to be present in similar numbers in Cubitermes spp. and other soil-feeding Termitinae (38, 46).
Another potential substrate for methanogenesis in the termite gut may be methanol. Methanomicrococcus blatticola, the only cultivated representative in the cluster of Methanosarcinaceae-related clones recovered from insect guts, is obligately methylotrophic; i.e., it uses H2 as an electron donor only in the presence of methanol or methylamines (43). Sequence similarities between C. orthognathus clones and M. blatticola range from 93.5 to 97.5%, which may allow careful speculations regarding a methylotrophic phenotype.
While the different methanogenic community structures of P1 versus the P3 and P4 sections of C. orthognathus may be caused by axial differences in the microenvironmental conditions, the diversity among the methanogenic populations within each section may reflect a radial organization of methanogens specifically adapted to their respective microhabitats. This is supported by the apparent location of the hydrogen sinks in the posterior hindgut compartments of C. orthognathus (P3-P4b) (41) and by the observation that different morphotypes of F420-fluorescent cells appear to be associated with the gut wall and the cuticular spines protruding into the lumen of the P4b compartment (Tholen and Brune, unpublished results). Functional adaptations to different ecological niches may also explain why Tokura et al. (49) found that the methanogens associated with gut epithelium of the wood-feeding R. speratus and the grass-feeding Hodotermopsis sjoestedti are phylogenetically different from those harbored by the intestinal flagellates (49).
Most-probable-number enumeration yielded highest numbers of methanogens in the posterior hindgut, i.e., in P3/4a and P4b/5 sections, but the low absolute numbers (2 × 106 cells per section [46]) are probably strongly biased by cultivation. It is important to consider that the proportions of methanogenic phylotypes among the archaeal populations in different gut sections reported in this study represent only relative numbers. These are based on PCR methods, which can be subject to method-inherent bias (for a review, see reference 51). In order to gain more information on the absolute abundance and physiological role of methanogens and other archaea in the individual gut segments, it will be necessary to determine their absolute numbers and their spatial distribution by a PCR-independent approach, e.g., by fluorescent in situ hybridization.
ACKNOWLEDGMENTS
This study was supported by a grant of the Deutsche Forschungsgemeinschaft within the program “Structural and Functional Analysis of Natural Microbial Communities” and by the Max-Planck Society.
We thank Bianca Wagner for excellent technical assistance and Ralf Conrad and Bernhard Schink for continuing support.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthelet M, Whyte L G, Greer C W. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiol Lett. 1996;138:17–22. doi: 10.1111/j.1574-6968.1996.tb08128.x. [DOI] [PubMed] [Google Scholar]
- 4.Bignell D E, Eggleton P. On the elevated intestinal pH of higher termites (Isoptera: Termitidae) Insectes Sociaux. 1995;42:57–69. [Google Scholar]
- 5.Bignell D E, Eggleton P, Nunes L, Thomas K L. Termites as mediators of carbon fluxes in tropical forests: budgets for carbon dioxide and methane emissions. In: Watt A B, Stork N E, Hunter M D, editors. Forests and insects. London, United Kingdom: Chapman and Hall; 1997. pp. 109–134. [Google Scholar]
- 6.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boone D R, Whitman W B, Rouviere P E. Diversity and taxonomy of methanogens. In: Ferry J G, editor. Methanogenesis. New York, N.Y: Chapman & Hall; 1993. pp. 35–80. [Google Scholar]
- 8.Brauman A, Kane M D, Labat M, Breznak J A. Genesis of acetate and methane by gut bacteria of nutritionally diverse termites. Science. 1992;257:1384–1387. doi: 10.1126/science.257.5075.1384. [DOI] [PubMed] [Google Scholar]
- 9.Breznak J A. Acetogenesis from carbon dioxide in termite guts. In: Drake H L, editor. Acetogenesis. New York, N.Y: Chapman & Hall; 1994. pp. 303–330. [Google Scholar]
- 10.Breznak J A. Ecology of prokaryotic microbes in the guts of wood- and litter-feeding termites. In: Abe T, Bignell D E, Higashi M, editors. Termites: evolution, sociality, symbiosis, ecology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 209–231. [Google Scholar]
- 11.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brune A. Termite guts: the world's smallest bioreactors. Trends Biotechnol. 1998;16:16–21. [Google Scholar]
- 13.Brune A, Friedrich M. Microecology of the termite gut: structure and function on a microscale. Curr Opin Microbiol. 2000;3:263–269. doi: 10.1016/s1369-5274(00)00087-4. [DOI] [PubMed] [Google Scholar]
- 14.Brune A, Kühl M. pH profiles of the extremely alkaline hindguts of soil-feeding termites (Isoptera: Termitidae) determined with microelectrodes. J Insect Physiol. 1996;42:1121–1127. [Google Scholar]
- 15.Buckley D H, Graber J R, Schmidt T M. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl Environ Microbiol. 1998;64:4333–4339. doi: 10.1128/aem.64.11.4333-4339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin K J, Lukow T, Conrad R. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl Environ Microbiol. 1999;65:2341–2349. doi: 10.1128/aem.65.6.2341-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLong E F. Everything in moderation: Archaea as ‘non-extremophiles’. Curr Opin Genet Dev. 1998;8:649–654. doi: 10.1016/s0959-437x(98)80032-4. [DOI] [PubMed] [Google Scholar]
- 18.Ebert A, Brune A. Hydrogen concentration profiles at the oxic-anoxic interface: a microsensor study of the hindgut of the wood-feeding lower termite Reticulitermes flavipes (Kollar) Appl Environ Microbiol. 1997;63:4039–4046. doi: 10.1128/aem.63.10.4039-4046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godon J J, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groβkopf R, Stubner S, Liesack W. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl Environ Microbiol. 1998;64:4983–4989. doi: 10.1128/aem.64.12.4983-4989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groβkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heck K L, Jr, Van Belle G, Simberloff D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology. 1975;56:1459–1461. [Google Scholar]
- 23.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurlbert S H. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 25.Kappler A, Brune A. Influence of gut alkalinity and oxygen status on mobilization and size-class distribution of humic acids in the hindgut of soil-feeding termites. Appl Soil Ecol. 1999;13:219–229. [Google Scholar]
- 26.Karner M B, DeLong E F, Karl D M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 27.Leadbetter J R, Breznak J A. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl Environ Microbiol. 1996;62:3620–3631. doi: 10.1128/aem.62.10.3620-3631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leadbetter J R, Crosby L D, Breznak J A. Methanobrevibacter filiformis sp. nov., a filamentous methanogen from termite hindguts. Arch Microbiol. 1998;169:287–292. doi: 10.1007/s002030050574. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 30.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 31.Lueders T, Friedrich M. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl Environ Microbiol. 2000;66:2732–2742. doi: 10.1128/aem.66.7.2732-2742.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McInerney J O, Wilkinson M, Patching J W, Embley T M, Powell R. Recovery and phylogenetic analysis of novel archaeal rRNA sequences from a deep-sea deposit feeder. Appl Environ Microbiol. 1995;61:1646–1648. doi: 10.1128/aem.61.4.1646-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moré M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Ohkuma M, Kudo T. Phylogenetic analysis of the symbiotic intestinal microflora of the termite Cryptotermes domesticus. FEMS Microbiol Lett. 1998;164:389–395. [Google Scholar]
- 33b.Ohkuma M, Noda S, Horikoshi K, Kudo T. Phylogeny of symbiotic methanogens in the gut of the termite Reticulitermes speratus. FEMS Microbiol Lett. 1995;134:45–50. doi: 10.1111/j.1574-6968.1995.tb07912.x. [DOI] [PubMed] [Google Scholar]
- 34.Ohkuma M, Noda S, Kudo T. Phylogenetic relationships of symbiotic methanogens in diverse termites. FEMS Microbiol Lett. 1999;171:147–153. doi: 10.1111/j.1574-6968.1999.tb13425.x. [DOI] [PubMed] [Google Scholar]
- 35.Osborn A M, Moore E R B, Timmis K N. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 36.Ramakrishnan B, Lueders T, Conrad R, Friedrich M. Effect of soil aggregate size on methanogenesis and archaeal community structure in anoxic rice field soil. FEMS Microbiol Ecol. 2000;32:261–270. doi: 10.1111/j.1574-6941.2000.tb00719.x. [DOI] [PubMed] [Google Scholar]
- 37.Rotthauwe J H, Witzel K P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouland C, Brauman A, Labat M, Lepage M. Nutritional factors affecting methane emission from termites. Chemosphere. 1993;26:617–622. [Google Scholar]
- 39.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Sanderson M G. Biomass of termites and their emissions of methane and carbon dioxide: a global database. Glob Biogeochem Cycles. 1996;10:543–557. [Google Scholar]
- 41.Schmitt-Wagner D, Brune A. Hydrogen profiles and localization of methanogenic activities in the highly compartmentalized hindgut of soil-feeding higher termites (Cubitermes spp.) Appl Environ Microbiol. 1999;65:4490–4496. doi: 10.1128/aem.65.10.4490-4496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinzato N, Matsumoto T, Yamaoka I, Oshima T, Yamagishi A. Phylogenetic diversity of symbiotic methanogens living in the hindgut of the lower termite Reticulitermes speratus analyzed by PCR and in situ hybridization. Appl Environ Microbiol. 1999;65:837–840. doi: 10.1128/aem.65.2.837-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprenger W W, van Belzen M C, Rosenberg J, Hackstein J H, Keltjens J T. Methanomicrococcus blatticola gen. nov., sp. nov., a methanol- and methylamine-reducing methanogen from the hindgut of the cockroach Periplaneta americana. Int J Syst Evol Microbiol. 2000;50:1989–1999. doi: 10.1099/00207713-50-6-1989. [DOI] [PubMed] [Google Scholar]
- 44.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 45.Suzuki M T, Rappe M S, Giovannoni S J. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tholen A, Brune A. Localization and in situ activities of homoacetogenic bacteria in the highly compartmentalized hindgut of soil-feeding higher termites (Cubitermes spp.) Appl Environ Microbiol. 1999;65:4497–4505. doi: 10.1128/aem.65.10.4497-4505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tholen A, Brune A. Impact of oxygen on metabolic fluxes and in situ rates of reductive acetogenesis in the hindgut of the wood-feeding termite Reticulitermes flavipes. Environ Microbiol. 2000;2:436–449. doi: 10.1046/j.1462-2920.2000.00127.x. [DOI] [PubMed] [Google Scholar]
- 48.Tholen A, Schink B, Brune A. The gut microflora of Reticulitermes flavipes, its relation to oxygen, and evidence for oxygen-dependent acetogenesis by the most abundant Enterococcus sp. FEMS Microbiol Ecol. 1997;24:137–149. [Google Scholar]
- 49.Tokura M, Ohkuma M, Kudo T. Molecular phylogeny of methanogens associated with flagellated protists in the gut and with the gut epithelium of termites. FEMS Microbiol Ecol. 2000;33:233–240. doi: 10.1111/j.1574-6941.2000.tb00745.x. [DOI] [PubMed] [Google Scholar]
- 50.Van Der Maarel M J E C, Artz R R E, Haanstra R, Forney L J. Association of marine archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol. 1998;64:2894–2898. doi: 10.1128/aem.64.8.2894-2898.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Wintzingerode F, Goebel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 52.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]